Abstract
Purpose
Targeting of the HER2 protein in human breast cancer represents a major advance in oncology, but relies on measurements of total HER2 protein and not HER2 signaling network activation. We utilized reverse phase protein microarrays (RPMAs) to measure total and phosphorylated HER2 in the context of HER family signaling to understand correlations between phosphorylated and total levels of HER2 and downstream signaling activity.
Experimental Design
Three independent study sets, comprising a total of 415 individual patient samples from flash frozen core biopsy samples and FFPE surgical and core samples, were analyzed via RPMA. The phosphorylation and total levels of the HER receptor family proteins and downstream signaling molecules were measured in laser capture microdissected (LCM) enriched tumor epithelium from 127 frozen pre-treatment core biopsy samples and whole tissue lysates from 288 FFPE samples and these results were compared to FISH and IHC.
Results
RPMA measurements of total HER2 were highly concordant (> 90% all sets) with FISH and/or IHC data, as was phosphorylation of HER2 in the FISH/IHC+ population. Phosphorylation analysis of HER family signaling identified HER2 activation in some FISH/IHC- tumors and, identical to that seen with FISH/IHC+ tumors, the HER2 activation was concordant with EGFR and HER3 phosphorylation and downstream signaling endpoint activation.
Conclusions
Molecular profiling of HER2 signaling of a large cohort of human breast cancer specimens using a quantitative and sensitive functional pathway activation mapping technique reveals IHC-/FISH-/pHER2+ tumors with HER2 pathway activation independent of total HER2 levels and functional signaling through HER3 and EGFR.
Keywords: HER2, breast cancer, reverse-phase protein microarrays, cell signaling, phosphorylated HER2
Introduction
The expression of HER family receptor tyrosine kinases has major biological impact on the pathogenesis of many solid tumors and is an important driving component of signal transduction networks that are deregulated in many cancers (1–3). Trastuzumab therapy offers significant disease-free and overall survival advantages in the metastatic as well as adjuvant settings for HER2 overexpressing breast cancer patients (4–7). However, recent findings suggest that the benefit of adjuvant trastuzumab may not be limited to patients with Her2 gene amplification (8). Hence, there is an emerging need for new additional diagnostic tests that allow the identification of new breast cancer patient subgroups that may also benefit from HER2-directed therapy.
Currently, HER2 status is routinely determined by immunohistochemistry (IHC), often with additional fluorescence in situ hybridization (FISH) to verify equivocal IHC results (9,10). Although IHC is the predominant method for assessing HER2, this assay could produce false positive or negative outcomes due to inter-operator variability (11–17). Moreover, IHC and FISH do not provide a quantitative measurement of total HER2 protein levels, nor do they provide data reflecting the activation status of the HER2 protein or the downstream signaling network itself, both of which may limit their effectiveness in patient selection for HER2-targeted therapeutics.
Several research groups have reported that the activation/phosphorylation status of HER2 protein provides significant information on breast cancer survival as compared to total HER2 (18–22). In this context, the reverse phase protein microarray (RPMA) represents a powerful tool for measuring and mapping the protein signaling architecture from clinical samples allowing both relative and absolute protein quantifications (23,24).
We utilized RPMA analysis and assessed its determination of HER2 expression and activation in formalin-fixed and paraffin-embedded (FFPE) and frozen breast cancer tissue samples. We evaluated these study sets in independent laboratories, and examined the relationship between a quantitative assay (RPMA) that measures both total and phosphorylation levels of HER2 and downstream HER family receptor-driven signaling with HER2 levels as measured by FDA-approved assays such as IHC and FISH. This analysis provides a starting point to understand the relationships between the quantitative measurement of important receptor tyrosine kinase levels in clinical samples, their relative levels of activation and measurements of downstream signaling.
Materials and Methods
Patient Samples and Tissue Processing
For the analysis of frozen tissue specimens, a total of 127 pretreatment breast cancer biopsy specimens collected in the ISPY-1 TRIAL (CALGB 150007/150012, ACRIN 6657) were subjected to laser capture microdissection (LCM) to enrich for tumor epithelium as described (25,26). All patients provided IRB-approved informed consent prior to specimen collection. For both data sets (frozen and FFPE tissue), central laboratory measurements of HER2 status (central IHC and/or FISH) were used for comparisons (Table 1). HercepTest™ (Dako, Glostrup, Denmark) was used for IHC measurements and Pathvysion® (Abbott Molecular, Wiesbaden, Germany) was used for FISH measurements of HER2 status in FFPE material. The anti-HER2 antibody (clone CB11, 1:100 diluton) (Biogenex, Fremont, CA) and the Tricolor HER2/TopoII/CEP17 probe kit (Abbott Molecular, Abbott Park, IL) was used for central IHC and FISH measurements in the frozen tissue samples, respectively. For the frozen tumor study set, a single pathologist was responsible for all IHC and FISH determinants. For the FFPE analysis, each tissue sample was reviewed by 2 independent pathologists.
Table 1.
HER2 Measurements for Patients in FFPE and Frozen Tissue Study Sets
| HER2 Status-FFPE Specimens | ||||||
|---|---|---|---|---|---|---|
| IHC- (IHC=0; IHC=1+; IHC=2+/FISH-) |
IHC=2+/FISH+ | IHC=3+ | ||||
| Surgical Specimen Training Set, n=73 | 57 (78%) | 10 (14%) | 6 (8%) | |||
| Surgical Specimen Validation Set, n=125 | 83 (66%) | 15 (12%) | 27 (22%) | |||
| Core Biopsy Training Set, n=31 | 18 (58%) | 2 (6%) | 11 (36%) | |||
| Core Biopsy Validation Set, n=59 | 36 (61%) | 6 (10%) | 17 (29%) | |||
| HER2 Status-Frozen Specimens | ||||||
| IHC- | IHC Indeterminate | IHC+ | FISH- | FISH Borderline | FISH+ | |
| Frozen, Microdissected Biopsy Set, n=127† | 96 (76%) | 2 (2%) | 28 (22%) | 43 (70%) | 2 (3%) | 17 (27%) |
FISH data available for n=62 cases; HER2 IHC data available for n=126 cases; 1 case between the two data sets is non-overlapping. FFPE=Formalin-fixed, paraffin-embedded
A total of 288 FFPE breast cancer samples diagnosed between 2000 and 2010 were included in this study (198 surgical specimens and 90 core biopsies). These were further divided into training sets (73 and 31, respectively) and validation sets (125 and 59, respectively). Protein extraction was performed as described (27,28). Only sections comprised of at least 85% tumor cells were analyzed. This investigation was approved by the Human Investigations Committee of the Technical University of Munich, Germany (project number: 2056/08). All patients gave informed consent.
