Abstract
The microbiota plays a fundamental role in the induction, education and function of the host immune system. In return, the host immune system has evolved multiple means by which to maintain its symbiotic relationship with the microbiota. The maintenance of this dialogue allows the induction of protective responses to pathogens and the utilization of regulatory pathways involved in the sustained tolerance to innocuous antigens. The ability of microbes to set the immunological tone of tissues, both locally and systemically, requires tonic sensing of microbes and complex feedback loops between innate and adaptive components of the immune system. In this review, we will highlight the dominant cellular mediators of these interactions and discuss emerging themes associated with our current understanding of the homeostatic immunological dialogue between the host and its microbiota.
Multicellular organisms exist as meta-organisms comprised of both the macroscopic host and its symbiotic commensal microbiota. With an estimated composition of 100 trillion cells, microbes colonizing humans outnumber host cells and express more unique genes than their host’s genome (Ley et al., 2006). These complex communities of microbes that include bacteria, fungi, protozoa and viruses play a fundamental role in controlling host physiology at large. Of late, the field of immunology has been transformed by the growing understanding of the fundamental role of the microbiota in the induction, education and function of the mammalian immune system.
The development of defined arms of the immune system and more particularly those associated with adaptive immunity has coincided with the acquisition of a complex microbiota, supporting the idea that a large fraction of this complex system evolved in order to maintain a symbiotic relationship with the microbiota. As such the immune system plays a fundamental role in shaping and preserving the ecology of the microbiota. In turn, and as discussed in this review, the microbiota promote and calibrate all aspects of the immune system. Indeed, commensals play a fundamental role in both the training of the immune system and its functional tuning, thereby acting as adjuvants to the immune system as a whole (Figure 1). Such dialogue can be revealed in patients with primary immuno-deficiencies that not only display increased ecological permissiveness but also often suffer from infections caused by normal constituents of the microbiota (Puel et al., 2010; Smeekens et al., 2014). Indeed, although members of the microbiota are often referred to as commensals, the symbiosis (persistent interaction) between the microbiota and its mammalian host encompasses various forms of relationships, and how members of the microbiota interact with their host can be highly contextual with the same microbe developing as mutualist, commensal or parasite according to the genetic landscape, nutritional status or co-infection of its host.
Figure 1. The microbiota plays a fundamental role in the induction, education and function of the mammalian immune system.
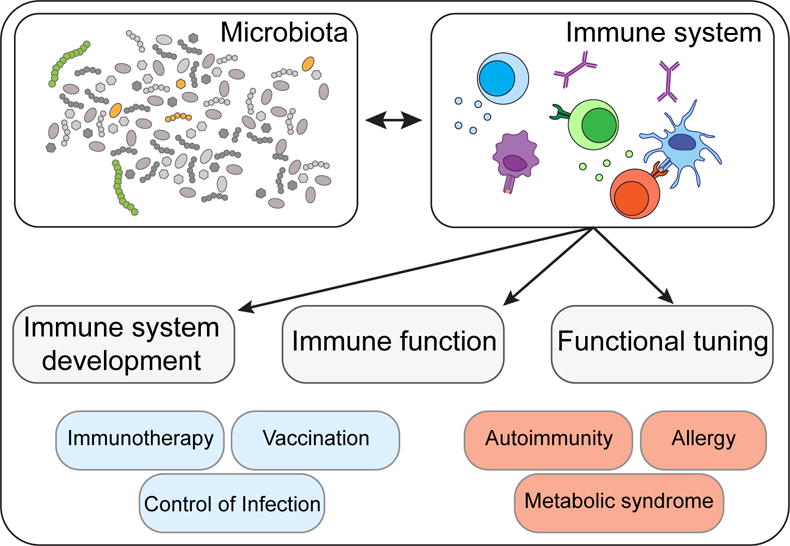
Equally, evolution of the mammalian immune system has coincided with the acquisition of a complex microbiota, demonstrating a symbiotic relationship between the host immune system and its commensal microbiota. A self-reinforcing, dynamic dialogue ensures that commensal colonization occurs as a state of mutualism, the breakdown of which can result in chronic inflammatory disorders, including autoimmunity, allergies and metabolic syndromes. Conversely, selective modulation of the microbiota presents immense therapeutic potential for bolstering tumor immunotherapy, vaccination and resistance to antibiotic-resistant microbes.
Microbial surveys unveiled the remarkable partitioning of commensals within the human body with each tissue and microenvironment hosting unique microbial communities (Belkaid and Naik, 2013). Although it is now clear that gut resident commensals can control both the local milieu and the systemic threshold of activation of cells and tissues, microbes residing in the lung, skin or other barrier sites also contribute to the local regulation of tissues. Acquisition of a complex immune system and its reliance on the microbiota also came at a price. Pathologies that increasingly affect humans such as allergies, autoimmune and inflammatory disorders all arise from a failure to control misdirected immune responses against self, environmental antigens and/or the microbiota. Alteration of the composition and function of the microbiota as a result of antibiotic usage, diet alteration and recent elimination of constitutive partners such as helminthic worms, is believed to have transformed our microbial allies into potential liabilities (Figure 1). The basal tuning of the immune system associated with constant sensing of microbes implies that subtle changes in this conditioning may have long-term consequences on the capacity of the host to mount systemic immune responses and develop inflammatory diseases. Furthermore, growing evidence supports a key role for early encounters between microbes and the neonate in setting the stage for the immune system in the long term. Altered early dialogue between the host immune system and the microbiota have been recently shown to have profound and long-term implications for the development of inflammatory diseases (reviewed in (Arrieta et al., 2014))
While the vast majority of host/microbe encounters are those resulting from the symbiotic relationship with the microbiota, most of what we understand today about the function of the immune system came from the exploration of inflammatory settings and responses to pathogenic microbes. The characteristics of this class of immunity to commensals has only recently begun to be explored and a remarkable property of the various modules of innate and effector cells engaged in this homeostatic dialogue is their ability to reinforce each other’s function.
As the impact of the microbiota in the etiology of diseases has been recently reviewed, in this review we will focus our discussion on the exploration of the homeostatic relationship between the host immune system and the microbiota, and discuss how the adjuvant effect of these microbes, and their ability to calibrate the threshold of activation of cells and tissues promote responses to infection, vaccines and tumor immunotherapy.
Control of epithelial cells by the microbiota
A central strategy utilized by the host to maintain its homeostatic relationship with the microbiota is to minimize contact between microorganisms and the epithelial cell surface, thereby limiting tissue inflammation and microbial translocation. In the gastrointestinal tract, home to the largest density of commensals, this segregation is accomplished by the combined action of epithelial cells, mucus, IgA, antimicrobial peptides and immune cells. Intestinal mucus production provides a primary shield, by limiting contact between the microbiota and host tissue and preventing microbial translocation (McGuckin et al., 2011). In addition to the production of mucus by goblet cells, all intestinal epithelial cell lineages can produce antimicrobial peptides that play a significant role in limiting exposure to the commensal microbiota (Hooper and Macpherson, 2010). These proteins can exert antimicrobial functions including enzymatic attack of the bacterial cell wall, or disruption of the bacterial inner membrane (Hooper and Macpherson, 2010). Some of these molecules, such as α-defensins, are constitutively expressed by epithelial cells, while in other cases engagement of pattern recognition receptors (PRRs) by commensal-derived products is required for their production (Hooper and Macpherson, 2010). One of the best-characterized mucosal antimicrobial peptides is RegIIIγ, which is expressed soon after birth or following colonization of germ-free mice (Cash et al., 2006). Production of this lectin is tightly controlled by the flora in a MyD88-dependent manner and has a direct microbicidal effect on gram-positive bacteria (Brandl et al., 2007; Cash et al., 2006; Ismail et al., 2011; Mukherjee et al., 2009). Accumulation of antimicrobial peptides in the mucus contributes to the maintenance of the segregation between the microbiota, a separation referred to as the “demilitarized zone” (Vaishnava et al., 2011). In part, these responses are controlled by direct sensing of microbes or microbe-derived products by epithelial cells that integrate multiple signals in order to ensure barrier integrity and tissue homeostasis including those from Toll-like receptors (Rakoff-Nahoum et al., 2004), Nod-like receptors (Elinav et al., 2011) and short chain fatty acid receptors (Macia et al., 2015). Additionally, intestinal epithelial cells are also influenced indirectly by the microbiota, via cytokine production by innate and adaptive immune cells induced by microbial colonization (reviewed in (Thaiss et al., 2016).
