Abstract
Recent technological advances have made it possible to decode DNA methylomes at single-base-pair resolution under various physiological conditions. Many aberrant or differentially methylated sites have been discovered, but the mechanisms by which changes in DNA methylation lead to observed phenotypes, such as cancer, remain elusive. The classical view of methylation-mediated protein-DNA interactions is that only proteins with a methyl-CpG binding domain (MBD) can interact with methylated DNA. However, evidence is emerging to suggest that transcription factors lacking a MBD can also interact with methylated DNA. The identification of these proteins and the elucidation of their characteristics and the biological consequences of methylation-dependent transcription factor-DNA interactions are important stepping stones towards a mechanistic understanding of methylation-mediated biological processes, which have crucial implications for human development and disease.
DNA methylation, one of the best-studied epigenetic marks in eukaryotes, is a biological process in which a methyl group is covalently added to a cytosine, yielding 5-methylcytosine (5mC)1–3 (BOX 1). The methylation process is carried out by a set of enzymes called DNA methyltransferases (DNMTs)4, which are encoded in many genomes, from bacteria to plants and mammals5,6. The evolutionary conservation of these enzymes suggests that DNA methylation provides a selective advantage to the organism. However, the percentage of methylated cytosine varies substantially across species. For example, vertebrates and plants often have a high percentage of methylated CpG dinucleotides outside CpG islands, whereas invertebrates typically exhibit intermediate levels or no methylation7,8. With the development of more sensitive methodological approaches, such as methylated DNA immunoprecipitation followed by bisulfite sequencing (MeDIP–BS-seq) — which sequences bisulfite-converted DNA species after enrichment for methylated DNA fragments using an anti-5mC antibody — some genomes previously considered not to have any DNA methylation (for example, that of Drosophila melanogaster) have now been found to be methylated at a limited number of cytosines9–11. In most animals, DNA is methylated predominantly at CpG dinucleotides, whereas in plants and fungi, a large fraction of DNA methylation also occurs at CHG or CHH (where H can be any nucleotide but G)12–15. That said, it was recently discovered that a small fraction of non-CpG methylation also occurs in animals (BOX 1).
Box 1. Non-CpG methylation and methylcytosine derivatives.
Methylated CpH
Cytosine methylation is usually considered to only occur at CpG sites. Recent advances in genome-wide single-nucleotide sequencing have led to a re-examination of this concept. Interestingly, non-CpG methylation (that is, CpH; where H can be any nucleotide but G) was observed in mammalian stem cells and neuronal cells27,28,124. A recent deep-sequencing survey on 18 human tissues revealed an unexpected presence of methylation at non-CpG sites in almost all tissues tested125. Several lines of evidence suggest that non-CpG methylation might be functional. First, the flanking sequences of the methylated CpH (mCpH) showed similar motifs to 5′-TNCA(C/G)125 (where N can be any nucleotide). Second, the position of DNA methylation is highly conserved across different cell types27. Third, gene expression level is negatively correlated with the methylation level in the gene body125. To understand the biological functions of the modification, identification of the proteins that interact with these modifications would be a crucial step.
One such protein is methyl-CpG-binding protein 2 (MeCP2), which is known to interact with mCpG sites and negatively regulates gene expression. Superimposing MeCP2 chromatin immunoprecipitation followed by sequencing (ChIP–seq) and mCpH profiles in neurons showed an enrichment of mCpH around the binding peak of MeCP2. MeCP2 ChIP followed by bisulfite sequencing (ChIP– BS-seq) confirmed its ability to bind mCpH sites in vivo124. In vitro electrophoretic mobility shift assays (EMSAs) demonstrated a direct interaction between MeCP2 and mCpH. The relative affinity of MeCP2 with mCpA is similar to that with mCpG126,127. However, the affinity of MeCP2 for mCpT and mCpC is markedly lower than for mCpA and mCpG126,127.
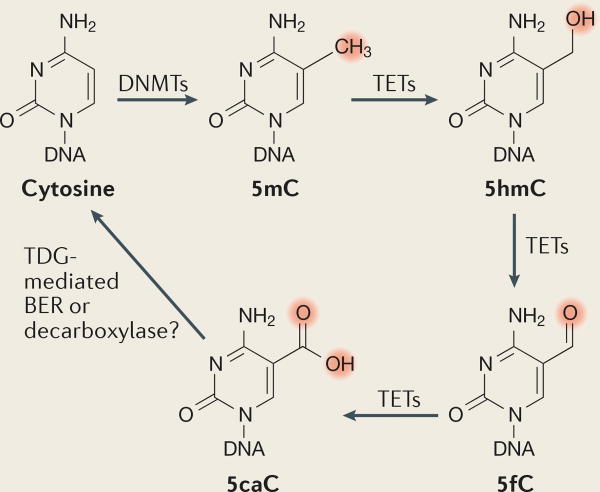
Methylcytosine derivatives
It is well known that DNA methyltransferases (DNMTs) are the enzymes responsible for cytosine methylation, although it long remained elusive which enzymes could reverse DNA methylation in metazoans. In 2009, it was discovered that DNA demethylation might be a multistep process that involves TET (ten-eleven translocation) methylcytosine dioxygenase enzymes that convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC)128,129 (see the figure). These enzymes can further oxidize 5hmC to 5-formylcytosine (5fC) and to 5-carboxylcytosine (5caC)130,131. Thymine-DNA glycosylase (TDG)-mediated base excision repair (BER) of 5fC and 5caC can regenerate unmethylated cytosines132,133.
One important question is whether these oxidized derivatives of 5mC are simply the intermediate products of the demethylation process, or whether they have a functional role themselves. Genome-wide sequencing approaches have generated 5hmC, 5fC and 5caC profiles and have revealed the distribution of these modifications across the genome130,131,134–137. The modification levels for these three derivatives are substantially lower than the mCpG levels. For example, the level of 5hmC (that is, 5hmCG/CG) varies from 1% to 30% depending on the cell type27,134,135,138, whereas the levels of 5fC and 5caC range from 8% to 10% (REFS 139,140). In comparison, the methylation level for mCpG typically ranges from 80% to 90% (REF. 28).
These modifications are not randomly distributed in the genome, but show a preference for certain genomic regions. For example, 5hmC is enriched at distal regulatory elements, such as enhancers and DNase I hypersensitivity sites134, 5fC is enriched in poised enhancers137 and a large fraction of 5fC sites are located in intragenic regions with a particular enrichment in exons141. By contrast, 5caC was found to be preferentially enriched at major satellite repeats136. Interestingly, different modifications showed distinct patterns surrounding protein-DNA binding sites135.
To understand the function of these modifications, researchers have started to identify proteins that interact with these modifications using various techniques, including mass spectrometry-based approaches51,142. For example, MeCP2 was recently found to bind to 5hmC127,143–145, and the binding affinity seems to be context-dependent. The binding of MeCP2 to 5hmCG, 5hmCC and 5hmCT is substantially weaker than their corresponding methylated probes. However, the conversion of mCpA to 5hmCA does not alter the high affinity binding to MeCP2 (REFS 127,143–145). Interestingly, the binding of these readers is often modification-specific and cell type-specific51,142. For example, THAP domain-containing protein 11 (THA11), a transcriptional repressor that plays a central part in embryogenesis, was identified as a brain-specific 5hmC reader51. In addition, a number of forkhead box proteins (FOXK1, FOXK2, FOXP1, FOXP4 and FOXI3) were found to interact with 5fC142. The dynamic nature of such interactions suggests specific and complex biological roles for these modifications. We expect that more proteins remain to be discovered because these studies only used one or two DNA probes, and because the binding of many proteins could depend on the sequence context surrounding the modifications.
DNA methylation has a critical role as a means to control gene expression; for example, during development to ensure X-chromosome inactivation or genomic imprinting16,17 through various mechanisms. Furthermore, aberrant DNA methylation is a hallmark of many diseases, including various types of cancers18. Indeed, abnormal gains in methylation in normally unmethylated CpG islands have been linked to the inactivation of tumour suppressor genes19–21. Such abnormal promoter CpG island methylation is emerging as a potential biomarker for cancer detection, diagnosis and prognosis22,23. More recently, DNA methylation has also been implicated in non-cancerous diseases, such as schizophrenia24 and autism spectrum disorders25,26.
Thanks to rapid technological advances, especially the range of techniques based on deep sequencing, it is now possible to monitor the dynamics of the DNA methylome at single-nucleotide resolution27–29. These developments have provided new insights into how the epigenome is shaped and how it regulates different biological processes, such as cellular differentiation and cancer development. For instance, comparing methylation profiles under different physiological conditions revealed tissue-specific or disease-specific differentially methylated regions30–32, suggesting that the role of DNA methylation in gene regulation is multifaceted and goes beyond simple repression of gene expression.
