Significance
The Niemann–Pick C1 (NPC1) protein is responsible for transporting LDL-derived cholesterol out of late endosomes. Mutations in NPC1 lead to the fatal Niemann–Pick Type C disease. We present here an improved structure of an NPC1 protein at 3.3 Å and decipher details of its C-terminal luminal domain (CTD), which could not be resolved in previous structures. In particular, a loop stabilized by a pair of disulfide bonds in the CTD binds to the N-terminal domain through a loop–loop interaction. We show that this interaction is important for cholesterol transport in cultured cells. Together, our data provide insights related to the molecular mechanism of NPC1 activity and Niemann–Pick Type C disease.
Keywords: cholesterol transport, Niemann–Pick type C disease, cysteine-rich domain, sterol-sensing domain, crystal structure
Abstract
Niemann–Pick C1 (NPC1) and NPC2 proteins are indispensable for the export of LDL-derived cholesterol from late endosomes. Mutations in these proteins result in Niemann–Pick type C disease, a lysosomal storage disease. Despite recent reports of the NPC1 structure depicting its overall architecture, the function of its C-terminal luminal domain (CTD) remains poorly understood even though 45% of NPC disease-causing mutations are in this domain. Here, we report a crystal structure at 3.3 Å resolution of NPC1* (residues 314–1,278), which—in contrast to previous lower resolution structures—features the entire CTD well resolved. Notably, all eight cysteines of the CTD form four disulfide bonds, one of which (C909–C914) enforces a specific loop that in turn mediates an interaction with a loop of the N-terminal domain (NTD). Importantly, this loop and its interaction with the NTD were not observed in any previous structures due to the lower resolution. Our mutagenesis experiments highlight the physiological relevance of the CTD–NTD interaction, which might function to keep the NTD in the proper orientation for receiving cholesterol from NPC2. Additionally, this structure allows us to more precisely map all of the disease-causing mutations, allowing future molecular insights into the pathogenesis of NPC disease.
Cholesterol is essential for the growth and viability of mammalian cells and serves as the precursor for steroid hormones, bile acids, and vitamin D. Cholesterol molecules, free and esterified, are packaged in liver or intestinal cells via a few specific proteins into lipoprotein particles, each of which contains a mixture of hundreds of free and esterified cholesterol and other lipid molecules. After secretion of lipoprotein particles at the basolateral membrane and their entry into the vasculature, they are then available to all other cells in the body via endocytotic uptake using specific lipoprotein receptors (1); once in the late endosomes, lipoprotein particles are disassembled and esterified cholesterol molecules are converted into free cholesterol by acid lipase (2). Because cholesterol is almost water-insoluble, efficient transport within the aqueous environment of the endosomal lumen to the endosomal membrane, followed by its unidirectional translocation across the hydrophobic bilayer of the endosomal membrane, is mediated by protein. Two proteins, the soluble NPC2 (Niemann–Pick type C protein 2) (3) and the integral membrane NPC1 (4), are required for cholesterol transport across the 8-nm glycocalyx (5) and for cholesterol exit from the endosomal membrane. Insight into the function of these two proteins comes from NPC disease, a fatal lysosomal storage disease, due to accumulation of cholesterol and other lipids in lysosomes within many organs (6). Besides its function in LDL-derived cholesterol export, NPC1 has also been identified as a receptor for Ebola virus in late endosomes (7, 8).
Human NPC1, consisting of 1,278 amino acid residues, includes three luminal domains [N-terminal domain (NTD), middle luminal domain (MLD), C-terminal domain (CTD, also called cysteine-rich domain)] and 13 transmembrane helices (TMs) (9). The TM3-7 segment, termed sterol-sensing domain (SSD), is highly conserved within a subclass of membrane proteins in the cholesterol metabolism pathway (10, 11). Biochemical and cell biological studies showed that small molecules such as U18666A and posaconazole can block cholesterol export from lysosomes and that a point mutation (P691S) in the NPC1-SSD can prevent binding of these ligands (12–16). Additionally, binding of some of these ligands is retained even after deletion of the NTD (13, 15), which contains a classical cholesterol-binding site (17, 18), suggesting that NPC1-SSD itself is able to bind multiple ligands of distinct structures.
Structural insights into NPC1 function have been reported by several groups since 2009: (i) An isolated N-terminal fragment of NPC1 presenting the NTD harbors a cholesterol molecule consistent with previous biochemical observations (19); (ii) a complex of the MLD and Ebola glycoprotein defines the site of Ebola virus attachment (20); (iii) a near-atomic X-ray structure of NPC1 (residues 314–1,278, termed NPC1*) shows that the SSD may indeed accommodate a cholesterol molecule (21); (iv) a cryo-EM structure of NPC1 at 4.4 Å presents the overall architecture of the full-length molecule (22); and (v) a complex of NPC1-MLD and NPC2 suggests a molecular mechanism of cholesterol transfer from NPC2 to the NTD of NPC1 (23). However, due to low resolution, our understanding of the structural details and function of the NPC1-CTD remain rudimentary. Intriguingly, almost half of the NPC disease-causing mutations are distributed in the CTD (9).
In this study, we present the crystal structure of NPC1* at 3.3 Å resolution (Fig. 1A), with the entire CTD domain clearly defined with unambiguous sequence assignment. By docking this structure into the full-length cryo-EM NPC1 structure, it reveals a detailed interface between the CTD and NTD. Disruption by mutagenesis of this interface blocks cholesterol transfer from the late endosome to ER.
Fig. 1.