Tonic control of hematopoiesis and innate immunity by the microbiota
Despite the anatomical separation of the microbiota and host immune system that limits systemic dispersion of commensal microbes, bacterial metabolites are detectable in peripheral tissues, following commensal colonization (Balmer et al., 2014b; Clarke et al., 2010b). Whether active transport mechanisms or passive diffusion underlie the systemic penetrance of microbial metabolites remains unclear. However, commensal metabolites and products reaching circulation impact development and as further discussed, the functional tuning of the host immune system. Indeed, experimental evidence suggests that the tonic sensing of commensal products or metabolites present in the blood stream can contribute to steady-state hematopoiesis (Maslowski et al., 2009; Shi et al., 2011). Over 30 years ago, antibiotic treatment was shown to reduce the frequency of granulocyte-macrophage colony formation and germ-free animals display a global defect in innate immune cells (Goris et al., 1985; Tada et al., 1996). The microbiota can impact both yolk sac- and stem cell-derived myeloid cell development (Balmer et al., 2014a; Khosravi et al., 2014) and the size of the bone marrow myeloid cell pool correlates with the complexity of the intestinal microbiota (Balmer et al., 2014a). Tonic sensing of minute concentrations of LPS, a setting that can reflect microbiota fluctuation, drives CCR2-dependent emigration of monocytes from the bone marrow (Shi and Pamer, 2011). Bone marrow mesenchymal stem cells (MSCs) and their progeny can rapidly express MCP1 in response to circulating TLR ligands thereby promoting monocyte trafficking into the bloodstream (Shi and Pamer, 2011). In vivo administration of NOD1 ligands to germ-free mice can also restore the numbers of hematopoietic stem cells and precursors to the levels displayed by specific pathogen-free animals. Mesenchymal stem cells can also sense NOD1 ligands, thereby expressing factors with known effects on hematopoiesis such as interleukin-7, Flt3L, stem cell factor, ThPO, and IL-6 (Iwamura et al., 2017). Such control may also occur in early life as antibiotic treatment of murine dams reduces their levels of IL-17 and G-CSF, events proposed to account for the reduced numbers of neutrophils and granulocyte/macrophage-restricted progenitor cells observed in neonates (Deshmukh et al., 2014).
Products of bacterial metabolism can also influence hematopoiesis and hematopoietic cell education (Figure 2). Mammals rely on bacteria to break down indigestible dietary components such as fiber, and one prominent class of metabolites resulting from this process are short chain fatty acids (SCFA), ubiquitous bacterial fermentation products (Cummings et al., 1987). In addition to providing an energy source for enterocytes, SCFA activate G protein-coupled receptors expressed by epithelial and hematopoietic cells, and can inhibit histone deacetylase (HDACs)(Macia et al., 2015; Maslowski et al., 2009). As such, SCFA are commensal-derived products that can act directly on diverse immune cells and tune their function (Rooks and Garrett, 2016). Elevation of circulating SCFA following increase in dietary fiber, or direct treatment with propionate, led to alterations in hematopoiesis characterized by enhanced generation of macrophage and dendritic cell (DC) precursors and subsequent seeding of the lungs by DCs with high phagocytic capacity (Trompette et al., 2014). In addition to a role for the microbiota in controlling the output and, potentially, the function of cells leaving the bone marrow, the microbiota continues to influence hematopoietic cells within tissues. For instance, in the gut, SCFA can alter the gene expression profile of local macrophages via HDAC inhibition (Chang et al., 2014; Singh et al., 2014). Antibiotic treatment of mice can influence alveolar macrophage polarization via outgrowth of a commensal fungal species and elevated circulating levels of prostaglandin E2 (Kim et al., 2014). The microbiota also control the homeostatic replenishment of monocyte-derived macrophages in the intestinal mucosa (Bain et al., 2014). The level of circulating innate cells, such as basophil, is also influenced by the microbiota, and tonic sensing of TLR ligands can promotes neutrophil “ageing” (Hill et al., 2012; Zhang et al., 2015).
Figure 2. Microbiota controls hematopoiesis and systemic hematopoietic cell education.
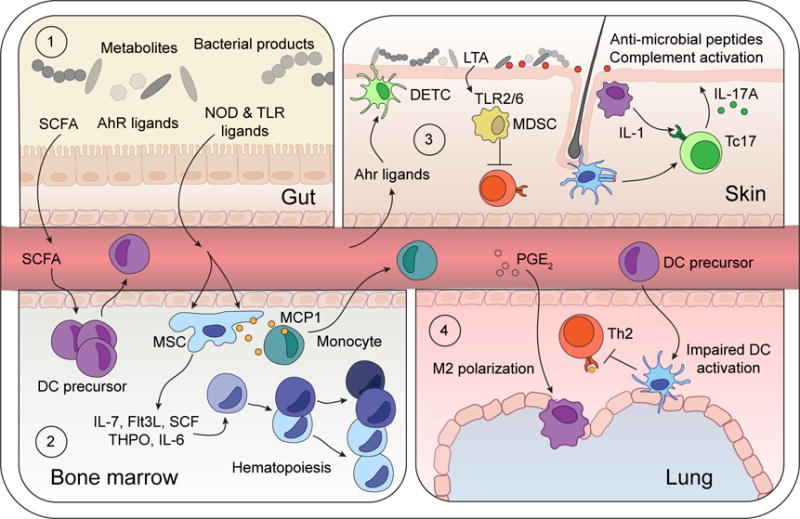
(1) Microbiota-derived TLR and NOD ligands and metabolites (SCFA and AhR ligands) can act directly on local intestinal tissue cells, but also penetrate beyond the mucosa, into circulation to tune immune cells in peripheral tissues.
(2) SCFA promote DC precursor activation and release into bloodstream. Microbiota-derived NOD1 ligands activate mesenchymal stem cells (MSC) to produce hematopoietic growth factors (IL-7, SCF, THPO, Flt3L, IL-6), and commensal-derived TLR ligands trigger MCP1 production by MSC and their progeny, thereby promoting monocyte exit into the bloodstream.
(3) Diet-derived AhR-ligands promote continued tissue residency of Dendritic Epidermal T cells (DETC) within the skin epidermis. Local skin commensals engage local immune responses. Lipoteichoic acid (LTA) derived from S. aureus limits T cell activation by TLR2/6-dependent generation of myeloid-derived suppressor cells (MDSC). Conversely, colonization with S. epidermidis promotes cognate CD8+ T cell seeding of the epidermis, licensing of cytokine production by myeloid-derived IL-1, and engagement of antimicrobial pathways by keratinocytes.
(4) SCFA-elicited DC precursors traffic to the lung and limit Th2 cell reactivation to dampen experimental asthma. Antibiotic-dependent dysbiosis elicits PGE2 production which promotes M2 polarization of alveolar macrophages.
The aryl hydrocarbon receptor (AhR) is a ligand-dependent transcription factor that senses both xenobiotic and endogenous ligands, including microbiota-derived factors. Catabolism of tryptophan-containing dietary components to indole-derivatives by commensal bacteria plays a key role in bolstering AhR-mediated homeostasis at barrier surfaces (Stockinger et al., 2014). Within the gastrointestinal tract, AhR-activation promotes IL-22 production by group 3 innate lymphoid cells (ILC) and Th17 cells (Kiss et al., 2011; Qiu et al., 2012; Veldhoen et al., 2008; Zelante et al., 2013), and the maintenance of intra-epithelial lymphocytes (Li et al., 2011). Commensal-catabolized indole-derivatives from gestationally-colonized mothers promoted increased ILC3 and mononuclear phagocyte frequencies in their offspring (Gomez de Aguero et al., 2016). Maternal antibodies mediated some of these effects, potentially by retention of AhR-ligands to promote transmission during pregnancy. Dietary-derived AhR-ligands promote maintenance of intra-epithelial lymphocytes within the gut, but also within the skin epidermis, demonstrating a vast systemic reach of these microbiota-conditioned metabolites (Li et al., 2011).
Non-classical lymphocytes interact with the microbiota
Innate lymphocytes, including ILC, γδT cells, MAITs, NKT or NK cells often reside in peripheral tissues, and as such, are well positioned to coordinate the interaction of the host with its microbiota (Figure 3). In further support for a role in these cells in setting the stage for host-microbiota interactions, non-classical lymphocytes have been shown to be enriched at barrier sites during early life (Gensollen et al., 2016), a time of extraordinary and dynamic exposure to the microbiota.
Figure 3. Commensal colonization differentially regulates innate and innate-like lymphoid cells.
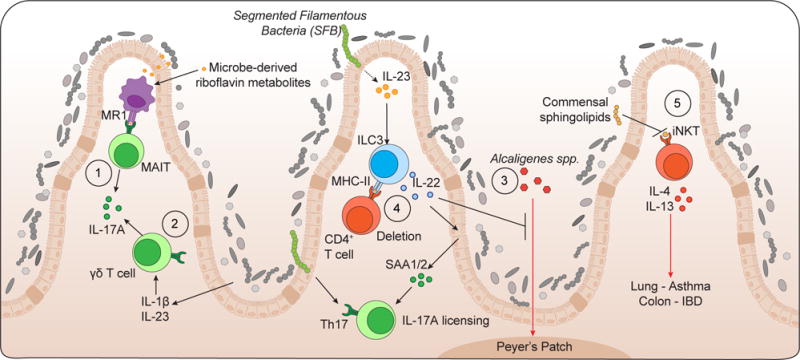
(1) Microbe-derived riboflavin metabolites promote development of mucosal-associated invariant T (MAIT) cells.
