Abstract
Antibody 34E4 catalyzes the conversion of benzisoxazoles to salicylonitriles with high rates and multiple turnovers. The crystal structure of its complex with the benzimidazolium hapten at 2.5-Å resolution shows that a combination of hydrogen bonding, π stacking, and van der Waals interactions is exploited to position both the base, GluH50, and the substrate for efficient proton transfer. Suboptimal placement of the catalytic carboxylate, as observed in the 2.8-Å structure of the GluH50Asp variant, results in substantially reduced catalytic efficiency. In addition to imposing high positional order on the transition state, the antibody pocket provides a highly structured microenvironment for the reaction in which the carboxylate base is activated through partial desolvation, and the highly polarizable transition state is stabilized by dispersion interactions with the aromatic residue TrpL91 and solvation of the leaving group oxygen by external water. The enzyme-like efficiency of general base catalysis in this system directly reflects the original hapten design, in which a charged guanidinium moiety was strategically used to elicit an accurately positioned functional group in an appropriate reaction environment and suggests that even larger catalytic effects may be achievable by extending this approach to the induction of acid-base pairs capable of bifunctional catalysis.
Keywords: crystal structure, base catalysis, proton transfer, medium effects, orientation effects
Proton abstraction from carbon constitutes a fundamental process catalyzed by numerous enzymes, including isomerases, epimerases, racemases, lyases, and some synthases (1). Although heterolytic C—H bond cleavage is a kinetically and thermodynamically demanding reaction, deprotonation catalysts rank among the most efficient enzymes known. For example, triosephosphate isomerase, ketosteroid isomerase, and fumarase operate at the diffusion limit (2). The mechanisms by which proteins that contain only weak acids and bases accelerate proton transfers by such enormous factors has long intrigued biochemists (3). Crystal structures and biochemical experiments of such enzymes have highlighted the importance of precisely positioned functional groups with optimized pKas (3–5), but the precise origins of their high activities remain controversial (6).
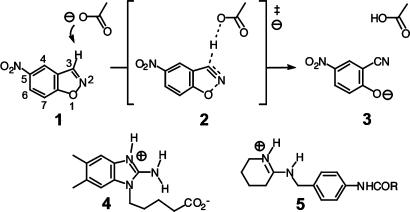
Valuable insight into the factors that contribute to efficient proton abstraction can be gained from the base-promoted Kemp elimination (1 → 3, Fig. 1), a widely used model system that is sensitive to base strength and solvent environment (7–9). This reaction is also susceptible to catalysis by antibodies (10, 11), albumins (12–14), polyethyleneimine “synzymes” (15, 16), organic hosts (17), cationic vesicles (18), and even natural coals (19). Antibody 34E4, which was raised against the cationic 2-aminobenzimidazolium derivative 4, is particularly effective in this regard (11). It promotes the decomposition of benzisoxazole 1 with >103 turnovers per active site and achieves a rate acceleration of 106 over background (11). A glutamate at position H50, elicited in response to the cationic hapten, serves as the catalytic base (20). The effective molarity (EM) of this residue is >50,000 M. This value, which corresponds to the concentration of a carboxylate base that would be needed in the absence of antibody to achieve the same rate, is several orders of magnitude larger than EMs typically obtained in intramolecular model systems for general base catalysis (21, 22).
Fig. 1.
Base-catalyzed Kemp elimination of 5-nitrobenzisoxazole (1). In this example, acetate is the base that abstracts the C3 proton from the substrate. Antibodies 34E4 and 4B2 were raised against haptens 4 and 5, respectively, to elicit a carboxylate group as a base. Both of these antibodies catalyze the Kemp elimination.
Although high effective molarities are often interpreted as the entropic advantage of precisely aligning reacting groups, the extreme solvent sensitivity of the acetate-promoted decomposition of benzisoxazoles raises the possibility that medium effects also contribute appreciably in 34E4. In fact, GluH50 is activated relative to acetate, as judged by the elevated pKa of its conjugate acid (≈6.0) (11), presumably because it is embedded in a relatively apolar active site. Based on the finding that the Kemp elimination can be effectively catalyzed by serum albumins and synzymes containing solvent-insensitive amine bases, it has even been argued (12, 15, 16) that medium effects are likely to be the dominant factor in 34E4 catalysis, with little importance attributed to the precise positioning of the catalytic base. Countering this conclusion, mutagenesis studies (20), as well as the unexpectedly low activity of benzisoxazoles containing 6- and 7-nitro substituents (23), have provided evidence that orientation effects play a major role in 34E4 catalysis.