Array Printing and Analysis
RPMA printing and analysis for the frozen tissue specimens was performed as described (25,26). For the FFPE tissue study, RPMAs were printed, stained and analyzed as described (29). Total protein levels were assessed in each sample by staining with Sypro Ruby Protein Blot Stain (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions for both data sets. Antibody staining intensities were quantified using the MicroVigene v3.5.0.0 software package (Vigenetech, Carlisle, MA).
Array Immunostaining
HER-related signaling pathway activation in tumor cells was evaluated by staining the arrays with various antibodies targeting key proteins in the HER signal transduction cascade (HER2, pHER2(Y1248), pHER3(Y1289), pEGFR(Y1086), pEGFR(Y1148), pEGFR(Y1173), pEGFR(Y992), pSHC(Y317), pFAK(Y576/Y577), pSTAT5(Y694)). Primary antibody dilutions and distributors are listed in Supplementary Table S1. Before use on RPMAs, antibody specificity was confirmed by Western blot analyses as previously described (26,30).
Quantification of HER2 protein expression by RPMA
For the quantification of HER2 protein carried out for FFPE specimens, purified recombinant HER2 protein (8.8 pg/nl start concentration; #PKSP011; Biaffin, Kassel, Germany) was printed alongside the patient samples in a six-point dilution curve. A signal intensity vs. concentration curve was plotted and HER2 concentrations were determined for each sample by interpolation of the array signal intensity to this standard concentration curve as previously described (30).
Western Blotting
Microdissected tumor epithelial cells from pretreatment specimens of selected patients were lysed in SDS sample buffer and resolved on 4–20% gradient Tris-Glycine polyacrylamide gels (Invitrogen), transferred to PVDF membrane, and probed with pHER2(Y1248) and β-actin antibodies (Cell Signaling; Supplementary Table S1).
Unsupervised Hierarchical Clustering and Statistical Analysis
Unsupervised hierarchical clustering of the FFPE data set was performed using Cluster and Tree View software (31). Subsequent to log transformation and center to median correction, average hierarchical clustering was performed. Cluster mapping for frozen tissue data was performed using the Ward method for two-way unsupervised hierarchical clustering in JMP v5.1 (SAS Institute, Cary, NC). Receiver operator characterization (ROC) analysis for determining positive RPMA staining thresholds for total and pHER2 in frozen LCM tissues was performed in JMP v5.1. T-tests and Wilcoxon Rank Sum tests and ANOVA analyses were performed using R v2.13.2. GraphPad Prism v5.02 (GraphPad Software, La Jolla, CA) was used to generate graphs.
Results
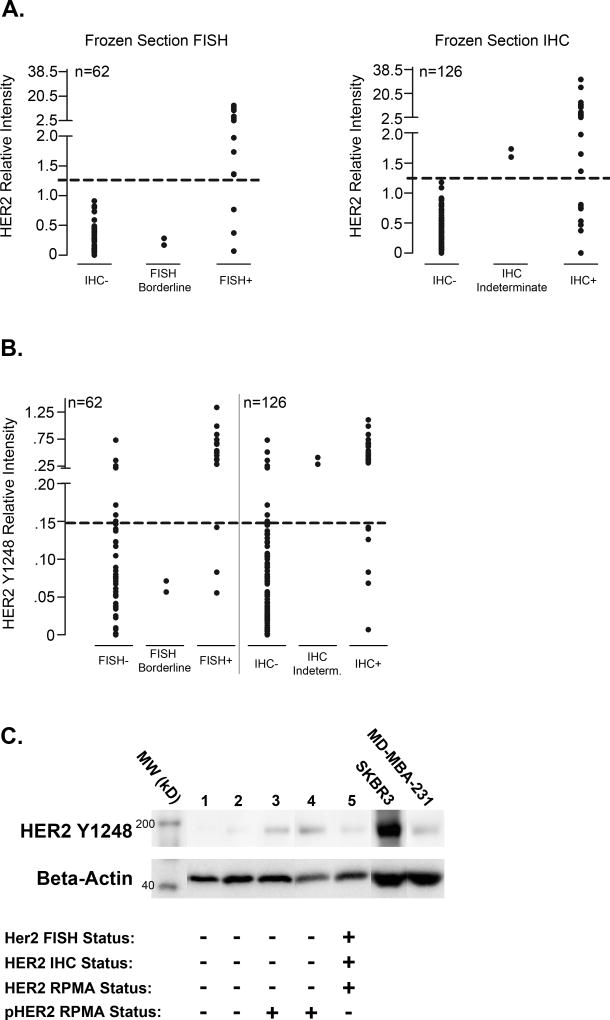
We first sought to understand the concordance of our total HER2 measurements in frozen tumor tissues by RPMA with current FDA-approved IHC and FISH methods. Table 1 shows the HER2 status distributions for the frozen tissue specimens based on central IHC and FISH measurements of HER2 status in the I-SPY 1 TRIAL. 22% of these cases were HER2-positive (HER2+) by central IHC (28/126) and 27% (17/62) were HER2+ by central FISH analysis (Table 1), which correlates well with rates of HER2 overexpression/amplification observed in the general population (32,33). A threshold level of relative staining intensity for total HER2 protein by RPMA analysis was selected based on ROC analysis, which minimized false-negative and false-positive results when compared with IHC- or FISH- populations (Figure 1A). An overall concordance of 95% with central FISH measurements and 94% with central IHC measurements was observed. These data demonstrate that RPMA-based continuous variable measurements of total HER2 protein levels in frozen tissues have excellent concordance with traditional IHC and FISH assessments.
Figure 1. Correlation of RPMA measurements of HER2 and pHER2 protein levels with IHC and FISH results for frozen breast tumor tissue specimens and identification of a HER2-/pHER2+ subpopulation of tumors.