(2) Commensal-induced cytokines IL-1β and IL-23 promote IL-17A production from γδT cells.
(3) ILC3-derived IL-22 prevents translocation of Alcaligenes spp. to lymphoid architecture and promotes epithelial cell production of serum amyloid A (SAA)1/2 to license IL-17A production from Th17 cells.
(4) ILC3-expressing MHC-II delete activated commensal-specific CD4+ T cells.
(5) Early life colonization limits iNKT expansion and pathogenic function within the colonic lamina propria and lungs.
ILC are enriched at mucosal and barrier sites (Tait Wojno and Artis, 2016). While several subsets of ILC have been identified, particular attention has been given to ILC3 and their interaction with the microbiota. Among other factors, these cells produce IL-22, a cytokine of central importance in the maintenance of tissue immunity and physiology via its pleiotropic action in promoting antimicrobial peptide production, enhancing epithelial regeneration, increasing mucus production and regulating wound repair (Kilian et al., 2017). Production of IL-22 by ILC can promote the containment of specific members of the microbiota community residing in mucosal lymphoid structures, such as bacteria of the Alcaligenes genus (Qiu et al., 2013; Sonnenberg et al., 2012). Although some reports propose that the development of these cells is independent of signals derived from the microbiota, their phenotype and functional capacity can evolve to accommodate physiologic alterations in the intestinal environment (Gury-BenAri et al., 2016; Satoh-Takayama et al., 2008). How the microbiota affect the turnover of ILC3 remains unclear but recent evidence support the idea that defined commensals may preferentially impact this subset. For instance, SFB colonization can promote IL-22 production by ILC3 in an IL-23 dependent manner (Atarashi et al., 2015; Sano et al., 2015). ILC3 have also been proposed to regulate immune reactivity to the microbiota as MHC-II expressing ILC3 can interact with and delete CD4+ T cells specific for commensal antigens (Hepworth et al., 2015; Hepworth et al., 2013). The conditions in which ILC3 promotes commensal specific responses (SFB) versus regulate them (deletion) remains unclear at this stage.
γδT cells are highly represented at sites highly exposed to microbial products or metabolites, such as the liver (Kabelitz and Dechanet-Merville, 2015). The nature of the antigens recognized by γδT cells remains largely unknown, but these cells are believed to have a prominent role in recognition of lipid antigens, a point of particular relevance for potential recognition of commensal derived products. In addition to TCR-mediated responses, dermal γδT cells can also be directly activated in response to cytokines such as IL-1 and IL-23 (Gray et al., 2011; Sumaria et al., 2011) that can be both promoted by the action of the microbiota on tissues. Indeed, microbiota colonization drives the expansion of γδT cells at barrier sites in an IL-1 and/or IL23 dependent manner (Duan et al., 2010; Naik et al., 2012; Paget et al., 2015). A recent report also links the ability of the microbiota to maintain the homeostasis of liver-resident γδT-17 cells via lipid antigen/CD1d-dependent pathways (Li et al., 2017).
Non-polymorphic MHC representatives, referred to as non-classical MHC exist for both class I and class II proteins, the most diverse being those related to the MHC class I molecules (Adams and Luoma, 2013). The ability of non-classical MHC molecules to present antigens with specific chemical modifications or amino acid motifs, which can be derived from a large constituency of the microbiota, place them as likely candidates for the constitutive sensing and recognition of microbiota-derived antigens or metabolites. MAIT cells are a population restricted by the non-polymorphic and highly evolutionarily conserved MHC-Ib molecule, MHC class I-related protein 1 (MR1) (Adams and Luoma, 2013). These cells are broadly reactive to bacterial and yeast species, because of their ability to recognize metabolic intermediates of the microbial riboflavin synthesis pathway (Kjer-Nielsen et al., 2012). The development of MAIT cells is highly dependent on the microbiota as these cells are absent in germ free mice (Treiner et al., 2003).
A dialogue between non-classical lymphocytes and the microbiota can also impede accumulation of potentially pathogenic effectors. Invariant natural killer T (NKT) cells recognize endogenous and microbe-derived lipid antigens in the context of CD1d, a MHC class-I like molecule. Notably, commensal colonization at a critical window in early life restricted invariant NKT cell accumulation in the lung and colonic lamina propria. Direct recognition of microbe-derived sphingolipids altered the long-term susceptibility to develop iNKT-mediated inflammatory disorders at these barrier tissues (An et al., 2014; Olszak et al., 2012). As previously discussed, maternal microbial colonization during gestation increases ILC3 population size in offspring (Gomez de Aguero et al., 2016) further supporting the idea that importance of non-classical lymphocytes in mediating interaction with the microbiota may be more relevant to early life than in adults, in which adaptive immunity is likely to dominate in controlling host microbiota interactions.
Adaptive immunity to the microbiota
Our understanding of the complex interactions between the gut microbiota and the host immune system has challenged the original perception that, under steady state conditions, microbes composing the microbiota were ignored by the adaptive immune system. One defining characteristic of adaptive responses to the microbiota is that their development and establishment occurs in the complete absence of inflammation, a process that could be referred to as ‘homeostatic immunity’. These adaptive responses are fundamental in containing the microbiota in its physiological niche and highly entwined with tissue physiology contributing to the maintenance of tissue integrity and regulation as a whole.
The most studied form of homeostatic immunity to the microbiota is that associated with IgA responses (Figure 4). IgA (maternal IgA, innate and adaptive IgA) is the most abundant secreted isotype in mammals and play a fundamental role in shaping early interactions with the microbiota as well as in maintaining microbiota diversity and compartmentalization through life (Kawamoto et al., 2014; Sutherland et al., 2016). Secretory IgA can be produced in both T independent (referred to as innate) and T cell dependent manners (referred to as adaptive). While innate IgA can exclude microorganisms from the gut, those are not as potent as adaptive IgA in shaping mutualistic relationship with the microbiota (Kawamoto et al., 2014; Sutherland et al., 2016).
Figure 4. Commensal colonization promotes effector and regulatory T cell responses.
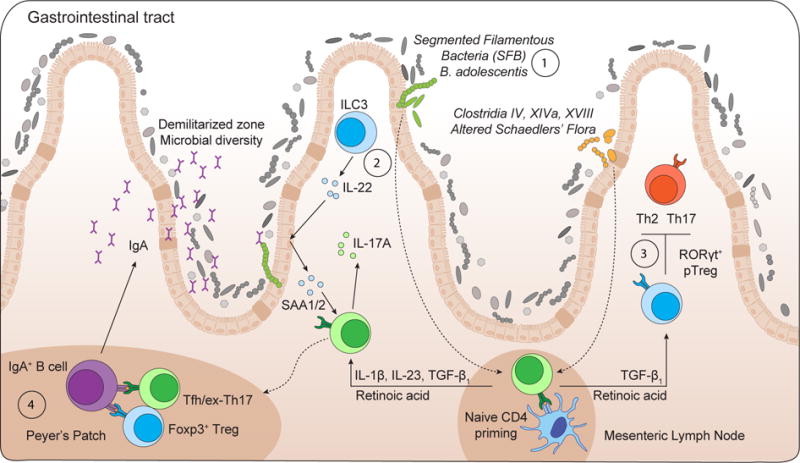
(1) Intestinal colonization by Segmented Filamentous Bacteria (SFB) or Bifidobacterium adolescentis promotes local Th17 cell differentiation.
(2) SFB colonization drives an ILC3/IL-22/SAA1/2 axis that licenses IL-17A production from ROR0γt+ Th17 cells within the terminal ileum.
(3) Clostridia colonization promotes RORγt+ Foxp3+ pTreg cell accumulation, which in turn, limit colonic Th2 and Th17 cell responses.
(4) Foxp3+ Treg cells and ex-Th17/Tfh cells localize in the Peyer’s patches and promote B cell class-switch and production of IgA, which fosters a diverse microbiota and ensures commensal compartmentalization from the intestinal epithelium.
Commensal-specific IgA responses are produced with the help of intestinal dendritic cells able to sample commensals associated with the intestinal epithelium and interact with B and T cells in the Peyer’s patches to produce IgA specific for commensal-derived antigens (Macpherson and Uhr, 2004). Within germinal centers of the Peyer’s patch, B cells preferentially undergo class-switch from IgM production to that of IgA. This preferential production appears due to the unique microenvironment of the Peyer’s patch, with enrichment of chemokines, cytokines and dietary factors (retinoic acid) required for the survival and proliferation of recently class-switched B cells (Haynes et al., 2007; Suzuki et al., 2010; Wang et al., 2011).