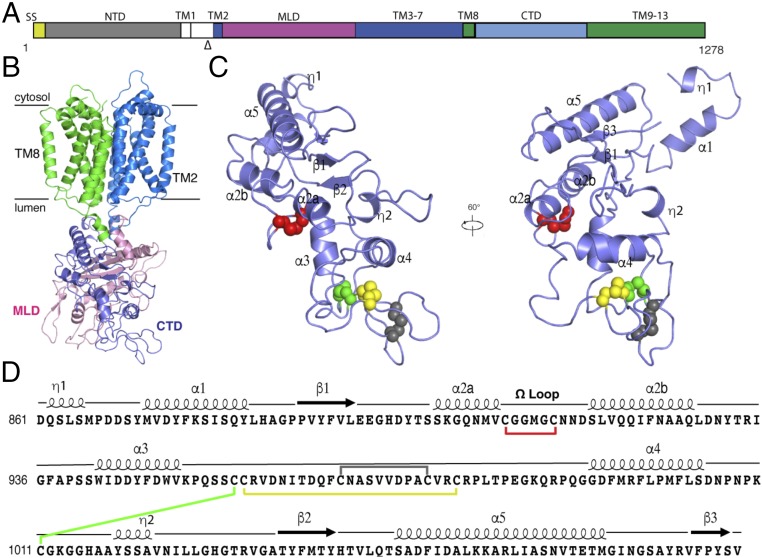
Crystal structure of the NPC1 protein and structural details of its CTD. (A) Schematic representation of NPC1 domains with the part after the proteinase K site (indicated by “Δ”) crystallized here in color. (B) The overall structure of NPC1* with TM2–7 colored blue, TM8-13 in green, MLD in pink, and CTD in light purple. (C) Structure of the NPC1-CTD with the secondary elements labeled and the cysteine side-chain atoms involved in disulfide bonds shown as balls with different colors. (D) Primary structure of the CTD, with the major structural elements indicated above the sequence and disulfide bonds indicated by colored lines (colored as in C).
Results
We expressed and purified the NPC1* (residues 314–1,278) as previously published (21). A previous photo-crosslinking study showed itraconazole binds to NPC1 in vitro (15). To stabilize the protein during the purification process, we supplemented each buffer with 10 μM itraconazole except for the initial buffer for resuspending cells. The protein after gel filtration was concentrated to 10 mg/mL and incubated with an additional 100 μM itraconazole on ice for 1 h before crystallization.
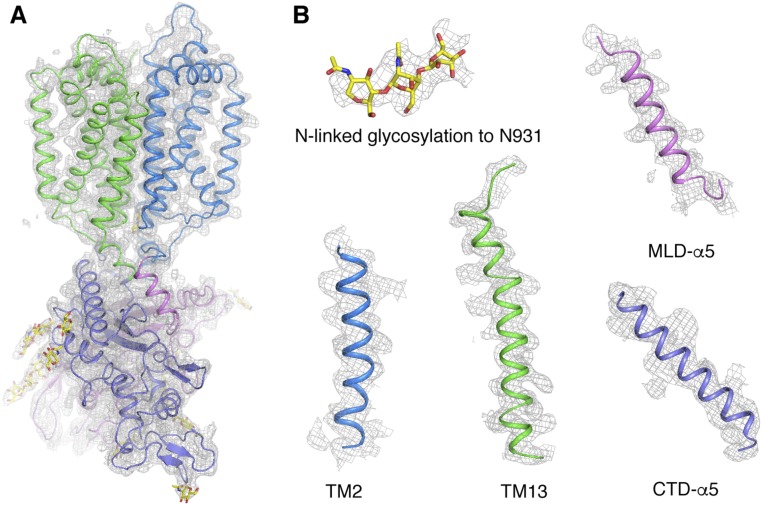
Crystals significantly larger than those grown without itraconazole appeared after 3 d. We determined the structure of the NPC1* molecule at a resolution of 3.3 Å (Fig. 1B) by molecular replacement using the previously determined NPC1* structure as an initial search model. Although the electron density of itraconazole is not observed in the electron density map, the quality of the whole map has been considerably improved and the entire CTD is well resolved and completely built. After several rounds of refinement, the final Rfree and Rwork reached 30.5% and 25.3%, respectively (Fig. S1 and Table S1). Using this improved model, our previously published 3.6 Å resolution structure has been rerefined with current Rfree 23.5% and Rwork 21.4% and has been updated in the Protein Data Bank (PDB ID code 5U73).
Fig. S1.
The 2Fo-2Fc electron density of NPC1*. (A) An overall view of the 2Fo-2Fc electron density map, contoured at 1.0 σ, in one unit cell. (B) A representative view of the 2Fo-Fc electron density map at various regions, contoured at 1.0 σ.
Table S1.
Data collection and refinement statistics
| Parameter | Native |
| Data collection | |
| Wavelength, Å | 0.98 |
| Space group | C2221 |
| Unit cell, Å and °, a, b, c; α, β, γ | 174.87, 222.11, 107.85; 90, 90, 90 |
| Resolution, Å | 50 ∼ 3.33 (3.42 ∼ 3.33)a |
| Rmerge | 0.147 (>1) |
| I/σI | 8.9 (1.2) |
| Completeness,% | 97.7 (98.7) |
| Redundancy | 3.7 (3.6) |
| Refinement | |
| Resolution, Å | 50 ∼ 3.33 |
| No. of reflections | 29,807 |
| Rwork/Rfree,b % | 24.62/29.88 |
| Molecules in ASU | 1 |
| No. atoms protein/ligand | 7,081/230 |
| B factors protein/ligand | 101.2/140.5 |
| rmsd in | |
| Bond distances, Å | 0.005 |
| Bond angles, ° | 1.982 |
| MolProbity score | 2.33 99th percentile |
Values in parentheses are for the highest resolution shell.
Rfree was calculated with 5% of the reflections selected in the thin shell.
The amino acid sequence of residues 334–1,255, including TM2–13, the MLD, and the CTD, could be unequivocally assigned. Only the loop between TM3 and TM4 containing residues 642–649 and the loop between TM7 and TM8 containing residues 800–813 are disordered. Compared with the previously reported X-ray NPC1* and cryo-EM NPC1 structures, this model is almost complete and significantly more accurate, providing a much improved atomic snapshot of this protein.