Despite the fast accumulating profiles of DNA methylomes in various biological processes and species, the interpretation of these data sets often falls short of providing a mechanistic understanding of the dynamic changes in DNA methylation levels. It still remains a challenge to establish the causality between DNA methylation and physiological outcomes in the epigenetic field. In our view, the first step towards a mechanistic understanding of the DNA methylome is to determine the protein–DNA interactions associated with the dynamics of the DNA methylome. In other words, the identification of DNA methylation ‘readers’ and ‘effectors’, which translate methylation signals into biological actions, will be crucial to decipher the epigenetic ‘code’ of methylation-mediated biological processes.
In this Analysis article, we review the discovery of a new class of methylated-DNA-binding proteins, namely transcription factors (TFs), and the approaches used to discover these interactions. We focus on the interaction partners with methylated CpG sites in mammals, with a brief discussion of other methylation derivatives (BOX 1). We then summarize the specific properties of methylation-dependent interactions between TFs and DNA, and discuss the causal relationship between TF–DNA interactions and DNA methylation, before concluding with an overview of potential biological consequences of methylation-dependent protein–DNA interactions.
Readers of methylated DNA
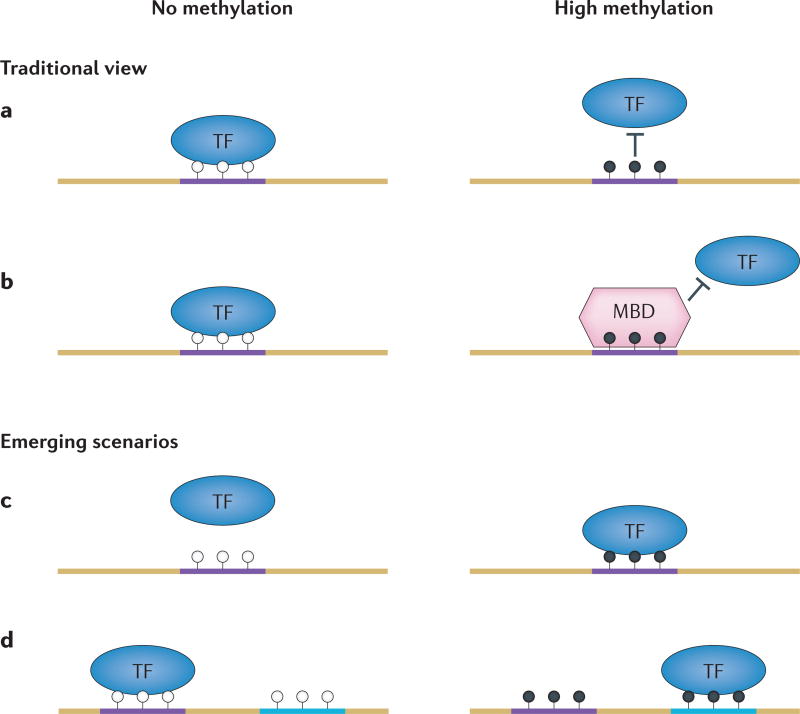
The classical view of methylation-mediated protein– DNA interactions is that only proteins with a methyl-CpG (mCpG)-binding domain (MBD) can recognize and bind to methylated CpG dinucleotides3,6,33–35 (FIG. 1). The MBD protein family has five known members in mammals, including MeCP2 (methyl-CpG-binding protein 2), MBD1, MBD2, MBD3 and MBD4. Except for MBD3, which does not bind to methylated DNA, all MBD proteins bind to methylated DNA in a non-sequence-specific manner36,37. Comparison of MBD proteins from different species showed conservation and divergence in terms of the number of MBD genes and the composition of the MBD domains6,38. Interestingly, the extent of genomic methylation generally correlates with the number of MBD proteins in a species6. Dysfunction of MBD proteins is associated with human diseases. For example, mutations in the gene encoding MeCP2 cause the neurodevelopmental disorder Rett syndrome39,40.
Figure 1. Interaction modes between proteins and DNA.
a,b | The traditional view of the protein–DNA interaction patterns. Transcription factors (TFs) usually bind to non-methylated DNA motifs (open circles, left panels) in open chromatin regions (part a). However, such interactions can be directly disrupted by methylation on the CpG sites in the motifs (filled circles, right panels). Alternatively, methyl-CpG binding-domain (MBD) proteins can be recruited to the methylated DNA motifs and compete off TFs through their higher affinity to the mCpG site in a sequence-independent fashion (part b). c,d | Newly emerging scenarios for protein–DNA interactions. DNA methylation could create a new binding site for TFs (part c). TFs may be able to recognize different sequences with or without DNA methylation (part d).
Over the past 15 years, evidence has emerged that suggests that some TFs lacking MBDs are able to interact with methylated DNA23–27 (FIG. 1). Unlike MBD proteins, a handful of mammalian TFs were found to possess sequence-dependent mCpG-binding activity in a few studies. For example, the transcriptional regulator Kaiso, which contains POZ (pox virus and zinc-finger) and zinc-finger domains, was found to bind to a specific methylated sequence with its C2H2 zinc-finger domains41. In other studies, the basic leucine zipper (bZIP) CCAAT/enhancer-binding protein-α (CEBPα)42, the zinc-finger protein ZFP57 and its cofactor KRAB-associated protein 1 (KAP1; also known as TIF1β)43,44 were shown to interact with specific methylated sequences. In addition to mammalian proteins, a bZIP herpesvirus protein, Zta, was found to bind to methylated regulatory elements and control the epigenetic landscape during the latency-to-lytic phase transition in infected mammalian cells45. Two amino acids, one cysteine and one serine, were found to interact with 5mC45. In rice, the nuclear protein MVBP (methylated VBE-binding protein) was shown to bind to a rice tungro bacilliform virus promoter region only when the promoter was methylated46.
As the discovery of these mCpG-binding proteins was often serendipitous, whether TFs represent a new class of DNA methylation readers and, potentially, effectors, and whether sequence-specific mCpG-dependent binding activity is a widespread phenomenon or merely an exception, remained questionable. In addition, recent large-scale analyses of gene expression profiles and DNA methylomes showed that a substantial portion of DNA methylation sites is positively correlated with gene expression31. This finding may result from the high levels of DNA methylation in the gene body of highly expressed genes; however, it also raises the possibility that some TFs bind to methylated regulatory elements and activate gene expression. It should be noted that DNA methylation of a promoter or an enhancer has been shown to be correlated with increased transcription of a target gene47,49, although most of the evidence showing a positive correlation between methylation and expression seems to result from methylation downstream of the transcription start site. Intrigued by these observations, several research groups have conducted unbiased, high-throughput screens to search for such a correlation in higher eukaryotes (TABLE 1).
Table 1.
Comparison of high-throughput approaches for mCpG reader discovery
| Method | Bait | Prey | Advantage | Disadvantage | Refs |
|---|---|---|---|---|---|
| MS/MS | Generic DNA sequences | Nuclear extracts | A comprehensive survey against nuclear proteins; tissue-specific interactions can be detected | Uses generic DNA probes with no sequence specificity; limited to high abundant proteins | 51,142 |
| Protein microarray | Sequence-specific DNA motifs | ∼1,500 human TF proteins | A comprehensive survey against the entire TF repertoire and is not limited to protein abundance in cells | Limited to a few hundred DNA probes | 53 |
| DNA microarray | Individual proteins | All possible combinations of 8–10-nucleotide-long DNA sequences | Accurate mapping of binding consensus | Candidate approach, prior knowledge required | 56 |
| ChIP–BS-seq | Antibodies against TF of interest | TF–DNA complexes in cultured cells | Genome-wide survey for in vivo binding events | Limited by antibody quality and availability | 57,58 |
ChIP–BS-seq, chromatin immunoprecipitation followed by bisulfite sequencing; MS/MS, tandem mass spectrometry; T F, transcription factor.
