Abstract
METTL3 catalyzes the formation of N6-methyl-adenosine (m6A) which has important roles in regulating various biological processes. However, the in vivo function of Mettl3 remains largely unknown in mammals. Here we generated germ cell-specific Mettl3 knockout mice and demonstrated that Mettl3 was essential for male fertility and spermatogenesis. The ablation of Mettl3 in germ cells severely inhibited spermatogonial differentiation and blocked the initiation of meiosis. Transcriptome and m6A profiling analysis revealed that genes functioning in spermatogenesis had altered profiles of expression and alternative splicing. Our findings provide novel insights into the function and regulatory mechanisms of Mettl3-mediated m6A modification in spermatogenesis and reproduction in mammals.
Keywords: Mettl3, m6A, spermatogenesis, spermatogonial differentiation, meiosis, alternative splicing
Introduction
Over 100 types of RNA modifications have been identified in mRNAs and non-coding RNAs, among which the N6-methyl-adenosine (m6A) is the most prevalent mRNA modification in eukaryotes1. m6A formation is catalyzed by the RNA methyltransferase complex containing methyltransferase like 3 (METTL3), METTL14 and Wilms' tumor 1-associating protein (WTAP)2,3,4,5,6. m6A is a reversible modification that can be erased by demethylases, fat-mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5)7,8, and recognized by YTH-domain containing family 'reader' proteins9,10,11,12,13. m6A modification mediates a variety of RNA processing steps, and thus regulates mRNA splicing11, mRNA stability9, translation efficiency10,12,13,14,15, microRNA processing16,17 and XIST-mediated transcriptional repression18. Recent studies revealed m6A modulation of mRNA expression to be involved in obesity19, the circadian clock20, and the DNA damage response21. Additionally, it also plays important roles in a variety of biological processes including pluripotency maintenance of stem cells22,23,24, the maternal-to-zygotic transition25, sex determination26,27 and plant shoot stem cell fate determination28. As the key 'writer' of m6A modification, Mettl3 has been demonstrated to modulate neuronal functions and sex determination in Drosophila and regulate early embryonic development and stem cell pluripotency in mice22,23,24,26,27. However, due to the early lethality of Mettl3 knockout mice24, the in vivo biological functions of Mettl3-mediated m6A modification in mammalian tissue development remain to be elucidated.
Here, we generated germ cell conditional Mettl3 knockout mice and found that Mettl3 regulates spermatogonial differentiation and meiosis and is essential for male fertility and spermatogenesis. Mechanistically, we found that METTL3-mediated m6A modification regulates the alternative splicing of genes functioning in spermatogenesis and the global gene expression pattern in testes.
Results
Mettl3 is essential for male fertility and spermatogonial differentiation during spermatogenesis
To explore the function of Mettl3 in mouse spermatogenesis, we first used immunostaining to examine the expression of METTL3 in the mouse testis at 6 and 12 days post-partum (P6 and P12). METTL3 was expressed in both germ cells and somatic cells during testis development (Supplementary information, Figure S1A). Furthermore, METTL3 expression was relatively higher in undifferentiated spermatogonia that expressed PLZF (promyelocytic leukemia zinc-finger protein) at P6 (Supplementary information, Figure S1B). To investigate the function of Mettl3-mediated m6A modification in gametogenesis in mice, we specifically knocked out the Mettl3 gene in the germ cells. Using CRISPR-Cas9 system-assisted homologous recombination, two loxp sites were inserted into the intron 1 and intron 4 of the Mettl3 gene to generate mice carrying the floxed allele (Mettl3flox/+) (Supplementary information, Figure S1C). To disrupt the Mettl3 gene in germ cells, the Mettl3flox/+ mice were further mated with the Vasa-Cre mice that specifically expressed the Cre recombinase in germ cells driven by a Vasa promoter as early as embryonic day 15.5 (E15.5)29 (Supplementary information, Figure S1C). Six genotypes of Mettl3 allele, including Mettl3+/+, Mettl3flox/+, Mettl3flox/flox, Vasa-Cre, Mettl3flox/+Vasa-Cre and Mettl3flox/−Vasa-Cre, were obtained and verified by PCR and Sanger sequencing (Supplementary information, Figure S1D-S1H). The Mettl3flox/−Vasa-Cre mice showed specific deletion of exons 2∼4 and loss of METTL3 expression in PLZF-positive spermatogonia, confirming the conditional knockout of Mettl3 (referred as Mettl3cKO) (Figure 1A and Supplementary information, Figure S1E, S1H). The Mettl3+/+, Mettl3flox/+, Mettl3flox/flox, Vasa-Cre and Mettl3flox/+Vasa-Cre mice were healthy and phenotypically normal, and were used as control in the following experiments (referred as Mettl3Ctrl).
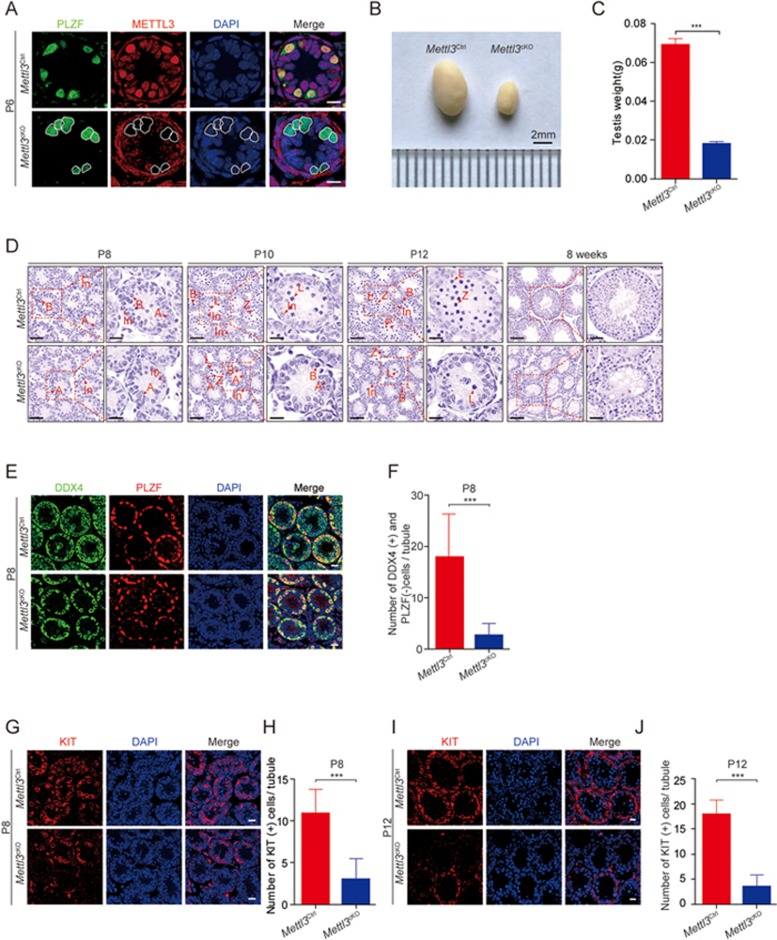
Figure 1.
