Abstract
PSI-BLAST is an iterative program to search a database for proteins with distant similarity to a query sequence. We investigated over a dozen modifications to the methods used in PSI-BLAST, with the goal of improving accuracy in finding true positive matches. To evaluate performance we used a set of 103 queries for which the true positives in yeast had been annotated by human experts, and a popular measure of retrieval accuracy (ROC) that can be normalized to take on values between 0 (worst) and 1 (best). The modifications we consider novel improve the ROC score from 0.758 ± 0.005 to 0.895 ± 0.003. This does not include the benefits from four modifications we included in the ‘baseline’ version, even though they were not implemented in PSI-BLAST version 2.0. The improvement in accuracy was confirmed on a small second test set. This test involved analyzing three protein families with curated lists of true positives from the non-redundant protein database. The modification that accounts for the majority of the improvement is the use, for each database sequence, of a position-specific scoring system tuned to that sequence’s amino acid composition. The use of composition-based statistics is particularly beneficial for large-scale automated applications of PSI-BLAST.
INTRODUCTION
BLAST and PSI-BLAST are widely used programs for detecting sequence similarities, including subtle ones, in searches of protein sequence databases (1,2). PSI-BLAST’s basic strategy is to construct a multiple alignment from the output of a BLAST protein database similarity search, abstract a position-specific score matrix from this multiple alignment, and search the database anew using the score matrix as query. The process may be iterated many times, as new significant similarities are found.
The ways in which PSI-BLAST estimates the statistical significance of the similarities it finds, and constructs position-specific score matrices from multiple alignments, are amenable to a variety of modifications. Some of these involve simple parameter adjustments, but others are based on deeper theoretical considerations. We sought to investigate the effect of a large number of proposed refinements on PSI-BLAST’s sensitivity to distant sequence relationships, with the aim of incorporating any that proved beneficial. To this end, we constructed a test set of query sequences, and annotated for each those yeast sequences that were related. We tested various versions of PSI-BLAST with a protocol described in the next section. To evaluate the performance of PSI-BLAST versions, we pooled the output from all queries, ranked by E-value, and plotted false positives versus true positives.
The outlined procedure was used to test over a dozen ideas for improving PSI-BLAST search accuracy. Among these, fewer than half proved of value. Nevertheless, PSI-BLAST version 2.2.1, which incorporates all useful changes found, is substantially more accurate than the original program. Among the modifications tested are four that we considered ‘known’ and could reasonably have been implemented in the original PSI-BLAST (version 2.0.1) or were published elsewhere (3). We included these four improvements first in what we call the ‘baseline’ version of PSI-BLAST, and in what follows we do not count the improvement that they yield. The remaining modifications were added, usually one at a time, in what seemed to us a plausible order. Among the modifications we included in the baseline version of PSI-BLAST are: (i) the ability to run the Smith–Waterman algorithm on all alignments reported, and (ii) the filtering of database sequences for the presense of ‘low-entropy’ segments of restricted amino acid composition. The modification that yields most of the improvement above the baseline version is the use of statistics tuned to the composition of the query and database sequences to evaluate the significance of local alignments.
THE TEST SET AND SEARCH PROTOCOL
We used as a primary test set some queries originally constructed for the purpose of detecting proteins with widespread regulatory and signal-transduction domains in the yeast Saccharomyces cerevisiae and the nematode worm Caenorhabditis elegans (4). Lists of true positives in yeast were delineated and curated by human experts (L.A. and E.V.K.).
An earlier version of this query set with 105 sequences was used to test IMPALA in Schäffer et al. (5) and called the aravind105 set. For the tests reported here we used 103 queries of which 91 are common to the aravind105 set. The changes in the new query test set are due to: 12 useful queries added after the IMPALA project started; nine queries dropped due to having no apparent true positives in yeast; five queries dropped due to overlaps with other queries and/or difficulties in identifying all true positives because of the PSI-BLAST corruption problem discussed below. Even though the methods presented here largely solve the PSI-BLAST problems that made some annotations difficult, we did not reintroduce the problematic query sequences. Starting with the copy of the yeast proteome used by Schäffer et al. (5) and containing 6406 proteins, we dropped 65 proteins that were identical or nearly identical to substrings of proteins remaining in the yeast database to avoid double counting what were really single true positives.
At the start of this project, the 103 lists of true positives included 918 total matches. As a result of testing with hundreds of PSI-BLAST versions we identified 87 more true positives, giving a total of 1005 for a mean of 9.8/query (range: 2–123). Improved software is expected to detect new true positives, and the ability to add these to the original true positive set is important. Therefore, we chose to use match lists developed and curated in house in lieu of fixed external ones. The 103 queries, copy of the yeast database, and lists of true positives are available from ftp.ncbi.nlm.nih.gov under directory pub/impala/blasttest.
For our 103 queries, we had comprehensive lists of true positives only for the yeast database. However, PSI-BLAST achieves greater sensitivity to distant relationships when it constructs its score matrices from larger and more diverse sets of related sequences (6). If one is ultimately interested in relationships between a query and the members of a restricted database, such as the proteins in PDB (7), it pays to search a comprehensive sequence database with PSI-BLAST through several iterations, save the resulting position-specific score matrix (PSSM) as a ‘checkpoint’, and then restart using the checkpoint matrix to search the database of interest. We have used this general protocol in evaluating PSI-BLAST search accuracy. To evaluate each version of PSI-BLAST, we first compare every query sequence to the NCBI nr protein sequence database (8) (frozen on 2 February 2000 for consistency across tests) and save the matrix computed after the fifth PSI-BLAST iteration, or when the program stops because no further significant similarities are found. After this, each saved PSSM is compared to the yeast database, and the program’s accuracy in detecting distant relationships is evaluated through an analysis of the pooled search outputs.
EVALUATING PSI-BLAST SEARCH ACCURACY
Given a classification of (query sequence, database sequence) pairs as related or unrelated, it is useful to have an objective criterion for comparing the performance of database search programs. Eliding for the moment the precision of reported E-values, one may treat them simply as a means of ordering database search results, and then plot the number of false versus the number of true relationships found as E-values increase. Such a procedure yields a ‘sensitivity curve’ such as those shown in Figures 1–4. In general, one prefers to find as many true positives as possible before a given number of false positives, so the farther to the right the sensitivity curve lies the better. A problem in assessing the relative merit of different search procedures is that their sensitivity curves are high-dimensional objects subject to a variety of reasonable linear orderings (9–13). A measure of search ‘accuracy’ derived from sensitivity curves that has gained fairly wide use is the truncated receiver operating characteristic (ROC) (13). Let T be total number of true positives available to be found. For a fixed number of false positives n, the quantity ROCn is the proportion of the area in the rectangle [0,T][0,n] that lies to the left of the sensitivity curve. ROCn is a proportion, so its values lie between 0 (worst) and 1 (best). See Appendix A for more information.
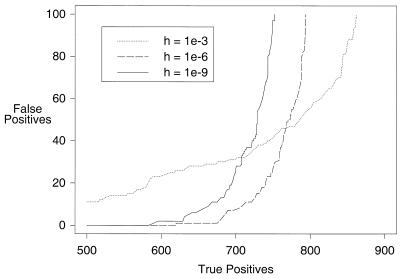
Figure 1.
Sensitivity curves of baseline program at three settings of the threshold parameter for including matching sequences in the PSI-BLAST multiple alignment. The versions compared are FWh10–3, FWh10–6 and FWh10–9. The sensitivity curve for FWh10–3 crosses the others because at this setting for h, three queries yield substantially corrupted results, while many other queries show improved search accuracy. The ROC100 scores for FWh10–9, FWh10–6 and FW10–3 are 0.713 ± 0.005, 0.758 ± 0.005 and 0.721 ± 0.020, respectively.