Antigen presenting cells from the lamina propria loaded with microbial antigens may also traffic to the mesenteric lymph node, but do not circulate further, allowing the induction of a mucosal compartmentalized IgA response (Macpherson and Uhr, 2004). Within the intestinal lamina propria, B cells secrete soluble IgA, which are subsequently transcytosed across epithelial cells (Sutherland et al., 2016) thereby controlling host-commensal interaction by both impacting commensal gene expression (Peterson et al., 2007) and preventing adhesion of commensal bacteria to the epithelial surfaces (Boullier et al., 2009; Fernandez et al., 2003; Hornquist and Lycke, 1995; Martinoli et al., 2007; Wade and Wade, 2008; Wei et al., 2011). Of note, as previously discussed, various modules of homeostatic immunity to the microbiota reinforce each other’s function. For instance, Foxp3+ Treg cells contribute to the diversification of gut microbiota via suppression of inflammation and regulation of IgA selection in Peyer’s patches (Kawamoto et al., 2014). The microbiota also promotes B cell receptor editing within the lamina propria upon colonization (Wesemann et al., 2013). Diversified and selected IgA contribute to the maintenance of a balanced microbiota, which in turn promote the expansion of Treg cells, induction of germinal centers, and IgA responses in the gut in a feed forward regulatory loop (Kawamoto et al., 2014). One remarkable feature of mucosal IgA responses is that these cells lack classical memory characteristics and are able to respond to flux in commensal microbiota composition allowing the mucosal immune system to respond to a changing microbiota (Hapfelmeier et al., 2010).
A high frequency of effector and memory T cells reside in tissues that are constitutively colonized by commensals. Notably, at steady state, most Th17 and Th1 cells are found at barrier sites such as the gastrointestinal (GI) tract or the skin and develop in response to cues derived from the microbiota (Figure 4). In germ-free mice, or mice treated with broad-spectrum antibiotics, Th17 cell frequencies are severely reduced within the gut-associated lymphoid tissue (GALT) or within the skin compartment (Atarashi et al., 2008; Hall et al., 2008; Ivanov et al., 2008; Naik et al., 2012). In a manner similar to IgA responses, T cell responses to the microbiota may not be long lived. An evolving pool of specificities within the T cell compartment may allow for the maintenance of tolerance and barrier function in the face of variable commensal populations. Of note, despite the extraordinary number of potential antigens expressed by the microbiota, only a handful of microbiota derived antigens have been identified thus far (Cong et al., 2009; Yang et al., 2014). Th17 cells play an important role in the maintenance of tissue physiology. The signature cytokines of Th17 cells, IL-17A, IL-17F and IL-22 can promote the production of antimicrobial peptides by epithelial cells and reinforce epithelial cell tight junctions (Weaver et al., 2013). Furthermore, Th17 cells can also promote IgA production within the gut associated lymphoid tissue (GALT) (Hirota et al., 2013). Failure to maintain the Th17 cell lineage in the gut, as observed during HIV or SIV infection is associated with microbial translocation (Klatt et al., 2013).
In addition to the pleiotropic effects of conserved microbial ligands or metabolites in the education and function of the immune system, it is now becoming clear that defined microbes or groups of bacteria can dominantly influence the immune system under steady state. In the language of ecological systems these organisms with paramount effect are termed ‘keystone species’. Segmented Filamentous Bacteria (SFB) represent a prototype of a keystone species in the GI tract. This spore-forming Gram-positive anaerobic bacteria colonizes the terminal ileum of mice and has a dominant effect on the mucosal immune system by promoting the accumulation of both Th17 and Th1 cells in the small intestine and driving the production of IgA (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009; Talham et al., 1999; Umesaki et al., 1999). The host mount a potent SFB-specific Th17 response, a phenomenon that has been associated both with its ability promote protection against gastrointestinal pathogens (Ivanov et al., 2009), but also mediate profound systemic effects in the context of inflammation (Lee et al., 2011; Wu et al., 2010; Yang et al., 2014). In contrast to most commensals that reside outside of the “demilitarized zone”, SFB interacts closely with the mucosal tissue via tight adhesion to Peyer’s Patches and epithelial cells, inducing cytoskeletal reorganization in these cells at the site of contact (Ivanov et al., 2008; Sczesnak et al., 2011). This intimate contact with epithelial cells, a property shared by a minority of commensal organisms, is believed to account for the ability of defined microbes to heighten tissue immunity (Atarashi et al., 2015; Ivanov et al., 2009), indeed mutant strains of Citrobacter rodentium and E. coli defective in adhesion also failed to induce these responses (Atarashi et al., 2015). A recent study identified Bifidobacterium adolescentis, a human symbiont, as a microbe that can also promote Th17 cell accumulation in intestine of monocolonized mice (Tan et al., 2016). Of note, B. adolescentis elicited a transcriptional program distinct from that of SFB, supporting the idea that the induction of Th17 cells, a subset of lymphocyte central to the homeostasis of barrier sites, can occur via distinct and/or overlapping routes (Tan et al., 2016). The skin microbiota also controls the accumulation of IL-17 producing T cells within the skin compartment (Naik et al., 2012). As for the gut, defined skin microbes display a high degree of specialization in their capacity to modulate distinct branches of the adaptive immune system. Colonization of mice with defined isolates of S. epidermidis, but not other members of the human or mouse microbiota, induces S. epidermidis specific IL-17A+ CD8+ T cells that home to the epidermis (Naik et al., 2015). In this compartment, S. epidermidis-specific T cells, via their ability to produce IL-17, promote AMP production by keratinocytes thereby promoting heterologous protection against fungal infections (Naik et al., 2015).
How tissue resident dendritic cell positioning and specialization could account for the capacity of the host to respond and regulate defined aspects of its relationship with the microbiota and in particular adaptive T cell responses to commensals or mutualistic microbes has only begun to be addressed. While the Peyer’s patches are equipped with cells specialized in the capture of microbes, how microbes and/or microbial antigens are captured in the lamina propria remains incompletely understood. This process is believed to result, at least in part, from direct capture of microbes by DC (Chieppa et al., 2006; Niess et al., 2005; Rescigno et al., 2001) but could also result from capture of microbial outer membrane vesicles (OMVs) (Shen et al., 2012). While in the gut Th17 have been proposed to depend upon DC co-expressing CD11b and CD103 (dependent on Notch2 and IRF4) (Lewis et al., 2011; Persson et al., 2013; Schlitzer et al., 2013), induction of SFB specific Th17 cells depends on gut resident CX3CR1 expressing cells (Panea et al., 2015). Cognate responses to S. epidermidis in the skin depend exclusively on tissue resident antigen presenting cells with CD103+DCs required for the induction of IL-17+CD8+T cells responses against S. epidermidis and production of IL-17 by these cells licensed within the skin by IL-1 producing CD11b+DCs (Naik et al., 2015). While much more remains to be learned about these processes, we could speculate that the ability of resident microbes to induce homeostatic effector T cell responses may perhaps rely on their capacity to exclusively utilize the endogenous network of APCs without active recruitment of new APC populations.
The presence of defined microbes with a keystone role on the immune system raises an intriguing possibility. Although optimal control of host metabolism and physiology may rely on complex and redundant populations of microbes, we could speculate that a more limited number of microbes may act as dominant adjuvants of immune responses. Under steady state conditions these ambivalent members of the microbiota such as E. coli, SFB or Tritrichomonas are maintained in check by the immune system but could be co-opted by the host to control invasive microbes.
While the differentiation of lymphocytes can occur in the absence of live microbes, the acquisition of effector function in the tissue is under the tight control of local cues provided by the action of resident microbes (Figure 4). This functional licensing by the microbiota promote its function as a potent tissue rheostat. Within the skin, the ability of the microbiota to promote local production of IL-1α is associated with the licensing of cutaneous αβ and γδT cells to release cytokines (Naik et al., 2012). Within the gut, RORγt-expressing CD4+ T cells specific for SFB accumulate within the mucosa upon SFB colonization, but robust IL-17A production is restricted to the ileum, where SFB makes direct contact with the epithelium. Such contact induces epithelial-cell production of serum amyloid A proteins 1 and 2 (SAA1/2), which promote local IL-17A production by RORγt-expressing T cells (Sano et al., 2015). Serum amyloid A is also a carrier of both high-density lipoprotein and retinol (Derebe et al., 2014), and as such can potentially deliver these molecules to antigen presenting cells providing another amplification mechanism for the induction of IL-17 responses within tissues. The ability of commensals to control tissue immunity may also have consequences for the maintenance of memory responses to pathogens or vaccines. For instance, the tonic action of the microbiota on tissue immunity could promote the formation of a niche within the tissue microenvironment that results in the enhanced recruitment, survival and maturation of both T and B cells. Absence of commensals is associated with decreased levels of T cell chemoattractants, as well as the survival factors IL-7 and IL-15 (Fink et al., 2012; Jiang et al., 2013; Vonarbourg et al., 2010). Another systemic control mediated by the microbiota occurs via the manipulation of the metabolic landscape. For instance, fatty acids that are regulated by gut commensals are also important in the development, survival and function of memory T cells (Cui et al., 2015; Martin et al., 2007; Pearce et al., 2009; van der Windt et al., 2013). Some of the local stimulatory effects of the microbiota can be attributed as previously discussed to dominant microbial derived signals such as ligands for TLRs or NODs. For instances, dendritic cells that reside in the lamina propria of the intestine are poised to respond to flagellin by rapidly expressing chemokines, antimicrobial peptides and cytokines involved in the initiation of immune responses (Kinnebrew et al., 2012; Uematsu et al., 2008; Uematsu et al., 2006) and Unmethylated cytosine phosphate guanosine (CpG) dinucleotides, which are abundant within the prokaryotic DNA of intestinal flora, can contribute to intestinal homeostasis under steady state conditions (Hall et al., 2008).