Mettl3 is essential for male fertility and spermatogonial differentiation during spermatogenesis. (A) Confocal immunofluorescence detection of METTL3 by staining of the Mettl3Ctrl and Mettl3cKO testes at postnatal day 6 (P6). PLZF was co-stained to indicate the location of the undifferentiated spermatogonia. The DNA was stained with DAPI. White circles denote the Mettl3 null spermatogonia. Scale bar, 10 μm. (B) Morphological analysis of the 8-week-old Mettl3Ctrl and Mettl3cKO testes. Scale bar, 2 mm. (C) Testis weight of the 8-week-old Mettl3Ctrl and Mettl3cKO mice. Student's t-test, error bars indicate standard error of measurement (SEM). ***P < 0.001, n = 8. (D) Hematoxylin eosin (H&E) staining of Mettl3Ctrl and Mettl3cKO testes at postnatal day 8 (P8), postnatal day 10 (P10), postnatal day 12 (P12) and 8 weeks showed that the spermatogonial differentiation was inhibited in Mettl3 knockout testes. Red arrows indicate the representative stages of the spermatocytes. A, type A spermatogonia; In, intermediate spermatogonia; B, type B spermatogonia; L, leptotene spermatocytes; Z, zygotene spermatocytes; P, pachytene spermatocytes. Left panel, P8, P10, P12, scale bar, 20 μm; 8 weeks, scale bar, 100 μm. Right panel, P8, P10, P12, scale bar, 5 μm; 8 weeks, scale bar, 20 μm. (E) Immunofluorescence co-staining of PLZF and DDX4 in Mettl3Ctrl and Mettl3cKO testes at P8. Scale bar, 20 μm. (F) Statistics results of DDX4-positive but PLZF-negative cells in Mettl3Ctrl and Mettl3cKO testes at P8. At least 100 tubules were counted from 3 different mice. Student's t-test, error bars indicate SEM. ***P < 0.001. (G) Immunofluorescence staining of KIT in Mettl3Ctrl and Mettl3cKO testes at P8. Scale bar, 20 μm. (H) Statistics of Kit-positive cells in Mettl3Ctrl and Mettl3cKO testes at P8. At least 100 tubules were counted from three different mice. Student's t-test, error bars indicate SEM. ***P < 0.001. (I) Immunofluorescence staining of KIT in Mettl3Ctrl and Mettl3cKO testes at P12. Scale bar, 20 μm. (J) Statistics of KIT-positive cells in Mettl3Ctrl and Mettl3cKO testes at P12. At least 100 tubules were counted from 3 different mice. Student's t-test, error bars indicate SEM. ***P < 0.001.
Next, we analyzed the phenotypes of the Mettl3cKO mice. The Mettl3cKO mice were normal in growth, but were completely infertile and had much smaller testes with 80% reduction in testis weight at 8 weeks (Figure 1B and 1C). Rare differentiated cell types of spermatocytes were observed in the Mettl3cKO seminiferous tubules at P8, P10 and P12, and no spermatids were observed at 8 weeks by hematoxylin and eosin (H&E) staining, indicating a severe defect in spermatogenesis in the Mettl3cKO mice (Figure 1D). Co-staining of the germ cell marker DDX4 and the undifferentiated spermatogonia marker PLZF showed the presence of undifferentiated spermatogonia that were normal, but the number of germ cells was significantly reduced in the Mettl3cKO testes at P8, P10 and P12 (Figure 1E, 1F and Supplementary information, Figure S1I-S1L). We next traced advanced stages of spermatogonial differentiation using the differentiated spermatogonial cell marker KIT at P8, the initial stage of spermatogonial differentiation, and P12. KIT-positive cells were significantly reduced in the Mettl3cKO testes at both P8 and P12 (Figure 1G-1J). Collectively, these results demonstrated that Mettl3 is essential for male fertility in mice and its deletion leads to defects in the initiation of spermatogonial differentiation.
Mettl3 deletion causes a severe defect in meiosis during spermatogenesis
Meiosis, as the key feature of spermatogenesis, initiates around P10 and spermatocytes enter meiotic prophase around P12. To investigate the function of Mettl3 in meiosis, we examined the expression of meiotic marker genes at P10 and P12 by immunostaining. STRA8, a marker of meiosis initiation, showed a much-reduced expression level in Mettl3cKO testes at P10 and P12. Both the ratio of STRA8-positive seminiferous tubules and the number of STRA8-expressing cells per tubule were significantly reduced in Mettl3cKO testes compared to Mettl3Ctrl testes at P10 and P12 (Figure 2A). The number of cells expressing the synaptonemal complex component SYCP3 in Mettl3cKO testes was also much less than that in Mettl3Ctrl testes (Figure 2B). We next traced the progression of meiotic prophase that can be divided into leptotene, zygotene, pachytene stages according to the distribution patterns of SYCP3 and γH2AX in spermatocyte nuclear spreads. Pachytene-stage spermatocytes were only detectable in Mettl3Ctrl mice and not in Mettl3cKO mice, while leptotene- and zygotene/zygotene-like-stage spermatocytes were observed in both Mettl3Ctrl and Mettl3cKO mice (Figure 2C-2E and Supplementary information, Figure S2A). Mettl3Ctrl testes had around 29%, 31% and 40% of spermatocytes at the leptotene, zygotene and pachytene stages, respectively, whereas 84% of spermatocytes in Mettl3cKO testes were in the zygotene/zygotene-like stage but none were in pachytene (Figure 2F).
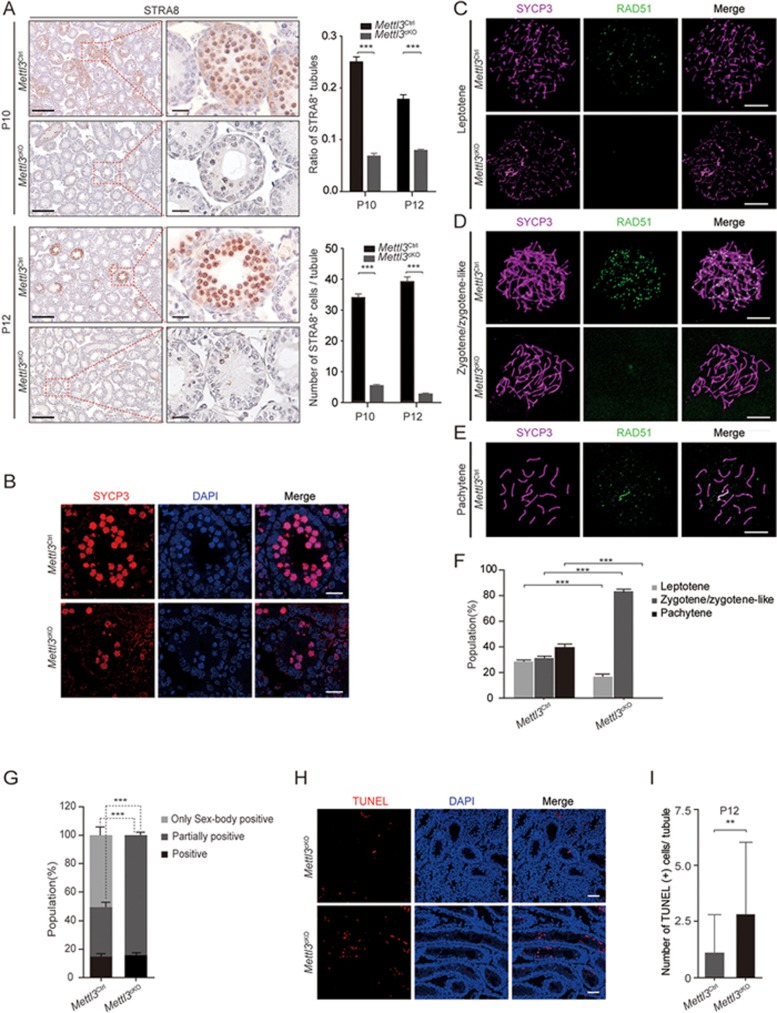
Figure 2.