To gain further insight into these mechanistic issues, crystal structures of the 34E4 antibody and a variant in which the active-site base has been repositioned by mutagenesis were determined in complex with hapten 4. The structural data confirm the importance of precise positional ordering of the catalytic base in this system and illuminate at the atomic level how medium effects likely augment catalytic efficiency in the active site.
Materials and Methods
34E4 Fab Preparation, Crystallization, and Data Collection. Wild-type 34E4 and the GluH50Asp variant were produced and purified as chimeric murine–human Fabs, as described (20). Crystallization experiments were performed by the sitting drop vapor diffusion method at 22.5°C. The Fabs, concentrated to 14 mg/ml, were crystallized in the presence of 3-fold molar excess of hapten under similar conditions [1.5 and 1.7 M (NH4)2HPO4, respectively/0.5 M NaCl/0.1 M imidazole HCl (pH 8.0)]. Both complexes crystallized in space group P3221 with similar unit cell dimensions and four molecules in the asymmetric unit (Table 1). For data collection, the crystals were flash-cooled to 110 K by using 20–25% glycerol as cryoprotectant. Data were collected at the Advanced Light Source, Lawrence Berkeley National Laboratory (Berkeley, CA) and processed and scaled with hkl2000 (24) (Table 1).
Table 1. Data collection and refinement statistics.
| 34E4-4 | 34E4 EH5OD-4 | |
|---|---|---|
| Space group | P3221 | P3221 |
| Unit cell dimensions, Å | a = b = 165.2, c = 152.6 | a = b = 163.0, c = 151.2 |
| Resolution range, Å | 50.0-2.5 (2.54-2.50)* | 50.0-2.8 (2.85-2.80)* |
| Observations | 191,280 | 135,576 |
| Unique reflections | 81,375 | 56,867 |
| Completeness, % | 97.6 (98.4)* | 98.6 (98.5)* |
| Rsym† | 0.07 (0.45)* | 0.09 (0.44)* |
| Mean I/σ | 14.7 (1.8)* | 8.2 (1.5)* |
| Rcryst‡ / Rfree§ | 0.20/0.24 | 0.22/0.25 |
| No. of refined protein atoms/hapten atoms/water molecules/chloride ions | 13,464/76/425/1 | 13,460/76/287/1 |
| RMSD¶ bond length, Å/angle, ° | 0.009/1.4 | 0.008/1.4 |
| Average B values protein/hapten/water/chloride, Å2 | 40.4/35.8/35.7/30.8 | 32.2/32.4/21.3/24.9 |
| Ramachandran plot most favored/additionally allowed/generously allowed/disallowed∥ (%) | 88.8/10.4/0.3/0.5 | 85.6/13.4/0.5/0.5 |
Highest-resolution shell.
Rsym = ΣhklΣi|Ii(hkl) - 〈I(hkl)〉|/ΣhklΣi Ii(hkl)
Rcryst = Σhkl||Fc(hkl)| - |Fo(hkl)||/Σhkl|Fo(hkl)|.
Rfree is calculated in the same manner as Rcryst but from 5% of the data that was not used for refinement.
RMSD, rms deviation.
ThrL51, SerL93, and their corresponding residues in Fabs AB, CD, and EF are the only residues in a disallowed region, but they both have well defined electron density. ThrL51 is in a γ turn, as commonly observed in other antibody structures (46).
Structure Determination and Refinement. The structure of 34E4 was determined by molecular replacement by using merlot (25) to rapidly screen ≈250 Fab structures for potential solutions to the rotation function. Among the 10 top solutions, the murine ribonucleotide-binding antibody Jel103 (26) was found to be the best model by using the program amore (27) [correlation coefficient (CC) of 27.3 and Rcryst = 51.6% compared with the next peak with a CC of 16.0 and Rcryst = 55.1%]. The model was refined by alternating cycles of model building with the program o (28) and refinement with cns (29). The hapten could easily be built into unambiguous difference electron density maps for both complex structures. During refinement, noncrystallographic symmetry (NCS) restraints of 300 kcal/mol were applied to all main-chain atoms in the four Fab molecules in the asymmetric unit, except for some loop regions. The final statistics of both structures are shown in Table 1. The quality of the structures was analyzed by using the programs molprobity (30), what if (31), and procheck (32).