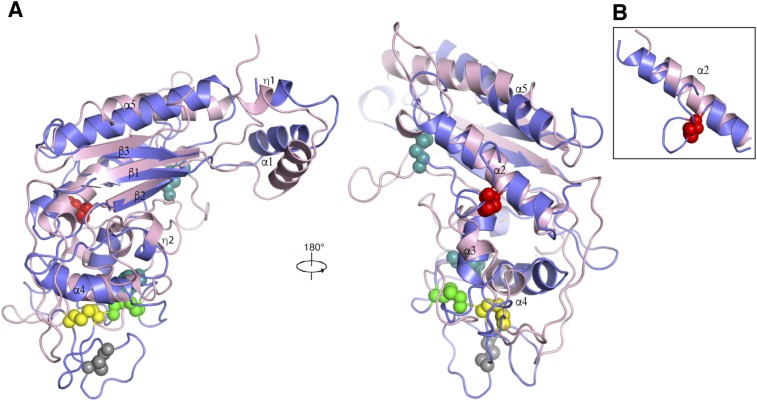
The CTD (residues 861–1,083) consists of three β strands surrounded by five α-helices (Fig. 1 C and D). As suggested by its other name, “cysteine-rich domain” (24), the CTD contains eight cysteines. All cysteine residues form four pairs of disulfide bonds to stabilize the loops on the tip of this domain (Fig. 1 C and D). In contrast, the MLD only contains two pairs of disulfide bonds, although it shares a similar fold with the CTD (Fig. S2A). Interestingly, Cys909 and Cys914 form a disulfide bond creating a loop (residue 909–917, termed “Ω loop”), which helps to break one long α-helix into two shorter α-helices (α2a and α2b) (Fig. 1 C and D) and is not observed in the analogous α-helix of the MLD (Fig. S2B).
Fig. S2.
Structural comparison of the MLD and the CTD. (A) Structures of the MLD (pink) and the CTD (light purple) are superimposed with the major elements of two domains indicated; disulfide bonds of the CTD are shown as balls with different colors as in Fig. 1C; disulfide bonds of the MLD are shown as balls in teal. (B) Close-up view of α2 of the both domains.
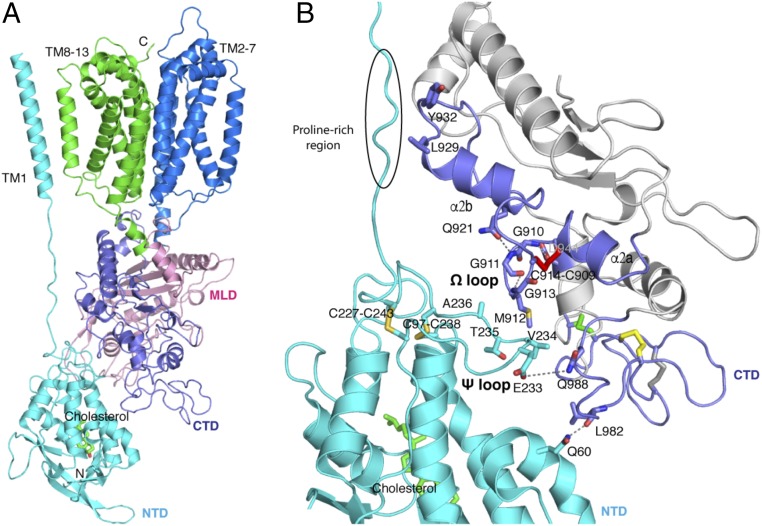
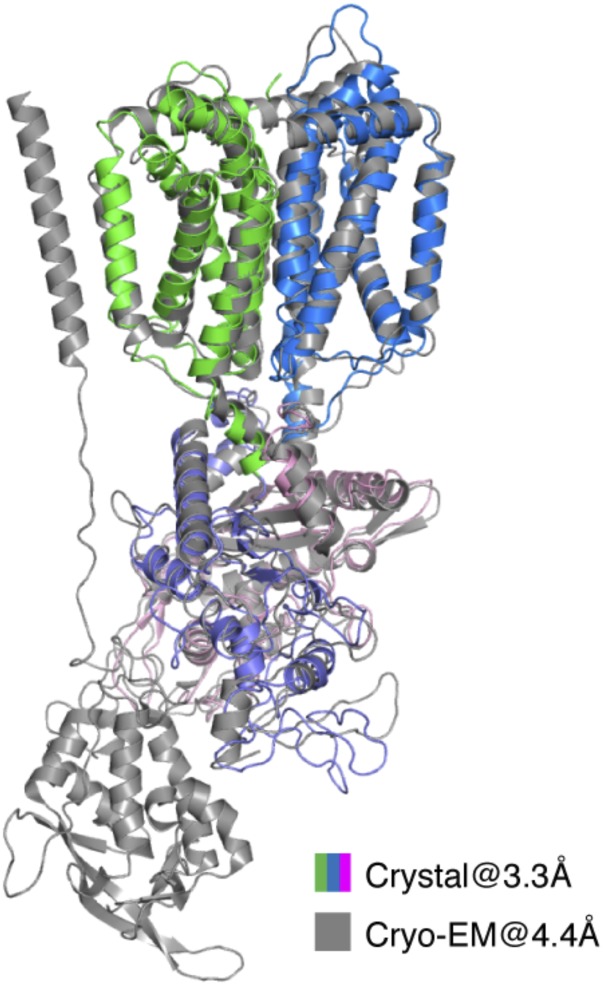
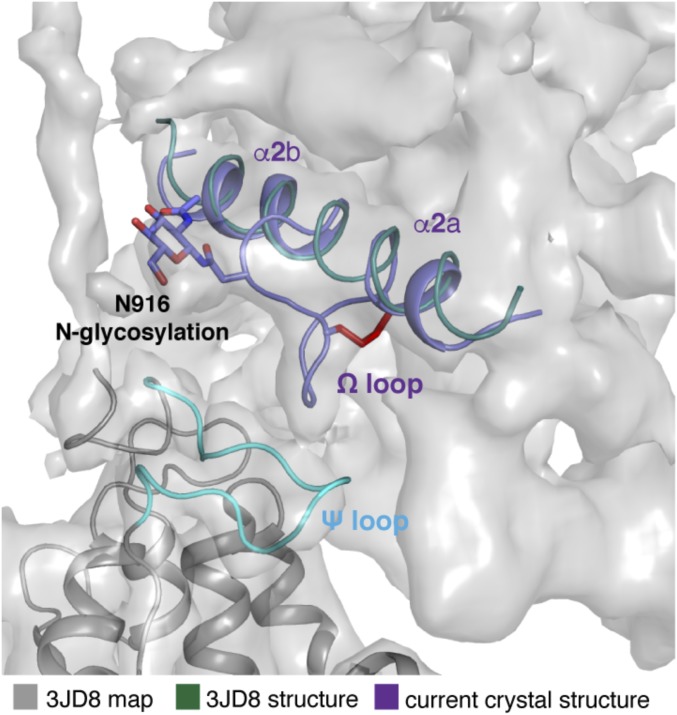
To determine the function of this special Ω loop, we docked the NPC1* structure into the full-length cryo-EM NPC1 map (Fig. 2A). In our docking results, the majority of the TMs and MLD from NPC1* can be aligned well with the cryo-EM NPC1 structure (Fig. S3) with an rmsd of 2.0 Å (over 538 residues), which allows us to confidently analyze the interactions between the NTD and CTD. Τhe Ω loop also can be observed in the cryo-EM map, but due to the limited resolution, this loop had been omitted in the cryo-EM structure (PDB ID code 3JD8) (Fig. S4). With 410 Å2, the CTD-NTD interface is larger than that of the MLD-NTD interface (∼260 Å2), suggesting that the CTD may modulate the NTD in a more pronounced manner than the MLD. Fig. 2B shows the details of the interaction between the CTD and the NTD. The Ω loop, containing three glycines (G910, G911, G913) and a methionine (M912), features a hydrophobic environment prone for association with V234, T235, and A236 in the adjacent loop from the NTD (residues 227–238, termed “Ψ loop”) (Fig. 2B). The main chain of G911 and G913 can form a hydrogen bond to satisfy the constraints of the disulfide bond in the Ω loop. Notably, two pairs of disulfide bonds (C97–C238 and C227–C243) also fix the Ψ loop. The compact loop–loop interface is enabled by the triple glycines of the Ω loop, generating an interface without spatial clashes.
Fig. 2.