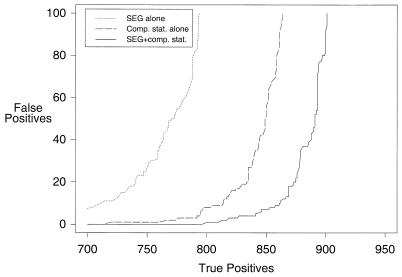
Figure 4.
Sensitivity curves showing the benefits of restricted score rescaling and of tuning the pseudocount parameter and the purging percentage. The versions compared are FWSDh0.005 and FWSDMb9p94h0.005.
The upper bound on false positives in calculating ROC values has a practical justification. Programs such as BLAST (1,2) report similarities only above a fairly high threshold score, so only a limited number of false positives appear in the output. Moreover, a researcher performing a database search generally is unwilling to analyze many weak matches, most of which are false positives, in the hope of finding a few more true positives. For comprehensive sequence database searches with a single query, Gribskov and Robinson (13) have recommended ROC50 as a figure of merit. Since we use pooled results from approximately 100 searches here, this would appear to translate into a recommendation of ROC5000. Because the database we ultimately search consists of only 6341 proteins and contains a mean of <10 true relationships per query, diminishing returns have a much earlier onset. After a total of about 100 pooled false positives have been observed, the number of additional false positives found per additional true positive escalates beyond a level most researchers would consider fruitful for detailed analysis. Thus, the figure of merit we will report for our pooled results is ROC100.
When analyzing database search programs, it is useful not only to compare their ROC values, but also to know when these values are significantly different. We approximate the ‘random’ distribution of ROC values for a given program through bootstrap resampling of the false positive database search results it returns. As described in Appendix A, this distribution should be approximately normal, so that we need estimate only its standard deviation, which can be calculated analytically.
As we investigate various methods for improving PSI-BLAST, we in essence are exploring the high-dimensional space of different possible combinations of methods for constructing PSSMs. Due to the combinatorial explosion, it would be impractical to consider all points in this space and find a version of PSI-BLAST that is on average optimal. Accordingly, we generally content ourselves with examining one proposed change at a time, and accepting it if it produces a program with improved search accuracy. Therefore we claim only that the version of PSI-BLAST we finally produce is a significant improvement over the baseline version of the program.
THE ‘BASELINE’ PSI-BLAST PROGRAM
To keep track of the various versions of PSI-BLAST we will be studying, we will use a shorthand to describe modifications to the original program. Single upper case letters will refer to modifications that we programmed so that they may be easily turned on or off in our test versions. Lower case letters will refer to changes in a numerical parameter, and will be followed by a numerical argument. In the released code, we do not enable the user to turn on/off each option individually because most combinations are of interest only in testing. As modifications to PSI-BLAST are described, letters signifying these changes will be introduced, but for reference purposes the meanings of all these codes are collected in Table 1.
Table 1. Abbreviations for modifications of BLAST and PSI-BLAST.
| F |
Filtering of database sequences with the SEG
program |
| W | Construction of final alignments with the Smith–Waterman algorithm |
| S | Composition-based statistics |
| R | Reversed sequence score normalization |
| D | Dispersed method for inferring amino acid frequencies from gaps |
| C | Concentrated method for inferring amino acid frequencies from gaps |
| M | Restricted score rescaling |
| bx | Pseudocount parameter (default 10) |
| px | Purging percentage (default 98) |
| hx | E-value threshold for inclusion in PSI-BLAST multiple alignment |
The ‘baseline’ version of PSI-BLAST with which we will begin our analysis is modified in four ways from that described by Altschul et al. (2). Because the first two changes are incorporated in all versions of PSI-BLAST we will study, neither will be assigned a letter for program designation. The other two changes will be assigned a letter because we will consider the effect of suppressing them.
Estimation of statistical parameters
BLAST and PSI-BLAST E-values are calculated using statistical and edge-effect parameters (3). These parameters cannot be calculated analytically, but must be estimated by prior random simulation for any amino acid substitution matrices and gap costs to be supported. BLAST release 2.2.1 employs parameter values estimated by a more accurate procedure (3,14) than that used for earlier releases (15).
Numerical precision and amino acid pair frequency ratios
The scores sij in the PAM (16,17) and BLOSUM (18) amino acid substitution matrices are constructed using the formula
sij = log rij 1
where rij is the ratio of the estimated frequency with which the amino acids i and j are aligned due to evolutionary descent, to the frequency with which they would be aligned by chance. All local alignment substitution matrices are implicitly of this form, with the base of the logarithm simply providing a scale factor (19,20).
For convenience, scores constructed using equation 1 are generally rounded to the nearest integer. However, as described below, we will have occasion to change, sometimes by small degrees, the scale of the matrices employed by BLAST and PSI-BLAST. Integral substitution scores of the usual scale discard too much precision for our purposes, so our new baseline version of BLAST and PSI-BLAST represents substitution matrices internally by the floating point rij rather than by the integral sij. This provides the programs with two opportunities for greater precision. [Note that the rij used in constructing substitution matrices generally do not appear explicitly in the literature. For the PAM matrices (16,17) we inferred these values from other published data, while for the BLOSUM matrices (18) we obtained these values from the authors, S. Henikoff and J. G. Henikoff.]
First, as described below, we re-evaluate all database sequences that participate in an alignment with good initial score. This is done using an integral score matrix but, since the rij are available, we employ a scaled-up version of the standard matrix. For greater precision, the log-odds scores to the usual scale derived from equation 1 are multiplied by 32 before rounding, as are any gap costs. (The usual scale varies among matrices.) To avoid confusion, ‘raw’ alignment scores are returned to the usual scale before printing.
Secondly, the availability of the rij gives PSI-BLAST an opportunity for greater precision in constructing amino acid frequency ratios for individual columns. In contrast to the frequency ratio Ri = Qi/Pi for amino acid i implied by equations 3–5 of Altschul et al. (2), where Qi and Pi are the predicted and background frequencies, respectively, for amino acid i, the baseline PSI-BLAST now derives this ratio using the equation:
![]() 2
2
Here, as defined and discussed further by Altschul et al. (2), fi is the weighted observed frequency of amino acid i for the column in question, α reflects the effective number of independent observations for that column, and β is the pseudocount parameter, which balances the relative importance given to data from the multiple alignment, and prior information on amino acid mutation propensities implicit in the reference substitution matrix. The ratio rij here replaces the earlier eλuSij. The earlier formulation was inferior both because the sij in general have low precision, and because the scale parameter λu was derived using standard amino acid background frequencies Pi (21) as opposed to the frequencies implicit in the original construction of the substitution matrix.
Sequence filtering
Alignments involving ‘low entropy’ protein segments with highly restricted or biased amino acid composition generally are not of interest, and BLAST has traditionally filtered such segments out of query sequences using the SEG program (22,23). However, because PSI-BLAST PSSMs are constructed from the output of database searches, it has been possible for low-entropy segments from database sequences to strongly influence the scores in these PSSMs, which are then aligned with unfiltered segments of other database sequences in further rounds of PSI-BLAST searching. Accordingly, we apply SEG filtering in our baseline program to database sequences as opposed to the query sequence in the original PSI-BLAST program; this change is designated with the letter ‘F’. For SEG filtering of the database we use a window-size of 10, a trigger complexity of 1.8 and an extension complexity of 2.1; this compares with the default values of 12, 2.2 and 2.5, respectively. The SEG parameters were tuned by testing them on all proteins in Mycoplasma genitalium plus some problematic queries that arose in studies by Y.I.W. We considered SEG parameter values to be safe if we could run all queries for five iterations without obvious signs of PSSM corruption, such as a program crash or massive increase in the size of the output file. We tried to find the lowest safe setting of the window and trigger-complexity, while keeping the difference between extension complexity and trigger complexity at 0.3 as suggested by the developer (J. Wootton, personal communication). It is not surprising that one can use more permissive SEG parameters for filtering the database because the composition-based statistics also help correct for composition biases. Applying SEG filtering to both query and database sequences appears to be overkill, and reduces search accuracy (data not shown).