High-throughput reader discovery
Tandem mass spectrometry
One systematic approach for the discovery of mCpG-binding proteins is based on tandem mass spectrometry (MS/MS)50,51. A recent study used a generic DNA sequence harbouring an mCpG site to pull down interacting proteins from nuclear extracts of cultured cells51. Proteins bound to methylated DNA sequences were then identified by MS/MS. Based on this approach, 19 proteins were identified that interact preferentially with the methylated DNA probe rather than the non-methylated counterpart in mouse embryonic stem (ES) cell nuclear extracts. Besides the known MBD proteins (such as MeCP2, MBD1 and MBD4), many TFs (such as MHC class II regulatory factor RFX1, zinc-finger homeobox 3 (ZFHX3), lysine-specific histone demethylase 1A (LSD1), zinc-finger and BTB domain-containing protein 44 (ZBT44) and thymocyte nuclear protein 1 (THYN1; also known as THY28), and the Krüppel-like factors (for example, KLF2, KLF4 and KLF5)), were identified as new mCpG-binding proteins (TABLE 2). The authors applied the same approach to neuronal progenitor cells and found that a large and distinct set of proteins showed preferential binding to mCpG sites, suggesting that the interaction with mCpG is dynamic and thus varies under different physiological conditions. A similar approach was also used to identify nucleosome-interacting proteins that are affected by DNA methylation50. Although proteins from cell extracts are in a more native state in the MS/MS-based approach, the DNA probes used in this approach are typically generic and, therefore, sequence specificity of observed interactions remains elusive.
Table 2.
Representative mCpG-binding transcription factors
| Protein Name* |
DNA-binding domain‡ |
Canonical motif | Methylated motif |
In vivo evidence? |
Pioneer TF? |
Refs |
|---|---|---|---|---|---|---|
| Kaiso | Zinc-finger | TCCTGCNA | TCTmCGmCGAGA | Yes | Yes | 41,50,51 |
| ZFP57 | Zinc-finger | Unknown | TGCmCGC | Yes | No | 43,60 |
| GATAD2A | Zinc-finger | Unknown | Unknown | No | No | 50,51 |
| GATAD2B | Zinc-finger | Unknown | Unknown | No | No | 50,51 |
| RFX5 | Other | HYRDBVMCH | Unknown | Yes | No | 50,53 |
| KLF4 | Zinc-finger | GCCMCRCCC | CCmCGCC | Yes | Yes | 51,53 |
| HOXA5 | Homeobox | CCYCATTAKTGN | Non-specific | No | No | 51,53 |
| CEBPα | Zinc-finger | TTKCNYMA | mCGTCA | Yes | No | 42,56 |
| CEBPβ | Zinc-finger | TTKCNYMA | TTGmCGYMA | Yes | No | 56 |
| ZBTB40 | Zinc-finger | Unknown | Unknown | No | No | 50 |
| ZBTB9 | Zinc-finger | Unknown | Unknown | No | No | 50 |
| ZHX1 | Zinc-finger homeobox | Unknown | Unknown | No | No | 50 |
| ZHX2 | Zinc-finger homeobox | Unknown | Unknown | No | No | 50 |
| ZHX3 | Zinc-finger homeobox | Unknown | Unknown | No | No | 50 |
| HOMEZ | Homeobox | YTCGYYY | Unknown | No | No | 50 |
| FOXA1 | Forkhead | TRTTTGYTYWN | Unknown | No | Yes | 50 |
| GZF1 | Zinc-finger | TGCGCKTMTATA | Unknown | No | No | 51 |
| KLF10 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| KLF2 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| KLF3 | Zinc-finger | CAGGGTGTG | Unknown | No | No | 51 |
| KLF5 | Zinc-finger | YYMCDCCC | Unknown | No | No | 51 |
| RREB1 | Zinc-finger | CCCCAAACMMCCCC | Unknown | No | No | 51 |
| SALL2 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| ZBTB4 | Zinc-finger | CNNTCACTGGNA | Unknown | No | No | 51 |
| ZBTB44 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| ZCHC8 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| ZFP597 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| ZN513 | Zinc-finger | NNAACATCTGGA | Unknown | No | No | 51 |
| ZNF710 | Zinc-finger | Unknown | Unknown | No | No | 51 |
| ZFHX2 | Zinc-finger homeobox | Unknown | Unknown | No | No | 51 |
| ZFHX3 | Zinc-finger homeobox | ATTAAYTRCAC | Unknown | No | No | 51 |
| ZFHX4 | Zinc-finger homeobox | Unknown | Unknown | No | No | 51 |
| DLX1 | Homeobox | NTGNNNTAATTANY | Unknown | No | No | 51 |
| DLX5 | Homeobox | NNRGYAATTRNYK | Unknown | No | No | 51 |
| DLX6 | Homeobox | YAATTA | Unknown | No | No | 51 |
| HOXB8 | Homeobox | NNNNGYAATTAATANW | Unknown | No | No | 51 |
| HOXB9 | Homeobox | NRRGCMATAAAA | Unknown | No | No | 51 |
| MEIS1 | Homeobox | NNNTGACAG | Unknown | No | No | 51 |
| PBX1 | Homeobox | ATCAATCAW | Unknown | No | No | 51 |
| PBX3 | Homeobox | Unknown | Unknown | No | No | 51 |
| BACH1 | bZIP | NNSATGAGTCATGNT | Unknown | Yes | No | 51 |
| TFCP2 | CP2 | DWCYRGH | Unknown | No | No | 51 |
| UBIP1 | CP2 | SCAGYB | Unknown | No | No | 51 |
| FOXK1 | Forkhead | AATGTAAACAAA | Unknown | No | Yes | 51 |
| FOXK2 | Forkhead | Unknown | Unknown | No | Yes | 51 |
| RFX4 | Other | CCNTAGCAACS | Unknown | No | No | 51 |
| RFXAP | Other | Unknown | Unknown | No | No | 51 |
| DIDO1 | Zinc-finger | Unknown | GCAGmCGAGC | No | No | 53 |
| FEZF2 | Zinc-finger | Unknown | SYmCGCC | No | No | 53 |
| GATA3 | Zinc-finger | NNGATARNG | Non-specific | No | Yes | 53 |
| GATA4 | Zinc-finger | AGATADMAGGGA | AAAmCGCTTCC | No | Yes | 53 |
| PF21A | Zinc-finger | Unknown | Unknown | No | No | 53 |
| PPARγ | Zinc-finger | NNWGRGGTCAAAGGTCA | Unknown | No | No | 53 |
| RN138 | Zinc-finger | Unknown | Unknown | No | No | 53 |
| RXRA | Zinc-finger | NNNNNTGACCCC | TCmCGVN | No | No | 53 |
| SCAPE | Zinc-finger | Unknown | Non-specific | No | No | 53 |
| ZCHC7 | Zinc-finger | Unknown | BKmCGDS | No | No | 53 |
| ZKSC5 | Zinc-finger | Unknown | Unknown | No | No | 53 |
| ZMYM3 | Zinc-finger | TTTGAAA | GAmCGTC | No | No | 53 |
| ZN114 | Zinc-finger | Unknown | Non-specific | No | No | 53 |
| ZNF22 | Zinc-finger | HYDCCYMCD | Unknown | No | No | 53 |
| ZNF28 | Zinc-finger | Unknown | TTTAmCGTGCAG | No | No | 53 |
| ZN416 | Zinc-finger | Unknown | Unknown | No | No | 53 |
| ZN461 | Zinc-finger | Unknown | VHmCGHM | No | No | 53 |
| ZN695 | Zinc-finger | Unknown | DNmCGCY | No | No | 53 |
| CERS4 | Homeobox | Unknown | Unknown | No | No | 53 |
| CRX | Homeobox | YNNNTAATCYSMN | CCCmCGTAA | No | No | 53 |
| HOXA9 | Homeobox | NCGGYCATWAAAWTANW | Unknown | No | No | 53 |
| TGIF1 | Homeobox | AGCTGTCANNA | RVmCGMM | No | No | 53 |
| ATF6β | bZIP | Unknown | Unknown | No | No | 53 |
| E2F3 | E2F TDP | GGCGGGN | Non-specific | No | No | 53 |
| E2F6 | E2F TDP | CNTTTCNT | Unknown | Yes | No | 53 |
| FOXC1 | Forkhead | GTAAATAAACA | HVmCGBS | No | Yes | 53 |
| ARNT2 | HLH | Unknown | AAAmCGCTTCC | No | No | 53 |
| NPAS2 | HLH | VCAMRTR | AAACmCGGCTC | No | No | 53 |
| ARI3B | Other | HWTAWW | AAAmCGCTTCC | No | No | 53 |
| PMS1 | Other | Unknown | ATGAmCGTCAC | No | No | 53 |
| RBPJ | Other | CGTGGGAA | AAACmCGAGAAC | No | No | 53 |
| SMAD4 | Other | GKSRKKCAGMCANCY | NCmCGGG | No | No | 53 |
| SUB1 | Other | Unknown | Unknown | No | No | 53 |
| AP2α | Other | GCCNNNRGS | GTCAmCGCCC | No | No | 53 |
AP2α, activating enhancer-binding protein 2α; ARI3B, AT-rich interactive domain-containing protein 3B; ARNT2, aryl hydrocarbon receptor nuclear translocator 2; ATF6β, activating transcription factor 6β; B, any nucleotide except A; bZIP, basic leucine zipper; CEBPα, CCAAT/enhancer-binding protein-α; CERS4, ceramide synthase 4; CP2, CCAAT box binding protein 2; CRX, cone-rod homeobox; D, any nucleotide except C; DIDO1, death-inducer obliterator 1; FEZF2, Fez family zinc-finger protein 2; FOXA1, forkhead box A1; GATA3, GATA-binding factor 3; GATAD2A, GATA zinc-finger domain-containing protein 2A; GZF1, GDNF-inducible zinc-finger protein 1; H, any nucleotide except G; HLH, helix-loop-helix; HOMEZ, homeobox and leucine zipper-containing protein; HOXA5, homeobox protein A5; K, G or T; KLF4, Krüppel-like factor 4; M, A or C; m, methyl; N, any nucleotide; NA, not available; NPAS2, neuronal PAS domain-containing protein 2; PBX1, pre-B cell leukaemia transcription factor 1; PF21A, PHD finger protein 21A; PPARγ, peroxisome proliferator-activated receptor-γ; R, A or G; RFXAP, regulatory factor X-associated protein; RN138, RING finger protein 138; RREB, Ras-responsive element-binding protein 1; RXRα, retinoic acid receptor RXRα S, G or C; SALL2, sal-like protein 2; SCAPE, S phase cyclin A-associated protein in the endoplasmic reticulum; SMAD4, mothers against decapentaplegic homologue 4; TDP, transcription factor E2F dimerization partner; TF, transcription factor; TFCP2, α-globin transcription factor CP2; UBIP1, upstream-binding protein 1; V, any nucleotide except T; W, A or T; Y, C or T; ZBTB40, zinc-finger and BTB domain-containing protein 44; ZCHC, zinc-finger CCHC domain-containing protein 8; ZFHX2, zinc-finger homeobox protein 2; ZFP57, zinc-finger protein 57; ZHX1, zinc-fingers and homeoboxes protein 1; ZKSC5, zinc-finger protein with KRAB and SCAN domains 5; ZMYM3, zinc-finger MYM-type protein 3.