The interface of NPC1-CTD and NPC1-NTD. (A) Docking of NPC1* to cryo-EM NPC1 structure (PDB ID code 3JD8); the NTD and TM1 from EM structure are presented with the 3.3 Å NPC1* crystal structure; cholesterol molecule is indicated as stick. (B) Close-up view of the interaction between the NPC1-CTD and NPC1-NTD; residues involved in this interaction are labeled and hydrogen bonds are indicated by dashed lines. The disulfide bonds are indicated by sticks and colored as Fig. 1 C and D; the proline-rich region is indicated; the CTD is colored in gray except for the light purple region that interacts with the NTD.
Fig. S3.
Structural comparison of NPC1* crystal structure and cryo-EM structure of full-length NPC1.
Fig. S4.
Comparison of NPC1* crystal structure and NPC1 cryo-EM structure (PDB ID code 3JD8). The structure of NPC1* is colored in purple, and the cryo-EM structure is colored in cyan; the map of cryo-EM (EMD-6640) is shown as gray surface at the 5.0 σ level.
In addition to the loop–loop interaction, we identify a secondary interface between the NTD and the CTD. In this interface, the side chain of E233 interacts with the side chain of Q988, and the amide group of Q60 forms a hydrogen bond with the main chain of L982 (Fig. 2B). Furthermore, L929 and Y932 may form hydrophobic contacts with the proline-rich region to connect the NTD and TM1 (Fig. 2B). Because the proline-rich region cannot be confidently built due to the limited resolution of the cryo-EM map in this region, we do not further discuss this region in the current paper.
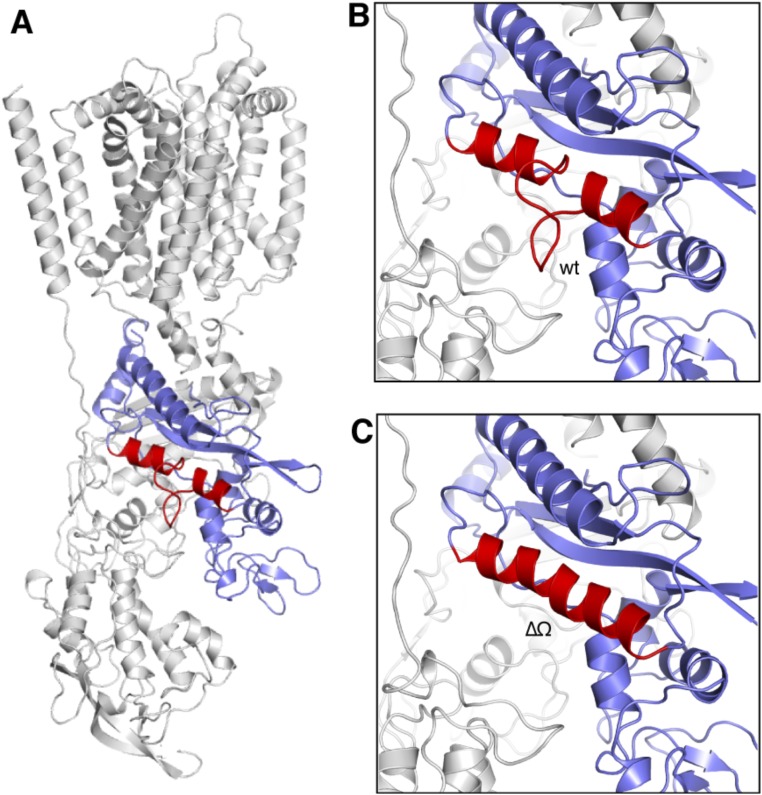
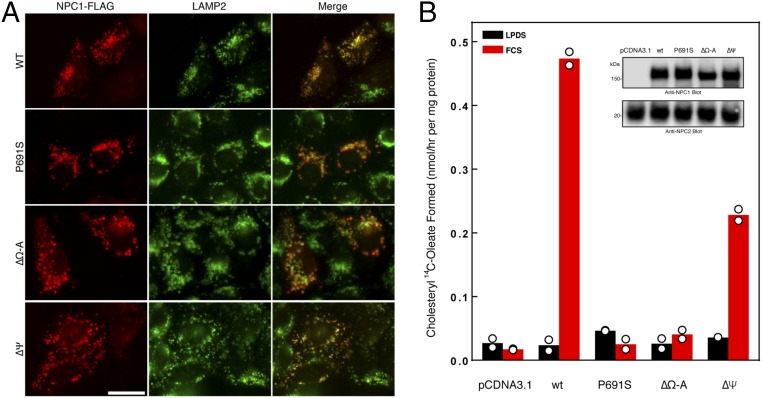
In our previous study, we proposed an NTD-SSD insertion mechanism to explain how cholesterol is transferred from the lysosomal lumen to the membrane domain (21, 23). In light of the structure, we suspect that the Ω loop–Ψ loop interaction might modulate the NTD-SSD insertion to facilitate cholesterol transport. To test this hypothesis, we generated two constructs: one termed NPC1ΔΩ-A, with residues 909–917 replaced by one alanine residue, allowing α2a and α2b to form one continuous alpha-helix (Fig. S5), and the other termed NPC1ΔΨ, with a deletion of residues 230–234. We then performed a cholesterol esterification assay by transfecting wild-type NPC1 (termed NPC1-WT), NPC1-P691S, NPC1ΔΩ-A, or NPC1ΔΨ individually to NPC1−/− 10–3 cells according to a previously published protocol (14). NPC1ΔΩ-A and NPC1ΔΨ are expressed well and correctly localized to lysosomes like NPC1-WT and NPC1-P691S (Fig. 3A). It is essential to establish the localization of the mutants, as many NPC1 mutations lead to misfolding and ER-associated degradation (25, 26). Although correctly localized, NPC1ΔΩ-A blocks cholesterol transport out of late endosomes, as does the NPC1-P691S mutant; NPC1ΔΨ loses 50% activity compared with NPC1-WT (Fig. 3B). This result supports our hypothesis that the Ω loop–Ψ loop interaction is physiologically important for the NPC1-mediated cholesterol export process.