Results
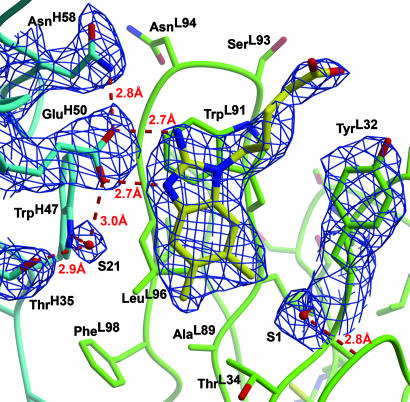
34E4 Fab Crystal Structures. The x-ray structures of wild-type Fab 34E4 and the GluH50Asp variant, both complexed with the benzimidazolium hapten 4, were determined at 2.5- and 2.8-Å resolution, respectively (Table 1). The Fab 34E4 structures resemble other antibodies in their general features (33). The occurrence of four noncrystallographic symmetry-related molecules, designated Fab LH, AB, CD, and EF, respectively, allowed any potential artifacts arising from crystal packing to be assessed and ruled out, while providing independent corroboration of key active-site features. The overall structure of the 34E4 GluH50Asp mutant is essentially unchanged from that of the wild-type Fab, with an overall rms deviation of 0.4 Å for Cα atoms and 1.5 Å for all atoms. In all eight 34E4 hapten complexes, well defined density for the active-site residues and the ligand was observed in 3Fo – 2Fc electron density maps (Fig. 2).
Fig. 2.
Antibody-combining site of 34E4 bound to hapten. The heavy and light chains are colored in blue and green, respectively. Two of the active-site water molecules are designated S1 and S21. The 3Fo–2Fc σA-weighted electron density map around the hapten and key active-site residues is contoured at 1.3σ. Hydrogen bonds are shown as broken lines. TrpL91 forms a cation–π interaction with the guanidinium moiety of the hapten. CDR H3 is omitted for clarity.
Hapten Recognition. The benzimidazolium hapten binds snugly in a deep cavity that is 4 Å wide, 6 Å long, and 10 Å deep (Fig. 3 and 4). Approximately 97% of its aromatic surface is buried (34) in the binding pocket, which results in a nearly complete sequestration from the aqueous medium. The light and heavy chains contribute equal numbers of contacts to the benzimidazolium moiety (49.5% and 50.5% by VH and VL, respectively). More specifically, complementarity-determining regions (CDR) H3 (39.1%) and L3 (33.5%) provide the majority of the contacts, whereas L1 (17.0%), H2 (8.6%), and H1 (1.8%) make minor contributions. Three ordered water molecules are found within the active site, but they do not contact the hapten directly. Water molecule S21 interacts with GluH50, TrpH47, and ThrH35, whereas water molecules S1 and S192 satisfy buried backbone carbonyl oxygens and amide protons through hydrogen bonding.
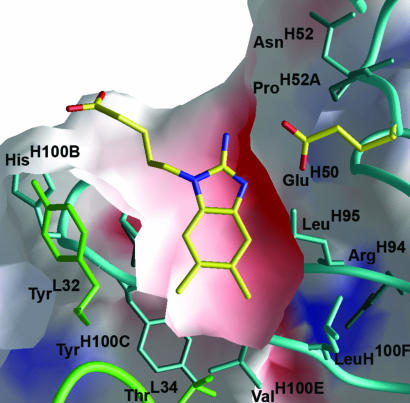
Fig. 3.
Electrostatic and shape complementarity of the hapten in the antigen-binding site. A slice through the center of the binding site is shown between the CDR H3 below (in cyan) and CDR L3 above (not shown). Negative and positive potentials are colored red and blue. The catalytic base GluH50 is highlighted.
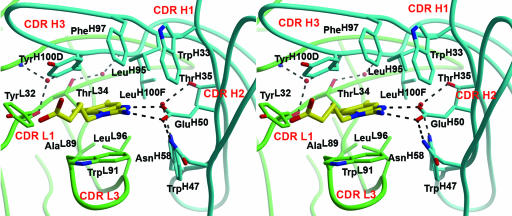
Fig. 4.