Although most of what is known today about the cross talk between the immune system and microbes arise from the exploration of the bacterial component of the flora, other microbes such as fungi, virus or protozoans can control host immunity. For instance, a common enteric RNA virus can replace the immune-promoting function of commensal bacteria in the intestine (Kernbauer et al., 2014). Mono-colonization of germ-free mice with Murine norovirus (MNV) also suppressed the expansion of group 2 innate lymphoid cells observed in the absence of bacteria and promote type I interferon (IFN) signature (Kernbauer et al., 2014). The protozoan Tritrichomonas musculis, a common member of the murine microbiota, activates the host epithelial inflammasome to induce IL-18 release (Chudnovskiy et al., 2016). Upon T. musculis colonization, epithelial-derived IL-18 promotes dendritic cell-driven Th1 and Th17 immunity thereby altering the capacity of the tissue to respond to infections (Chudnovskiy et al., 2016). Tuft cells, which are taste-chemosensory epithelial cells, accumulate during colonization with a commensal protozoan or a nematode and represent the primary source of interleukin-25 post colonization, which indirectly induces tuft cell expansion (Howitt et al., 2016; von Moltke et al., 2016).
Control of immune reactivity to the microbiota
Maintenance of tissue homeostasis is imperative to host survival. This fundamental process relies on a complex and coordinated set of innate and adaptive responses that calibrates responses against self, food, commensals and pathogens. To this end, specialized populations of cells have to integrate local cues such as defined metabolites, cytokines, or hormones allowing the induction and contraction of responses in a way that preserve the physiological and functional requirements of each tissue.
In the gut, failure to regulate the formidable challenge represented by the exposure to the microbiota, food derived antigens, metabolites and pathogens can lead to severe pathological outcomes ranging from Inflammatory Bowel Diseases (IBD) to metabolic syndromes (Blumberg and Powrie, 2012). Regulatory T cells play a dominant role in the control of this complex equilibrium. These cells maintain both peripheral and mucosal homeostasis throughout the lifespan of the host and disruption of the homeostasis of these cells in the GI tract results in loss of oral tolerance and development of aberrant effector responses in the gut (Josefowicz et al., 2012b; Mucida et al., 2005; Worbs et al., 2006). Although Foxp3+ Treg can arise as differentiated cells in the thymus (tTreg), the gut-associated lymphoid tissue is a privileged site for the peripheral induction of Treg cells in response to oral antigens (Coombes et al., 2007; Mucida et al., 2005; Sun et al., 2007) (Figure 4). A proportion of peripherally-induced Treg (pTreg) in the colonic tissue have been shown to be specific for antigens derived from the commensal microbiota (Lathrop et al., 2011) and optimal maintenance of tolerance to commensal and environmental antigens is believed to require the combined effect of both tTreg and pTreg (Cebula et al., 2013; Josefowicz et al., 2012a; Lathrop et al., 2011). The specialized property of the gut-associated lymphoid tissue to induce Treg can be at least in part explained by the presence of defined populations of mucosal antigen-presenting cells, and in particular those expressing CD103, which are endowed with the capacity to produce factors involved in the induction of Treg such as the cytokine TGF-β and the vitamin A metabolite retinoic acid (RA)(Coombes et al., 2007; Esterhazy et al., 2016; Loschko et al., 2016; Mucida et al., 2007; Sun et al., 2007).
Peripherally-induced Treg are enriched in the colon and a fraction express RORγt under the control of the microbiota (Ohnmacht et al., 2015; Sefik et al., 2015; Yang et al., 2016). A large proportion of these RORγt+ Treg cells express IL-10 and high levels of CTLA-4 and can efficiently restrain aberrant type 2 or type 17 responses (Ohnmacht et al., 2015; Sefik et al., 2015; Yang et al., 2016). Within the gut compartment, another fraction of Treg cell express the transcription factor GATA3 (independently of the microbiota) that upregulates the expression of Foxp3 and IL-33R/ST2 in a feed forward manner that promotes Treg cell fitness (Schiering et al., 2014; Wohlfert et al., 2011). The remaining Treg cells are unaffected by the absence of gut microbiota, but are absent in germ free mice fed an antigen free diet (Kim et al., 2016). The relative contribution of these various flavors of Treg cell subsets to the maintenance of tolerance to the microbiota remains unclear, but is likely to be highly contextual and dependent on the microbial community, host developmental stage and the inflammatory status of the host.
The skin contains one of the highest frequencies of Foxp3+ Treg cells within the body (Belkaid, 2002; Suffia et al., 2006); in mice with defects in the Foxp3 gene, such as scurfy mice, this compartment is one of the primary targets of severe inflammation. In the skin of both mice and human, Foxp3+ Treg cells reside in the dermis and a large fraction of these cells can be found in close proximity to hair follicles (Sanchez Rodriguez et al., 2014), appendage structures that serve as a natural habitat for skin resident microbes. Of interest, the skin of neonates is populated by a wave of activated Treg cells at a time that coincides with post-natal hair-follicle morphogenesis (Scharschmidt et al., 2015). This observation raises the possibility that early occupation of the hair follicle by microbes may be coupled with the induction of regulatory responses aimed at limiting aberrant reactivity against these constitutive partners. Indeed, association of S. epidermidis to neonate (but not adult) skin is associated with the induction of S. epidermidis-specific Foxp3+ Treg cells that subsequently limit aberrant responses against this microbe in the context of tissue damage (Scharschmidt et al., 2015).
Induction of regulatory responses by the microbiota
Commensals themselves control the induction of regulatory responses. Notably, the establishment of oral tolerance – the active suppression of inflammatory responses to food and other orally ingested antigens – cannot be induced in the absence of gut flora derived signals (Kiyono et al., 1982; Sudo et al., 1997; Wannemuehler et al., 1982; Weiner et al., 2011). Although the control of immunological tolerance by commensals is likely to be achieved via multiple and redundant mechanisms (Weiner et al., 2011), most of what we understand today about the impact of commensals on regulatory responses relates to their impact on Foxp3+ Treg cells. Notably, peripherally-induced Treg cells are reduced under germ free conditions (Geuking et al., 2011; Ohnmacht et al., 2015; Sefik et al., 2015; Yang et al., 2016). Furthermore, commensals can also control luminal antigen sampling by mucosal DCs and promote the induction of lamina propria resident macrophages associated with local expansion of Treg cells (Chieppa et al., 2006) (Niess and Adler, 2010). The ability of gut resident macrophages to sense the microbiota is also associated with IL-1β release that in turn promotes the accumulation of Th17 cells (Shaw et al., 2012) and production of GM-CSF by ILC3 (Mortha et al., 2014). Such GM-CSF production reinforces the ability of mucosal antigen presenting cell to produce both IL-10 and retinoic acid, thereby promoting local immune regulation and Treg cell induction (Mortha et al., 2014).
The first demonstration that a single molecule derived from a microbial symbiont could promote regulatory responses was provided by the identification of the polysaccharide A (PSA), produced by Bacteroides fragilis (Mazmanian et al., 2005). B. fragilis, via PSA expression, can protect mice from experimental colitis induced by Helicobacter hepaticus (Mazmanian et al., 2008). This protective activity was associated with the capacity of PSA to induce and expand IL-10 producing Treg cells via interaction of PSA with TLR2 (Mazmanian et al., 2008; O’Mahony et al., 2008; Ochoa-Reparaz et al., 2010; Round et al., 2011). The discovery of a link between defined members of the microbiota and the induction of regulatory cells able to limit mucosal inflammation and promote tolerance led to a rational approach for the identification of the next generation of probiotics with superior capacity to induce Treg cells.