Mettl3 deletion causes a severe defect in meiosis during spermatogenesis. (A) Immunohistochemical staining of STRA8 in Mettl3Ctrl and Mettl3cKO testes at P10 and P12. Left panel, scale bar, 100 μm. Right panel, scale bar, 5 μm. Statistics of the ratio of STRA8-positive tubule in Mettl3Ctrl and Mettl3cKO testes at P10 and P12. At least 750 tubules were counted from 3 different mice. Student's t-test, error bars indicate SEM. ***P < 0.001. Statistics of the number of STRA8-positive cells per tubule in Mettl3Ctrl and Mettl3cKO testes at P10 and P12. At least 50 tubules were counted from 3 different mice. Student's t-test, error bars indicate SEM. ***P < 0.001. (B) Immunofluorescence staining of SYCP3 in Mettl3Ctrl and Mettl3cKO testes at P12. DNA was stained with DAPI. Scale bar, 20 μm. (C-E) Immunofluorescence staining for detection of SYCP3 and RAD51 on nuclear surface spreads of leptotene (C), zygotene-like (D) and pachytene (E) spermatocytes from Mettl3Ctrl and Mettl3cKO testes at P12. Scale bar, 5 μm. (F) Statistics of the proportion of leptotene, zygotene/zygotene-like, pachytene spermatocytes in Mettl3Ctrl and Mettl3cKO testes at P12. At least 1 000 cells were counted from 3 different mice. Student's t-test, error bars indicate SEM. ***P < 0.001. (G) Statistics of the proportion of γ-H2AX expression patterns including only sex-body positive, partially positive and positive in Mettl3Ctrl and Mettl3cKO spermatocytes at P12. At least 550 cells were counted from 3 different mice. Student's t-test, error bars indicate SEM. ***P < 0.001. (H) Immunofluorescence staining of TUNEL in Mettl3Ctrl and Mettl3cKO testes at P12. The DNA was stained with DAPI. Scale bar, 50 μm. (I) Statistics of number of TUNEL-positive cells per tubule in Mettl3Ctrl and Mettl3cKO testes at P12. At least 50 tubules were counted from 3 different mice. Student's t-test, error bars indicate SEM. **P < 0.01.
To confirm the blockage of meiosis at the zygotene/zygotene-like stage, we examined testes for the presence of additional marker proteins. We found that the synapsis marker SYCP1 was also not detected in Mettl3cKO spermatocytes (Supplementary information, Figure S2B). In contrast to the much greater number of foci of the DSB repair protein RAD51 at the zygotene/zygotene-like stage in Mettl3Ctrl spermatocytes, very few RAD51 foci were detected in Mettl3cKO spermatocytes, indicating defective DSB repair during meiosis upon Mettl3 knockout (Figure 2C-2E). γH2AX expression is strong at the leptotene stage (referred to as positive), gradually decreases around the zygotene stage (referred to as partial positive), and the protein is enriched in the sex-body at the pachytene stage (referred as only sex-body positive). While all three expression patterns could be seen in nuclear spreads of Mettl3Ctrl spermatocytes, the only sex-body-positive pattern was not observed in Mettl3cKO nuclear spreads (Figure 2G and Supplementary information, Figure S2C). Spermatocytes blocked in meiosis will undergo apoptosis; this was detectable by TUNEL assays showing a significantly increased apoptotic signal in Mettl3cKO testes at P12 (Figure 2H and 2I). These results demonstrate that Mettl3-deficient spermatocytes are unable to reach the pachytene stage of meiotic prophase and that Mettl3 is required for normal meiosis.
Mettl3 deletion alters expression pattern of spermatogenesis-related genes
To systematically illuminate gene expression changes resulting from the Mettl3 conditional knockout in spermatogenesis, we compared the transcriptome between Mettl3Ctrl and Mettl3cKO testes at P6 and P12 through high-throughput RNA sequencing. The correlation analysis showed that the expression of the majority of genes in testes was not influenced by Mettl3 deletion (Supplementary information, Figure S3A, S3B and Table S4). We found that a total of 258 genes were differentially expressed in the Mettl3cKO testes at P6. These included 157 down-regulated genes that were significantly enriched in genes required for the regulation of spermatogenesis including the meiotic cell cycle and synaptonemal complex assembly (Supplementary information, Figure S3C). We found that 941 genes were differentially expressed in Mettl3cKO testes at P12. These included 699 down-regulated genes that were mainly enriched in genes required for spermatogenesis and regulation of meiosis (Figure 3A and Supplementary information, Table S4). We next examined expression patterns of 50 genes specifically expressed in germ cells according to a previous study30. Unsupervised cluster analysis showed that the Mettl3cKO expression pattern at P12 was much more similar to Mettl3Ctrl at P6 than Mettl3Ctrl at P12, consistent with the severely impaired spermatogonial differentiation observed in Mettl3cKO testes (Figures 3B, 3C and Supplementary information, Table S4). Genes with reported functions in spermatogenesis31,32,33 were mapped into an interaction network to illustrate regulationary relationships by Pathway Studio (Figure 3D). The genes significantly down-regulated in Mettl3cKO testes included 9 genes involved in spermatogonial stem cell maintenance (Dazl, Ddx4, Plzf, Pax7, Nanos2, Id4, Pou3f1, Taf4b, Bcl6); 6 genes in spermatogonial differentiation (Sohlh1, Sohlh2, Neurog3, Kit, Dmrt1, Sox3); 8 genes in cell cycle (Ccna1, Ccna2, Ccnb1, Ccnb2, Ccnb3, Ccne2, Cdk1, Rad51); and 9 genes in meiosis (Stra8, Sycp1, Sycp3, Spo11, Rad18, Dmc1, Rec8, Mlh1, Smc1b) (Figure 3E, 3F and Supplementary information, Figure S3D). Taken together, this indicate that the Mettl3 deficiency globally influenced the expression pattern of spermatogenesis-related genes regulating multiple biological processes including spermatogonial stem cell maintenance, differentiation and meiosis.