A. RPMA measurements of HER2 protein expression in frozen, microdissected breast tumor epithelium were plotted against reported FISH status (left panel) or IHC results (right panel). A threshold intensity value that generated no false-positive values in either data set is shown as a dashed line. B. pHER2 relative intensity values measured by RPMA are plotted against FISH status (left) or IHC status (right) for frozen, microdissected tissue samples. A tentative threshold pHER2 intensity value of 0.15 is indicated by a dashed line. C. Immunoblot validation of pHER2 RPMA relative expression levels in frozen, microdissected tissues. Samples of microdissected tumor cells from 4 HER2- tumors (1–4) and one HER2+ tumor (5) were probed with antibody against pHER2(Y1248) to verify relative protein expression level differences measured in the same cases by RPMA. SKBR3 and MD-MBA-231 cell lysates were run as high and low level expression controls, respectively, and serve as an internal control for antibody specificity. Immunoblotting of beta-actin was used for normalization of protein loading. Central FISH and IHC status for HER2 as well as HER2 status by RPMA measurements for each tumor are shown. Immunoblot images are cropped for clarity.
We next used RPMA analysis to measure levels of phospho-HER2(Y1248) (pHER2) protein in our frozen sample set to determine total HER2 and pHER2 protein correlations as well as concordance between pHER2 and IHC and FISH-based HER2 assessments. In a similar manner to the threshold intensity determination for total HER2, we again used ROC analysis to determine an optimal threshold value for pHER2 positivity in the context of the IHC and FISH data. In this study set, 82% (14/17) of FISH+ tumors and 79% (22/28) of the IHC+ tumors exhibited above-threshold relative levels of pHER2 expression, suggesting that the majority of the HER2+ tumors in the frozen study set also exhibited activation of the receptor (Figure 1B). We also found that a subset of the HER2-negative (HER2-) tumors, 16% (7/44) and 8% (8/96) of the FISH and IHC- tumors, respectively, demonstrated pHER2 levels that were above threshold (Figure 1B). All but one of the 8 HER2 IHC-/pHER2+ tumors were also central FISH-. The remaining patient had central IHC data only (IHC-) due to a larger central IHC data set than FISH for the frozen tissue set. We were able to confirm our pHER2 RPMA relative measurements from frozen tumors in a small subset of cases by immunoblotting (Figure 1C). These results provide evidence for activation of the HER2 receptor in the absence of HER2 overexpression (Figure 1B), with HER2 activation levels as high (as a population) as IHC/FISH+ patients.
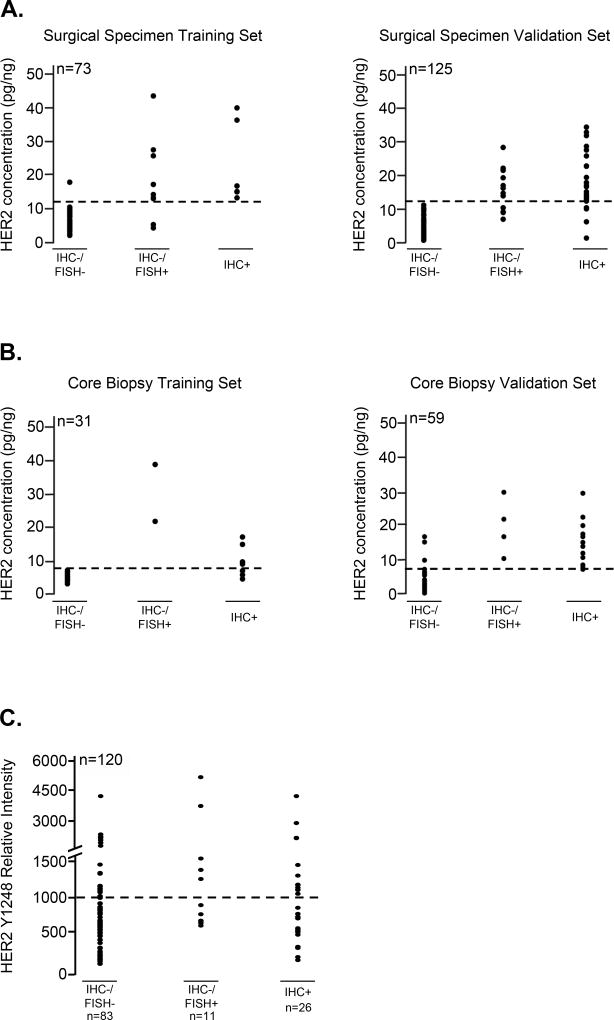
FFPE tissues are used in hospitals worldwide for diagnostic determination of HER2 expression. Therefore, we wanted to determine whether the small HER2-/pHER2+ subpopulation of patients found in frozen tumors could also be found in FFPE tissues. We identified an independent study set of 288 FFPE specimens which included both core biopsies (N=90) and surgical specimens (N=198). Overall, 29% (58/198) of the FFPE surgical specimen population was determined to be HER2+ by a 3+ IHC score or a 2+ IHC score and a FISH/CEP17 ratio >2.2 (Table 1). In the core biopsy set of FFPE specimens (N=90), 40% were HER2+ (36/90) (Table 1). Both of these figures trended slightly higher than estimates for HER2 positivity in the general population of breast cancers (15–25%) (32,33). Using RPMA, we assessed HER2 protein concentration in each FFPE patient sample. Array-based HER2 concentration thresholds were established for training sets of surgical specimens (N=73; HER2 threshold: 12 pg HER2/ng protein) and core biopsies (N=31; HER2 threshold: 7 pg HER2/ng protein) separately, to optimally distinguish IHC+ (HER2 IHC=2+/FISH+; IHC=3+) from IHC- (HER2 IHC=0 or 1+; IHC=2+/FISH-) tumor samples in each set. In FFPE tissues, IHC and RPMA-based HER2 assessment showed a concordance of 96% for surgical and 84% for core biopsy training specimens (Figure 2 A, B). Application of these RPMA-derived thresholds for total HER2 in the surgical validation set revealed an overall concordance between RPMA and IHC of 92% (Figure 2A, B). All surgical samples in this set determined as HER2- by IHC were also classified as HER2- by RPMA, and 8% (10 of 125) of IHC+ samples were determined as HER2- by RPMA. HER2 determination in the core biopsy validation set demonstrated an overall concordance of 93%. Only 3 of 59 (5%) samples that were scored HER2+ by IHC were negative using RPMA, and 1 of 59 (2%) samples classified negative by IHC showed a positive RPMA HER2 status (Figure 2A, B). Assessment of pHER2 levels in the surgical validation study set revealed pHER2 expression in 46% (17/37) of the IHC/FISH+ HER2 cases and in 26.5% (22/83) of the IHC/FISH- subset of samples (Figure 2C). While the range of pHER2 values in the FISH- and FISH+ group were nearly identical, there was a statistically significant difference in the median values (664 RU vs. 986 RU, respectively) between the two populations (p=0.024 by Chi-square testing). These data successfully confirm the presence of a similar HER2-/pHER2+ subpopulation of samples in our FFPE study set. In order to more fully validate the concordance between HER2 and pHER2 levels in the FFPE set, tumor cells were independently lysed from 3 sequential FFPE sections and printed to generate 3 separate lysates (one from each section) on 2 separate sets of arrays and then probed for total HER2 and phospho-HER2 (Y1248). The results (Supplementary Figure S2), clearly demonstrate excellent inter- and intra-array reproducibility of the HER2 and pHER2 FFPE data. The pHER2 measurements obtained by RPMA in the frozen and FFPE samples were generated by two independent laboratories at different times, using different instrumentation and methods. While the data gathered can be compared within an individual study set, in keeping with other immunoassays, the absence of bridging cases and controls between the study sets and differences in operating conditions and procedures prevented the direct comparison of data between study sets as there are scalar differences in the data output.