Stereoview of the antibody-combining site of the 34E4–hapten complex. Hydrogen bonds are shown as broken lines, and heavy and light chains are colored in blue and green, respectively. The hapten is sandwiched between two aromatic residues TrpL91 and TyrH100D. GluH50 forms a bidentate salt bridge to the guanidinium moiety of the hapten. The CDRs of the light and heavy chains are labeled L1–L3 and H1–H3, respectively.
The binding pocket is highly complementary to the hapten, with respect to both shape [Sc parameter of 0.87 (35)] and charge, as illustrated in Fig. 3. Key features include parallel π stacking of the protonated benzimidazole ring of 4 with the aromatic side chain of TrpL91 of CDR L3 (Fig. 4). The ensuing cation–π interactions presumably contribute to the high affinity of the complex (Kd ≈ 1 nM) (36). Additional van der Waals contacts are provided by TyrL32, LeuL96, TrpH33, LeuH95, PheH97, TyrH100D, LeuH100F, and the methyl group of ThrL34. The highly hydrophobic collar surrounding the ligand is interrupted only by GluH50, the negatively charged catalytic base, which forms a bidentate salt bridge (2.7 Å) with the guanidinium group of 4 (Fig. 2). As expected, the pentanoic acid linker is exposed to the solvent.
Carboxylate Positioning. In the wild-type antibody, the carboxylate side chain of GluH50 is anchored within the active site by hydrogen bond interactions with AsnH58 (2.8 Å) and water molecule S21 (3.0 Å), which is further positioned by ThrH35 and TrpH47 (Figs. 2 and 4). These interactions constrain the GluH50 carboxylate to a conformation that is optimally suited for formation of the bidentate salt bridge with the benzimidazolium hapten. The more basic syn lone pairs of the carboxylate (37) are directed toward the ligand.
Replacement of the catalytic glutamate with aspartate leads to significant losses in hapten affinity (150-fold) and catalytic activity (30-fold) without altering the apparent pKa of the functional group (20). The structural perturbations caused by removal of a methylene unit from the side chain of the base are localized to this residue and its immediate environment (Fig. 5). The conformation of AspH50 itself varies somewhat in each of the four noncrystallographic symmetry-related active sites (Fig. 5), suggesting enhanced conformational mobility compared with glutamate in the wild-type antibody. Although the AspH50 carboxylate still contacts the hapten, it does not form the bidentate salt bridge. Instead, AspH50 interacts via its Oδ2 atom with the protonated N3 atom of 4 in a monodentate fashion, with the Oδ2-N3 distance in the mutant complexes increased somewhat (2.7–3.1 Å) compared with the wild-type complexes (2.6–2.7 Å). The other oxygen (Oδ1) of AspH50 forms a hydrogen bond with the AsnH58 side chain as in the parent antibody.
Fig. 5.
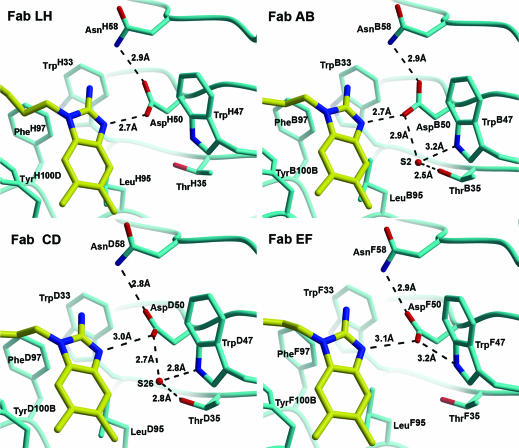
Plasticity of the catalytic base in the active site of the 34E4 GluH50Asp mutant. The proton-abstracting Oδ2ofAspH50 is positioned further away from the hapten compared with the analogous oxygen Oε1 in the wild-type 34E4 (Fig. 2). The syn electron pair of this aspartate oxygen is variably disposed at α = 135° and 155° in Fabs LH and AB, respectively. In Fabs CD and EF, the aspartate carboxylate is rotated out of the ligand plane, which precludes a linear N3—H—O bond in the hapten–antibody complex. Only two of the complexes show convincing electron density for the active-site water (S2 in AB, and S26 in CD) that correspond to S21 in the wild-type Fab LH.