Fig. S5.
Structural prediction of NPC1ΔΩ-A. (A) The localization of Ω loop in the NPC1 (red), with the CTD colored in light purple. (B) Close-up view of CTD-α5 structure in the NPC1-WT (red). (C) Close-up view of CTD-α5 model in the NPC1ΔΩ-A(red).
Fig. 3.
Specific residues of the NPC1-CTD are required for cholesterol esterification. (A) Confocal immunofluorescence microscopy analysis of the localization of NPC1ΔΩ-A, NPC1Δψ, and LAMP2 proteins in NPC1−/− 10–3 cells 24 h after transfection as described in Experimental Procedures. (Scale bar, 20 µm.) (B) Cholesterol esterification: After labeling as described in Experimental Procedures, cells were harvested for measurement of their content of cholesteryl [14C]oleate and [14C]triglycerides. Each value is the mean of duplicate incubations with individual values shown. The cellular content of [14C]triglycerides in all transfected cell lines did not differ significantly in cells treated with LPDS (8–11 nmol·h–1·mg–1) or FCS (13–15 nmol·h–1·mg–1). The immunoblot analysis of whole cell extracts (6 µg) from the indicated transfection shown below was done using 0.5 µg/mL NPC1 antibody and 2 µg/mL NPC2 antibody as a loading control.
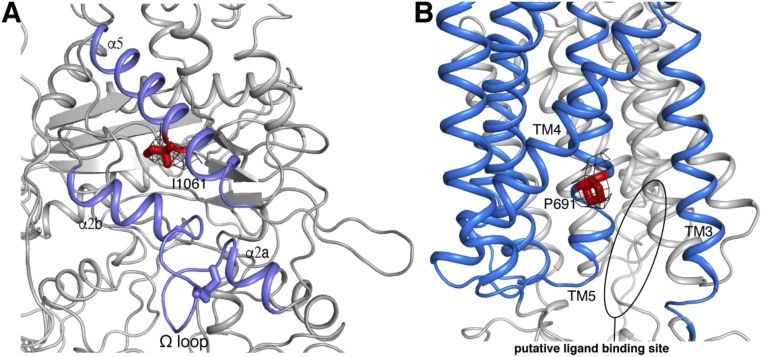
Our structure provides an improved atomic model for an understanding of NPC-causing mutations. The most frequent mutation, I1061T, present in 15–20% of all disease alleles (27, 28), is in the α5 helix of the CTD (Fig. 4A). This mutation does lead to protein mislocalization and degradation due to misfolding (25). Our structural analysis suggests that I1061T would not disrupt the interaction between the α5 and α2b (Fig. 4A), which could explain why overexpression of NPC1-I1061T in NPC1−/− cells still retains late endosome localization and rescues the NPC1-deficient phenotype (25). In addition, previous cross-linking studies showed that mutation P691S in human, or P692S in mouse, abolishes binding of different ligands to the SSD (12, 14–16). In our structure, P691 is located in the TM5 helix and faces the SSD pocket that accommodates cholesterol or other small molecules (Fig. 4B).
Fig. 4.
The specific location of I1061 and P691 in NPC1. (A) The location of I1061. Close-up view of 2Fo-Fc density map of I1061 (black mesh) contoured at σ level 1.0. (B) The location of P691. Close-up view of 2Fo-Fc density map of P691 (black mesh) contoured at σ level 1.0. The putative ligand-binding pocket of the SSD is indicated by a black oval.