Figure 2. Correlation of RPMA measurements of HER2 and pHER2 protein levels with IHC and FISH results for FFPE breast tumor tissue specimens and identification of a HER2-/pHER2+ subpopulation of tumors.
A, B. HER2 concentration thresholds optimally distinguishing HER2 IHC+ (HER2 IHC score 3+; 2+/FISH+) from HER2 IHC- (HER2 IHC score 0; 1+; 2+/FISH-) tumor samples were established within training sets for FFPE surgical specimens (A) (left panel; HER2 concentration threshold 12pg HER2/ng protein), as well as core biopsies (B) (left panel; HER2 concentration threshold 7pg HER2/ng protein). Threshold values were validated using independent tissue samples for each study set (right panels). Thresholds for positive and negative samples are marked by horizontal dashed lines. C. pHER2 relative intensity values measured by RPMA are plotted against HER2 IHC status for the FFPE surgical validation study set. A tentative threshold intensity value of 1000RU is indicated by a dashed line. RU=relative units.
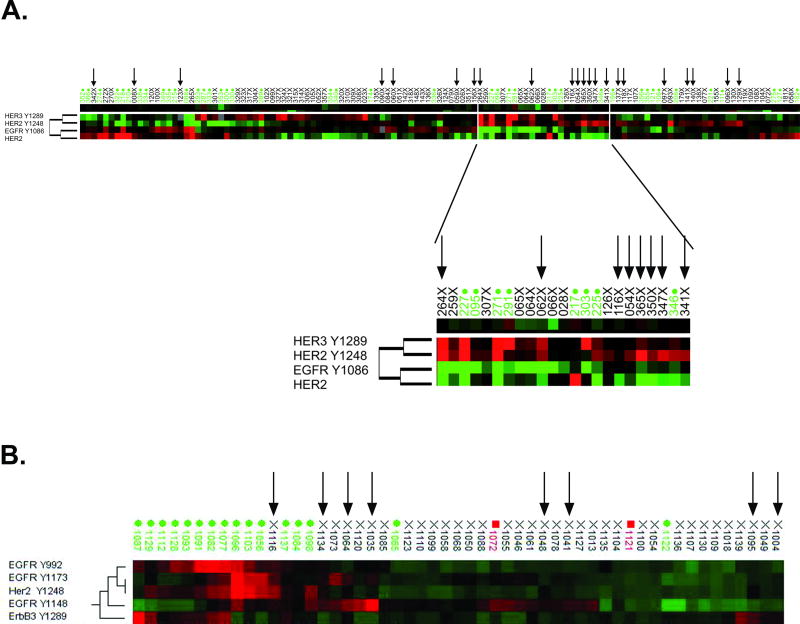
Based on these results, we explored whether activated/phosphorylated HER2, regardless of IHC/FISH status, produced a functional signal through the known dimerization-mediated phosphorylation of other HER family members. Unsupervised hierarchical clustering of the FFPE surgical validation study set revealed a subgroup of mostly IHC-/FISH- patients characterized by low relative expression of total HER2 but higher relative levels of pHER2 and pHER3 (Figure 3A, inset). The statistically confirmed correlation of pHER2 and pHER3 (Spearman correlation coefficient rs=0.705, p=0.0003) suggests that HER-based signaling in this subgroup of HER2-low tumors may occur via heterodimerization between HER2 and HER3 receptor molecules. Clustering analysis of the frozen microdissected tissues with known IHC/FISH status revealed that the IHC-/FISH-/pHER2+ cases (Figure 3B, black arrows) often showed concomitant high relative activation of EGFR at various sites compared to the IHC-/FISH-/pHER2- population (Figure 3B). Specifically, the phosphorylation levels of pHER2(Y1248), pEGFR(Y1173) and pEGFR(Y1148) were elevated (p=0.00002, p=0.01 and p=0.07, respectively) in the IHC-/FISH-/pHER2+ population compared to the IHC-/FISH-/pHER2- population. Compared to the IHC-/FISH-/pHER2- group, the IHC+/FISH+/pHER2+ cohort exhibited co-activation of HER2, EGFR and HER3 (p=0.00007 for pHER2(Y1248); p=0.00002 for pEGFR(Y992); p=0.02 for pEGFR(Y1173); p=0.004 for pHER3(Y1289)).
Figure 3. Co-activation of various HER family receptors with activation of HER2.
Unsupervised hierarchical clustering of various activated HER family relative expression levels in FFPE surgical validation specimens (A) or frozen, microdissected breast tumor tissues (B). Green case labels indicate IHC+/FISH+, black indicate IHC-/FISH-, red indicate IHC-/FISH borderline tumors (horizontal axis). Endpoints examined are clustered on the vertical axis. Black arrows indicate IHC-/FISH-/pHER2+ tumors. Within the heatmap, red color represents higher levels of relative activity/expression; black represents intermediate levels and green represents lower levels of relative activity/expression.