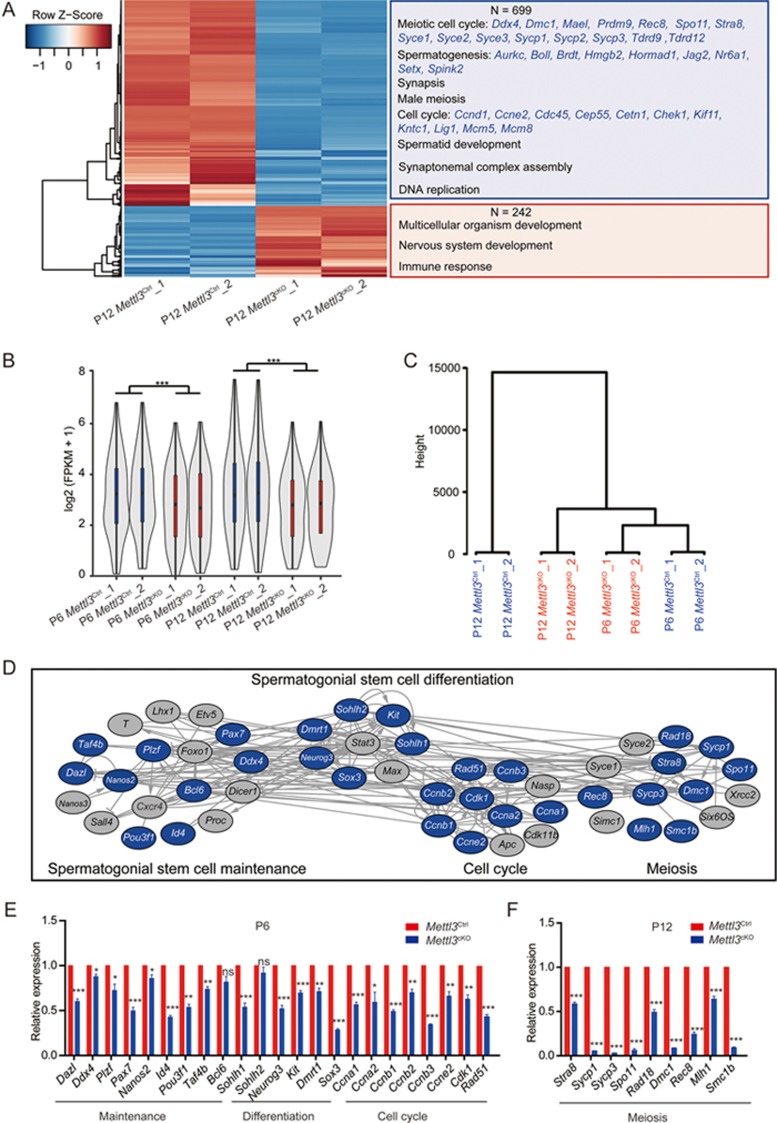
Figure 3.
Mettl3 deletion alters expression pattern of genes involved in mouse spermatogenesis. (A) Heatmap showing the differentially expressed genes between Mettl3Ctrl and Mettl3cKO testes at P12 and their function enrichment. Two-fold expression difference and P = 0.05 as the cutoff. The enriched Gene Ontology (GO) terms of the biological process category in down or upregulated genes in Mettl3cKO samples were shown on the right panels. (B) Violin plot of expression levels of germ cell specifically expressed genes in Mettl3Ctrl and Mettl3cKO samples at P6 and P12. The log2-transformed expression level (FPKM) was used for drawing the plot. *** means the P < 10e-5, as calculated by the Wilcoxon signed-rank paired test. (C) The unsupervised hierarchical cluster analysis for the expression of germ cell specifically expressed genes in Mettl3Ctrl and Mettl3cKO samples at P6 and P12. (D) The interaction network showing genes involved in spermatogenesis regulation. The downregulated genes in Mettl3cKO testes which had been validated by qRT-PCR marked in blue. The regulation relationships were produced by pathway studio; the positive, negative regulation among these genes was marked as arrow, T-head respectively, while the regulation with unknown effects between two genes were connected directly. (E, F) qRT-PCR validation of downregulated genes involved in spermatogenesis in Mettl3cKO testes at P6 (E) and P12 (F). Student's t-test was performed and data was shown as mean ± SEM. of three independent experiments. *P < 0.05, *P < 0.01, **P < 0.001; ns, no significance.
Mettl3-mediated m6A regulates alternative splicing of genes functioning in spermatogenesis
Previous studies have shown that m6A could influence biological process through regulating alternative splicing events11,19. We therefore employed rMATS software to analyze the differential alternative splicing events between Mettl3Ctrl and Mettl3cKO transcriptomes34. Among the differential splicing events, we mainly detected exon skipping to be enriched, consistent with previous studies11,19 (Figure 4A and Supplementary information, Table S5). Gene ontology analysis revealed that the differentially spliced genes between Mettl3Ctrl and Mettl3cKO samples at P6 were enriched in those for DNA replication and cell division processes whereas those at P12 were enriched for spermatogenesis (Supplementary information, Figure S4A).
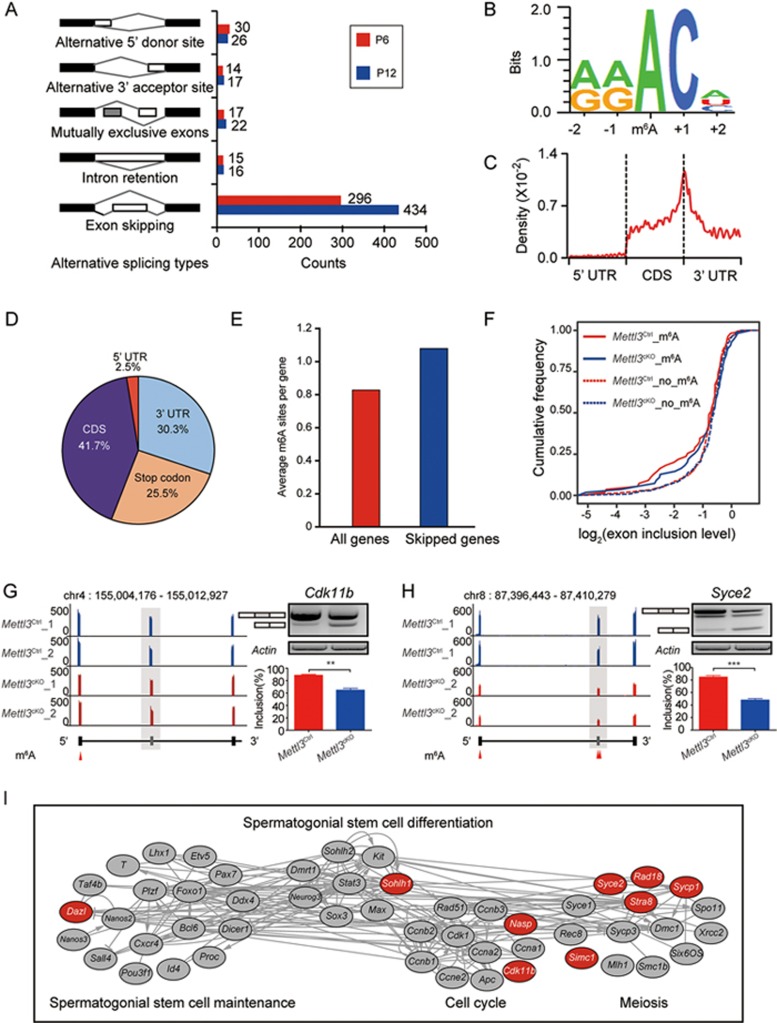
Figure 4.