Smith–Waterman alignments
Database search heuristics such as FASTA (24) and BLAST (1,2) are used in place of the Smith–Waterman local alignment algorithm (25) primarily for the purpose of speed. These heuristics may completely miss some significant alignments, and may produce non-optimal alignments for some sequence similarities they find. At a small price in speed it is possible to avoid the second of these problems in BLAST. One simply runs the full Smith–Waterman algorithm on the small fraction of database sequences the heuristic algorithm chooses to report. We have implemented this option, and represent it with the letter ‘W’; thus the baseline version of PSI-BLAST is designated FW.
THE CHOICE OF THRESHOLD PARAMETER AND THE PROBLEM OF CORRUPTION
A major potential problem for users of PSI-BLAST is that of PSSM ‘corruption’. At each iteration, PSI-BLAST constructs a multiple alignment, from which it then abstracts a PSSM. If a sequence S that is unrelated to the original query sequence Q is included in the multiple alignment, then the resulting PSSM will tend to produce highly significant alignments to sequences related to S as well as those related to Q. Such a PSSM is said to have been corrupted, and the search results from further iterations are unreliable (26). For the purpose of analysis, we shall say that the PSSM produced by one of our query sequences after five rounds of comparison to the nr database has become corrupted if it produces a false positive alignment with E-value ≤10–4 when compared to the yeast database.
With PSI-BLAST, a single corrupted query can yield many false positives with very low E-values. Because we consider pooled results, the sensitivity curves and corresponding ROC values may be greatly affected. One might argue that this penalizes corruption too heavily, and conclude that program evaluation should instead be conducted on a query-by-query basis (12). However, for researchers using PSI-BLAST on a large scale, even a small percentage of corrupted queries can be a major problem, and should therefore weigh heavily in any evaluation. First, the results of a corrupted search can be almost completely meaningless, and this casts considerable doubt on the reliability of results from the large majority of searches that are uncorrupted. Secondly, a corrupted search can consume a great quantity of computing time, exhaust all virtual memory causing a crash, or produce a huge volume of bogus output, limiting the applicability of PSI-BLAST to large-scale, automatic annotation projects.
One may attempt to avoid PSSM corruption by setting to a sufficiently low value the parameter h, which defines the maximum E-value a similarity may have for inclusion in the multiple alignment. We use the letter ‘h’, followed by its setting, in designating versions of PSI-BLAST. For most queries, the threshold h = 10–3 is sufficient to avoid corruption with the baseline program FW, but a small percentage yield corrupted PSSMs at this and even much lower values of h. Among our 103 query sequences, three become corrupted with h = 10–3, one with h = 10–6 and none at h = 10–9. The corresponding sensitivity curves are shown in Figure 1. Although one may avoid most corruption by lowering h sufficiently, one pays a price in search accuracy for the majority of queries that do not get corrupted. Among the thresholds considered, the highest ROC100 score, 0.758 ± 0.005, is for FWh10–6. Several of the refinements described below influence PSI-BLAST accuracy to a large degree by suppressing the appearance of false positives with low E-values in the iterated searches of the nr database, thereby allowing the value of the h parameter to be raised.
COMPOSITION-BASED STATISTICS
Our most important refinement is to compute the statistical significance of a match by taking into account the composition of the query and database sequences. The statistical significance of a local alignment produced by BLAST is assessed with an E-value, calculated using the formula:
E = Kmne–λS 3
where m and n are the effective lengths of the query sequence and database, S is the nominal score of the alignment, and λ and K are statistical parameters dependent upon the scoring system used and the composition of the sequences being compared (2). Because it enters equation 3 exponentially, the scale parameter λ is the far more important.
For alignments that are not allowed to contain gaps, the parameters λu and Ku (where the subscript indicates an ungapped scoring system) may be calculated analytically (19,27), but for gapped alignments λg and Kg must be estimated (3,14,15,28–33). BLAST employs estimates of λg and Kg precomputed from the comparison of a large number of ‘random protein sequences’ (3,15), generated using standard amino acid frequencies (21).
Occasionally, the amino acid compositions of a particular query and matching database sequence pair imply a λ′g substantially different from the precomputed λg, rendering unreliable the estimates of statistical significance based upon λg for any alignments between these sequences. Most often this is due to a similar, slightly biased amino acid composition shared by the sequences, yielding a λ′g < λg. Using the standard λg then results in a smaller E-value than is justified, sometimes off by several orders of magnitude. The same problem can arise with the PSI-BLAST comparison of PSSMs to protein sequences.
It is not practical to estimate statistical parameters by random simulation for each (query sequence, database sequence) pair for BLAST, or each (PSSM, database sequence) pair for PSI-BLAST. Thus we adopt the rescaling strategy introduced by the IMPALA program (5), an inverse form of PSI-BLAST. In brief, for a given pairwise comparison, the ungapped scale parameter λ′u can be calculated analytically (19). By rescaling the substitution scores, this parameter may be forced to equal λu, the ungapped scale parameter for a reference scoring system in the context of standard amino acid frequencies. Allowing gaps, with specified costs, changes the scale parameter for this reference scoring system to the pre-estimated λg. We simply assume the same holds for our rescaled substitution scores in the context of non-standard amino acid frequencies (2,5). This assumption is supported by random simulation (2,5). More details on the rescaling method are given in Appendix B.
It would be possible to rescale each PSSM for each database sequence, but this would slow down the program unduly. We rescale PSSMs and then recalculate alignments only for those pairwise comparisons that have yielded near-significant alignments after a first pass that employs scores scaled assuming a standard amino acid composition. Versions of PSI-BLAST that employ these composition-based statistics will be designated with the letter ‘S’.
With PSI-BLAST, it is important to use the rescaling strategy outlined here for the initial BLAST search as well as any subsequent PSI-BLAST searches, because corruption can occur at any stage. As a result, the alignment scores produced by the initial BLAST search in general do not correspond precisely to those implied by a standard substitution matrix, and even the same alignment involving two different database sequences can receive slightly different scores because of differing amino acid compositions.
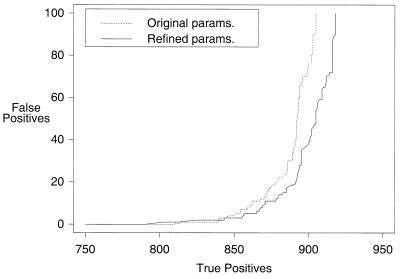
The gain in PSI-BLAST accuracy from the use of composition-based statistics is substantial. Whereas the baseline PSI-BLAST program FWh10–6 yields one corrupted PSSM and achieves a ROC100 of 0.758 ± 0.005, with composition-based statistics the program FWSh0.002 yields no corrupted PSSMs, and the ROC100 rises to 0.879 ± 0.003 (Fig. 2). Composition-based statistics alone go a long way toward removing the need for SEG filtering of database sequences. For our test set, the program WSh0.0002 yielded no corruption and achieved a ROC100 of 0.838 ± 0.003 (Fig. 2).
Figure 2.
Sensitivity curves comparing the effects of adding filtering of the database and composition-based statistics. The versions compared are FWh10–6, WSh0.0002 and FWSh0.002.