Oral administration of a mixture of 46 strains of Clostridia derived from conventional mice or a mixture of 17 stains isolated from a healthy human induces Treg cell in the colon of mice (Atarashi et al., 2013; Atarashi et al., 2011). Optimal induction of Treg relies on the synergistic effect of this consortium while individual species have a modest effect on the immune system (Atarashi et al., 2013; Atarashi et al., 2011). Notably, strains that fall within cluster IV, XIVa and XVIII of Clostridia promote the accumulation of Treg cells in the colon characterized by high levels of CTLA-4 and IL-10 and preferential expression of RORγt (Atarashi et al., 2013; Atarashi et al., 2011; Ohnmacht et al., 2015). The mechanism by which these Clostridia strains promote regulatory response remains unclear but is proposed to rely on their ability to create a TGF-β rich environment and through SCFA production (Atarashi et al., 2011; Narushima et al., 2014). Notably, SCFA regulate the size and function of the regulatory T cell network by promoting the induction and fitness of regulatory T cells in the colonic environment (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013). The action of SCFA on Treg cell induction appears to occur both through HDAC inhibition and Gpr43 activation on both T cells and dendritic cells (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013). Several other microbes have shown to increase the frequency of Treg cells in the colon (Faith et al., 2014; Geuking et al., 2011). Altogether these findings reveal a major role for the microbiota in shaping the repertoire, number and activation of Treg cells and in the maintenance of host-microbe mutualism at barrier sites.
It is unclear to what extent the microbiota resident at other barrier surfaces such as the skin or the lung can also contribute to the establishment of a milieu compatible with the induction of regulatory responses. Neonatal colonization of the skin with S. epidermidis can promote accumulation of commensal-specific Foxp3+ Treg cells (Scharschmidt et al., 2015). However, skin microbes are not essential for the seeding of dermal Treg cells (Naik et al., 2012), supporting the idea that while commensals may control Treg specificities they are not required for the establishment their niche. How commensal colonization affects the repertoire, function and tTreg/pTreg ratio of cutaneous Treg cells remains unclear. However, cutaneous microbes may also be able to promote regulatory responses in adult mice, as exposure to a lysate of Vitreoscilla filiformis can promote the accumulation of Treg cells within the skin (Volz et al., 2014). Further, secreted products from S. epidermidis promotes IL-10 production by human dendritic cells (Laborel-Preneron et al., 2015) and skin exposure to TLR2-6 binding S. aureus derived lipopeptides can potently suppress immune responses via the induction of regulatory myeloid cells (Skabytska et al., 2014). The microbiota can also limit Thymic strmal lymphopoietin (TSLP) expression in mice with a defective skin barrier (Yockey et al., 2013). Based on the fundamental role of Treg cells and regulatory network in maintaining tissue homeostasis, it is likely that a large fraction of any given microbiota or products at all barrier sites may have evolved to favor regulatory responses.
Commensal derived products can also act by controlling directly or indirectly the function of inflammatory cells. For example, recognition of the commensal derived metabolites SCFA by innate immune cells is critical for the regulation of inflammation (Maslowski et al., 2009). Further, during acute mucosal infection, encounter of inflammatory monocytes with the microbiota in the gastrointestinal tract promotes their production of the lipid mediator PGE2 that in turn limits the level of activation of tissue damaging neutrophils (Grainger et al., 2013). Although most of what is known today about the regulatory properties of the microbiota arise from the exploration of the bacterial component of the flora, other microbes such as fungi and virus are likely to promote similar or complementary aspects of the regulatory network. In the gastrointestinal tract, experimentally, interaction of commensal fungi with the C-type lectin receptor Dectin-1 was able to prevent inflammation in the context of acute mucosal injury (Iliev et al., 2012). Much remains to be learned about the molecular basis of host-microbe interactions within individual barrier sites and the overall dynamic of effector versus regulatory responses required to maintain or restore host-microbiota homeostasis.
Adjuvant properties of the microbiota: role in infection, vaccine and tumor immunotherapy
The ability of the microbiota to control all aspects of immunity ranging from its development to its fine tuning within tissue makes these partners remarkable allies in the control of infections caused by acquired pathogens or by members of the resident microbiota themselves. Tissues that are natural habitats of the microbiota such as the skin, the GI tract or the lung are also the portals by which pathogens access the host and often the primary site of infections. This implies that the initial encounter of pathogens with the immune system occurs in an environment conditioned and regulated by its endogenous microbiota. Members of the microbiota can directly and dynamically interact with pathogens (or contextual pathogens) and immune cells and the results of this interaction can define the pathogenesis and outcome of a given infection. Early studies have identified significant impairment of host immune responses, and in particular Th17 and Th1 cell responses, to pathogens in mice treated with antibiotics or raised under germ free conditions (Cebra, 1999; Hall et al., 2008; Ivanov et al., 2009; Mazmanian et al., 2005; Naik et al., 2012). The adjuvant effect of the microbiota allows the control of parasitic, viral or bacterial infection (Hall et al., 2008; Ivanov et al., 2009) not only at the site colonized by microbes but also distally via the ability of the microbiota to calibrate systemic immunity. For example, the microbiota prime intestinal resident macrophages for rapid IL-1-β activation (Franchi et al., 2012). Antibiotic treatment also impaired adaptive and innate responses following exposure to systemic viral infections (Abt et al., 2012). Genome wide transcriptional profiling of macrophages from antibiotic treated mice revealed a broad decrease of genes associated with antiviral immunity (Abt et al., 2012). Commensal derived peptidoglycan found in the serum can also improve the killing of Streptococcus pneumonia and Staphylococcus aureus by bone-marrow derived neutrophils in a NOD1 dependent manner (Clarke et al., 2010a). Constitutive antibody responses to the microbiota not only provide protection against gut microbiota translocation but also against infections with unrelated pathogenic bacteria via recognition of conserved outer-membrane molecules (Zeng et al., 2016). In this context, commensal specific antibodies have been shown to also cross-react with HIV-1 (Jeffries et al., 2016; Trama et al., 2014), supporting the idea that pre-existing commensal-specific antibodies could influence responses to a diverse range of microbes. The microbiota may also control the tissue milieu via the maintenance of a pool of commensal reactive cells endowed with the ability to provide a heterologous adjuvant effect. Indeed, previous infection and transient breach in barrier can lead to microbial translocation, a phenomenon that can lead to the induction of commensal-specific T cells with a heightened inflammatory potential (Hand et al., 2012). Because of the extraordinary number of antigens expressed by the host microbiota, primary exposure to a pathogen is likely to occur in the context of a much broader recall response against commensal bacteria.
A protective effect for the microbiota has also been revealed in clinical and experimental settings in which broad antibiotic treatment allow the domination of intestinal microbiota by drug resistant microbes such as Vancomycin Resistant Enterococcus (VRE) or Clostirudium difficile (Buffie and Pamer, 2013; Murray, 2000). Infections caused by multidrug-resistant organisms are on the rise and have developed into endemic and epidemic situations worldwide (Gupta et al., 2011). Harnessing the microbiota to combat these infections represents an important therapeutic avenue with the most spectacular results obtained thus far in the context of Clostridium difficile colitis (van Nood et al., 2013). During this recurrent infection, transfer of a microbiota from healthy donors eradicated the infection with a remarkable efficiency (van Nood et al., 2013). Subsequent work has utilized rational approaches to develop cocktails of microbes endowed with superior ability to compete with these invasive species (Buffie et al., 2015). While colonization resistance plays an important role in the control of these pathogens, the adjuvant effect of the microbiota on the immune system also contributes to the control of these life threatening microbes (Pamer, 2016).
Antibiotic treatment abrogates Th1 and Th17 response to oral vaccine in mice (Hall et al., 2008). Further, optimal antibody responses to the seasonal trivalent influenza vaccine (TIV) vaccine, as well as to the polio vaccine (IPOL) require the presence of gut commensals (Oh et al., 2014). Recent studies have suggested that the gut microbiota could influence vaccine efficacy in human and non-human primates (Valdez et al., 2014). For instance, a study using Cynomolgus macaques supports the idea that a stable and more diverse gut microbiota correlates with a better response to vaccination and subsequent immunity (Seekatz et al., 2013). This finding is consistent with another study involving a small human cohort, in which the majority of individuals that responded to vaccination had greater community richness and diversity amongst their gut microbiota (Eloe-Fadrosh et al., 2013). While these studies remain limited in number and in scope, their findings raise some intriguing possibilities. In the context of future vaccine trials, it may be important to stratify individuals based on their microbiota profile and more importantly, microbiota metabolism as well as host genetic influences. Such lines of research should provide insight into the link between host genetic variation in shaping both immunity and the composition of the human microbiome (Blekhman et al., 2015; Goodrich et al., 2014), and provide a starting point towards understanding factors that predict vaccine success.