Mettl3-mediated m6A regulates alternative splicing of genes involved in spermatogenesis. (A) Schematics and statistics of the five types of alternative splicing events in Mettl3Ctrl and Mettl3cKO testes at P6 and P12. The differential alternative splicing events were identified by rMATS and the FDR = 0.05 were used as cutoff. Each group sample included two replicates. (B) The logo plot for bases adjacent to m6A sites. All 12 516 putative m6A residues were used to enrich the m6A motif and the plot was produced by WebLogo. (C) Distribution of m6A sites across the length of mRNA transcripts. (D) Transcriptome-wide distribution of m6A sites. Pie charts showing the percentage of m6A sites in distinct RNA sequence types: 5′ UTR, CDS, Stop codon and 3′ UTR. (E) The average m6A site frequency per gene in all genes and in differentially spliced genes in Mettl3Ctrl testes at P12. (F) The cumulative frequency curve for the alternative exon inclusion ratio in Mettl3Ctrl and Mettl3cKO testes at P12. The exon inclusion level was calculated by alternative exon coverage divided by constitutive exon coverage and transformed by log2. The exons containing m6A sites in Mettl3cKO testes showed lower coverage level than in Mettl3Ctrl testes (P < 0.01, Wilcoxon signed-rank paired test). No significant difference was exhibited between exons without m6A modification. (G, H) Distribution of RNA-seq reads of Cdk11b (G) and Syce2 (H) in Mettl3Ctrl and Mettl3cKO testes samples, the alternative exon of each gene was marked in grey and shadow. The m6A site location was marked as a red triangle. The exon skipping event was validated by RT-PCR (right panel). (I) The interaction network showing genes involved in spermatogenesis regulation. The genes containing alternative exons which had been validated by RT-PCR were marked in red. The regulation relationships were produced by Pathway Studio; the positive, negative regulation among these genes was marked as arrow, T-head respectively, while the regulation with unknown effects between two genes was connected directly.
We then detected the m6A sites and explored their distribution in P12 testes of wild-type mice through m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP) and sequencing (miCLIP-seq). A total of 12 516 putative m6A residues within 5 092 genes were identified in testes (Supplementary information, Table S6). These sites were enriched in the m6A consensus motif (Figure 4B) and were predominantly distributed in the CDS and 3′ UTR regions, especially near stop codon regions (Figure 4C and 4D). Notably, the average m6A site number was higher in genes with skipped exons (Figure 4E). An alternative exon inclusion level measurement also revealed that genes containing m6A had a lower inclusion level in Mettl3cKO samples compared to that in Mettl3Ctrl samples whereas genes without m6A modification did not show this tendency (Figure 4F and Supplementary information, Figure S4B). These results are consistent with previous reports showing the role of m6A modification in promoting exon inclusion11,19.
Next, we examined differential splicing events between Mettl3Ctrl and Mettl3cKO samples in spermatogenesis-related genes. Our findings showed that 9 genes had significantly decreased exon inclusion levels in Mettl3cKO testes (Figure 4G, 4H and Supplementary information, Figure S4C-S4I). The m6A-RIP-qPCR analysis was used to validate the presence of m6A residues in alternative exons or neighboring regions, and the results supported occurrence of alternative splicing events regulated by m6A (Supplementary information, Figure S4J-S4Q). For example, Dazl, essential for spermatogenesis, had a shorter transcript with loss of 17 amino acids next to its RNA recognition motif in Mettl3cKO testes (Supplementary information, Figure S4C). Similar splicing events were also found in Sohlh1 regulating spermatogonial stem cell differentiation35,36,37, and Nasp- and Cdk11b-regulating cell division38,39,40,41 (Figure 4I and Supplementary information, Figure S4D, S4E). Taken together, these data revealed that Mettl3-mediated m6A modification regulates the alternative splicing of genes that function in spermatogenesis.
Discussion
m6A is the most abundant modification in RNA molecules in eukaryotes and is catalyzed by one of the 'writer' proteins, METTL32,3,4,5,6. Although several biological functions of Mettl3-mediated m6A methylation have been revealed in Drosophila, mouse early embryos and embryonic stem cells22,23,24,26,27, the in vivo function in mammalian tissues has remained elusive due to the early lethality of Mettl3-deficient mice24. Here, we presented a number of findings demonstrating the significance of m6A in male fertility and spermatogenesis: (1) conditional deletion of Mettl3 in germ cells led to defective spermatogonial differentiation and meiosis, demonstrating an essential role of METTL3-mediated m6A modification in male fertility and spermatogenesis (Figures 1 and 2). (2) Transcriptome analysis and qRT-PCR assays revealed that deletion of Mettl3 resulted in downregulation of several spermatogenesis-related genes (Figure 3). (3) The Mettl3 deficiency dysregulated alternative splicing of spermatogenesis-related genes (Figure 4). Thus, our study provides evidence that m6A plays a significant role in regulating spermatogenesis by affecting the expression and splicing of spermatogenesis-related genes.
Previous studies have shown that deletion of Alkbh5, one of the m6A demethylases, led to impaired fertility with compromised production of meiotic metaphase-stage spermatocytes8, suggesting a regulatory role of m6A in spermatogenesis. However, the underlying mechanism remains unknown. Compared to the phenotypes of Alkbh5 knockout mice, the defects in Mettl3cKO mice arise much earlier and are more severe, suggesting a less redundant and moreover, indispensable function of METTL3-catalyzed m6A formation in tissue development. The essential role of both the m6A demethylase and methyltransferase in spermatogenesis also suggest that the overall balance of RNA m6A modification is key to maintain the differentiation program during spermatogenesis. Highly dynamic developmental processes, such as the spermatogenesis, require dynamic gene expression involving multiple layers of transcriptional regulation. Hence both demethylase- and methyltransferase-catalyzed m6A modifications play an important role in normal development.
Spermatogenesis is a complicated process involving dynamic spermatogonial differentiation coupled with meiosis. DNA methylation and demethylation, histone modification, together with lncRNAs, microRNAs, piRNA and many transcriptional factors have been shown to participate in regulating spermatogenic progression by modulating gene expression42,43,44,45,46,47,48. Alternative splicing is another important mechanism to regulate gene expression, and a recent report showed that Bcas2, an important alternative splicing factor, functions in spermatogonia and is involved in the transition to meiosis through regulating alternative mRNA splicing49.
Recent studies have implicated an essential regulatory role of m6A in mRNA splicing4,11,16,19,24,26,27,50,51,52,53,54. First, m6A alters the local structure in mRNA to facilitate the binding of some HNRNP proteins51,53. Alarcon et al.16 revealed that modulation of METTL3 and HNRNPA2B1 causes similar changes to alternative splicing; Second, the m6A reader YTHDC1 promotes SRSF3 but antagonizes SRSF10 binding to mRNAs and affects mRNA splicing11. An m6A-modified Sxl intron regulates the adjacent exon splicing and controls Drosophila sex determination26,27,52. Moreover, Ye et al.54 demonstrated an essential role of m6A in regulating replication transcription activator pre-mRNA splicing in Kaposi's sarcoma-associated herpesvirus.
Nevertheless, the recent study of Ke et al.55 showed that only 99 exons from ∼2 000 alternatively spliced cassette exons containing m6A have a significantly changed inclusion level upon Mettl3 depletion using Quantas software with FDR < 5% and ΔPSI ≥ 0.1, raising a question regarding the role of m6A in the regulation of alternative splicing. However, many factors may affect the callings of alternative splicing, such as sequencing depth, the analysis pipeline and parameters56,57,58. We further analyzed the RNA-seq datasets19,24,50,51 used in the Ke et al. study55 to detect the changed alternative splicing events by rMATS34, a popular software used in alternative splicing analysis with FDR < 0.05. The number of changed alternative splicing events varied from dozens to thousands in different tissues or cell types, indicating that m6A regulates alternative splicing events in a tissue-dependent manner (Supplementary information, Figure S4R). Given that one mRNA can be expressed by alternative splicing according to different regulatory programs, mild alternative splicing events can be sufficient to induce important functional changes19,59. Future systematic analysis of m6A single-site resolution of pre-mRNAs will be able to give straight forward answers how m6A regulates mRNA splicing.