Other approaches to accounting for amino acid composition are possible when evaluating statistical significance. Mott (33) has derived an empirical formula for estimating statistical parameters that relies upon sequence composition. Employing this formula would require slightly less execution time than our rescaling procedure, but the statistics it produces may be somewhat less accurate (3).
Karplus et al. (34) suggest subtracting from the optimal local alignment score the score obtained from the best local alignment of the query with a reversed copy of the database sequence. We designate subtracting the reverse alignment score by the letter ‘R’; this procedure has the advantage that it accounts for higher-order statistical sequence properties beyond that merely of composition. A disadvantage is that the reversed alignment score introduces some random noise. To get a handle on this effect, one may make the rough assumption that after suitable normalization the scores for locally aligning a random query sequence to a database sequence and its reversed copy are independent random variables X1 and X2 that follow a standard extreme value distribution. Some calculus then shows that X1 – X2 has a probability density function with right-hand tail asymptotically equal to e–x, the same as the right-hand tail for X1 (K. Karplus, personal communication). However, the extreme value distribution is positively skewed with mean equal to Euler’s constant γ ≈ 0.577, so subtracting the reversed alignment score X2 tends to dilute statistical significance. The mean change in score corresponds to a multiplicative factor of eγ ≈ 1.8 in significance. Incorporating the reversed-alignment score adjustment procedure decreased the ROC100 score from 0.879 ± 0.003 for FWSh0.002 to 0.872 ± 0.003 for FWSRh0.01 (where the ROC score was maximum), and we have therefore not adopted this method. The R option does provide added protection against corruption arising from sequences with certain anomalous statistical properties, and may be useful in some contexts.
AMINO ACID FREQUENCIES FOR COLUMNS INCLUDING GAPS
To calculate the weighted observed frequencies for the 20 amino acids in a given column of a multiple alignment (hereafter called simply the observed frequencies), the baseline PSI-BLAST program considers only sequences of the multiple alignment actually containing a residue in that column, and ignores any sequences with a gap there (2). This choice, however, may throw away important information.
To test whether integrating information from sequences with gaps in a column could improve the sensitivity of the PSSMs constructed by PSI-BLAST, we implemented the following. When calculating sequence weights for a given column (2), include all sequences that participate in the column, whether with a residue or null character (gap). A null character is treated initially as if it were simply a twenty-first amino acid. After each sequence is assigned a weight, one is faced in general with a non-zero observed frequency for the null character. However, to calculate substitution scores for the given column, the observed frequencies of the 20 amino acids should sum to 1 (2), and thus the frequency for the null character must somehow be distributed among the amino acids. Two alternative ways to do this, which we call respectively the dispersed (letter ‘D’) and concentrated (letter ‘C’) methods, distribute the null character frequency in proportion to the standard background frequencies of all 20 amino acids, and in proportion to the observed frequencies of those amino acids present in the column in question. The dispersed method captures the idea that gaps in a given position should imply a degree of indifference as to which amino acids might occur there in related proteins, while the concentrated method involves no such assumption.
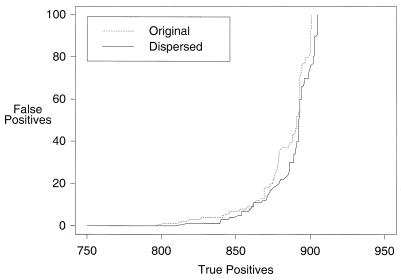
When added to FWS, the dispersed method yielded a small but not statistically significant improvement in PSI-BLAST accuracy, partially by better suppressing the appearance of false positives at low E-values, and thereby allowing the h parameter to be raised. Furthermore, with the dispersed method at h = 0.005, the pseudocount parameter (see below) could vary over a wide range of values without inducing corruption, whereas for the same pseudocount parameter range some corruption occurred with the concentrated or original method even at h = 0.002. The ROC100 value for FWSDh0.005 was 0.884 ± 0.002, in contrast to 0.878 ± 0.003 for FWSCh0.002 and 0.879 ± 0.003 for FWSh0.002 (Fig. 3). We adopted the dispersed method for further tests.
Figure 3.
Sensitivity curves showing the benefit of the ‘dispersed’ method for columns with gaps in the multiple alignment. The versions compared are FWSh0.002 and FWSDh0.005.
DISCORDANT SEQUENCE COMPOSITIONS AND RESTRICTED SCORE RESCALING
Sometimes when two sequences, or a sequence and a PSSM are compared, the corresponding ungapped scale parameter λ′u before score rescaling is greater than the parameter λu for sequences with standard amino acid composition. This is a rarer event than the case of main concern above, in which λ′u < λu, and the departure from the standard parameter tends to be of much smaller magnitude (5). Larger than standard values for λ′u usually result from sequences with discordant rather than similar amino compositions. If λ′u > λu, using the standard statistical parameters, without rescaling the substitution scores as described above, results in an overestimate of E-values. Nevertheless, there are several reasons it may be desirable not to rescale the substitution scores in this case.
First, discordant compositions alone provide some evidence against biological relatedness, so rescaling is more likely to yield a chance high-scoring and now statistically significant false positive than a newly statistically significant true positive. Secondly, the amino acid composition of some proteins has a mosaic structure, which effectively implies different λs appropriate for the comparison of different regions. Scaling scores to account for globally discordant compositions may yield exaggerated significance estimates for local alignments involving regions with more typical compositions. Thus, it may be desirable to rescale substitutions scores only when λ′u < λu, and not when λ′u > λu. We designate such restricted rescaling with the letter ‘M’.
Cases where λ′u > λu are relatively rare (5), so including restricted rescaling changed the ROC100 score only insignificantly, from 0.8837 ± 0.0025 for FWSDh0.005 to 0.8845 ± 0.0021 for FWSDMh0.005. We adopted this feature because it renders the program less susceptible to producing spurious results in the cases discussed above.
We have slightly changed two of PSI-BLAST’s default parameters. The program’s sequence weighting scheme (2,35) may somewhat overweight closely related sequences. Thus the original version of PSI-BLAST purged from its multiple alignment all but one copy of aligned sequence segments ≥98% identical, and this percentage has been changed to 94, indicated by ‘p94’. Also, the pseudocount parameter (2), which balances multiple alignment data with prior knowledge of amino acid substitutability, and originally set empirically to 10, has been changed to 9, indicated by ‘b9’. The best values for these parameters appear to change slightly from one version of PSI-BLAST to another, and so we have attempted to optimize them only as a final step. Combined with the other changes discussed above, the refined parameters yield a ROC100 of 0.895 ± 0.003 for FWSDMb9p94h0.005 (Fig. 4).
IDEAS THAT DID NOT WORK
We have discussed several modifications to PSI-BLAST that in combination significantly improve search accuracy. We also tested a substantial number of modifications that did not yield improved results, several of which we describe briefly here. Most of these modifications were motivated by appealing theoretical ideas, so it is possible that some refinement or combination of these approaches could still prove fruitful.
Adaptive pseudocount parameters
The range of evolutionary divergence optimally detected by a given amino acid substitution matrix is perhaps best captured by the matrix’s relative entropy (20,36), and the concept of average relative entropy is easily extended to PSSMs that are constructed using log odds ratios. A scoring system whose relative entropy is too high will tend to detect only closely related sequences, whereas one whose relative entropy is too low is optimized for detecting relationships so distant that they are not distinguishable from chance in any case. As a result, most substitution matrices used in database searches are tuned to distant but still detectable evolutionary distances (16–18). PSI-BLAST’s pseudocount parameter affects the average relative entropy of the PSSMs the program produces, with a higher value of the parameter yielding lower average relative entropies. Thus it may be advisable to choose the pseudocount parameter adaptively, so as to produce PSSMs with a ‘target’ average relative entropy near that associated with the most effective amino acid substitution matrices (34). However, our implementation of adaptive pseudocounts degraded search accuracy. It may be possible to parse PSI-BLAST multiple alignments so as to determine those regions most important for detecting distant relationships, and base average relative entropy calculations on those regions alone.