TFs are sorted by assay type performed (see reference indicated); the TFs identified by multiple studies are ranked on top.
The DNA-binding domains that are found only in a small number of TFs are denoted as ‘Other’.
Functional protein microarray
Functional protein microarrays have been used as a powerful tool to profile protein–DNA interactions in the past52. A comprehensive examination of sequence-specific mCpG-binding activities was conducted by sequentially probing a human protein microarray containing 1,321 TFs and 210 cofactors with 154 DNA motifs that each carried at least one mCpG site53. To identify human TFs that preferentially bind to methylated DNA motifs, each methylated motif was mixed with its unlabelled and unmethylated counterpart in tenfold excess in the binding assays. This competition assay ensures that the identified interactions are indeed methylation-dependent, rather than due to CpG-flanking sequences. Of the 154 methylated motifs examined, 150 showed strong binding signals to at least one protein on the microarray. In total, 41 TFs and 6 cofactors were found to bind to at least one methylated sequence. Most of these factors were found to bind to only a few methylated sequences, suggesting that the interactions are not only methylation-dependent but also sequence-specific. Interestingly, the factors that showed binding activity to methylated sequences were widespread among various TF subfamilies, such as zf-C2H2, homeobox, bHLH (basic helix–loop–helix), forkhead, bZIP and HMG (high-mobility group) box. Many of these factors are known to be involved in tissue development or have been associated with cancer. A subsequent validation assay showed that some of these TFs indeed bind to methylated DNA in vivo and regulate gene expression53.
DNA microarray
DNA (or protein-binding) microarray technology has been used to determine the binding specificity of TFs54,55. A double-stranded DNA microarray, typically comprising 40,000 unique DNA sequences that cover all possible combinations of 8–10-nucleotide-long DNA sequences that could constitute a binding motif, is incubated with a purified TF so that its binding preference can be accurately determined. In a recent study, the bacterial DNMT SssI was used to methylate the CpG sites of the sequences on the array56, followed by individual probing with eight purified proteins containing bZIP domains. By comparing the binding profiles of each protein obtained on the methylated and unmethylated microarrays, proteins that preferentially bind to specific sequences were determined. Among the eight bZIP proteins, CEBPα and CEBPβ were found to specifically bind to a methylated sequence56. This approach enables a large amount of DNA sequences to be surveyed for protein–DNA interactions; accurate sequence specificity can, therefore, be determined for a given protein. However, prior knowledge of a candidate TF is required because it can be cumbersome to survey an entire TF family. Therefore, this approach is ideally used for fine-mapping sequence specificity of a previously identified mCpG-binding protein.
ChIP–BS-seq
To determine methylation-dependent protein-DNA interactions in vivo, chromatin immunoprecipitation followed by bisulfite sequencing (ChIP–BS-seq) is an ideal approach. ChIP is first performed to obtain the DNA sequences that are bound by a protein of interest, and then the methylation level is sequentially determined using BS-seq. This approach was developed recently to determine the crosstalk between histone modifications and DNA methylation57–59. However, it requires prior knowledge of mCpG-binding proteins and the availability of antibodies that are directed against the proteins of interest. For example, after KLF4 was determined to bind to mCpG sites, ChIP–bisulfite conversion followed by PCR was used to validate the methylated DNA–protein interactions in vivo53. It is important to note that ChIP–BS-seq is the only approach that does not use naked DNA fragments to identify the TFs that bind to methylated DNA. Therefore, TFs identified using the other methods may not necessarily recognize mCpGs in vivo, and further studies are needed to dissect the functionality of the interactions.
Methylated DNA–TF interactions in vivo
Several studies have demonstrated that methylated DNA–TF interactions can occur in a cellular context (in vivo). For example, ZFP57 and its cofactor KAP1 were shown to bind selectively to nine DNA-methylated alleles of imprinting control regions (ICRs) in ES cells43. In another study, ZFP57 binding sites were mapped in hybrid ES cells, and ZFP57 was found to interact with the methylated parental-origin allele60. Similarly, Kaiso was shown to bind to the methylated promoter of the MTA2 gene in HeLa cells61. Another study, which used a quantitative ChIP–PCR assay, demonstrated that Kaiso binds to the methylated promoters of CDKN2A, MGMT and HIC1 in both HCT116 and Colo320 human colon cancer cell lines62. The finding that Kaiso binds to the methylated promoter of CDKN2A was recently reproduced in an independent study63. By contrast, a different study discovered that Kaiso was not associated with highly methylated promoters in GM12878 lympho-blastoid cells or in K562 human myeloid leukaemia cell lines64. Of note, this observation does not necessarily rule out the possibility that Kaiso binds to methylated DNA motifs in other cell types; rather, it suggests that methylation-dependent TF–DNA interactions may be cell type-specific. That is, some TFs might bind to methylated DNA motifs in certain cell types but not in others, presumably owing to variations in accessibility to methylated motifs in different cell types and/or dynamics of the DNA methylomes during differentiation and development.
Although these studies may suggest that some TFs can bind to methylated DNA in vivo, one important question remains: how prevalent are methylated-DNA–TF interactions in a given genome? For example, the studies showing that Kaiso binds to methylated DNA in vivo58–60 were focused on a few genes or genomic regions rather than genome-wide surveys. To determine to what extent these TFs interact with methylated loci in cells, we globally evaluated the accessibility of highly methylated regions in the H1 human ES cell line. Integration of the DNA accessibility data obtained by mapping DNase I hypersensitivity sites (DHSs)65 and the DNA methylome data obtained from the same cell type28 revealed that numerous open chromatin regions (that is, accessible regions) indeed contain highly methylated CpG sites. Overall, 258,188 DHS peaks were determined in the H1 human ES cell line by The ENCODE Consortium. By superimposing the DHS peaks with the DNA methylome of the H1 cells determined by whole-genome BS-seq28, we calculated the average methylation level (m) of CpG sites within a DHS peak, defined as:
| (1) |
where N is the number of CpG sites within a peak, and mi is the methylation level for CpG site i. Overall, 77,124 (29.9%) of the 258,188 DHSs detected in H1 cells had an average methylation level greater than 80% at CpG sites (FIG. 2a), suggesting that many methylated CpG sites are accessible to TFs.