Recent evidence implies that the capacity of commensals to calibrate systemic immunity has profound consequences in the context of tumor therapy. Total body irradiation, used in defined settings of immunotherapy and bone marrow transplantation, is associated with gut damage and microbial translocation, providing an adjuvant effect to the anti-tumoral T cells (Paulos et al., 2007). Cyclophosphamide (CTX), a clinically important cancer drug, also leads to intestinal damage, microbial translocation and subsequent induction of Th17, Th1 and γδT responses that, collectively, contribute to the anti-tumoral response (Daillere et al., 2016; Viaud et al., 2013). During treatment, translocation of Enterococcus hirae increases the intratumoral CD8/Treg ratio while Barnesiella intestinihominis accumulates in the colon and promotes tumor infiltration of IFN-γ-producing γδT (Daillere et al., 2016). Th1 cell memory responses against these bacteria predicted longer progression-free survival in advanced lung and ovarian cancer patients treated with chemo-immunotherapy (Daillere et al., 2016). Disruption of the gut flora via antibiotic treatment or in germ free mice also impairs the capacity of the host to control subcutaneous tumors during immunotherapy (Iida et al., 2013). In these experimental settings, the protective effect results from the capacity of the commensal derived ligands to control the status of activation of tumor myeloid cells and more particularly their level of TNF-α and reactive oxygen species both associated with optimal tumor control (Iida et al., 2013). Remarkably, tumor control was also associated with the presence of defined commensal species such as Alistipes shahii (Iida et al., 2013).
Cancer therapy has been transformed by the advent of immune checkpoint blockade, which targets regulatory pathways in T cells to enhance antitumor immune responses. Remarkably, experimental and clinical evidence support a role for the microbiota in controlling response to treatment. Experimentally, the antitumor effects of CTLA-4 blockade depend on distinct Bacteroides species (Vetizou et al., 2015). In mice and patients, T cell responses specific for B. thetaiotaomicron or B. fragilis are associated with the efficacy of CTLA-4 blockade (Vetizou et al., 2015). Fecal microbial transplantation from humans to mice confirmed that treatment of melanoma patients with antibodies against CTLA-4 favored the outgrowth of B. fragilis with anti-cancer properties (Vetizou et al., 2015). In mice, Bifidobacterium alone improved melanoma control to the same degree as programmed cell death protein 1 ligand 1 (PD-L1)-specific antibody therapy, and combination treatment nearly abolished tumor outgrowth. This effect was associated with dendritic cell activation leading to enhanced CD8+ T cell priming and accumulation in the tumor microenvironment (Sivan et al., 2015). Thus, commensals, and more particularly defined member of the microbiota, can control various aspects of immunity associated with anti-tumoral responses, a phenomenon that has profound clinical implications.
Concluding remarks
As discussed in this review, all aspect of immunity can be directly or indirectly controlled by the microbiota. However much remains to be learned about the mechanisms utilized by these microbes in the control of local and systemic immunity. Further, how communities of microbes, including those involving virus, fungi and protozoan cooperate and influence each other’s to sustain immune health remains virtually unknown. Much also remains to be learned about the quality of homeostatic immunity to the microbiota. As discussed in this review, this class of immunity appears to be orchestrated by self-reinforcing modules of innate and adaptive immunity and aimed at reinforcing microbiota containment, barrier immunity, and tissue repair in a manner uncoupled from inflammation. A complex regulation that remains to be explored via the integration of ecology, genomic, microbiology and immunological approaches.
While the identification of novel molecular determinants and the underlying mechanisms of microbiota host interaction remains in its infancy, such lines of investigation bear enormous potential. Because the microbiota has co-evolved with its host to finely tune the unique requirements of the gut, these microbes may produce highly adapted tissue-specific adjuvants. Uncovering these pathways and the microbiota-derived molecules (Donia and Fischbach, 2015; Medema and Fischbach, 2015) involved in these processes may allow for the development of novel classes of adjuvants with potent potential for the control of infection as well as for the promotion of anti-tumoral responses. Further, manipulation of microbe function or composition, via diet alteration or microbiota-engraftment, may soon become a viable and therapeutic approach to enhance the efficacy of cancer therapy or to combat life threatening antibiotic-resistant pathogens. Understanding the factors controlling the fundamental dialogue between microbes and the immune system and gradual integration of this dialogue with the nervous and hormonal system will represent an important challenge for the scientific community but also a necessary step in our quest for the next generation of therapeutics.
Acknowledgments
We would like to apologize to our colleagues whose work we could not cite because of space constrains. This work was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases (NIAID). We thank Jacqueline Kehr, Dr. Nicholas Collins and Dr. Seong-Ji Han for editorial comments and help with figure design.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013;31:529–561. doi: 10.1146/annurev-immunol-032712-095912. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, Deroos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013 doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Frontiers in immunology. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer ML, Schurch CM, Saito Y, Geuking MB, Li H, Cuenca M, Kovtonyuk LV, McCoy KD, Hapfelmeier S, Ochsenbein AF, et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol. 2014a;193:5273–5283. doi: 10.4049/jimmunol.1400762. [DOI] [PubMed] [Google Scholar]
- Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, Grieco A, Van Vlierberghe H, Fahrner R, Patuto N, et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014b;6:237ra266. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nature immunology. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Piccirilo AC, Mendez S, Shevack E, Sacks DL. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D, Clark AG. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boullier S, Tanguy M, Kadaoui KA, Caubet C, Sansonetti P, Corthesy B, Phalipon A. Secretory IgA-mediated neutralization of Shigella flexneri prevents intestinal tissue destruction by down-regulating inflammatory circuits. Journal of Immunology. 2009;183:5879–5885. doi: 10.4049/jimmunol.0901838. [DOI] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. The American journal of clinical nutrition. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, Bry L, Kraj P, Kisielow P, Ignatowicz L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell. 2016;167:444–456. e414. doi: 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine. 2010a;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010b;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-{beta}- and retinoic acid-dependent mechanism. J Exp Med. 2007 doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell. 2015;161:750–761. doi: 10.1016/j.cell.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CP, Flament C, Lepage P, Roberti MP, et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Derebe MG, Zlatkov CM, Gattu S, Ruhn KA, Vaishnava S, Diehl GE, MacMillan JB, Williams NS, Hooper LV. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. Elife. 2014;3:e03206. doi: 10.7554/eLife.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, Kolls JK, Weiser JN, Worthen GS. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell host & microbe. 2010;7:140–150. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloe-Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, Fraser CM. Impact of oral typhoid vaccination on the human gut microbiota and correlations with s. Typhi-specific immunological responses. PLoS One. 2013;8:e62026. doi: 10.1371/journal.pone.0062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterhazy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat Immunol. 2016;17:545–555. doi: 10.1038/ni.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MI, Pedron T, Tournebize R, Olivo-Marin JC, Sansonetti PJ, Phalipon A. Anti-inflammatory role for intracellular dimeric immunoglobulin a by neutralization of lipopolysaccharide in epithelial cells. Immunity. 2003;18:739–749. doi: 10.1016/s1074-7613(03)00122-5. [DOI] [PubMed] [Google Scholar]
- Fink LN, Metzdorff SB, Zeuthen LH, Nellemann C, Kristensen MB, Licht TR, Frokiaer H. Establishment of tolerance to commensal bacteria requires a complex microbiota and is accompanied by decreased intestinal chemokine expression. Am J Physiol Gastrointest Liver Physiol. 2012;302:G55–65. doi: 10.1152/ajpgi.00428.2010. [DOI] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Nunez G. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nature immunology. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013 doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris H, de Boer F, van der Waaij D. Myelopoiesis in experimentally contaminated specific-pathogen-free and germfree mice during oral administration of polymyxin. Infect Immun. 1985;50:437–441. doi: 10.1128/iai.50.2.437-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, Legrand F, Koo LY, Brenchley JM, Fraser ID, Belkaid Y. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat Med. 2013;19:713–721. doi: 10.1038/nm.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Suzuki K, Cyster JG. Cutting edge: Identification of a motile IL-17-producing gammadelta T cell population in the dermis. J Immunol. 2011;186:6091–6095. doi: 10.4049/jimmunol.1100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell. 2016;166:1231–1246. e1213. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA Limits Regulatory T Cell Conversion and Is a Natural Adjuvant of Intestinal Immune Responses. Immunity. 2008 doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, Maynard CL, Elson CO, 3rd, Belkaid Y. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Hornquist E, Lycke N. Cholera toxin increases T lymphocyte responses to unrelated antigens. Advances in experimental medicine and biology. 1995;371B:1507–1512. [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, Ruhn KA, Hou B, DeFranco AL, Yarovinsky F, Hooper LV. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamura C, Bouladoux N, Belkaid Y, Sher A, Jankovic D. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 2017;129:171–176. doi: 10.1182/blood-2016-06-723742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries TL, Jr, Sacha CR, Pollara J, Himes J, Jaeger FH, Dennison SM, McGuire E, Kunz E, Eudailey JA, Trama AM, et al. The function and affinity maturation of HIV-1 gp120-specific monoclonal antibodies derived from colostral B cells. Mucosal Immunol. 2016;9:414–427. doi: 10.1038/mi.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang X, Zeng B, Liu L, Tardivel A, Wei H, Han J, MacDonald HR, Tschopp J, Tian Z, Zhou R. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J Exp Med. 2013;210:2465–2476. doi: 10.1084/jem.20122490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012a;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012b;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabelitz D, Dechanet-Merville J. Editorial: “Recent Advances in Gamma/Delta T Cell Biology: New Ligands, New Functions, and New Translational Perspectives”. Frontiers in immunology. 2015;6:371. doi: 10.3389/fimmu.2015.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian E, Valentina D, Andrea C. IL-17 and IL-22 in immunity: driving protection and pathology. Eur J Immunol. 2017 doi: 10.1002/eji.201646723. [DOI] [PubMed] [Google Scholar]
- Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, Lee JY, Lee M, Surh CD. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351:858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2) Cell Host Microbe. 2014;15:95–102. doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Kiyono H, McGhee JR, Wannemuehler MJ, Michalek SM. Lack of oral tolerance in C3H/HeJ mice. The Journal of experimental medicine. 1982;155:605–610. doi: 10.1084/jem.155.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends in microbiology. 2013;21:6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborel-Preneron E, Bianchi P, Boralevi F, Lehours P, Fraysse F, Morice-Picard F, Sugai M, Sato’o Y, Badiou C, Lina G, et al. Effects of the Staphylococcus aureus and Staphylococcus epidermidis Secretomes Isolated from the Skin Microbiota of Atopic Children on CD4+ T Cell Activation. PLoS One. 2015;10:e0141067. doi: 10.1371/journal.pone.0141067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, Klinakis A, Charo IF, Jung S, Gommerman JL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Li F, Hao X, Chen Y, Bai L, Gao X, Lian Z, Wei H, Sun R, Tian Z. The microbiota maintain homeostasis of liver-resident gammadeltaT-17 cells in a lipid antigen/CD1d-dependent manner. Nature communications. 2017;7:13839. doi: 10.1038/ncomms13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Loschko J, Schreiber HA, Rieke GJ, Esterhazy D, Meredith MM, Pedicord VA, Yao KH, Caballero S, Pamer EG, Mucida D, Nussenzweig MC. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J Exp Med. 2016;213:517–534. doi: 10.1084/jem.20160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Partch CL, Lehotzky RE, Whitham CV, Chu H, Bevins CL, Gardner KH, Hooper LV. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J Biol Chem. 2009;284:4881–4888. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE. Vancomycin-resistant enterococcal infections. The New England journal of medicine. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narushima S, Sugiura Y, Oshima K, Atarashi K, Hattori M, Suematsu M, Honda K. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes. 2014;5:333–339. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- O’Mahony C, Scully P, O’Mahony D, Murphy S, O’Brien F, Lyons A, Sherlock G, MacSharry J, Kiely B, Shanahan F, O’Mahony L. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget C, Chow MT, Gherardin NA, Beavis PA, Uldrich AP, Duret H, Hassane M, Souza-Fonseca-Guimaraes F, Mogilenko DA, Staumont-Salle D, et al. CD3bright signals on gammadelta T cells identify IL-17A-producing Vgamma6Vdelta1+ T cells. Immunol Cell Biol. 2015;93:198–212. doi: 10.1038/icb.2014.94. [DOI] [PubMed] [Google Scholar]
- Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscso B, Gowda K, Hohl TM, Bogunovic M, Ivanov II. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, Palmer DC, Boni A, Muranski P, Yu Z, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, Gudjonsson S, Hakansson U, Reizis B, Kotarsky K, Agace WW. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Puel A, Doffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, Cobat A, Ouachee-Chardin M, Toulon A, Bustamante J, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Rodriguez R, Pauli ML, Neuhaus IM, Yu SS, Arron ST, Harris HW, Yang SH, Anthony BA, Sverdrup FM, Krow-Lucal E, et al. Memory regulatory T cells reside in human skin. J Clin Invest. 2014;124:1027–1036. doi: 10.1172/JCI72932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014 doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AW, See P, Shin A, Wasan PS, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell host & microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seekatz AM, Panda A, Rasko DA, Toapanta FR, Eloe-Fadrosh EA, Khan AQ, Liu Z, Shipley ST, Detolla LJ, Sztein MB, Fraser CM. Differential response of the cynomolgus macaque gut microbiota to Shigella infection. PLoS One. 2013;8:e64212. doi: 10.1371/journal.pone.0064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Jia T, Mendez-Ferrer S, Hohl TM, Serbina NV, Lipuma L, Leiner I, Li MO, Frenette PS, Pamer EG. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabytska Y, Wolbing F, Gunther C, Koberle M, Kaesler S, Chen KM, Guenova E, Demircioglu D, Kempf WE, Volz T, et al. Cutaneous innate immune sensing of Toll-like receptor 2–6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity. 2014;41:762–775. doi: 10.1016/j.immuni.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Smeekens SP, Huttenhower C, Riza A, van de Veerdonk FL, Zeeuwen PL, Schalkwijk J, van der Meer JW, Xavier RJ, Netea MG, Gevers D. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. Journal of innate immunity. 2014;6:253–262. doi: 10.1159/000351912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159:1739–1745. [PubMed] [Google Scholar]
- Suffia IJ, Reckling SK, Piccirillo CA, Goldszmid RS, Belkaid Y. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J Exp Med. 2006;203:777–788. doi: 10.1084/jem.20052056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumaria N, Roediger B, Ng LG, Qin J, Pinto R, Cavanagh LL, Shklovskaya E, Fazekas de St Groth B, Triccas JA, Weninger W. Cutaneous immunosurveillance by self-renewing dermal gammadelta T cells. J Exp Med. 2011;208:505–518. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland DB, Suzuki K, Fagarasan S. Fostering of advanced mutualism with gut microbiota by Immunoglobulin A. Immunol Rev. 2016;270:20–31. doi: 10.1111/imr.12384. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Maruya M, Kawamoto S, Sitnik K, Kitamura H, Agace WW, Fagarasan S. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity. 2010;33:71–83. doi: 10.1016/j.immuni.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Tada T, Yamamura S, Kuwano Y, Abo T. Level of myelopoiesis in the bone marrow is influenced by intestinal flora. Cell Immunol. 1996;173:155–161. doi: 10.1006/cimm.1996.0261. [DOI] [PubMed] [Google Scholar]
- Tait Wojno ED, Artis D. Emerging concepts and future challenges in innate lymphoid cell biology. J Exp Med. 2016;213:2229–2248. doi: 10.1084/jem.20160525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A. 2016;113:E8141–E8150. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- Trama AM, Moody MA, Alam SM, Jaeger FH, Lockwood B, Parks R, Lloyd KE, Stolarchuk C, Scearce R, Foulger A, et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16:215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nature immunology. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M, Kato H, Sougawa N, Matsui H, Kuwata H, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez Y, Brown EM, Finlay BB. Influence of the microbiota on vaccine effectiveness. Trends Immunol. 2014;35:526–537. doi: 10.1016/j.it.2014.07.003. [DOI] [PubMed] [Google Scholar]
- van der Windt GJ, O’Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz T, Skabytska Y, Guenova E, Chen KM, Frick JS, Kirschning CJ, Kaesler S, Rocken M, Biedermann T. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J Invest Dermatol. 2014;134:96–104. doi: 10.1038/jid.2013.291. [DOI] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529:221–225. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Holscher C, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade TK, Wade WF. Variable gene family usage of protective and non-protective anti-Vibrio cholerae O1 LPS antibody heavy chains. Microbiol Immunol. 2008;52:611–620. doi: 10.1111/j.1348-0421.2008.00078.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Cho B, Suzuki K, Xu Y, Green JA, An J, Cyster JG. Follicular dendritic cells help establish follicle identity and promote B cell retention in germinal centers. J Exp Med. 2011;208:2497–2510. doi: 10.1084/jem.20111449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannemuehler MJ, Kiyono H, Babb JL, Michalek SM, McGhee JR. Lipopolysaccharide (LPS) regulation of the immune response: LPS converts germfree mice to sensitivity to oral tolerance induction. J Immunol. 1982;129:959–965. [PubMed] [Google Scholar]
- Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nature Immunology. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Weiner HL, da Cunha AP, Quintana F, Wu H. Oral tolerance. Immunol Rev. 2011;241:241–259. doi: 10.1111/j.1600-065X.2011.01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, Panchakshari RA, Rodig SJ, Kepler TB, Alt FW. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J Clin Invest. 2011 doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, Forster R, Pabst O. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. The Journal of experimental medicine. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Fohse L, Prinz I, Pezoldt J, Suerbaum S, et al. Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9:444–457. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey LJ, Demehri S, Turkoz M, Turkoz A, Ahern PP, Jassim O, Manivasagam S, Kearney JF, Gordon JI, Kopan R. The absence of a microbiota enhances TSLP expression in mice with defective skin barrier but does not affect the severity of their allergic inflammation. J Invest Dermatol. 2013;133:2714–2721. doi: 10.1038/jid.2013.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, Burk RD, Kunisaki Y, Jang JE, Scheiermann C, et al. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525:528–532. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]