Discussion
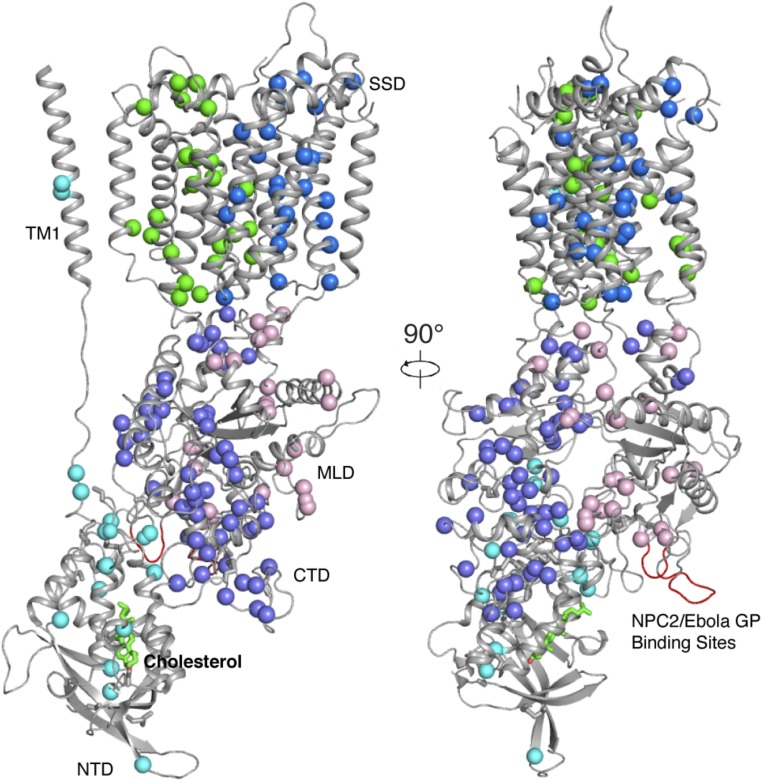
NPC disease is associated with mutations either in the NPC1 (95% of families) or the NPC2 gene. The improved resolution of the structure in this study allows us to map all of the reported NPC-causing mutations, which distribute broadly over the NPC1 protein (Fig. 5). In the transmembrane domain, the mutations only locate to the TMs but not the loops that connect the TMs. Notably, there is no mutation identified on the NPC2 or Ebola glycoprotein interacting site of the MLD (Fig. 5). It is possible that such mutations are lethal for embryonic development so that they will not be identified in patients. Among the three luminal domains, the CTD carries most disease-causing mutations, many of which result in protein misfolding and degradation in the ER (9). This abundance of mutations in the CTD may be explained by the decreased stability of this domain, as the CTD may require more disulfide bonds than the MLD and the NTD for proper folding (Fig. S2).
Fig. 5.
The distribution of NPC-causing mutations. The overall structure rotated by 90° with balls at each mutated residue. Color code for mutations: cyan, NTD and TM1; blue, TMs 2–7; pink, MLD; green, TMs 8–13; and light purple, CTD.
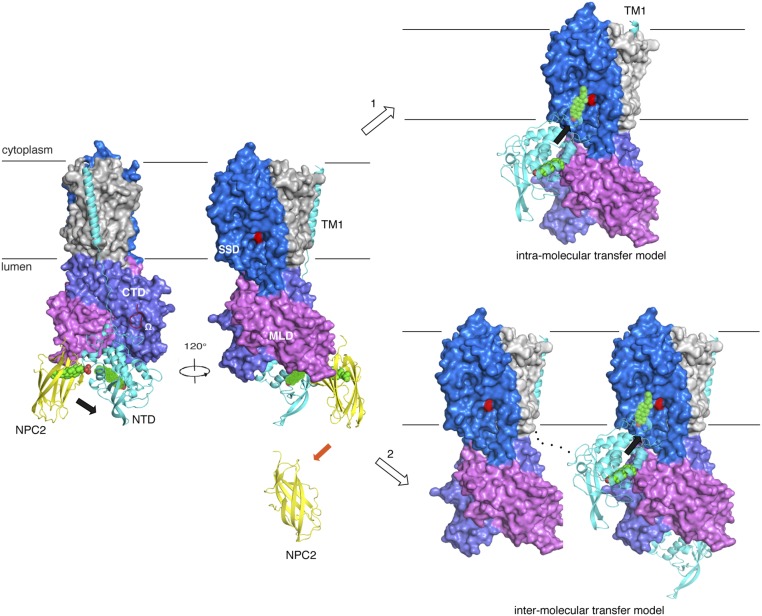
Our previous and current structural observations support the “hydrophobic hand-off” cholesterol transfer model (Fig. 6). In brief, binding of cholesterol in the endosomal lumen to NPC2 induces conformational changes that facilitate its interaction with the NPC1-MLD (23). The NTD, unlike the MLD and CTD, is not buried in the ∼8 nm-thick glycocalyx, and its orientation is crucial to NPC2 for delivering cholesterol into the lysosomal lumen. Because the Ω loop–Ψ loop interaction is the major contributor to the NTD–CTD interface and larger than any other interface between the NTD and the MLD, we suggest that without the Ω loop, the NTD may not be in the favorable position to receive cholesterol from NPC2. A cholesterol esterification assay, showing that NPC1 without the Ω loop or the Ψ loop can prevent cholesterol transport out of the late endosomes, supports this hypothesis. Alternatively, or in addition, the regulation of the NTD could be affected by Lamp (lysosomal-associated membrane protein) proteins (29).
Fig. 6.
Models of NPC2–NPC1-mediated cholesterol transfer. The Ω loop (red) might keep the NTD in the proper orientation for receiving the cholesterol from NPC2 when NPC2 binds to the MLD. Cholesterol (green balls) is transferred from NPC2 (yellow) to NPC1-NTD (cyan) when NPC2 binds to NPC1-MLD (magenta). After this transfer, the NTD could reorient across the glycocalyx to dock in the gap between the SSD (blue) and the MLD for delivering the cholesterol to the SSD. The mechanism of this step is still unclear. Two possibilities exist: (1) The first is intramolecular transfer. Because the NTD is far away from the SSD, a conformational change is required for intramolecular transfer. The long linker between TM1 (cyan) and TM2 (on the edge of the SSD) might induce movement of TM1 to allow the proximity between NTD to SSD in one NPC1 molecule to trigger this transfer. (2) The other is intermolecular transfer. The NTD of one NPC1 molecule could insert into another NPC1-SSD without movement of TM1 and without any large conformational change. Black arrows indicate the transfer orientation of the cholesterol molecule. The P691 in the SSD cavity is indicated in red.