Sequence weight scaling
In calculating the ‘observed’ residue frequencies for a given column of a multiple alignment, it is important to weight the raw counts of amino acids appearing in the column to correct for correlations among the sequences included in the alignment (2). Many sequence weighting schemes have been proposed, of varying sophistication and ease of implementation (35,37–46). PSI-BLAST implements a slight modification (2) of the method of Henikoff and Henikoff (35), because it is simple and rapidly computable. However, this method has no strong mathematical foundation, and the weights it produces may tend to either overweight or underweight highly correlated sequences. We introduced an extra exponent parameter into the weighting scheme, transforming the weights w1,w2, w3, … to cw1P,cw2P, cw3P, …, where the coefficient c is chosen so the new weights sum to 1. As the exponent p was varied about 1, no clear improvement in PSI-BLAST performance could be observed. We measured the performance of exponents 0.9, 0.95, 1.05, 1.10, as possible enhancement to FWSDMb9p94h0.005. The ROC100 scores were 0.892, 0.892, 0.894, 0.890, all slightly lower than 0.895 for FWSDMb9p94h0.005. It is an open problem to provide theoretical justification for why p = 1.0 should perform near optimally.
Window-based amino acid composition calculations
Because amino acid composition may vary through a protein, one may argue that an alignment’s statistical significance should be assessed based upon the ‘local’ composition near the sequence segments involved in the alignment. Accordingly, we implemented a procedure whereby the amino acid compositions used for rescaling scores for a particular alignment are drawn from the sequence segments constituting the alignment, plus a window of fixed length to each side of each segment, although shorter if the end of the protein is encountered. This procedure was executed in lieu of the restricted score rescaling discussed in the previous section. A Bayesian variation involved assuming a prior standard background amino acid composition (21), which entered the amino acid frequency calculation in the form of pseudocounts. For window lengths 50, 100 and 200, and using Bayesian amino acid pseudocounts or not, we observed no improvement in accuracy using an earlier implementation of composition-based statistics. We remeasured the performance of windows of 100 and 200 without pseudocounts as a modification to FWSDMb9p94h0.005. The ROC100 score decreased significantly from 0.895 to 0.852 for window length 100 and to 0.866 for window length 200.
Generalized affine gap costs
Most popular sequence alignment and database search programs employ affine gap costs, which charge a fixed penalty for the existence of a gap, and an additional penalty for each residue within a gap (47–50). Altschul (51) has suggested permitting a gap to leave an arbitrary and in general different number of residues in both sequences unaligned; the cost of a gap involving x and y residues in the two sequences, where x ≤ y, would be a + b(y – x) + cx. We implemented such generalized affine gap costs for use with BLAST and PSI-BLAST, and explored a variety of plausible settings for the gap cost parameters, but did not observe any improvement in PSI-BLAST accuracy vis a vis traditional affine gap costs.
TESTING PSI-BLAST VERSIONS WITH AN INDEPENDENT QUERY SET
To confirm that the refinements to PSI-BLAST described above yield improved accuracy in detecting distant relationships, we tested several versions of PSI-BLAST on their ability to detect relationships within three diverse protein families whose members within the nr database of 2 February 2000 have been delineated. For each family we employed two separate queries, and we pooled the results from the six runs to generate ROC scores. Each query was compared to the nr database through five rounds of PSI-BLAST searching, and the search results returned on a final sixth round against nr were evaluated.
The three protein domain families employed in this test were: (i) DHH phosphoesterases (52), a family of predicted phosphoesterases with a broad spectrum of substrates; (ii) POZ domains (53,54), a family of eukaryotic and viral domains involved in specific protein–protein interactions; (iii) metallo-β-lactamase domain proteins (55), containing a metal-dependent hydrolase domain particularly abundant in archaea, but also present in a wide variety of bacterial and eukaryotic proteins. A list of family members, and the frozen nr database are available upon request. The query sequences used respectively to seek members of these families have gi numbers 4982166 (positions 6–297) and 2498554 (7–317); 482321 (50–169) and 2315751 (95–205); and 115023 (36–257) and 1172877 (536–778).
Compared to the development query set used for refining PSI-BLAST, this set contained many fewer queries, but many more true positives per query. The aggregate point of diminishing returns appeared to occur near 50 false positives, so we used ROC50 scores to compare versions of PSI-BLAST. Those versions we tested were FWh10–6, WSh0.0002, FWSh0.002, FWSDh0.005, and FWSDMb9p94h0.005. The results appear in Table 2 along with ROC100 scores from our development query set. There is broad agreement on relative PSI-BLAST version accuracy between the two sets of queries. For the development set, the use of the dispersed method to score columns containing gaps led to a statistically non-significant increase in accuracy, while for the test set it led to a statistically non-significant decrease. However, there is strong agreement that FWSDMb9p94h0.005 is far superior to the baseline version of PSI-BLAST with which we began.
Table 2. Search accuracy of PSI-BLAST, measured using development and test query sets.
| PSI-BLAST version |
Aggregate ROC100 score
for score for development query set versus yeast |
Aggregate ROC50 test query set
versus nr |
| FWh10–6 | 0.758 ± 0.005 | 0.615 ± 0.004 |
| WSh0.0002 | 0.838 ± 0.003 | 0.839 ± 0.004 |
| FWSh0.002 | 0.879 ± 0.003 | 0.906 ± 0.003 |
| FWSDh0.005 | 0.884 ± 0.002 | 0.902 ± 0.002 |
| FWSDMb9p94h0.005 | 0.895 ± 0.003 | 0.910 ± 0.003 |
THE ACCURACY OF BLAST DATABASE SEARCHES
Most of the refinements discussed above concern the construction of PSSMs, and are therefore applicable to PSI-BLAST but not to simple BLAST searches. The exceptions are composition-based statistics, and restricted score rescaling. To test whether these refinements led to improved BLAST as well as PSI-BLAST accuracy, we compared both our development and test query sets directly to their respective target databases using the programs FW, FWS and FWSM. For the development set, the ROC100 scores were 0.529 ± 0.003, 0.522 ± 0.003 and 0.525 ± 0.003, respectively. For the test set, the ROC50 scores were 0.118 ± 0.002, 0.112 ± 0.002 and 0.113 ± 0.003, respectively.
Composition-based statistics do not appear to improve BLAST search accuracy, even when restricted score rescaling is also incorporated. One reason for the discrepancy in these results with those for PSI-BLAST is that a rare unrelated database sequence with an anomalously low E-value has only a minor effect on the BLAST ROC score, but by corrupting the resulting PSSM it can have a major negative effect on that for PSI-BLAST. Some users may find it desirable to use composition-based statistics and restricted score rescaling even for BLAST, to avoid rare spuriously low E-values. These changes have the disadvantage of frequently returning different scores for identical alignments, due to the varying compositions of database sequences, and of rarely returning scores that correspond precisely to those implied by the standard, unscaled substitution matrix used. For these reasons, we recommend the changes described above primarily in the context of PSI-BLAST.
We note that using our testing protocol, the ROC100 for the development set improves from ∼0.53 using one round of searching with BLAST to 0.89 using six rounds (five versus nr and one versus yeast) of searching with PSI-BLAST. This reinforces the conclusions of previous studies (56–60) that most biologists who still commonly use only BLAST for protein queries would benefit substantially by switching to PSI-BLAST.