The stereoelectronic interactions between the catalytic base and the hapten are also less ideal in the mutant than in the parent antibody (Fig. 5). In all four copies of the wild-type complex, the ligand N3—H forms a linear hydrogen bond with the carboxylate Oε1 of the catalytic glutamate. Importantly, the angle between the ligand N3—H and the carboxylate C—Oε1 bond vectors in the four complexes is 115°, 115°, 110°, and 120°, which are all close to the 120° optimum for syn orbital interactions. In contrast, the corresponding angle is 135° and 155° in the LH and AB active sites of the GluH50Asp mutant, respectively, whereas in the active sites of Fabs CD and EF, the carboxylate groups of AspD50 and AspF50 are rotated ≈30° out of the ligand plane, precluding a linear N3—H—Oδ2 bond. In Fab EF, the AspF50 conformation is stabilized by a hydrogen bond (3.2 Å) to Nε1 of TrpF47 instead of the S21 water molecule. Interestingly, this water molecule, which has strong electron density in all four Fab wild-type complexes, has good electron density only in Fab AB (S2), very weak density in Fab CD (S26), and no density in Fab LH, indicative of comparatively weak binding to the mutant antibody (Fig. 5).
Discussion
The positively charged benzimidazolium hapten 4 was designed to elicit a reaction chamber for the Kemp elimination in an antibody-combining site containing (i) a carboxylate group for proton abstraction and (ii) aromatic groups for transition state stabilization (11). The structural data show that these design goals have been exceptionally well realized in the active site of antibody 34E4. The positively charged guanidinium moiety of the hapten forms a bidentate salt bridge with the catalytic base GluH50 (Fig. 2). At the same time, this moiety is sandwiched between two aromatic side chains (Fig. 4) in an active-site pocket that exhibits high overall shape complementarity (Fig. 3). Although the factors that contribute to catalytic efficiency are highly interdependent and hence difficult to quantify individually, these structural details provide a molecular basis for qualitatively assessing the roles of medium and orientation effects in 34E4 catalysis.
Transfer from water to aprotic dipolar solvents is known to increase the base strength and reactivity of carboxylate anions dramatically. For instance, the pKa of acetic acid is 22.3 in acetonitrile vs. 4.75 in water (38). This type of nonspecific medium effect is largely responsible for the enormous solvent-induced rate accelerations observed for Kemp eliminations promoted by carboxylate bases (9). Loss of hydrogen bonding in the low dielectric environment of the antibody active site was similarly expected to activate the catalytic carboxylate of 34E4 (11). The structure of the antibody confirms that GluH50 resides in a relatively hydrophobic pocket. However, its carboxylate group is clearly not “dry” (39) but is stabilized in its anionic form by hydrogen bond interactions with the amide side chain of AsnH58 and an ordered water molecule. These interactions explain the relatively modest 10-fold increase in base strength of the GluH50 carboxylate (ΔpKa ≈ 1.25) relative to a normal carboxylic acid in aqueous solution. Because even a single hydrogen bonding interaction inhibits solvent catalysis of the related decarboxylation of 3-carboxybenzisoxazoles (9), we conclude that destabilization of the carboxylate anion through desolvation is unlikely to be a major determinant of the large rate accelerations achieved by 34E4.
The highly polarizable transition state of the Kemp elimination can also be stabilized by dispersion interactions with solvent. This situation represents another type of medium effect that has been invoked to explain, in part, the modest solvent sensitivity of the amine-catalyzed reaction (9), as well as the catalytic efficacy of synzymes (15, 16) and a macrocyclic host (17). In 34E4, the polarizable aromatic side chain of TrpL91 is potentially well suited to mediate such interactions. However, the small catalytic benefit provided by dispersion forces in diverse model systems (9, 17, 40) suggests that their contribution to the overall efficiency of 34E4 is relatively minor.
In contrast, juxtaposition of the substrate and the catalytic base appears to be a significant factor in catalysis by this antibody. The catalytic glutamate, GluH50, adopts a fixed conformation in all four independent active sites, buttressed by its interactions with AsnH58 and a water molecule (S21 in Fab LH, Fig. 2). Its carboxylate is directed toward the bound ligand in an orientation that would allow the more basic syn orbital (37) to be poised for proton abstraction from substrate. The observed degree of organization is consistent with biochemical experiments that showed GluH50 to be comparable to a typical enzymatic base in its effectiveness (20).