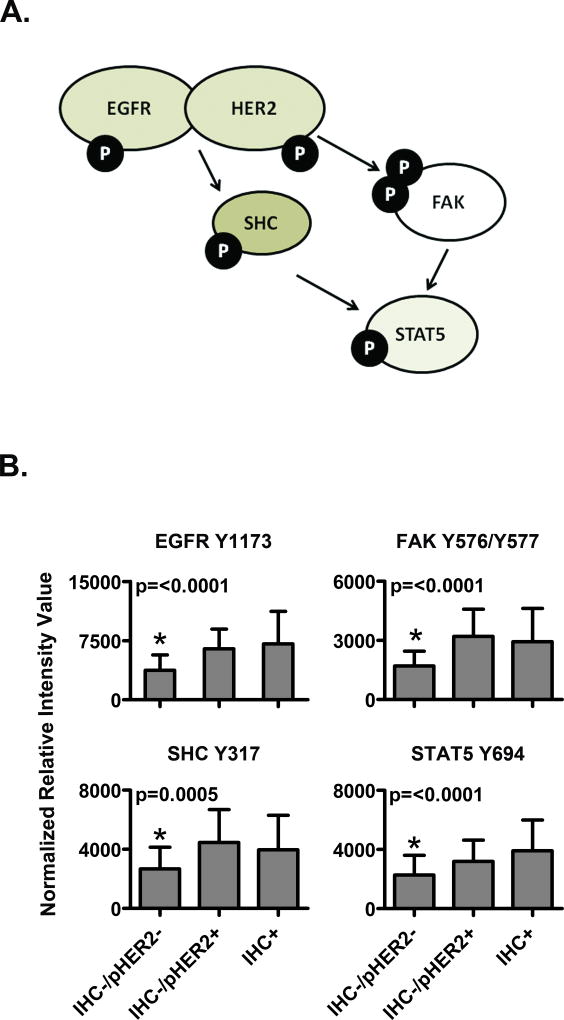
We also looked for further evidence of intact HER2-directed signaling in our HER2-/pHER2+ patients. For our frozen-LCM study set, in addition to HER2 receptor family activation analysis, we also measured phosphorylation of SHC, STAT5, and FAK, which are downstream cytoplasmic signaling molecules known to be linked to HER2 signaling, We used central IHC data to represent HER2 status in this analysis due to sample size considerations and its near complete redundancy with central FISH results available for this study set. As shown in Figure 4A, the level of phosphorylation of each of these signaling proteins was statistically indistinguishable between the IHC-/pHER2+ cohort and the IHC+/pHER2+group. Conversely, the IHC-/pHER2- group had statistically different and lower activation of these molecules (Fig. 4A). These data suggest that functional HER2 signaling occurs in the IHC-/pHER2+ subpopulation of samples.
Figure 4. The IHC-/pHER2+ subpopulation of frozen, microdissected tumors demonstrate full HER-signaling pathway activation.
(A) Pathway diagram of selected HER signaling pathway downstream proteins measured in frozen-LCM tissues. (B) Comparison of HER pathway activation levels of the proteins displayed in (A) between the IHC-/pHER2- cohort (N=87), IHC-/pHER2+ cohort (N=9) and IHC+/pHER2+ cohort (N=28) P-value results for ANOVA multiple mean comparisons are shown, with statistically different cohorts marked by an asterisk (*).
Discussion
Currently, IHC and FISH are most commonly used for HER2 assessment and the selection of patients for treatment with trastuzumab (34). However, these methods provide only semi-quantitative and granular measurement of the analyte, and are currently under increased scrutiny (10,16,17,35). The present study investigated the potential of RPMA technology to quantitatively measure HER2 protein levels in human breast cancer tissues and investigated the utility of functional mapping of the activation state of the HER family receptors in HER2+ and HER2- tissue samples.
We successfully demonstrated that RPMA-based measurements of total HER2 protein in three independent study sets of 415 individual patient samples had very strong concordance (>90% in all study sets) with IHC and/or FISH measurements in the same tissues (Figure 1, 2). This concordance was seen for both frozen and FFPE materials, surgical and biopsy specimens, and LCM-enriched isolates or whole tissue lysates from tissues selected for high (>85%) tumor content.
Our independent analyses of both frozen and FFPE tissues revealed a population of HER2- tumors that exhibit levels of pHER2 similar to that of HER2+ tumors. While the use of these different sets precluded a direct comparison between laboratories and tissue sets used, the novel identification of a cohort of women with HER2- breast cancer exhibiting HER2-activated signaling architecture was seen systemically across labs, cohorts and tissue input. This subpopulation of HER2- tumors represents a group of patients who could possibly benefit from treatment with HER2-targeted therapies such as trastuzumab, but would not be identified with IHC or FISH testing. Previous studies using a sensitive, but experimental immunoassay for HER2 and pHER2 measurement also found evidence of HER2 phosphorylation in a HER2- group of tumors, but their HER2 status determinations were not correlated back to approved FISH or IHC assays (18).
We observed much larger percentages of FISH or IHC+ tumors expressing high levels of pHER2 in our frozen, microdissected tissues compared to the FFPE study set, with approximately 80% of the HER2+ frozen tumors also containing high relative amounts of pHER2. One possible explanation for this is that the LCM process, which enriches for tumor cells, allows for activated HER2 receptor molecules to be more readily detected within a more homogeneous background. However, the FFPE tissue study set was selected on the basis of highly abundant tumor cells. Because formalin penetrates tissue very slowly (~0.1mm/hr) (36), an alternative explanation for the reduced HER2 phosphorylation in the HER2+ FFPE tissue is the loss or degradation of phosphate moieties during the fixation process (37–39), and indicates that pHER2 may be more reliably measured in frozen tissues, although this would have to be determined in larger study sets.
We observed a difference in the optimal cut-points for HER2 expression between the FFPE surgical and biopsy specimens. Recent reports for some clinically important proteins such as ER have revealed substantial differences in the measurement obtained between surgical material and biopsy material (40). Some possible reasons for different results from core and surgical specimens may include differences in sampling because the core biopsy likely has a smaller number of tumor epithelial cells and may not reflect the overall levels of the analyte measured in a larger sampling of the entire tumor. Other contributing factors could be from differences produced by fixation artifact because core biopsy material will have fixed much faster than the surgical material, with the delay in formalin exposure of the center of a surgical specimen resulting in changes in analyte concentration due to analyte instability and production in tissue that is alive but metabolically dying (41). The difference in cut-off values used in our FFPE analysis is not based on RPMA technical issues, but likely on the underpinning IHC determinations because, even in the face of FFPE tissue fixation issues and potential differences between core biopsy and surgical material, our concordance between RPMA and IHC for the FFPE sample set was 93% and 92% in the core biopsy and surgical FFPE validation sets, respectively. This was only slightly less than that achieved in our LCM tumor samples that were from flash-frozen core biopsies which represent the best practice.