THE PRECISION OF BLAST AND PSI-BLAST E-VALUES
The ROC method we have used to compare various versions of BLAST and PSI-BLAST relies on reported E-values only to sort into one list the matches for different queries. The accuracy of E-values in describing statistical significance is relevant within the program only for setting the h parameter, whose values have in any case been chosen empirically. However, a user of BLAST or PSI-BLAST may wish to rely upon reported E-values in evaluating the potential relevance of similarities found. Accordingly, we test here how well the E-values returned by these programs reflect the distribution of scores produced by chance.
To obtain a large number of relatively independent query sequences, we selected 1000 Escherichia coli proteins from GenBank (8) with GI numbers ending in 1, 2 or 3. For BLAST, we compared these sequences to a ‘shuffled’ version of our yeast database, in which the letters of each sequence were randomly permuted. For PSI-BLAST, we compared each query to the unscrambled nr database through five iterations or until convergence, and then compared the resulting PSSM to the scrambled yeast database. Because we were unable to assess theoretically the magnitude of the effect restricted score rescaling has on random score distributions, we studied versions of BLAST and PSI-BLAST that include and exclude this feature.
Specifically, we tested versions of FWSDh0.005 and FWSDMb9p94h0.005 for PSI-BLAST, and the corresponding versions FWS and FWSM for BLAST. For the 1000 query sequences, we recorded the total number or alignments returned with E-value ≤ 1.0, 0.1 and 0.01, which by theory are expected to be near 1000, 100 and 10 respectively; the results are shown in Table 3. In all cases, the number of alignments observed at various E-values are within a factor of two of the number predicted. The reported E-values for BLAST appear slightly high (i.e. conservative), while those for the version of PSI-BLAST without restricted score rescaling appear slightly low.
Table 3. Precision of BLAST and PSI-BLAST E-values, measured with 1000 queries.
| BLAST or PSI-BLAST version |
Number of alignments with E-value
less than or equal to |
|
|
| 1.0 | 0.1 | 0.01 | |
| FWS | 759 | 60 | 10 |
| FWSM | 641 | 53 | 8 |
| FWSDh0.005 | 1179 | 132 | 18 |
| FWSDMb9p94h0.005 | 815 | 86 | 14 |
IMPLEMENTATION
We use the terms BLAST and PSI-BLAST throughout to refer to versions of the command-line program more precisely called blastpgp. There is also a Web-based version (http://www.ncbi.nlm.nih.gov/BLAST/, follow hyperlinks there), which differs primarily in how the options are set, and in that the user can control which matching sequences get used in constructing the PSI-BLAST PSSM. Since it is impossible to synchronize the public releases of the command-line blastpgp (because it is part of the larger NCBI software toolkit), the Web version and papers, we summarize how the modifications described herein are encapsulated in different numbered versions of the released code.
The improved statistical parameters λ and K were first made available in version 2.1.3. The new ‘edge-effect’ parameters (3) accounting for the fact that alignments are unlikely to begin near the ends of sequences, are in version 2.2.1. The improved precision and use of proper amino acid pair frequency ratios started in version 2.1.1. Version 2.1.1 made available Smith–Waterman alignments (W) as a command-line option only, but disallowed its use on the Web page because of worst-case running time. We evaluated the effect on search accuracy of turning off W on versions of PSI-BLAST. The resulting ROC100 scores for our development query set improve from 0.780 ± 0.005 for Fh10–6 to 0.884 ± 0.004 for FSDMb9p94h0.005.
Version 2.1.1 first made available an option that combines filtering of database sequences (F), a preliminary implementation of composition-based statistics (S) and restricted score rescaling (M), under the combined name ‘composition-based statistics’. Version 2.2.1 includes a better implementation of S. Version 2.1.1 and beyond implement the dispersed method for inferring frequencies from gaps (D), but it cannot be turned off. No released version implements reversed sequence score normalization (R). Among the three numerical parameters, the user can control the settings of the pseudocount parameter (b) and the E-value threshold for inclusion in a PSI-BLAST multiple alignment (h), but not the purging percentage (p). Versions 2.0.* used b10p98h0.001 by default. Version 2.1.1 changed the default to b7p98h0.002, which is conservative, since we showed that one can safely raise h to 0.005. Version 2.2.1 changes the default again to b9p94h0.005.
DISCUSSION AND CONCLUSION
PSI-BLAST is a widely-used extension to BLAST that permits iterative searching, and is particularly good at finding distant relationships. There have been several evaluations of PSI-BLAST accuracy published by other groups (56–60) which show there was substantial room for improvement in the accuracy of versions 2.0.*. A particularly frustrating limitation to using PSI-BLAST for large-scale automated protein analysis was that on a small, but certainly not negligible percentage of queries, false positives could enter the list of matches at one iteration with an E-value low enough to corrupt the PSSMs constructed for searching in subsequent iterations. In some cases the corruption got so bad that the program would exhaust all virtual memory and crash, or produce tens of megabytes of worthless output.
We have described a long list of modifications that were implemented in a large-scale attempt to improve PSI-BLAST accuracy in protein database searching. Of course, there are more ideas left to test, especially involving methods for generating the multiple alignments used to construct the PSI-BLAST PSSMs. However, the improvements described herein are substantial enough that it seemed desirable to suspend the testing of new ideas long enough to quantify these improvements and make them publicly available. The modifications that improved performance are retained in PSI-BLAST version 2.2.1. Most of these were introduced, at least in preliminary implementations in version 2.1.1. Version 2.1.1 appears to have nearly eliminated the corruption problem in PSI-BLAST.
The change that yielded the majority of the improvement is the use of ‘composition-based’ statistics to re-evaluate candidate alignments. Composition-based statistics take into account the letter frequencies in database sequences, and adjust the scale of the query PSSM accordingly. The use of composition-based statistics represents a more careful interpretation of the statistical theory behind BLAST (19,27), initially assessing the significance of a local alignment between two sequences, and then extending the results to database searching. The original BLAST implementation of that extension codified the questionable assumption that all database sequences should be treated as if they have the same average letter frequencies. This assumption is dangerous in the iterative context of PSI-BLAST, where allowing false positives to enter the set of matches used to construct a PSSM can lead to corruption. We implemented composition-based statistics efficiently by using them only to re-evaluate candidate matches identified using the average composition assumption.
Incorporating composition-based statistics substantially improved the accuracy of PSI-BLAST searches, primarily by decreasing the retrieval of false positives, and thereby suppressing the corruption of constructed PSSMs. This modification of PSI-BLAST is most important for large-scale searches, for example during genome annotation, and for all searches with compositionally-biased queries. However, our results are averaged over many cases, and for individual queries the use of composition-based statistics, or indeed any of the other refinements we have introduced, may degrade performance.
We measured the performance of our modifications on a set of 103 query sequences, with lists of true positives in yeast curated by human experts. On this set the ROC100 score improved from 0.758 ± 0.005 to 0.895 ± 0.003, even without accounting for four improvements implemented in the baseline version. Nevertheless the gap between our current best ROC100 score of 0.895 and the best possible score of 1.0 is substantial. More research is needed to identify new algorithmic and/or statistical refinements to narrow this gap further.
Our testing protocol involved saving a checkpoint PSSM after five PSI-BLAST rounds versus the nr database, and then comparing that checkpoint to an annotated yeast database. The choice of five rounds versus nr was based on early experiments which suggested that five initial rounds of searching produced results comparable to 10. We performed a post facto evaluation of this choice by saving the PSSM generated by PSI-BLAST version FWSDMb9p94h0.005 after one to 10 rounds versus nr. The resulting ROC100 scores were: 0.754, 0.829, 0.861, 0.886, 0.895, 0.896, 0.894, 0.893, 0.891 and 0.895. From round 5 through round 10, the scores move slightly up and down, but there is no corruption with any query in any round. We also recorded the number of queries that converged after each round versus nr, meaning that no new matches were found with an E-value ≤ h = 0.005. The numbers of converged queries out of 103 were: 2, 19, 27, 43, 55, 60, 67, 72, 75 and 78. These experiments suggest that for large-scale, automated applications, running PSI-BLAST for five to six rounds (which corresponds to saving the checkpoint computed after four to five rounds) may reveal most of the matches that could be found by running until convergence.