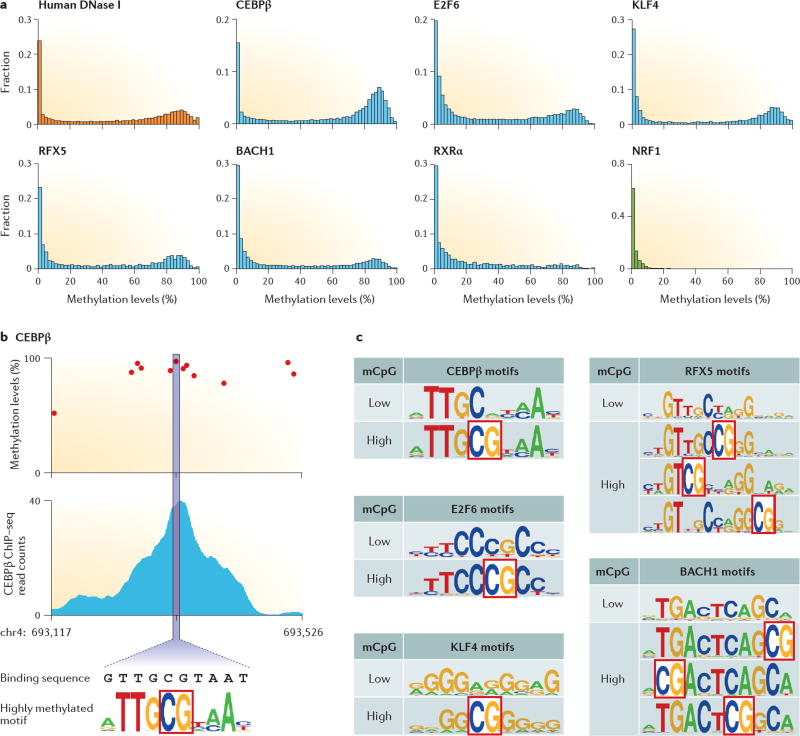
Figure 2. Methylated-DNA-TF interactions in vivo.
a | Integration of DNA methylome data28 with transcription factor (TF) chromatin immunoprecipitation followed by sequencing (ChIP–seq) data28 (see also Further Information) or with DNA accessibility data65 obtained from mapping DNase I hypersensitivity sites (DHSs) from the same cell lines suggests that some TFs bind to methylated DNA in vivo. All of the data, including DHS, ChIP–seq and DNA methylome, were obtained from the H1 embryonic stem cell line. The x axis shows the average methylation level (percentage) of the CpG sites within a DHS or TF ChIP–seq peak, and the y axis shows the fraction of peaks with a certain average methylation level. The methylation levels within each DHS peak show bimodal distribution, indicating that a large portion of highly methylated CpGs (mCpGs) are accessible to TFs. The methylation levels within the ChIP–seq peaks for five TFs (namely CCAAT/enhancer-binding protein-β (CEBPβ), E2F6, Krüppel-like factor 4 (KLF4), RFX5 and BACH1) also show a bimodal distribution, suggesting that they can interact with methylated DNA in vivo. Retinoic acid receptor RXRα shows no binding activity in highly methylated regions, similar to the negative control nuclear respiratory factor 1 (NRF1). b | We examined whether methylation occurred in the binding sites within the ChIP–seq binding peaks. DNA sequences within each ChIP–seq peak were extracted and grouped based on their average methylation level. The low methylation group contains peaks with a methylation level of <60% and the high methylation group contains peaks with a methylation level of >80%. MEME (multiple EM for motif elicitation) analysis was performed on each of the top 500 peaks with the highest ChIP–seq intensities for the two groups and the most significant motifs were identified. The motif was then used to scan the DNA sequence within each peak and the DNA segment with highest match score to the motif was recorded. The methylation level was examined for the CpG sites within the identified DNA segment. As illustrated with CEBPβ, a matched segment (for example, 5’-GTTGCGTAAT) containing a highly methylated CpG site in the middle was identified within a ChIP–seq peak. c | Motifs were identified separately for binding peaks with low-level (<60%) methylation and high-level (>80%) methylation. To obtain the most reliable methylated motifs, the DNA sequences that match to the MEME motif were further grouped based on the position of the mCpG, and the sequences with a methylation level of >80% were assembled to generate the methylated motif for each subgroup. The mCpG sites for each subgroup are outlined with red boxes.
We then examined whether the TFs listed in TABLE 2 could interact with methylated DNA in vivo. We obtained TF ChIP–seq data sets in H1 ES cells from The ENCODE Consortium, and uniform peaks were called using the Irreproducible Discovery Rate (IDR) method66. The ChIP–seq peaks were superimposed with the methylome data set and the average methylation levels within each ChIP–seq peak were calculated using the method described above. For each TF, we obtained the distribution of the methylation level for each ChIP–seq peak. Although the availability of ChIP–seq data sets was limited, six TFs (namely CEBPβ, E2F6, BACH1, RFX5, KLF4 (REF. 28) and retinoic acid receptor RXRα) had ChIP–seq data in H1 cells (TABLE 2). The DNA methylation levels within the ChIP–seq peaks showed a bimodal distribution for all TFs except RXRα, indicating that a substantial fraction of their binding sites are located in highly methylated regions (FIG. 2a). For example, of the 15,557 ChIP–seq peaks identified for CEBPβ, 6,675 (42.9%) had a methylation level greater than 80%. As a comparison, we selected two TFs (nuclear respiratory factor 1 (NRF1) and transcription initiation factor TFIID subunit 1 (TAF1)), which are known not to interact with methylated DNA (based on our current knowledge), as negative controls: neither NRF1 (FIG. 2a) nor TAF1 (Supplementary information S1 (figure)) showed a bimodal distribution, demonstrating that these TFs only bind to regions with low levels of methylation.
We further examined whether the methylated CpG sites located exactly at the TF binding sites (∼10–20 bp) within the ChIP–seq peaks (200–500 bp), using CEBPβ as an example (FIG. 2b). We first used the MEME (multiple EM for motif elicitation) algorithm to predict significantly enriched sequence motifs using the sequences of the ChIP–seq peaks that have a low methylation level67. The most significantly enriched motif did not contain a CpG site. Interestingly, when the same analysis was applied to those peaks that have high methylation levels, a significantly enriched motif containing a CpG site at position 4 was discovered (FIG. 2b). We next examined the methylation level of the CpG sites within the motif in each ChIP–seq peak (FIG. 2b). Among the 6,675 peaks with a high methylation level, 3,894 carried a highly methylated (>80%) CpG site within the enriched motifs. A motif could be reconstructed with these 3,894 binding peaks, which represented the methylated motif for CEBPβ (FIG. 2c). The same analysis was performed for the other four TFs. In summary, 25.0% (3,894 out of 15,557), 7.7% (1,103 out of 14,396), 5.2% (88 out of 1,695), 3.0% (115 out of 3,793) and 1.6% (186 out of 11,457) of binding sites were highly methylated for CEBPβ, E2F6, RFX5, KLF4 and BACH1, respectively (FIG. 2c). Note that this is a conservative estimate because we used a stringent definition of highly methylated sites (that is, >80%).
Taken together, the above analysis suggests that many TFs shown to bind methylated DNA in vitro are also able to interact with methylated DNA in vivo, although further in vivo genome-wide characterization of TF binding patterns and high-resolution DNA methylation analyses are needed to strengthen the evidence base. The list of TFs that interact with methylated DNA (TABLE 2) provides a foundation for further functional characterization of methylated DNA–TF interactions in various biological processes.
Features of methylated-DNA–protein interaction
Protein domains that interact with methylated DNA
Identification of the protein domains that recognize mCpG sites is important to characterize mCpG-dependent protein–DNA interactions. Such knowledge will enable the mutation of critical residues within these domains that abolish the mCpG-dependent binding activity of these proteins, while maintaining their ability to bind non-methylated DNA. Therefore, mutated proteins can be useful tools to dissect the physiological roles of mCpG-dependent protein–DNA interactions.
Besides the well-known MBDs, other protein domains seem to interact with mCpG sites. For example, the recent crystal structure of mouse ZFP57 in complex with a methylated DNA sequence demonstrated that its two zinc-fingers interact with methylated DNA, and that an arginine (Arg178), which is involved in hydrophobic interactions, plays a crucial part in mCpG binding44. A separate study suggested that an arginine and glutamate pair in KLF4 recognizes the mCpG site68. A structural comparison of MeCP2 and KLF4 indeed showed a common structural feature involving one arginine and one asparagine53. A global survey of methylated-DNA-binding proteins suggests that many other protein domains might also be able to interact with mCpG sites, including homeobox, HLH and E2F domains53.
There are currently no general rules of evolutionary conservation for the domains that interact with methylated sites, owing to a lack of data from multiple species. However, in a comparison of mCpG-binding proteins between humans and mice51,53, a few proteins such as KLF4 and homeobox A5 (HOXA5) were shown to bind mCpG sites in both species, which is indicative of the functional importance of methylation-dependent protein–DNA interactions.