In addition to the analysis of activated HER2 in our study sets, RPMA analysis allowed us to evaluate the activation status of other HER family receptors as well as downstream signaling endpoints in order to determine the network-based aspects of HER2 phosphorylation and signaling. Indeed, the HER2-/pHER2+ subgroup had levels of activated SHC, FAK and STAT5 proteins statistically higher than the HER2-/pHER2- group and indistinguishable from the levels seen in the HER2+/pHER2+ cohort (Figure 4). This result supports the postulate that the HER2 activation observed in the IHC/FISH- group is functional, and that the receptor directed signaling is transduced and results in a functional active network regardless of the underpinning total HER2 status. In our FFPE tissues, we found that a number of HER2- tumors with HER2 activation also displayed activation of HER3 (Figure 3A, inset). These results indicate a possible functional association between HER2 and HER3 in this patient subgroup. A recent study of early breast cancers described evidence for the detection of HER2/HER2 and HER2/HER3 homo- and heterodimers in situ using proximity ligation assays, which supports our current findings (42). By contrast, our results from frozen tissue analysis revealed more frequent co-activation of various EGFR phosphorylation sites in IHC-/FISH-/pHER2+ tumors compared to HER3. One straightforward, possible explanation for the different observations between the frozen and FFPE study sets is that several more, as well as different EGFR phosphorylation sites were measured in the frozen study set compared to the FFPE set. Earlier studies looking for predictors of trastuzumab efficacy found that a combination of EGFR and HER2 activation status was a better predictor of trastuzumab response over HER2 activation alone (22). EGFR phosphorylation may also be labile and could have been affected in the FFPE set, although this is speculative. Further analysis of a larger series of FFPE and frozen material whereby the same antibodies are used and the samples are arrayed and analyzed together at the same time would be necessary in order to more definitively understand these issues. These results further support the postulate that the pHER2 levels, independent of HER2 status, can produce an active biochemically linked network.
Our study revealed the presence of intact HER2 signaling in tumors from patients who were determined to be IHC and FISH- by central laboratory testing. This new cohort was initially characterized in frozen-LCM obtained tissue as a baseline “gold standard” input where tumor cellularity was maximized and normalized between tumor samples and minimized any pre-analytical issues arising from the FFPE tissue fixation/preservation. This finding was confirmed by immunoblotting with snap-frozen patient-matched tissue biopsies, and also in an independent FFPE core biopsy and surgical sample set. The use of multiple independent study sets, across two different laboratories was an important aspect of our analysis and points to the overall validity of the findings. The nature and clinical impact of this HER2 signaling as well as the entirety of the downstream signaling architecture itself remains to be elucidated by ongoing and future studies. Analysis of downstream HER2 and any associated HER signaling will require upfront sample enrichment techniques such as the LCM we used in our frozen core biopsy sets because past work has revealed inaccurate determination of signaling activation of ubiquitously expressed signaling proteins such as AKT, SHC, ERK, mTOR etc, that are not cell type specific (43,44).
Our findings could be clinically relevant, as studies from the recent NSABP B-31 adjuvant therapy clinical trial suggest that some breast cancer patients with HER2- tumors benefit from trastuzumab (8). It has been speculated that this result could stem from individuals with HER2- primary tumors having circulating tumor cells that are HER2+, or that trastuzumab may block other membrane receptor tyrosine kinases or block other pathways acting through AKT even if HER2 is not amplified. We provide an alternative explanation based on the activation status of the HER receptors. Using a quantitative, highly-sensitive protein array assay we identified a subgroup of HER2- breast cancer patients with levels of activated HER2 comparable to FISH+ tumors, and showed this activation is coincident with HER3 and EGFR activation and concomitant downstream signaling. This subgroup of breast cancer patients is not detectable by FISH or IHC, nor identified with intrinsic genomic subtypes or by mRNA expression (data not shown), and is thus excluded from trastuzumab treatment although these patients may respond to the drug. The activity of trastuzumab can also be affected by the expression of other HER family members as well as the activation of downstream effectors (45,46). For example, it has been shown that HER3 expression is increased after long-term trastuzumab treatment of HER2+ breast cancer cell lines that show primary resistance to trastuzumab (47). In this context, trastuzumab does not inhibit the dimerization of HER2 with other HER receptors such as EGFR and HER3. The monoclonal antibody pertuzumab, which targets the HER2 dimerization domain II and possibly prevents the formation of HER2/HER3 heterodimers (46), may be an alternative therapy option in these instances. Based on our results, we believe that retrospective analysis of phosphorylated HER2 levels in tissues from trials such as NSABP B-31, and neoadjuvant therapy trials such as ISPY-2 (www.ispy2.org) where HER2-directed therapies are being evaluated, would be justified.
While we have identified a cohort of tumors from patients with HER2 IHC/FISH- breast cancer that appear to have HER2 signaling profiles indistinguishable from IHC/FISH+ tumors, the true prevalence of this subgroup can only be established in larger study sets. Routine measurement of phosphorylated proteins such as HER2 would require across and between lab/platform standardization and control over any pre-analytical variables, such as tissue fixation, that could adversely impact precision and accuracy. Fortunately, new types of tissue fixatives and tissue processing methods (38,48–50) are being developed specifically to preserve labile analytes in the clinical setting so that the impact of these variables on routine analysis can be more effectively minimized. In the future, clinical trials will be needed to evaluate whether women with breast cancers which are HER2- but have high levels of pHER2 may benefit clinically from combinations of HER-targeted therapies. Quantitative analysis of the phosphorylation/activation levels of receptor tyrosine kinases, along with the analysis of activation of the linked downstream signal transduction network can potentially identify new patient cohorts that could benefit from molecular targeted inhibitors. Such patients may be missed by current testing methods that measure only the presence or absence of the drug target (e.g. total HER2) and thus, functional signaling analysis may provide new opportunities for personalized therapy if these measurements prove clinically useful.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
The identification and characterization of HER2-based protein signaling activation in breast tumors would be of critical importance in the clinical management of a significant number of breast cancer patients. Using a quantitative, highly-sensitive protein array assay we identified a subgroup of IHC and FISH HER2-negative breast cancer patients with levels of activated/phosphorylated HER2 comparable to IHC and FISH HER2-positive tumors that was accompanied by co-activation of HER2 binding partners as well as downstream pathway targets. This group of patients was not identified by current clinically-approved tests for HER2 and are currently excluded from trastuzumab treatment. Analysis of the phosphorylation/activation levels of receptor tyrosine kinases, along with the analysis of activation of the linked downstream signal transduction network can potentially identify new patient cohorts that could benefit from molecular targeted inhibitors, and thus, functional signaling analysis may provide new opportunities for personalized therapy if these measurements prove clinically useful.