APPENDIX A
Estimating the standard deviation of ROC scores
Fix a query sequence and consider any database search method that ranks the relatedness of database sequences to the query (e.g., by using E-values). For simplicity, classify each database sequence as either a true positive or a false positive with respect to the query. Let i = 1,2,3 … index the rank of the false positives, and let ti be the number of true positives ranked ahead of the ith false positive. For the Appendix, we assume for simplicity that there are no ties in rank, although ties can be accounted for easily in the estimates below, and were accounted for in the ROC scores and standard deviations reported above. The accuracy of the search method can be assessed by the ‘receiver operator characteristic’ for n false positives, defined as:
![]() 4
4
Here T is total number of true positives in
the database, and we used either n = 50
or 100 in the results reported. The sum alone in equation 4 we
denote ![]()
Given two search methods, resampling with a bootstrap (described in detail below) can assign an empirical p-value to the difference between their corresponding ROCn values. For each bootstrap sample, calculate the difference between the sample ROCn’s and then note the position of the actual difference within the bootstrap distribution. This appendix announces some simple analytic results that can be used to decide whether the difference between two ROCn values is statistically significant under bootstrap resampling. The mathematical details will be published elsewhere (Czabarka and Spouge, manuscript in preparation).
Two bootstrap resampling schemes could be used: either resample all sequences or resample only from the false positives, leaving true positives fixed. Usually, the set of true positives is well characterized, so the false positives generate the real ‘noise’ in the ROCn measurement. When only the false positive sequences are resampled, the analytic results simplify because the denominator (nT) in equation 4 remains constant. The following presents results on resampling only the false positive sequences.
Let F be the total number of false positives in the database. A bootstrap sample consists of F false positives sampled uniformly, independently and with replacement from the entire set of false positives. Let Fi be the (random) number of times that the false positive of retrieval rank i is resampled. Define the random variable Nn to be the smallest integer satisfying
![]() 5
5
i.e., the false positive ranked n in the resampling has original rank Nn.
The false positive sequences contributing to ROCn, for any reasonable choice of n, form a very small proportion of the database. Thus, the distribution of the ROCn under resampling can be approximated by taking each false positive in its rank order and sampling it Fi times, where Fi is chosen from a Poisson distribution with mean 1. One can stop when the total of the Poisson sample counts reaches n.
The equivalence of an experiment resampling false positives and an experiment sampling from a Poisson distribution implies that many important quantities concerning ROCn can be approximated analytically. For example, by definition, the resampled, non-normalized ROCn is:
![]() 6
6
The mean µ(Rn)
and the variance σ2(Rn) have analytic forms that can
be practically computed under all conditions (although they are
not shown here because they are too complex as formulas). Mathematical
theorems show that under typical conditions µ(Rn) ≈ Σ1 ≤ i ≤ n ti = ![]() and σ2(Rn) ≈ Σ1 ≤ i ≤ n (tn+1 – ti)2. Moreover,
the distribution of Rn is
close to normal, so the approximate standard deviation can be used
to provide an approximate p-value.
and σ2(Rn) ≈ Σ1 ≤ i ≤ n (tn+1 – ti)2. Moreover,
the distribution of Rn is
close to normal, so the approximate standard deviation can be used
to provide an approximate p-value.
These distributional results can be extended to differences between resampled ROCn’s, which is of use when comparing a pair of retrieval methods. Let us use the prime symbol (′) to denote quantities pertaining to a second search method. Then:
![]()
and
![]()
![]() 7
7
The summation condition Di = D′j indicates a sum over sequences common to the first n false positives in both retrieval lists. The notation expresses the condition that false positive Di in the first list is the same as false positive D′j in the second list.
The comparisons of experimental data above use the estimate σ2(Rn – R′n) ≈ σ2(Rn) + σ2(R′n) and ignore the final correlation term in equation 7, so that we can assign a standard deviation to each version rather than each pair. The omission of the third term, which is subtracted, leads to an overestimate of the standard deviation.
APPENDIX B
Matrix rescaling
The method of rescaling matrices can be be summarized as follows. Let λu be the ungapped scale parameter (19) for the reference substitution matrix (e.g., BLOSUM62) in the context of standard amino acid frequencies (21). Suppose that Q is the query sequence and Dinit is the initial matching database sequence.
(i) Multiply the gap costs by a scaling factor f, and divide λu by f. We currently set f = 32 for five extra bits of precision.
(ii) Compute the residue frequencies in Q.
(iii) If filtering (modification F) is turned on, let D be the output of filtering Dinit with SEG (currently using parameters 10, 1.8, 2.1). If F is off, let D = Dinit. Compute the residue frequencies for D, ignoring all segments that were replaced with X’s by SEG.
(iv) Given frequency ratios Rij for the current score matrix, compute scaled-up scores Sij as the nearest integer to log(Rij)/λu. Since λu has been previously divided by f, this will have the effect of multiplying each score by roughly f.
(v) Using the residue frequencies for Q and D and the scaled up matrix scores Sij compute a match-specific λ′u (19).
(vi) Let the ratio r = λ′u/λu. If restricted score rescaling (modification M) is turned on, change r to min(r,1).
(vii) For each position i, j in the matrix, compute the rescaled score S′ij as the nearest integer to r · log(Rij)/λu.
Since the resulting rescaled score matrix S′ and gap costs are both scaled up by a factor of f, we divide the final raw alignment scores by f before printing the output. Note that at steps 4 and 7, the scaling is done in floating point first, and the nearest integer score is computed at the end.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Drs Jorja and Steven Henikoff for providing the frequency ratios used to derive the BLOSUM score matrices. Thanks to two referees for some very helpful suggestions.
References
- 1.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S.F., Bundschuh,R., Olsen,R. and Hwa,T. (2001) The estimation of statistical parameters for local alignment score distributions. Nucleic Acids Res., 29, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chervitz S.A., Aravind,L., Sherlock,G., Ball,C.A., Koonin,E.V., Dwight,S.S., Harris,M.A., Dolinski,K., Mohr,S., Smith,T. et al. (1998) Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science, 282, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schäffer A.A., Wolf,Y.I., Ponting,C.P., Koonin,E.V., Aravind,L. and Altschul,S.F. (1999) IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics, 15, 1000–1011. [DOI] [PubMed] [Google Scholar]
- 6.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The protein data bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. et al. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamber D. (1975) The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J. Math. Psychol., 12, 387–415. [Google Scholar]
- 10.Swets J.A. (1988) Measuring the accuracy of diagnostic systems. Science, 240, 1285–1293. [DOI] [PubMed] [Google Scholar]
- 11.Wilbur W.J. (1992) An information measure of retrieval performance. Information Systems, 4, 283–298. [Google Scholar]
- 12.Pearson W.R. (1995) Comparison of methods for searching protein sequence databases. Protein Sci., 4, 1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gribskov M. and Robinson,N.L. (1996) Use of receiver operating characteristic (ROC) analysis to evaluate sequence matching. Comput. Chem., 20, 25–33. [DOI] [PubMed] [Google Scholar]
- 14.Olsen R., Bundschuh,R. and Hwa,T. (1999) Rapid assessment of extremal statistics for gapped local alignment. In Lengauer,T., Schneider,R., Bork,P., Brutlag,D., Glasgow,J., Mewes,H.-W. and Zimmer,R. (eds), Proceedings of the Seventh International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 211–222. [PubMed]
- 15.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 16.Dayhoff M.O., Schwartz,R.M. and Orcutt,B.C. (1978) A model of evolutionary change in proteins. In Dayhoff,M.O. (ed.), Atlas of Protein Sequence and Structure. National Biomedical Research Foundation, Washington, DC, Vol. 5, suppl. 3, pp. 345–352.