Sequence specificity
Notably, many proteins can bind both non-methylated and methylated sequences in a different sequence context. For example, CEBPα is known to bind a particular sequence element, 5′-TGACGTCA42. However, when the CpG is methylated, CEBPα can effectively recognize half of the motif: 5′-mCGTCA42. Similarly, although KLF4 recognizes a non-methylated canonical motif of 5′-TTTACGCC, it has been demonstrated that KLF4 specifically recognizes a 5′-TCCmCGCCC motif only when the CpG is methylated53. If the methylation status of these two sequences is exchanged, KLF4 loses the ability to bind to either sequence53. Indeed, for many newly discovered methylated DNA-binding proteins, the methylated motifs differ from the non-methylated motifs53. Therefore, it is reasonable to speculate that 5mC might represent the fifth nucleotide that further fine-tunes the specificity of protein–DNA interactions; that is, 5mC acts as an additional regulatory layer to remove, create and/or change TF binding sites (FIG. 1).
Several recent studies have started to provide the structural basis for the altered sequence specificity due to DNA methylation. Both in vitro DNase I digestion assays and structural studies indicate that methylation has a profound impact on DNA structure and shape (for an in-depth review see REF. 69). Adding a methyl group to the cytosine could affect the local DNA shape, as evidenced by the altered DNase I digestion rate and patterns70,71. Similarly, based on a few reported crystal structures of double-stranded DNA fragments with 5mC bases68,72,73, the presence of a bulky methyl group in the major groove leads to a subtle widening of the major groove and a subtle narrowing of the minor groove. Consequently, 5mC can affect the access of a given TF to the affected motifs in both major and minor grooves in genomic DNA and thus change the sequence specificity of protein–DNA interactions.
Binding affinity
One important question is whether methylated-DNA–protein interactions have a similar binding affinity to the interactions between the same protein and a non-methylated DNA, or whether they are just labile interactions. Using an in vitro pulldown-coupled MS/MS approach, the relative affinity of protein–mCpG interactions can be estimated51. For example, for a particular sequence (5′-GGGCGTG), which was determined on the basis of the KLF4 ChIP– seq data sets74, KLF4 showed higher affinity when the cytosine in the motif was methylated compared with the unmethylated sequence51. The protein microarray approach53, which uses the concept of relative affinity to identify proteins that preferentially bind to methylated DNA, revealed proteins with strong fluorescent signals, which are expected to bind tighter to the methylated motif than to the unmethylated counterpart. The absolute binding affinity (that is, Kd values) can be measured by applying the oblique incidence reflectivity difference (OIRD), which is a real-time, label-free method to measure the kinetics of a binding event53,75. Three proteins (ZMYM3, AP2α and KLF4) were selected to determine the Kon and Koff values with their corresponding motifs in methylated forms. The deduced Kd values of ZMYM3, AP2α and KLF4 were determined as 460 nM, 399 nM and 479 nM, respectively. Importantly, no obvious affinity could be detected when these tested motifs were unmethylated. As a comparison, the Kd values of the short isoform of MBD2, MBD2b, for the same motifs ranged from 97 nM to 197 nM, suggesting that MBD-lacking TFs bind to methylated DNA motifs nearly as strongly as MBD2b.
Cis-regulatory elements
To better understand the physiological role of mCpG-binding proteins, it is important to determine which methylated regions in the genome can be specifically recognized by these proteins. Although MBD family proteins tend to bind to regions with a high methylation density (that is, high methylation level and high CpG density)76, it is interesting to examine whether the same is true for sequence-specific mCpG-binding proteins.
As protein-DNA interactions are dynamic, differentially methylated regions might be possible candidates for methylation-dependent interactions. Analysis of the methylomes obtained from 17 adult mouse tissues at single base-pair resolution showed that approximately 6.7% of the mouse genome is differentially methylated, mostly at distal cis-regulatory regions77. Another study discovered that regions with a low level of methylation, ranging from 10% to 50%, often occur at distal regulatory regions78; that is, regions that are enriched for enhancer marks, including high levels of histone H3 lysine 4 monomethylation (H3K4me1) as well as binding sites for p300 histone acetyltransferase and other regulatory factors. Similarly, extensive DNA methylation was found to coexist with active H3K27 acetylation (H3K27ac) marks in a large number of enhancers79. More importantly, the reduction of DNA methylation led to a decrease in H3K27ac marks, suggesting an active role of DNA methylation in regulating enhancer activity. Based on the analysis of KLF4 binding in ES cells, we also found that KLF4 binds to methylated enhancer regions53, which may suggest that sequence-specific mCpG-binding proteins interact preferentially with distal enhancer regions.
Cause or consequence?
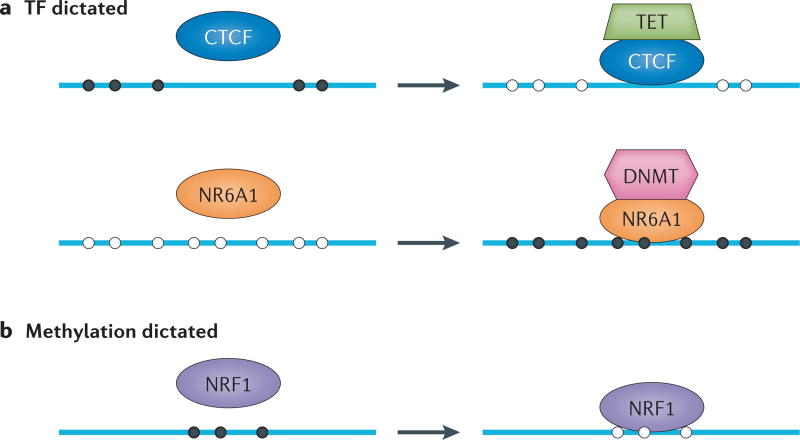
Although many proteins have been found to recognize methylated DNA, the causality between DNA methylation and TF binding is far from clear. On the one hand, DNA methylation could dictate the interaction between proteins and DNA, but on the other hand, the binding of certain proteins may affect the methylation of DNA. Recent studies suggest that both scenarios can occur in different contexts (FIG. 3).
Figure 3. Two action models between TF and methylated DNA interactions.
a | The binding of transcription factors (TFs) dictates the methylation status surrounding the binding sites. The filled circles represent methylated DNA and the open circles represent unmethylated DNA. The binding of the transcriptional repressor CTCF reduces the local methylation level, presumably by recruitment of the TET (ten-eleven translocation) enzymes, which can demethylate surrounding CpG sites. The interaction between CTCF and TET remains to be experimentally validated. Conversely, the binding of NR6A1 (nuclear receptor subfamily 6 group A member 1) induces DNA methylation by interacting with DNA methyltransferase (DNMT) proteins. b | The DNA methylation status dictates TF binding activity. NRF1 (nuclear respiratory factor 1) only binds to DNA when its consensus sequence is non-methylated.
Protein binding affects the DNA methylation status
The binding of methyltransferases or methylcytosine dioxygenases (for example, DNMTs and TETs (ten-eleven translocation proteins)) affects the status of DNA methylation, but recent studies suggest that many non-enzymatic proteins, such as TFs, could regulate the establishment and maintenance of the local DNA methylation levels in a sequence-specific fashion. One such regulator is the transcriptional repressor CTCF, which is known to have an essential role in imprinting control; that is, to achieve allele-specific gene regulation80. CTCF binds to the unmethylated ICRs in maternal alleles, which prevents distal enhancers from activating downstream genes81. By contrast, when the paternal ICR is methylated, CTCF cannot bind to the ICR, thus allowing the activation of downstream genes by distal enhancers. One study suggests that CTCF itself contributes to the maintenance of the non-methylated status of maternal ICRs, as maternally transmitted mutant ICRs in neonatal mice that harbour point mutations in CTCF binding sites acquire a heterogeneous degree of methylation82. Although the traditional view of imprinting control is that differential methylation leads to differential binding of CTCF, and thus yields allele-specific gene regulation, this study suggests that CTCF binding itself is necessary to maintain differential methylation of ICRs.
Moreover, a recent report confirmed that the binding of some proteins (for example, CTCF and RE1-silencing transcription factor (REST)) can affect local methylation patterns78. The authors first created a reporter construct with a CTCF-binding motif that was inserted into a genomic locus in mouse ES cells. Insertion of the binding site induced CTCF binding and resulted in a reduced methylation level in local genomic regions. A single-nucleotide mutation in the CTCF binding motif had no effect on the DNA methylation level. To test the effect in an endogenous setting, the authors generated a Rest−/− mouse ES cell line and, as expected, observed that the REST binding regions were highly methylated. Most importantly, they found that the methylation levels at these sites were much reduced after reintroduction of wild-type Rest into the cells. Altogether, these results support a model whereby the binding of certain proteins can directly affect DNA methylation levels (FIG. 3).