Acknowledgments
Grant Support:
The I-SPY 1 TRIAL (CALGB 150007 and 150012) was supported, in part, by grants from the National Cancer Institute (CA31946) to the Cancer and Leukemia Group B (Monica M. Bertagnolli, M.D., Chair) and to the CALGB Statistical Center (Daniel J. Sargent, Ph.D., CA33601). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
This research was also supported by NGFN (Nationale Genomforschungsnetz) Project of the BMBF (Bundesministerium für Bildung und Forschung), Grant #01GR0805 (KFB); College of Science, George Mason University (EFP, LAL); and Grant #CA45808, University of California, San Francisco (LE).
Footnotes
Conflicts of Interest:
Stock ownership in Theranostics Health, LLC: (JDW, VE, MP,LAL, EFP);
Uncompensated consultancy Theranostics Health, LLC: (JDW, VE, LAL, EFP);
Advisory role in Theranostics Health, LLC: (LAL, EFP)
References
- 1.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 5.Romond EH, Perez EA, Bryant J, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 6.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 7.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 8.Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358:1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 9.Roepman P, Horlings HM, Krijgsman O, Kok M, Bueno-de-Mesquita JM, Bender R, et al. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin Cancer Res. 2009;15:7003–7011. doi: 10.1158/1078-0432.CCR-09-0449. [DOI] [PubMed] [Google Scholar]
- 10.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 11.Arber DA. Effect of prolonged formalin fixation on the immunohistochemical reactivity of breast markers. Appl Immunohistochem Mol Morphol. 2002;10:183–186. doi: 10.1097/00129039-200206000-00015. [DOI] [PubMed] [Google Scholar]
- 12.Hashizume K, Hatanaka Y, Kamihara Y, Kato T, Hata S, Akashi S, et al. Interlaboratory comparison in HercepTest assessment of HER2 protein status in invasive breast carcinoma fixed with various formalin-based fixatives. Appl Immunohistochem Mol Morphol. 2003;11:339–344. doi: 10.1097/00129039-200312000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Selvarajan S, Bay BH, Choo A, Chuah KL, Sivaswaren CR, Tien SL, et al. Effect of fixation period on HER2/neu gene amplification detected by fluorescence in situ hybridization in invasive breast carcinoma. J Histochem Cytochem. 2002;50:1693–1696. doi: 10.1177/002215540205001215. [DOI] [PubMed] [Google Scholar]
- 14.Thomson TA, Hayes MM, Spinelli JJ, Hilland E, Sawrenko C, Phillips D, et al. HER-2/neu in breast cancer: interobserver variability and performance of immunohistochemistry with 4 antibodies compared with fluorescent in situ hybridization. Mod Pathol. 2001;14:1079–1086. doi: 10.1038/modpathol.3880440. [DOI] [PubMed] [Google Scholar]
- 15.Khoury T, Sait S, Hwang H, Chandrasekhar R, Wilding G, Tan D, et al. Delay to formalin fixation effect on breast biomarkers. Mod Pathol. 2009;22:1457–1467. doi: 10.1038/modpathol.2009.117. [DOI] [PubMed] [Google Scholar]
- 16.American Society of Clinical Oncology-College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer; American Society of Clinical Oncology-College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer, 4/2010 update. http://www.cap.org/apps/docs/committees/immunohistochemistry/clinical_notice.pdf.
- 17.Summary of ASCO/CAP HER2 Guideline Recommendations, 4/2011 update. http://www.cap.org/apps/docs/committees/immunohistochemistry/summary_of_recommendations.pdf.
- 18.Cicenas J, Urban P, Küng W, Vuaroqueaux V, Labuhn M, Wight E, et al. Phosphorylation of tyrosine 1248-ERBB2 measured by chemiluminescence-linked immunoassay is an independent predictor of poor prognosis in primary breast cancer patients. Eur J Cancer. 2006;42:636–645. doi: 10.1016/j.ejca.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Frogne T, Laenkholm AV, Lyng MB, Henriksen KL, Lykkesfeldt AE. Determination of HER2 phosphorylation at tyrosine 1221/1222 improves prediction of poor survival for breast cancer patients with hormone receptor positive tumors. Breast Cancer Res. 2009;11:R11. doi: 10.1186/bcr2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thor AD, Liu S, Edgerton S, Moore D2nd, Kasowitz KM, Benz CC, et al. Activation (tyrosine phosphorylation) of ErbB-2 (HER-2/neu): a study of incidence and correlation with outcome in breast cancer. J Clin Oncol. 2000;18:3230–3239. doi: 10.1200/JCO.2000.18.18.3230. [DOI] [PubMed] [Google Scholar]
- 21.DiGiovanna MP, Stern DF, Edgerton SM, Whalen SG, Moore D2nd, Thor AD. Relationship of epidermal growth factor receptor expression to ErbB-2 signaling activity and prognosis in breast cancer patients. J Clin Oncol. 2005;23:1152–1160. doi: 10.1200/JCO.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 22.Hudelist G, Kostler WJ, Czerwenka K, Kubista E, Attems J, Müller R, et al. Her-2/neu and EGFR tyrosine kinase activation predict the efficacy of trastuzumab-based therapy in patients with metastatic breast cancer. Int J Cancer. 2006;118:1126–1134. doi: 10.1002/ijc.21492. [DOI] [PubMed] [Google Scholar]
- 23.Liotta LA, Espina V, Mehta AI, Calvert V, Rosenblatt K, Geho D, et al. Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell. 2003;3:317–325. doi: 10.1016/s1535-6108(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 24.Wulfkuhle JD, Edmiston KH, Liotta LA, Petricoin EF3rd. Technology insight: pharmacoproteomics for cancer--promises of patient-tailored medicine using protein microarrays. Nat Clin Pract Oncol. 2006;3:256–268. doi: 10.1038/ncponc0485. [DOI] [PubMed] [Google Scholar]
- 25.Wulfkuhle JD, Aquino JA, Calvert VS, Fishman DA, Coukos G, Liotta LA, et al. Signal pathway profiling of ovarian cancer from human tissue specimens using reverse-phase protein microarrays. Proteomics. 2003;11:2085–2090. doi: 10.1002/pmic.200300591. [DOI] [PubMed] [Google Scholar]