Our findings demonstrated an additional mechanism for METTL3 in spermatogenesis by regulating the alternative splicing of mRNAs. We found that many important spermatogenesis-regulating genes, such as Sohlh1 and Dazl35,36,37,49, carried m6A modification and exhibited altered splicing pattern in Mettl3cKO mice. This indicates that the loss of m6A modification on these genes upon Mettl3 deletion indeed influenced their alternative splicing pattern and resulted in defective spermatogenesis.
Mettl3-mediated m6A modification was a prerequisite for the initiation of spermatogenesis from undifferentiated spermatogonia since both the initial spermatogonial differentiation and the initial meiosis were severely blocked upon Mettl3 disruption, as evidenced by the findings that KIT-positive cells were significantly reduced and spermatocytes could not reach the pachytene stage of meiotic prophase in Mettl3cKO testes. Thus, our findings provide strong evidence for deciphering the in vivo functions of METTL3-mediated m6A modification in mouse spermatogenesis.
Materials and Methods
Mouse
The mice used in this study were B6D2F1 (C57BL/6 × DBA2), DBA2 and C57BL6 strains. Specific pathogen-free-grade mice were purchased from Beijing Vital River laboratory animal center and housed in the animal facilities of the Institute of Zoology, Chinese Academy of Sciences. All animal experiments were carried out in accordance with the guidelines for the Use of Animals in Research issued by the Institute of Zoology, Chinese Academy of Sciences.
Mouse breeding
Mettl3flox/+ mice were generated by the CRISPR-Cas9 system-assisted homologous recombination. Vasa-Cre transgenic mice were a gift from professor Jia-Hao Sha, Nanjing Medical University. Mettl3flox/+Vasa-Cre mice were obtained by mating Mettl3flox/+ and Vasa-Cre mice and Mettl3flox/flox mice were obtained by mating Mettl3flox/+ and Mettl3flox/+ mice. Mettl3flox/+Vasa-Cre and Mettl3flox/flox were mated to generate Mettl3flox/−Vasa-Cre mice (Mettl3cKO).
Construction of gene targeting vector
The T7-Cas9 vector was from our lab. The T7-sgRNA vector was digested with BsaI enzyme (NEB) and the linearized vector was gel purified. A pair of oligonucleotides for each targeting site (Mettl3 intron 1 and intron 4) was annealed and ligated to linearized vector. All oligonucleotides were listed in the Supplementary information, Table S1.
In vitro preparation of sgRNAs, Cas9 mRNA and oligonucleotide DNAs for microinjection
All RNAs prepared for microinjection were in vitro transcribed. SgRNAs and Cas9 mRNA were transcribed by the HiScribe T7 In Vitro Transcription Kit (NEB). The Cas9 mRNA was capped with the m7G (5′) ppp (5′) G RNA Cap Structure Analog (NEB) by T7 transcriptase. All RNAs were dissolved in DEPC water and stored in −80 °C.
Oligonucleotides used for CRISPR-Cas9 system-assisted homologous recombination through embryo injection were synthetized in Integrated DNA Technologies. All oligonucleotides were listed in the Supplementary information, Table S1.
Intracytoplasmic RNA microinjection
The intracytoplasmic RNA microinjection was performed according to a previous report. Briefly, one-cell-stage embryos were collected at 0.5 day post coitum (dpc). Each embryo was microinjected with 25 ng/μl L-sgRNA, 25 ng/μl R-sgRNA, 100 ng/μl Cas9 mRNA, 50 ng/μl L-loxp oligonucleotide and 50 ng/μl R-loxp oligonucleotide into the cytoplasm. After microinjection, surviving embryos were implanted on the same day into the oviduct of pseudopregnant B6D2F1 female mice. Full-term pups were obtained by natural labor at 19.5 dpc.
Immunohistochemical analysis
For immunostaining analysis: testes were fixed in 4% paraformaldehyde overnight at 4 °C. Following dehydration, the samples were embedded in paraffin and sectioned (5 μm). After dewaxing and hydration, the sections were boiled in an antigen retrieval buffer (0.01 M citric acid/sodium citrate, pH 6.0) for 10 min using a microwave oven, washed in PBS three times and permeated in 0.2% Triton X-100 for 15 min. The sections were blocked with 5% BSA in PBS for 2 h at room temperature before adding primary antibodies. The samples were washed three times in PBS after incubating at 4 °C overnight and then incubated with secondary antibodies for 2 h at room temperature. After washing in PBS three times, the sections were incubated in 2 μg/ml of DAPI (Molecular Probes, D1306) for 10 min at room temperature and washed in PBS three times. Sections were mounted on slices with Dako Fluorescence Mounting Medium (Dako Canada, ON, Canada). The immunohistochemical analysis of STRA8 were imaged with a Leica Aperio VERSA 8 microscope (Leica Biosystems, Germany). The immunofluorescence staining was imaged with a laser scanning confocal microscope LSM780 (Carl Zeiss, Germany). All the antibodies were listed in the Supplementary information, Table S2.
Genotyping of mice
First, all mice were genotyped with the tail DNA. Two pair of primers were used to detect the loxp insertion into the Mettl3 intron 1 (L-loxp-forward and L-loxp-reverse) and intron 4 (R-loxp-forward and R-loxp-reverse). The product sizes were 222 bp and 335 bp with the loxp sequence insertion into Mettl3 intron 1 and intron 4, respectively; whereas the product sizes from WT were 182 bp and 295 bp, respectively. Cre recombinase was detected by the Vasa-Cre primers and the product of PCR was 240 bp. The PCR reaction conditions were as follows: 95 °C, 2 min; 95 °C, 30 s, 61 °C, 30 s, 72 °C, 30 s, 35 cycles; 72 °C, 5 min. Then the germ cells were used to confirm the deletion of Mettl3 with the primers of L-loxp-forward and R-loxp-reverse and the product of Mettl3 deletion was 318 bp whereas the WT product was 2 554 bp. The PCR reaction conditions were as follows: 95 °C, 5 min; 95 °C, 30 s, 61 °C, 30 s, 72 °C, 2.5 min, 35 cycles; 72 °C, 10 min. All primers are listed in the Supplementary information, Table S3.
Histological analysis
For histological analysis: testes from Mettl3Ctrl and Mettl3cKO male mouse were fixed in Modified Davidson's Fluid (Formaldehyde:Glacial acetic acid:Ethanol:H2O = 6:3:1:10) overnight at room temperature. The testes were dehydrated stepwise through an ethanol series (70%, 80%, 90%, 100% ethanol) and processed for paraffin embedding. 5 μm sections were cut with a Leica slicing machine (Leica RM2235, Leica Biosystems, Germany) and mounted on poly-D-lysine coated glass slices (Zhong Shan Golding Bridge biotechnology, Beijing, China). After dewaxing and hydration, the sections were stained with H&E using standard methods and imaged with a Leica Aperio VERSA 8 microscope (Leica Biosystems, Germany).
TUNEL analysis
The TUNEL assay was performed using the TUNEL BrightRed Apoptosis Detection Kit (Vazyme) according to the manufacturer's instructions. Briefly, sections were permeabilized by protein K and labeled with rTdT reaction mix for 1 h at 37 °C, reaction was stopped by 1× PBS. After washing in PBS, the sections were incubated in 2 μg/ml of DAPI (Molecular Probes, D1306) for 10 min at room temperature and washed in PBS. Sections were mounted on slices with Dako Fluorescence Mounting Medium (Dako Canada, ON, Canada). Images were obtained using a laser scanning confocal microscope LSM780 (Carl Zeiss).