Another study examined the methylation levels of hundreds of sequences that were individually inserted at the same genomic site in mouse ES cells83. Using this approach, the contribution of various sequence motifs to methylation levels could be quantified. They found that CpG density showed a negative correlation with methylation level, which is consistent with the previously established view that CpG islands are generally unmethylated. Interestingly, when the sequences of binding motifs were altered, overall methylation levels decreased83. This work suggests that protein binding has a general role in reducing DNA methylation levels, perhaps by preventing DNMT enzymes from gaining access to these sites, which is consistent with previous findings84.
As TFs have no enzymatic activity to methylate or demethylate a CpG dinucleotide, a possible model would be that these proteins provide sequence-specific guidance and recruit methyltransferases or methylcytosine dioxygenases to these specific sites (FIG. 3). A recent study showed that the nuclear receptor PPARγ (peroxisome proliferator-activated receptor-γ) recruits TET1, resulting in a reduced methylation level around its binding sites through the interaction with TET1 (REF. 85). The reverse can also happen. DNMTs have been found to form protein complexes with various TFs or chromatin modification enzymes. For instance, Sato et al.86 demonstrated that DNMT3A and DNMT3B interact with an orphan nuclear receptor, NR6A1 (nuclear receptor subfamily 6 group A member 1), and that this interaction induced the methylation of the OCT4 (also known as OCT3 and POU5F1) promoter carrying the NR6A1 binding site.
DNA methylation dictates protein–DNA interactions
It is well known that DNA methylation can affect the binding of some TFs87. The manipulation of the methylation status of DNA sequences has been shown (mostly in in vitro studies) to result in the differential binding of TFs, including E2F, AP2α, MYC and MYN88–95. Specifically, hypermethylation is often associated with a depletion of TF binding. Recently, a few studies examined the effect of DNA methylation on TF binding in vivo96,97.
Using a gene-editing approach, Domcke et al.97 generated a genetic deletion of three methyltransferases (Dnmt3a, Dnmt3b and Dnmt1) in a mouse ES cell line. A large number of novel binding sites for the TF NRF1 were created as a result of the triple knockout (TKO). These binding sites often correlated with novel DHSs in the TKO cells, which exhibited predominantly low methylation levels due to their generation in cells lacking DNMTs. Interestingly, novel NRF1 binding sites were hypermethylated in the wild-type cell line97, suggesting that the removal of DNA methylation in TKO cells generated new binding sites for NRF1. These new binding sites had poor sequence conservation, indicating that these sites are non-functional in the wild-type background. In an earlier study, the same group showed that CTCF was able to reduce the DNA methylation level near its binding sites78. In this work, the authors tested whether CTCF binding could affect NRF1 binding by reducing the methylation level of NRF1 binding sites. Reporter constructs harbouring an NRF1 binding motif and a CTCF motif were introduced into the ES cell line. Deletion of CTCF motifs within the construct led to hypermethylation and thus decreased NRF1 binding, suggesting that NRF1 binding in vivo depends on both methylation levels and co-occurring TFs, such as CTCF97.
The examples above represent the two major mechanisms by which protein–DNA interactions and DNA methylation influence each other (FIG. 3). Of note, these two mechanisms are not mutually exclusive. In some cases, the two mechanisms have been found to coexist for the same TFs. For example, although CTCF is known to change the local methylation status78, it has been shown that the binding of CTCF is also methylation sensitive96. Finally, it is worth noting that the crosstalk between TF– DNA interaction and DNA methylation is not restricted to the TFs whose binding motifs contain CpGs. Changes in DNA methylation are often associated with chromatin status, resulting in increased or decreased DNA accessibility 96,97. Differences in chromatin states will either create or eliminate TF binding sites and thus lead to differential TF binding. Although only approximately 25% of known TF binding motifs contain at least one CpG site98, through such indirect crosstalk mechanisms, the binding of TFs without CpGs in their binding motifs could also be influenced by DNA methylation.
Biological consequences
Activation or repression
Methylation in promoters is often considered the hallmark for gene repression99. However, large-scale analyses of gene expression profiles and DNA methylomes have revealed that a substantial proportion of DNA methylation sites is positively correlated with gene expression31. This analysis was performed on methylation sites located within 300 bp upstream from transcriptional start sites, which raises the possibility that methylation in promoters could also be positively correlated with increased transcription of a target gene31 (FIG. 4). Of course, whether these methylation sites fall exactly within the regulatory elements and whether they are recognized by TFs remains to be tested. Single-gene studies have also demonstrated that DNA methylation can activate gene expression47–49. For example, the sequence-specific DNA-binding protein RFX activates a methylated promoter49. Interestingly, this protein was previously shown to bind to methylated DNA47,51. Moreover, it was found that methylation at the 3′ end of the CpG island confers tissue-specific transcriptional activation during human ES cell differentiation100.
Figure 4. Possible biological consequences of methylated DNA–TF interactions.
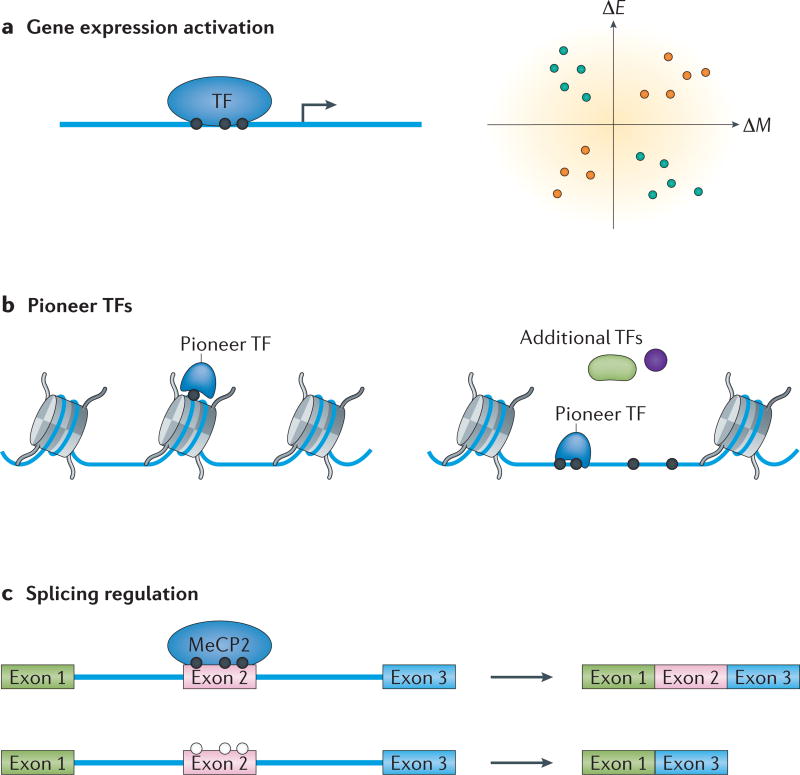
a | Some transcription factors (TFs) can bind to methylated DNA and activate gene expression. Genome-wide profiling of gene expression and DNA methylome data have revealed that many methylation sites positively correlate with gene expression. The orange and blue dots are the CpG sites with methylation levels that are positively or negatively correlated with gene expression, respectively. ΔE is the difference of gene expression, and ΔM is the difference in methylation level between two conditions. b | Pioneer TFs bind to DNA sequences wrapped around the nucleosomes. Consequently, other cofactors (such as chromatin remodelling enzymes and other TFs) are recruited to open up the chromatin regions. As condensed chromatin is often associated with DNA methylation, TFs that can bind to methylated DNA might be good candidates as pioneer TFs. c | Factors bound to exons or introns can affect the splicing activity. For example, MeCP2 (methyl-CpG-binding protein 2) binds to a methylated (filled circles) exon (Exon 2), which results in the inclusion of the exon. When the exon is not methylated (open circles), the MeCP2 does not bind, which leads to exclusion of the exon.
A recent comparative study of mouse retina and brain explicitly explored the possible role of methylation sites whose methylation levels were positively correlated with gene expression101. Among the differentially methylated regions located within 4 kb upstream of transcriptional start sites, approximately 47% showed a positive correlation with the expression of their putative target genes. These methylation regions are overrepresented in DHSs and are evolutionarily conserved, suggesting that these sites are likely to be functional101. More importantly, a distinct set of sequence motifs was discovered in these regions, suggesting that some TFs bind preferentially to these regions101.
Pioneer TFs
The human genome is not made of linear, naked DNA strands. Instead, it is mainly organized into two forms. One is heterochromatin (or condensed chromatin), in which DNA sequences and histones are highly condensed, and genes in these regions are inactive. The other form is euchromatin (or open chromatin), in which DNA sequences are largely accessible to TFs, and genes in these regions can be activated102,103. Chromatin organization is dynamic, and the different types of chromatin can change from one form to another during development or differentiation104 (FIG. 4).