- 26.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Wolff C, Malinowsky K, Berg D, Schragner K, Schuster T, Walch A, et al. Signalling networks associated with urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in breast cancer tissues: new insights from protein microarray analysis. J Pathol. 2011;223:54–63. doi: 10.1002/path.2791. [DOI] [PubMed] [Google Scholar]
- 28.Becker KF, Schott C, Hipp S, Metzger V, Porschewski P, Beck R, et al. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J Pathol. 2007;211:370–378. doi: 10.1002/path.2107. [DOI] [PubMed] [Google Scholar]
- 29.Berg D, Langer R, Tran K, Walch A, Schuster T, Bronger H, et al. Protein microarray-based comparison of HER2, estrogen receptor, and progesterone receptor status in core biopsies and surgical specimens from FFPE breast cancer tissues. Appl Immunohistochem Mol Morphol. 2011;19:300–305. doi: 10.1097/PAI.0b013e3182054f9f. [DOI] [PubMed] [Google Scholar]
- 30.Berg D, Hipp S, Malinowsky K, Malinowsky K, Böllner C, Becker KF. Molecular profiling of signalling pathways in formalin-fixed and paraffin-embedded cancer tissues. Eur J Cancer. 2010;46:47–55. doi: 10.1016/j.ejca.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 33.Schecter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, et al. The neu oncogene: and erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- 34.Press MF, Slamon DJ, Flom KJ, Park J, Zhou JY, Bernstein L. Evaluation of HER-2/neu gene amplification and overexpression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J Clin Oncol. 2002;20:3095–3105. doi: 10.1200/JCO.2002.09.094. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt C. How do you tell whether a breast cancer is HER2 positive? Ongoing studies keep debate in high gear. J Natl Cancer Inst. 2011;103:87–89. doi: 10.1093/jnci/djq557. [DOI] [PubMed] [Google Scholar]
- 36.Start RD, Layton CM, Cross SS, Smith JH. Reasessment of the rate of fixative diffusion. J Clin Pathol. 1992;45:1120–1121. doi: 10.1136/jcp.45.12.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics. 2008;10:1998–2018. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller C, Edmiston KH, Carpenter C, Gaffney E, Ryan C, Ward R, et al. One-step preservation of phosphoproteins and tissue morphology at room temperature for diagnostic and research specimens. PLoS One. 2011;6:e23780. doi: 10.1371/journal.pone.0023780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Espina V, Mueller C, Liotta LA. Phosphoprotein stability in clinical tissue and its relevance for reverse phase protein microarray technology. Methods Mol Biol. 2011;785:23–43. doi: 10.1007/978-1-61779-286-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann GB, Fahey VD, Feleppa F, Buchanan MR. Reliance on hormone receptor assays of surgical specimens may compromise outcome in patients with breast cancer. J Clin Oncol. 2005;23:5148–5154. doi: 10.1200/JCO.2005.02.076. [DOI] [PubMed] [Google Scholar]
- 41.Espina V, Mueller C, Edmiston K, Sciro M, Petricoin EF, Liotta LA. Tissue is alive: New technologies are needed to address the problems of protein biomarker pre-analytical variability. Proteomics Clin Appl. 2009;3:874–882. doi: 10.1002/prca.200800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spears M, Taylor KJ, Munro AF, Cunningham CA, Mallon EA, Twelves CJ, et al. In situ detection of HER2:HER2 and HER2:HER3 protein-protein interactions demonstrates prognostic significance in early breast cancer. Breast Cancer Res Treat. 2012;132:463–70. doi: 10.1007/s10549-011-1606-z. [DOI] [PubMed] [Google Scholar]
- 43.Wulfkuhle JD, Speer R, Pierobon M, Laird J, Espina V, Deng J, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008;7:1508–1517. doi: 10.1021/pr7008127. [DOI] [PubMed] [Google Scholar]
- 44.Silvestri A, Colobatti A, Calvert VS, Deng J, Mammano E, Belluco C, et al. Protein pathway biomarker analysis of human cancer reveals requirement for upfront cellular-enrichment processing. Lab Invest. 2010;90:787–796. doi: 10.1038/labinvest.2010.47. [DOI] [PubMed] [Google Scholar]
- 45.Smith BL, Chin D, Maltzman W, Crosby K, Hortobagyi GN, Bacus SS. The efficacy of Herceptin therapies is influenced by the expression of other erbB receptors, their ligands and the activation of downstream signalling proteins. Br J Cancer. 2004;91:1190–1194. doi: 10.1038/sj.bjc.6602090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koutras AK, Fountzilas G, Kalogeras KT, Starakis I, Iconomou G, Kalofonos HP. The upgraded role of HER3 and HER4 receptors in breast cancer. Crit Rev Oncol Hematol. 2009;74:73–78. doi: 10.1016/j.critrevonc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Narayan M, Wilken JA, Harris LN, Baron AT, Kimbler KD, Maihle NJ. Trastuzumab-induced HER reprogramming in "resistant" breast carcinoma cells. Cancer Res. 2009;69:2191–2194. doi: 10.1158/0008-5472.CAN-08-1056. [DOI] [PubMed] [Google Scholar]
- 48.Bellet V, Boissière F, Bibeau F, Desmetz C, Berthe ML, Rochaix P, et al. Proteomic analysis of RCL2 paraffin-embedded tissues. J Cell Mol Med. 2008;12:2027–36. doi: 10.1111/j.1582-4934.2008.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rountree CB, Van Kirk CA, You H, Ding W, Dang H, VanGuilder HD, et al. Clinical application for the preservation of phospho-proteins through in-situ tissue stabilization. Proteome Sci. 2010;8:61. doi: 10.1186/1477-5956-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viertler C, Groelz D, Gündisch S, Kashofer K, Reischauer B, Riegman PHJ, et al. A new technology for stabilization of biomolecules in tissues for combined histological and molecular analyses. J Mol Diagn. 2012 doi: 10.1016/j.jmoldx.2012.05.002. 28 June Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.