RNA extraction and qRT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen, 15596-018) from whole testes. After removing the genomic DNA with the RNase-Free DNase I Kit (Promega, M6101), 1 μg total RNA reverse-transcribed into cDNAs using the reverse Transcription System (Promega, A3500). qRT-PCR was performed using a SYBR Premix Ex Taq kit (TaKaRa, RR420A) on Agilent Stratagene Mx3005P. Relative gene expression was analyzed based on the 2−ΔΔCt method with Actin as internal control. At least three independent experiments were analyzed. All primers were listed in the Supplementary information, Table S3.
m6A-miCLIP-seq
Single-base resolution high-throughput sequencing of the mouse testes methylome was carried out according to previously reported methods60 with some modifications. Briefly, mRNA was purified using a Dynabeads® mRNA Purification Kit (Life Technologies, 61006) and fragmented into around 100 nt fragments using the RNA fragmentation reagent (Life Technologies, AM8740). 5 μg mRNA was incubated with 10 μg of anti-m6A antibody (Abcam, ab151230) in 450 μl immunoprecipitation buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 0.05% NP-40) and incubated at 4 °C for 2 h with gentle rotation. The mixture was transferred to a clear flat-bottom 96-well plate (Corning) on ice and irradiated three times with 0.15 J/cm2 at 254 nm in a CL-1000 Ultraviolet Crosslinker (UVP). After irradiation, the mixture was collected together and then immunoprecipitated by incubation with dynabeads protein A (Life Technologies, 10008D) at 4 °C by rotating for another 2 h. After extensive washing, end-repair and linker ligation was performed on-bead. Finally, the bound RNA fragments were eluted from the beads with 2 μg/μl proteinase K in PK buffer (100 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10 mM EDTA) by digestion at 55 °C for 20 min. RNA was isolated from the eluate by phenol-chloroform (Life Technologies, AM9730) extraction and ethanol precipitation. Purified RNA was reverse transcribed with Superscript III reverse transcriptase (Life Technologies, 18080093) according to the manufacturer's protocol as described previously18,61. cDNA was size-selected on a 6% TBE-Urea gel (Life Technologies, EC68655BOX) and regions corresponding to 70-100 nt were used for further analysis. Thus, purified cDNA was circularized with Circligase II (Epicentre, CL9021K) and re-linearized with BamHI (NEB, R0136M). Libraries were PCR amplified with Accuprime Supermix 1 enzyme for 18 cycles and size-selected on an 8% TBE gel (Life Technologies, EC62155BOX). Sequencing was carried out on an Illumina HiSeq 3000 platform according to the manufacturer's instructions.
m6A-RIP-qPCR
The procedure of m6A immunoprecipitation (m6A-RIP) was modified from previously reported methods23. In brief, purified mRNAs from P6/P12 mouse testes were digested by DNase I and then fragmented into ∼300 nt fragments by 30 s incubation at 94 °C in RNA fragmentation reagent (Life technologies, AM8740). This reaction was then stopped with stop solution (Life technologies, AM8740), followed by standard ethanol precipitation and collection. 12 μg anti-m6A polyclonal antibody (Synaptic Systems) was pre-incubated with 50 μl Dynabeads Protein A (Life technologies, 10013D) in IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl, pH 7.4) for 1 h at room temperature. After pre-heating at 75 °C for 5 min and chilling on ice immediately, 6 μg mRNAs were added to the above prepared antibody-beads mixture and incubated for 4 h at 4 °C by rotating. After extensive washing, bound RNAs were extracted by proteinase K digestion, phenol-chloroform (Life Technologies, AM9730) extraction and ethanol precipitation. The enrichment of m6A was quantified through qPCR as reported23. m6A-RIP-qPCR primers are listed in the Supplementary information, Table S3.
RNA-seq library preparation and data processing
Total RNA was extracted from cultured cells by TRIzol reagent (Invitrogen, 15596-018). For RNA-seq library construction, the PolyA+ tailed RNA purification was performed for each sample using a Dynabeads mRNA purification kit (Ambion, 61006). The cDNA library was generated with a KAPA Stranded mRNA-Seq Kit (KAPA, KR0960) protocol. Sequencing was performed on an Illumina HiSeq 3000 platform with 101 bp paired-end-sequencing reactions. The RNA-seq reads of each sample were mapped to the mouse mm9 genome assembly independently by the HISAT2 software using the annotated gene structures as templates. Default parameters of HISAT2 were used except with the options “--dta-cufflinks” and “--rna-strandness RF” opening62. Reads with unique genome location were retained for gene expression calculation using Cufflinks (version 2.0.2) with the option “--GTF”63. Genes with no less than 0.5 FPKM in at least one sample were used for the differentially expressed genes analysis.
Alternative splicing analysis
Reads with a unique genome location in SAM format were used for the splicing events identification. The AS events between the two sample groups were identified using rMATS version 3.2.534. Five types of AS events from RNA-seq data with two replicates were identified. In each rMATS run, the first group was compared to the second group to identify differentially spliced events with FDR less than 0.05. The alternative exon accumulation levels in each sample were calculated using methods previously described. RT-PCR validation of alternative splicing events were performed with cDNA reverse-transcribed from mouse testes RNA. Actin was used as an internal control. All primers are listed in the Supplementary information, Table S3.
Analysis of miCLIP-sequencing data
The method of pre-processing for reads was performed essentially as previously reported64. The adaptor was trimmed by fastx_clipper fastx_toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Low quality bases were filtered by fastq_filter.pl, a custom perl script from CLIP Tool Kit (CTK)65 and reads shorter than 24 nt discarded. After trimming, reads without a barcode marker sequence or within large fragment (length > 60 nt) are discarded to eliminate unspecific RNA tags. Based on the same criteria of a previously reported approach to process paired-end data, the forward reads were demultiplexed based on 5′ barcodes for individual replicates by fastq2collapse to remove PCR amplified reads and the reverse reads were reverse-complemented and processed like their forward mates. Finally, the random barcodes of remaining reads were stripped by stripBarcode.pl and moved to read headers for downstream processing by the CIMS pipeline.
Remaining reads were mapped to the mouse genome (version mm9) with BWA (v0.7.10)66 and allowed ≤ 0.06 error rate (substitutions, insertions or deletions) per read as online CTK Documentation (https://zhanglab.c2b2.columbia.edu/index.php/CTK_Documentation). To identify the m6A locus, the mode of mutation calling was performed as previously reported with minor modification60. For each mutation position, the coverage of a unique tag (k) and mutations (m) was determined by the CIMS.pl program67. The positions within k ≥ 15 and m/k ≤ 50% were kept and only mutation positions within the RRACH motif were identified as m6A for downstream analysis to exclude the potential m6Am modification60. The logo plot for bases adjacent to m6A was generated with WebLogo68.
Other tools
The unsupervised hierarchical cluster analysis performed by the hcluster function of the amap package in R. Point plots were produced by the smoothScatter functions of R. And the Pearson correlation coefficients shown in the figures. The heatmap was produced by the heatmap.2 function of R. Gene ontology analysis of differentially expressed genes was analyzed in DAVID69,70 and biological processes were selected based on P-values smaller than 0.05 and the figures were produced by ggplot2. The interaction relationship of spermatogenesis related genes was constructed by Pathway Studio71 based on the regulation reported in public reference and the network was showed by the Cytoscape72. The cumulative frequency curve and violin plot were also produced by ggplot2. The RNA-seq reads distribution were exhibited in IGV browser73.