- 17.Schwartz R.M. and Dayhoff,M.O. (1978) Matrices for detecting distant relationships. In Dayhoff,M.O. (ed.), Atlas of Protein Sequence and Structure. National Biomedical Research Foundation, Washington, DC, Vol. 5, suppl. 3, pp. 353–358.
- 18.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlin S. and Altschul,S.F. (1990) Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA, 87, 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F. (1991) Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol., 219, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson A.B. and Robinson,L.R. (1991) Distribution of glutamine and asparagine residues and their near neighbors in peptides and proteins. Proc. Natl Acad. Sci. USA, 88, 8880–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wootton J.C. and Federhen,S. (1993) Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem., 17, 149–163. [Google Scholar]
- 23.Altschul S.F., Boguski,M.S., Gish,W. and Wootton,J.C. (1994) Issues in searching molecular sequence databases. Nature Genet., 6, 119–129. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST – a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 27.Dembo A., Karlin,S. and Zeitouni,O. (1994) Limit distribution of maximal non-aligned two-sequence segmental score. Ann. Prob., 22, 2022–2039. [Google Scholar]
- 28.Smith T.F., Waterman,M.S. and Burks,C. (1985) The statistical distribution of nucleic acid similarities. Nucleic Acids Res., 13, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins J.F., Coulson,A.F.W. and Lyall,A. (1988) The significance of protein sequence similarities. Comput. Appl. Biosci., 4, 67–71. [DOI] [PubMed] [Google Scholar]
- 30.Mott R. (1992) Maximum-likelihood estimation of the statistical distribution of Smith-Waterman local sequence similarity scores. Bull. Math. Biol., 54, 59–75. [DOI] [PubMed] [Google Scholar]
- 31.Waterman M.S. and Vingron,M. (1994) Rapid and accurate estimates of statistical significance for sequence database searches. Proc. Natl Acad. Sci. USA, 91, 4625–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson W.R. (1998) Empirical statistical estimates for sequence similarity searches. J. Mol. Biol., 276, 71–84. [DOI] [PubMed] [Google Scholar]
- 33.Mott R. (2000) Accurate formula for P-values of gapped local sequence and profile alignments. J. Mol. Biol., 300, 649–659. [DOI] [PubMed] [Google Scholar]
- 34.Karplus K., Barrett,C. and Hughey,R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 35.Henikoff S. and Henikoff,J.G. (1994) Position-based sequence weights. J. Mol. Biol., 243, 574–578. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S.F. (1993) A protein alignment scoring system sensitive at all evolutionary distances. J. Mol. Evol., 36, 290–300. [DOI] [PubMed] [Google Scholar]
- 37.Altschul S.F., Carroll,R.J. and Lipman,D.J. (1989) Weights for data related by a tree. J. Mol. Biol., 207, 647–653. [DOI] [PubMed] [Google Scholar]
- 38.Sibbald P.R. and Argos,P. (1990) Weighting aligned protein or nucleic acid sequences to correct for unequal representation. J. Mol. Biol., 216, 813–818. [DOI] [PubMed] [Google Scholar]
- 39.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 40.Vingron M. and Sibbald,P.R. (1993) Weighting in sequence space: a comparison of methods in terms of generalized sequences. Proc. Natl Acad. Sci. USA, 90, 8777–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstein M., Sonnhammer,E.L.L. and Chothia,C. (1994) Volume changes in protein evolution. Appendix: A method to weight protein sequences to correct for unequal representation. J. Mol. Biol., 236, 1067–1078. [DOI] [PubMed] [Google Scholar]
- 42.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci., 10, 19–29. [DOI] [PubMed] [Google Scholar]
- 43.Eddy S.R., Mitchison,G. and Durbin,R. (1995) Maximum discrimination hidden Markov models of sequence consensus. J. Comput. Biol., 2, 9–23. [DOI] [PubMed] [Google Scholar]
- 44.Gotoh O. (1995) A weighting system and algorithm for aligning many phylogenetically related sequences. Comput. Appl. Biosci., 11, 543–551. [DOI] [PubMed] [Google Scholar]
- 45.Krogh A. and Mitchison,G. (1995) Maximum entropy weighting of aligned sequences of protein or DNA. In Rawlings,C., Clark,D., Altman,R., Hunter,L., Lengauer,T. and Wodak,S. (eds), Proceedings of the Third International Conference on Intelligent System for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 215–221. [PubMed]
- 46.Bailey T.L. and Gribskov,M. (1996) The megaprior heuristic for discovering protein sequence patterns. In States,D.J., Agarwal,P., Gaasterland,T., Hunter,L. and Smith,R. (eds), Proceedings of the Fourth International Conference on Intelligent System for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 15–24. [PubMed]
- 47.Gotoh O. (1982) An improved algorithm for matching biological sequences. J. Mol. Biol., 162, 705–708. [DOI] [PubMed] [Google Scholar]
- 48.Fitch W.M. and Smith,T.F. (1983) Optimal sequence alignments. Proc. Natl Acad. Sci. USA, 80, 1382–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul S.F. and Erickson,B.W. (1986) Optimal sequence alignment using affine gap costs. Bull. Math. Biol., 48, 603–616. [DOI] [PubMed] [Google Scholar]
- 50.Myers E.W. and Miller,W. (1988) Optimal alignments in linear space. Comput. Appl. Biosci., 4, 11–17. [DOI] [PubMed] [Google Scholar]
- 51.Altschul S.F. (1998) Generalized affine gap costs for protein sequence alignment. Proteins, 32, 88–96. [PubMed] [Google Scholar]
- 52.Aravind L. and Koonin,E.V. (1998) A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci., 23,17–19. [DOI] [PubMed] [Google Scholar]
- 53.Ahmad K.F., Engel,C.K. and Prive,G.G. (1998) Crystal structure of the BTB domain from PLZF. Proc. Natl Acad. Sci. USA, 95, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aravind L. and Koonin,E.V. (1999) Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol., 285, 1353–1361. [DOI] [PubMed] [Google Scholar]
- 55.Aravind L. (1999) An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol., 1, 69–91. [PubMed] [Google Scholar]
- 56.Park J., Karplus,K., Barrett,C., Hughey,R., Haussler,D., Hubbard,T. and Chothia,C. (1998) Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol., 284, 1201–1210. [DOI] [PubMed] [Google Scholar]
- 57.Muller A., MacCallum,R.M. and Sternberg,M.J.E. (1999) Benchmarking PSI-BLAST in genome annotations. J. Mol. Biol., 293, 1257–1271. [DOI] [PubMed] [Google Scholar]
- 58.Rychlewski L., Jaroszewski,L., Li,W.Z. and Godzik A. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information Protein Sci., 9, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sauder J.M., Arthur,J.W. and Dunbrack,R.L. (2000) Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins, 40, 6–22. [DOI] [PubMed] [Google Scholar]
- 60.Wallqvist A., Fukunishi,Y., Murphy,L.R., Fadel,A. and Levy,R.M. (2000) Iterative sequence/secondary structure search for protein homologs: comparison with amino acid sequence alignments and application to fold recognition in genome databases. Bioinformatics, 16, 988–1002. [DOI] [PubMed] [Google Scholar]