The importance of base positioning in wild-type 34E4 is underscored by structural studies of the GluH50Asp variant. Removal of a methylene group from the base substantially displaces the terminal carboxylate group, increasing its distance from the ligand (or substrate) by 0.1–0.5 Å and reorienting its syn orbital so that it is no longer optimally disposed for proton abstraction. Both factors are expected to be deleterious for catalysis. The existence of multiple conformations probably further diminishes the effectiveness of AspH50 as a base (Fig. 5). Together, these observations readily account for the 30-fold decrease in activity associated with mutation. More pronounced structural perturbation should reduce activity even further, as seen for the analogous Glu165Asp mutation of triose phosphate isomerase, where a 1-Å increase in the distance from the base to the bound intermediate analog and a switch from the syn to the anti orbital of the carboxylate results in 1,000-fold lower rates (41). That inactive GluH50Ala and GluH50Gly 34E4 variants cannot be chemically rescued by high concentrations of formate or acetate (20), i.e., by bases that are not covalently fixed within the active site, supports this conclusion.
Precise positioning of the catalytic base alone is not sufficient for efficient proton transfer. The substrate must also be held in place so that its acidic hydrogen is aligned with the base. Because substrate 1 is planar and similar in size and shape to hapten 4, the high shape complementarity of the 34E4-binding pocket guarantees that the substrate will be constrained to bind in an orientation that favors proton abstraction by GluH50 (22). The aromatic clamp would then severely restrict the movement of a benzisoxazole within the active site, so that it would lie roughly in the same plane as the carboxylate base, whereas steric barriers at the bottom and sides of the pocket would limit lateral displacement. Although 5-substituted benzisoxazoles can bind in an orientation similar to the benzimidazolium hapten, with their C3 proton poised for abstraction by GluH50, bulky substituents at the 6- and 7-positions would sterically clash with the walls of the active site. The ensuing misalignment would explain why 5,6-dinitro-, 5,7-dinitro-, and 6-nitrobenzisoxazoles are substantially poorer substrates than expected from their intrinsic reactivity (23).
Binding of the benzisoxazole substrate so as to juxtapose the acidic C3—H and the catalytic base simultaneously positions its ring oxygen near the mouth of the cavity where it can be solvated by external water. This orientation directly reflects the strategy used to couple the hapten to the carrier protein for immunization, because the leaving group oxygen corresponds to N1 of the hapten to which the pentanoic acid linker was attached (Fig. 1). Because negative charge is transferred from the carboxylate base to the leaving group oxygen as the Kemp elimination proceeds (7, 8, 42), interactions of the incipient phenoxide with bulk solvent would be expected to facilitate the reaction. However, this favorable effect is likely to be at least partially offset by the relatively hydrophobic nature of this region of the pocket (Fig. 3). Indeed, a Brønsted leaving group analysis has shown that the antibody-catalyzed reaction is more sensitive to electron-withdrawing substituents that stabilize the developing charge electronically than are the analogous reactions in water or acetonitrile (23). Unfortunately, attempts to increase activity by providing a dedicated general acid proximal to the leaving group oxygen (42), for example by replacing TyrL32 with a cationic lysine or arginine, have not yet been successful (20). The absence of such a group in 34E4 differentiates it from natural enzymes that typically exploit multiple functional groups to achieve their extraordinary efficiencies.
In assessing the relative importance of medium vs. orientation effects, it is instructive to compare 34E4 with a less-efficient catalyst of the Kemp elimination. Antibody 4B2, which was generated against compound 5 (43), also exploits a glutamate base [GluL34 (44)] with an elevated pKa to promote the decomposition of 1. However, GluL34 has been estimated to be 10,000 times less effective as a base than GluH50 in 34E4 (20). Although the carboxylate side chain of GluL34 is well ordered and directed into the active site where it forms a salt bridge with the cationic hapten, the hydrophobic 4B2-binding pocket is more than three times larger than that of 34E4 (≈8 Å × 8 Å × 12 Å) (44). As a consequence of its poor shape complementarity to planar 1, the probability of constraining the benzisoxazole substrate in a productive orientation relative to the catalytic base will be low. Simple placement of a carboxylate in a hydrophobic environment is obviously insufficient for efficient catalysis of the Kemp elimination. High structural complementarity between the antibody and the substrate in the transition state, as seen in 34E4, apparently sets a good catalyst apart from a more primitive one.