Quantification and statistical analysis
Statistical parameters including statistical analysis, statistical significance and n value are reported in the Figure legends. Statistical analyses were performed using Prism Software (GraphPad). For statistical comparison, Student's t-test was employed. A value of P < 0.05 was considered significant.
Data and software availability
The accession number for the sequencing data reported in this paper is NCBI GEO: GSE99773. This parent directory includes the following datasets: GEO: GSE99771 (RNA-seq) and GEO: GSE99772 (m6A-miCLIP-seq). The public RNA-seq datasets (GSE53249, GSE56010, GSE61997 and GSE37005) for alternative splicing events identification by rMATS were downloaded from GEO database.
Authors Contributions
WL, Y-GY and QZ conceived this project and supervised all the experiments and data analysis. Y-GY, WL, QZ, KX, G-HF, YY and B-FS analyzed the data and wrote the manuscript. YY performed m6A-miCLIP and m6A-RIP-qPCR assays. G-HF performed bioinformatics analysis with assistance from B-FS, Y-SC and X-JW. KX performed molecular biology, protein chemistry, cell culture and mouse experiments with assistance from J-QC, Y-FL, X-XZ, C-XW, L-YJ, CL, Z-YZ.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Dr Y-L Zhao from Beijing Institute of Genomics, Chinese Academy of Sciences for scientific suggestion. This work was supported by the National Natural Science Foundation of China (31422038, 31471395, 31625016 and 31670824), the National Basic Research Program of China (2014CB964800, 2016YFC0900300), CAS Strategic Priority Research Program (XDB14030300, QYZDY-SSW-SMC027/002, QYZDB-SSW-SMC022). We thank BIG sequencing core facility for sequencing.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Generation and characterization of Mettl3 conditional knockout mice.
Mettl3 deletion causes meiotic defect and abnormal spermatogenesis.
Global gene expression profiles and differential expression analyses between Mettl3Ctrl and Mettl3cKO Testes.
Mettl3-mediated m6A regulates alternative splicing of genes functioning in spermatogenesis.
Oligonucleotides used in this study.
Antibodies used in this study.
Primers used in this study.
Differentially expressed genes between Mettl3Ctrl and Mettl3cKO testes in differential stages and gene ontology enrichment analysis for these genes.
Differential exon skipping events identified between Mettl3Ctrl and Mettl3cKO testes in differential stages by rMATS.
The location of m6A sites identified in Mettl3Ctrl testes.
References
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Genet 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep 2014; 8:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014; 16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- Li A, Chen YS, Ping XL, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 2017; 27:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, et al. 5′ UTR m6A promotes cap-independent translation. Cell 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015; 162:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature 2015; 519:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014; 24:1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793–806. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017; 543:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilo F, Zhang F, Sancho A, et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 2015; 17:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, et al. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015; 16:289–301. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017; 542:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016; 540:301–304. [DOI] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 2016; 540:242–247. [DOI] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, et al. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell 2016; 38:186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dong J, Yan L, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 2017; 20:891–892. [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 2010; 11:124–136. [DOI] [PubMed] [Google Scholar]
- Song HW, Wilkinson MF. Transcriptional control of spermatogonial maintenance and differentiation. Semin Cell Dev Biol 2014; 30:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SR, Liu YX. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by sertoli cell signaling. Reproduction 2015; 149:R159–R167. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol 2006; 294:161–167. [DOI] [PubMed] [Google Scholar]
- Barrios F, Filipponi D, Campolo F, et al. SOHLH1 and SOHLH2 control Kit expression during postnatal male germ cell development. J Cell Sci 2012; 125:1455–1464. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Ahn HW, Chu T, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol 2012; 361:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseev OM, Bencic DC, Richardson RT, Widgren EE, O'Rand MG. Overexpression of the Linker histone-binding protein tNASP affects progression through the cell cycle. J Biol Chem 2003; 278:8846–8852. [DOI] [PubMed] [Google Scholar]
- Li T, Inoue A, Lahti JM, Kidd VJ. Failure to proliferate and mitotic arrest of CDK11(p110/p58)-null mutant mice at the blastocyst stage of embryonic cell development. Mol Cell Biol 2004; 24:3188–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RT, Alekseev O, Alekseev OM, O'Rand MG. Characterization of the NASP promoter in 3T3 fibroblasts and mouse spermatogenic cells. Gene 2006; 371:52–58. [DOI] [PubMed] [Google Scholar]
- Richardson RT, Alekseev OM, Grossman G, et al. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem 2006; 281:21526–21534. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431:96–99. [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 2007; 26:720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev Biol 2007; 307:368–379. [DOI] [PubMed] [Google Scholar]
- Huszar JM, Payne CJ. MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in mice. Biol Reprod 2013; 88:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WS, Falciatori I, Tam OH, et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev 2015; 29:1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhade VS, Dighe SN, Kataruka S, Rao MR. Mechanism of Wnt signaling induced down regulation of mrhl long non-coding RNA in mouse spermatogonial cells. Nucleic Acids Res 2016; 44:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataruka S, Akhade VS, Kayyar B, Rao MRS. Mrhl lncRNA mediates meiotic commitment of mouse spermatogonial cells by regulating Sox8 expression. Mol Cell Biol 2017; 37:pii:e00632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang F, Xu Q, et al. BCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosis. Nat Commun 2017; 8:14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L, Grozhik AV, Vedanayagam J, et al. The m6A pathway facilitates sex determination in Drosophila. Nat Commun 2017; 8:15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 2017; 10:6051–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen ER, Nilsen TW. Kaposi's sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. J Virol 2017, doi:10.1128/JVI.00466-17. [DOI] [PMC free article] [PubMed]
- Ke S, Pandya-Jones A, Saito Y, et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 2017; 31:990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan-Otto Attolini C, Pena V, Rossell D. Designing alternative splicing RNA-seq studies. Beyond generic guidelines. Bioinformatics 2015; 31:3631–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- Zyprych-Walczak J, Szabelska A, Handschuh L, et al. The impact of normalization methods on RNA-Seq data analysis. Biomed Res Int 2015; 2015:621690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkestein M, Laber S, McMurray F, et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun 2015; 6:6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 2014; 9:263–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 2017; 33:566–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 2014; 6:1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio-the analysis and navigation of molecular networks. Bioinformatics 2003; 19:2155–2157. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2007; 2:2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation and characterization of Mettl3 conditional knockout mice.
Mettl3 deletion causes meiotic defect and abnormal spermatogenesis.
Global gene expression profiles and differential expression analyses between Mettl3Ctrl and Mettl3cKO Testes.
Mettl3-mediated m6A regulates alternative splicing of genes functioning in spermatogenesis.
Oligonucleotides used in this study.
Antibodies used in this study.
Primers used in this study.
Differentially expressed genes between Mettl3Ctrl and Mettl3cKO testes in differential stages and gene ontology enrichment analysis for these genes.
Differential exon skipping events identified between Mettl3Ctrl and Mettl3cKO testes in differential stages by rMATS.
The location of m6A sites identified in Mettl3Ctrl testes.