After cholesterol is slipped from NPC2 to the NPC1-NTD, NPC2 changes its conformation to disassociate from the MLD-binding site (23), and the Ω loop–Ψ loop interaction might be weakened to allow the NTD to reach across the glycocalyx for delivering cholesterol to the SSD pocket in the membrane. How the NTD delivers cholesterol to the SSD in the glycocalyx is still a mystery. We suggest two possibilities for this step (Fig. 6). One is an intramolecular NPC1 transfer, whereby the long linker between TM1 and TM2, which is disordered in the full-length cryo-EM structure, might induce movement of TM1 to allow docking between the NTD and SSD pocket. Another is an intermolecular transfer, whereby the NTD of one NPC1 molecule could dock to an SSD pocket of a neighboring NPC1.
The functions of the NTD and the MLD have been well established by previous studies (18, 30, 31). Our higher resolution data revealed several hitherto unknown contacts between the CTD and NTD domains of NPC1 and allowed an atomic model for mapping NPC-causing mutations.
Experimental Procedures
Protein Expression and Purification.
Human NPC1 protein was expressed and purified as previously described (21). Briefly, the cells were disrupted by sonication in buffer A, containing 20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM PMSF, and 5 μg/mL each of leupeptin and aprotinin. After low-speed centrifugation, the resulting supernatant was incubated in buffer A with 1% (wt/vol) n-Dodecyl-β-d-malto-pyranoside (Anatrace) and 100 μM itraconazole (Sigma-Aldrich) for 2 h at 4 °C. The lysate was centrifuged again, and the supernatant was loaded onto a Ni2+-NTA affinity column (Qiagen). After washing twice with 20 mM Hepes, pH 7.0, 150 mM NaCl, 50 mM imidazole, 0.02% n-Dodecyl-β-d-maltopyranoside, and 10 μM itraconazole, the protein was eluted in 20 mM Hepes, pH 7.0, 150 mM NaCl, 300 mM imidazole, 0.02% n-Dodecyl-β-d-maltopyranoside, and 10 μM itraconazole and concentrated. The concentrated protein was incubated with 10 μg/mL proteinase K (Sigma-Aldrich) at room temperature for 15 min and purified by Superdex-200 size-exclusion chromatography (GE Healthcare) in a buffer containing 20 mM MES, pH 5.5, 150 mM NaCl, 0.06% (wt/vol) Cymal-6 (Anatrace), and 10 μM itraconazole. The peak fractions were collected and concentrated to 10 mg/mL for crystallization.
Crystallization.
Crystals were grown at 20 °C by the hanging-drop vapor-diffusion method. Before crystallization, protein was incubated with 100 μM itraconazole on ice for 1 h.
After optimization, crystals in the C2221 space group appeared in 3 d in well buffer containing 0.1 M sodium acetate, pH 5.5, 0.1 M NaCl, 10% (vol/vol) PEG4000, 6% (vol/vol) Jeffamine M-600, pH 7.0, and 6% (vol/vol) PEG P400. The crystals in space group C2221 have unit cell dimensions of a = 174.87 Å, b = 222.11 Å, and c = 107.85 Å. All crystals were flash-frozen in a liquid nitrogen stream with well buffer plus 20% Jeffamine M-600, pH 7.0, for cryoprotection.
Data Collection and Structure Determination.
The data were collected at the Advanced Photon Source beamline ID24-E at 100 K. The dataset was processed using XDS from the NECAT RAPD website (https://rapd.nec.aps.anl.gov/rapd/). The structure was solved by molecular replacement using Phaser from the CCP4 program suite (Collaborative Computational Project) with the previously reported NPC1 structure (PDB ID code 5I31) as search models. The initial model was built in Coot (32) manually. The structure was refined with PHENIX.REFINE (33) at 3.3 Å resolution. Model validation was performed with MolProbity (34). All figures were generated with PyMOL.
Cholesterol Esterification in 10–3 Cells.
The rate of incorporation of [14C]oleate into cholesteryl [14C]esters and [14C]triglycerides by monolayers of 10–3 cells was measured as described previously in detail (35).
Medium A is a 1:1 mixture of Ham's F-12 medium and DMEM containing 2.5 mM l-glutamine. Medium B is l-glutamine–free DMEM. All media contained 100 U/mL penicillin and 100 μg/mL streptomycin sulfate, unless noted otherwise in the figure legends.
On day 0, NPC1−/− 10–3 cells were set up in medium A with 5% FCS at 2.5 × 105 cells per 60-mm dish. On day 1, monolayers were washed once with Dulbecco’s PBS, switched to fresh medium A with 5% lipoprotein-deficient serum (LPDS), and then transfected with 2 µg DNA per dish with the indicated plasmids as described above. After incubation for 24 h, cells were washed once with PBS and switched to medium A with 5% LPDS containing 50 µM sodium compactin and 50 µM sodium mevalonate. On day 3, the cells received fresh medium B containing compactin and mevalonate in the presence of either 5% LPDS or 5% FCS as indicated. After incubation for 4 h at 37 °C, each cell monolayer was pulse-labeled for 2 h with 0.1 mM sodium [14C]oleate (18,412 dpm/nmol). The cells were then washed, and the lipids were extracted in hexane:isopropanol (3:2), separated on a silica gel G thin-layer chromatogram (developed in heptane:ethylether:acetic acid, 90:30:1), and quantified by scintillation counting. The amounts of cholesteryl [14C]oleate and [14C]triglycerides formed are expressed as nanomoles formed per hour per milligram cell protein. For reproducibility, each experiment was repeated at least three times, with similar results obtained.
Western Blot Analysis.
To verify the amount of NPC1 protein transfected into cells, Western blot analysis was performed. Cell lysates were subjected to electrophoresis in Bolt 4–12% Bis–Tris Plus gradient gels, after which the proteins were transferred to nitrocellulose filters. The filters were incubated at 4 °C overnight with either 0.5 µg/mL rabbit monoclonal IgG against amino acids 1,261–1,278 of human NPC1 (cat. no. 134113; Abcam) or 2 µg/mL mouse monoclonal IgG-13G4 against NPC2 (36). Bound antibodies were visualized by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientific) after a 1 h incubation with a 1:5,000 dilution of either donkey anti-mouse or goat anti-rabbit IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch). The images were scanned using an Odyssey FC Imager (Dual-Mode Imaging System; 2-min integration time) and analyzed using Image Studio ver. 5.0 (LI-COR).