Electrostatic stabilization of the transition state and preorganization of the active site are generally believed to largely account for the high efficiency of enzymes (45). The structure of 34E4 has revealed many similarities to biological counterparts that promote proton transfers. The 34E4 active site affords a highly structured microenvironment for the Kemp elimination in which noncovalent forces are effectively used to preorganize a carboxylate base and the benzisoxazole substrate for reaction and to shuttle charge from the base to bulk solvent as the reaction proceeds. The reactivity of the carboxylate and the stability of the transition state appear to be fine-tuned by the hydrophobic and polarizable residues that line the binding pocket. These attributes, which accurately reflect the stereoelectronic properties of the hapten used for immunization, demonstrate the feasibility of eliciting functional groups in antibody pockets in response to haptenic charge that are as effective as analogous groups in highly evolved enzymes. Nevertheless, the reliance of 34E4 on a single catalytic residue probably prevents it from achieving the rates of the most efficient enzymes. Development of strategies to install multiple functional groups, therefore, could afford even more potent catalysts for a wide range of important chemical transformations.
Acknowledgments
We thank the Advanced Light Source staff at beamlines 5.0.2 and 8.3.1 for their assistance, R. Stanfield and X. Dai for help with data collection, M. Elsliger for computational assistance, and X. Zhu for helpful discussions. This research was supported by National Institutes of Health Grant GM38273 (to I.A.W.), a Skaggs predoctoral fellowship (to E.W.D.), and the Eidgenössische Technische Hochschule Zurich (to D.H.). This is publication 17028-MB from The Scripps Research Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: CDR, complementarity-determining region.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1Y0L and 1Y18 for the 34E4 and the GluH50Asp variant complexes, respectively).
References
- 1.Walsh, C. (1979) Enzymatic Reaction Mechanisms (Freeman, New York).
- 2.Radzicka, A. & Wolfenden, R. (1995) Science 267, 90–93. [DOI] [PubMed] [Google Scholar]
- 3.Remington, S. J. (1992) Curr. Opin. Struct. Biol. 2, 730–735. [Google Scholar]
- 4.Ha, N. C., Choi, G., Choi, K. Y. & Oh, B. H. (2001) Curr. Opin. Struct. Biol. 11, 674–678. [DOI] [PubMed] [Google Scholar]
- 5.Alber, T., Banner, D. W., Bloomer, A. C., Petsko, G. A., Phillips, D., Rivers, P. S. & Wilson, I. A. (1981) Philos. Trans. R. Soc. London Ser. B 293, 159–171. [DOI] [PubMed] [Google Scholar]
- 6.Richard, J. P. & Amyes, T. L. (2001) Curr. Opin. Chem. Biol. 5, 626–633. [DOI] [PubMed] [Google Scholar]
- 7.Casey, M. L., Kemp, D. S., Paul, K. G. & Cox, D. D. (1973) J. Org. Chem. 38, 2294–2301. [Google Scholar]
- 8.Kemp, D. S. & Casey, M. L. (1973) J. Am. Chem. Soc. 95, 6670–6680. [Google Scholar]
- 9.Kemp, D. S., Cox, D. D. & Paul, K. G. (1975) J. Am. Chem. Soc. 97, 7312–7318. [Google Scholar]
- 10.Genre-Grandpierre, A., Tellier, C., Loirat, M.-J., Blanchard, D., Hodgson, D. R. W., Hollfelder, F. & Kirby, A. J. (1997) Bioorg. Med. Chem. Lett. 7, 2497–2502. [Google Scholar]
- 11.Thorn, S. N., Daniels, R. G., Auditor, M. T. & Hilvert, D. (1995) Nature 373, 228–230. [DOI] [PubMed] [Google Scholar]
- 12.Hollfelder, F., Kirby, A. J. & Tawfik, D. S. (1996) Nature 383, 60–63. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi, K., Thorn, S. N. & Hilvert, D. (1996) J. Am. Chem. Soc. 118, 8184–8185. [Google Scholar]
- 14.Hollfelder, F., Kirby, A. J., Tawfik, D. S., Kikuchi, K. & Hilvert, D. (2000) J. Am. Chem. Soc. 122, 1022–1029. [Google Scholar]
- 15.Hollfelder, F., Kirby, A. J. & Tawfik, D. S. (2001) J. Org. Chem. 66, 5866–5874. [DOI] [PubMed] [Google Scholar]
- 16.Hollfelder, F., Kirby, A. J. & Tawfik, D. S. (1997) J. Am. Chem. Soc. 119, 9578–9579. [Google Scholar]
- 17.Kennan, A. J. & Whitlock, H. W. (1996) J. Am. Chem. Soc. 118, 3027–3028. [Google Scholar]
- 18.Perez-Juste, J., Hollfelder, F., Kirby, A. J. & Engberts, J. B. (2000) Org. Lett. 2, 127–130. [DOI] [PubMed] [Google Scholar]
- 19.Shulman, H. & Keinan, E. (2000) Org. Lett. 2, 3747–3750. [DOI] [PubMed] [Google Scholar]
- 20.Seebeck, F. P. & Hilvert, D. (2005) J. Am. Chem. Soc. 127, 1307–1312. [DOI] [PubMed] [Google Scholar]
- 21.Kirby, A. J. (1980) Adv. Phys. Org. Chem. 17, 183–278. [Google Scholar]
- 22.Kirby, A. J. (1997) Acc. Chem. Res. 30, 290–296. [Google Scholar]
- 23.Hu, Y., Houk, K. N., Kikuchi, K., Hotta, K. & Hilvert, D. (2004) J. Am. Chem. Soc. 126, 8197–8205. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald, P. M. D. (1988) J. Appl. Crystallogr. 21, 273–278. [Google Scholar]
- 26.Pokkuluri, P. R., Bouthillier, F., Li, Y., Kuderova, A., Lee, J. & Cygler, M. (1994) J. Mol. Biol. 243, 283–297. [DOI] [PubMed] [Google Scholar]
- 27.Navaza, J. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 28.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 29.Brünger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 30.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S. & Richardson, D. C. (2003) Proteins 50, 437–450. [DOI] [PubMed] [Google Scholar]
- 31.Vriend, G. (1990) J. Mol. Graphics 8, 52–56. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 33.Amzel, L. M. & Poljak, R. J. (1979) Annu. Rev. Biochem. 48, 961–997. [DOI] [PubMed] [Google Scholar]
- 34.Sheriff, S., Hendrickson, W. A. & Smith, J. L. (1987) J. Mol. Biol. 197, 273–296. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence, M. C. & Colman, P. M. (1993) J. Mol. Biol. 234, 946–950. [DOI] [PubMed] [Google Scholar]
- 36.Dougherty, D. A. (1996) Science 271, 163–168. [DOI] [PubMed] [Google Scholar]
- 37.Gandour, R. D. (1981) Bioorg. Chem. 10, 169–176. [Google Scholar]
- 38.Kolthoff, I. M., Chantooni, M. K., Jr., & Bhowmik, S. (1968) J. Am. Chem. Soc. 90, 23–28. [Google Scholar]
- 39.Kemp, D. S. (1995) Nature 373, 196–197. [DOI] [PubMed] [Google Scholar]
- 40.Ngola, S. M. & Dougherty, D. A. (1996) J. Org. Chem. 61, 4355–4360. [DOI] [PubMed] [Google Scholar]
- 41.Joseph-McCarthy, D., Rost, L. E., Komives, E. A. & Petsko, G. A. (1994) Biochemistry 33, 2824–2829. [DOI] [PubMed] [Google Scholar]
- 42.Na, J., Houk, K. N. & Hilvert, D. (1996) J. Am. Chem. Soc. 118, 6462–6471. [Google Scholar]
- 43.Goncalves, O., Dintinger, T., Lebreton, J., Blanchard, D. & Tellier, C. (2000) Biochem. J. 346, 691–698. [PMC free article] [PubMed] [Google Scholar]
- 44.Golinelli-Pimpaneau, B., Goncalves, O., Dintinger, T., Blanchard, D., Knossow, M. & Tellier, C. (2000) Proc. Natl. Acad. Sci. USA 97, 9892–9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warshel, A. (1998) J. Biol. Chem. 273, 27035–27038. [DOI] [PubMed] [Google Scholar]
- 46.Arévalo, J. H., Stura, E. A., Taussig, M. J. & Wilson, I. A. (1993) J. Mol. Biol. 231, 103–118. [DOI] [PubMed] [Google Scholar]