Transfections and Confocal Microscopy.
On day 0, 10–3 cells were plated on glass coverslips in a 1:1 mixture of Ham’s F-12 medium and DMEM containing 2.5 mM l-glutamine with 5% FCS (Medium A) at 1.5 × 105 cells per 35 mm well in a six-well plate. On day 1, cells were transfected with 1 µg plasmid DNA per well using FuGENE HD (Promega) with one of the following plasmids: pCDNA3.1 empty vector, pCMV-NPC1-Flag-TEV-StrepTactin, pCMV-NPC1(P691S)-Flag-TEV-StrepTactin, or pCMV-NPC1(C909A, Δ910–917)-Flag-TEV-StrepTactin (ΔΩ-A). On day 2, cells were fixed in 3.7% formaldehyde in PBS for 15 min at room temperature and permeabilized in methanol for 10 min at −20 °C. After blocking with 1 mg/mL BSA in PBS, coverslips were incubated with 1 µg/mL rabbit monoclonal anti-FLAG IgG (Sigma) and 1 µg/mL mouse monoclonal anti–LAMP-2 IgG (clone H4B4; BD Pharmingen) followed by 6.7 µg/mL AlexaFluor 488-conjugated goat anti-rabbit and 6.7 µg/mL AlexaFluor 594-conjugated goat anti-mouse secondary antibodies (Invitrogen). Coverslips were mounted in Mowiol (EMD) solution (37), and fluorescence images were acquired using a Plan-Neofluar 40×/1.3 DIC objective (Zeiss), on an Axiovert 200 M microscope (Zeiss), using an Orca 285 camera (Hamamatsu) and the Openlab 4.0.2 software.
Acknowledgments
We thank Joseph L. Goldstein and Michael S. Brown for support and suggestions; Erik Debler for help with manuscript editing; Elias Coutavas for help with manuscript preparation and revision; and Joseph Fernandez and Henrik Molina (Proteomics Resource Center at The Rockefeller University, funded by the Leona M. and Harry B. Helmsley Charitable Trust) for mass spectrometry analyses. This work was supported by funds from the Howard Hughes Medical Institute (G.B., investigator) and by NIH Grants HL20948 and GM096070 (to J.S.). X.L. is the recipient of a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5U74).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711716114/-/DCSupplemental.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250:8487–8495. [PubMed] [Google Scholar]
- 3.Naureckiene S, et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 4.Carstea ED, et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 5.Neiss WF. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 1984;80:603–608. [PubMed] [Google Scholar]
- 6.Vanier MT. Niemann-Pick disease type C. Orphanet J Rare Dis. 2010;5:16. doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Côté M, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott C, Ioannou YA. The NPC1 protein: Structure implies function. Biochim Biophys Acta. 2004;1685:8–13. doi: 10.1016/j.bbalip.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 11.Kuwabara PE, Labouesse M. The sterol-sensing domain: Multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- 12.Ohgami N, et al. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc Natl Acad Sci USA. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgane K, Karaki F, Dodo K, Hashimoto Y. Discovery of oxysterol-derived pharmacological chaperones for NPC1: Implication for the existence of second sterol-binding site. Chem Biol. 2013;20:391–402. doi: 10.1016/j.chembiol.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Lu F, et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife. 2015;4:e12177. doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trinh MN, et al. Triazoles inhibit cholesterol export from lysosomes by binding to NPC1. Proc Natl Acad Sci USA. 2016;114:89–94. doi: 10.1073/pnas.1619571114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Head SA, et al. Simultaneous targeting of NPC1 and VDAC1 by itraconazole leads to synergistic inhibition of mTOR signaling and angiogenesis. ACS Chem Biol. 2017;12:174–182. doi: 10.1021/acschembio.6b00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Infante RE, et al. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J Biol Chem. 2008;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 18.Infante RE, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 19.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Ren J, Harlos K, Stuart DI. Structure of glycosylated NPC1 luminal domain C reveals insights into NPC2 and Ebola virus interactions. FEBS Lett. 2016;590:605–612. doi: 10.1002/1873-3468.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, et al. Structure of human Niemann-Pick C1 protein. Proc Natl Acad Sci USA. 2016;113:8212–8217. doi: 10.1073/pnas.1607795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong X, et al. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell. 2016;165:1467–1478. doi: 10.1016/j.cell.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Saha P, Li J, Blobel G, Pfeffer SR. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc Natl Acad Sci USA. 2016;113:10079–10084. doi: 10.1073/pnas.1611956113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greer WL, et al. Mutations in NPC1 highlight a conserved NPC1-specific cysteine-rich domain. Am J Hum Genet. 1999;65:1252–1260. doi: 10.1086/302620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelsthorpe ME, et al. Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem. 2008;283:8229–8236. doi: 10.1074/jbc.M708735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pipalia NH, et al. Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann-Pick C1 mutant cells. J Lipid Res. 2017;58:695–708. doi: 10.1194/jlr.M072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauniyar N, et al. Quantitative proteomics of human fibroblasts with I1061T mutation in Niemann-Pick C1 (NPC1) protein provides insights into the disease pathogenesis. Mol Cell Proteomics. 2015;14:1734–1749. doi: 10.1074/mcp.M114.045609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millat G, et al. Niemann-Pick C1 disease: The I1061T substitution is a frequent mutant allele in patients of western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet. 1999;65:1321–1329. doi: 10.1086/302626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Pfeffer SR. Lysosomal membrane glycoproteins bind cholesterol and contribute to lysosomal cholesterol export. Elife. 2016;5:e21635. doi: 10.7554/eLife.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deffieu MS, Pfeffer SR. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci USA. 2011;108:18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller EH, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 36.Wang ML, et al. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010;12:166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei JH, Seemann J. Induction of asymmetrical cell division to analyze spindle-dependent organelle partitioning using correlative microscopy techniques. Nat Protoc. 2009;4:1653–1662. doi: 10.1038/nprot.2009.160. [DOI] [PubMed] [Google Scholar]