Pioneer TFs are a unique subset of TFs that drive these chromatin changes. A typical characteristic of pioneer TFs is their ability to bind directly to heterochromatic DNA and recruit other factors to change the status to euchromatin to initiate transcription105,106. As DNA in heterochromatin is wrapped tightly around the nucleosomes and is often methylated, it is inaccessible to most TFs; pioneer TFs must possess special features to enable protein–DNA interactions. For example, a handful pioneer TFs (such as OCT4, SOX2 and KLF4) were shown to bind only partial motifs displayed on the nucleosome surface107.
It could be speculated that the ability to bind mCpG sites might prove a useful property for pioneer TFs. If a pioneer TF can interact with an mCpG site, such an interaction would provide an anchor point for the pioneer TF to open up the closed chromatin. Indeed, we observed a large overlap between known pioneer TFs and proteins that bind to methylated DNA (for example, forkhead box protein A (FOXA) and GATA families, which are the best-studied pioneer factors)106,108–110. Interestingly, several of their members showed the ability to bind to methylated DNA, including HOXA5, HOXA9, GATA3 and GATA4 (REF. 53). The mCpG-binding protein KLF4 was also shown to be a pioneer factor105,111. Although there is no simple assay to identify pioneer TFs, evidence that TFs are able to bind methylated DNA would provide a short list of candidate pioneer TFs for future tests. Notably, as methylation-binding proteins participate in multiple biological processes, including gene regulation and splicing regulation, not all methylation-binding proteins are likely to be pioneer factors. Yet, binding to an mCpG site is just one approach for a pioneer TF to access condensed chromatin. Other pioneer TFs might have alternative approaches such as binding to partial motifs.
Splicing regulation
Historically, RNA splicing was considered to be regulated only at the post-transcriptional level. On the basis of this idea, DNA methylation was not expected to have any substantial role in splicing regulation. However, it is now well-established that splicing occurs co-transcriptionally, which means that DNA modification could influence RNA splicing. In one study, the authors observed that the binding of CTCF in an exon region created a roadblock for RNA polymerase II elongation and thus promoted the inclusion of the exon112. Importantly, the binding of CTCF to the exon or intron was dependent on DNA methylation, suggesting that the methylation status surrounding the spliced exons could affect the inclusion level of these exons. Similarly, the mCpG-binding protein MeCP2 was found to play a part in regulating exon splicing113. In this case, a high methylation level led to MeCP2 binding to alternatively spliced exons, which resulted in exon inclusion (FIG. 4). The same trend was observed in a study of the brain methylome of honeybees114. A comparison of methylation levels between queen and worker bees revealed that intron-containing histone genes were highly methylated, whereas intronless histone genes were not methylated, suggesting that mCpG-binding proteins might play a part in splicing regulation. This observation is consistent with a global correlation analysis of DNA methylation and differential splicing events between the brain and the retina115. Although CTCF motifs were significantly enriched in differentially methylated regions associated with alternative splicing, other motifs were also enriched, suggesting that other TFs might also participate in splicing regulation. Interestingly, the methylation levels in some of the regions were positively associated with the inclusion level of the spliced exons, indicating that other mCpG-binding proteins are involved in regulating the splicing process.
Human diseases
Many studies have shown that aberrant DNA methylation is associated with various human diseases, including some types of cancer19–21. For example, profiling the DNA methylation status in the promoters of 272 glioblastoma tumours showed that a distinct subset of samples displayed hypermethylation at a large number of loci, a phenotype termed ‘CpG island methylator phenotype’ (REF. 116). However, the mechanism by which the altered epigenetic state causes disease remains elusive. A recent study analysed the effect of methylation-dependent protein–DNA interactions on gliomas117. The IDH genes (IDH1 and IDH2) encode isocitrate dehydrogenases, and mutations in these genes are among the most frequent found in diffuse gliomas118,119. Mutant IDH protein is a competitive inhibitor of hydroxylases, including the TET family of 5mC hydroxylases120–122. As a result, the IDH mutation leads to a remodelling of DNA methylation profiles. Specifically, owing to the interference with TET family proteins, the mutation causes the CpG island methylator phenotype116,123. IDH mutant gliomas have been shown to exhibit hypermethylation at CTCF binding sites, which leads to a reduction in CTCF binding; loss of CTCF in topologically associated domains removed the domain boundary and caused aberrant gene activation117.
Conclusions
Similar to genome-wide association studies (GWAS), the profiling of epigenomes (including DNA methylomes) has been extensively carried out under various physiological conditions and in many different biological systems. Transitioning to a post-epigenome era, it is time to elucidate the functional consequences of the observed changes in DNA methylation status and link these changes to phenotypes. Although the role of MBD proteins, as non-sequence-specific methylation readers, has been fairly well-studied, the biological functions of an emerging class of sequence-specific methylation readers and/or effectors remain elusive.
To fully understand the biological processes that are mediated by DNA methylation, many challenges and unanswered questions regarding the methylation readers and/or effectors remain to be tackled in future research. First, we need a more comprehensive catalogue of methylation readers and effectors. Although a few studies have provided more than 100 proteins that can interact with methylated DNA in humans and mice, more readers remain to be discovered in these and other species. An evolutionary conservation analysis of these proteins will provide critical insights into their functional importance. In addition, the identification of the readers for 5mC derivatives (BOX 1) will greatly facilitate the elucidation of their roles in epigenetics. Second, these newly identified methylation readers require more and detailed characterization. For example, it is imperative to understand whether these TFs actually interact with genomic DNA in vivo. As we demonstrated above, superimposing ChIP–seq and DNA methylome data sets can be an effective approach to validate mCpG-dependent DNA–TF interactions in vivo. Although more technically challenging, ChIP-coupled genome-wide BS-seq is a more direct approach to map the in v ivo protein–mCpG interactions. Another possible approach is to observe genome-wide changes in TF binding sites by perturbing DNA methylation; for example, by knocking out DNMTs or by pharmacologically removing DNA methylation. Finally, the physiological relevance of protein–mCpG interactions will need to be established. Given a lack of adequate assays or approaches, this could well be a daunting task. A methylation reader usually interacts with both methylated sites and unmethylated sites. Therefore, simply knocking down a methylation reader will not help reveal its role. Identification of the key residues that interact with mCpG sites and the effects of mutations of these residues will provide the next step to dissect the functional role of methylation readers.
Taken together, the notion that TFs may act as DNA methylation readers is an emerging concept supported by predominantly in vitro but also by emerging in vivo evidence. Of note, this new concept does not refute the conventional view that most TFs do not interact with methylated DNA. Instead, these two scenarios may well coexist in cells. Here, we have focused on this exciting and novel concept with a full awareness that it may apply only to a subset of TFs and to a subset of their binding sites.
Supplementary Material
Acknowledgments
The authors thank J. Wan, Y. Zhao and other laboratory members from the Zhu and Qian groups for their discussions. The authors are supported in part by the NIH (EY024580, EY023188 to J.Q. and GM111514 to H.Z.).
Glossary
- DNA methylation
A biological process in which a methyl group is covalently added to a cytosine
- DNA methyltransferases
(DNMTs). Enzymes that catalyse the transfer of a methyl group to DNA
- CpG islands
A segment of DNA with a high frequency of CpG dinucleotides that often overlaps with promoters
- Genomic imprinting
A phenomenon by which some genes are expressed in an allele-specific manner; that is, alleles inherited either from the father or the mother are expressed
- Deep sequencing
A next-generation sequencing approach (for example, RNA sequencing or bisulfite sequencing) with high coverage
- Methylome
The collection of methylation status in an entire genome
- Epigenome
The collection of chemical modifications added to DNA or histones of a given genome, which do not alter the genetic codes but can be inherited and lead to changes in the function of the genome
- Differentially methylated regions
Regions of DNA with significant differences in methylation levels between two physiological conditions (for example, disease versus healthy) different developmental stages or different tissues
- Kd
The dissociation constant Kd is defined by the Koff/Kon ratio, which has the unit of concentration
- Oblique incidence reflectivity difference
(OIRD). A form of polarization-modulated imaging ellipsometer for label-free, high-throughput detection of binding events on protein microarrays
- Kon and Koff
In a simple binding event Kon and Koff refer to the on-rate and off-rate constants, which have units of 1/(concentration time) and 1/time, respectively
- TETs
(Ten-eleven translocation proteins). The TET family of methylcytosine dioxygenases is made of TET1, TET2, TET3 and TET4, which catalyse the conversion of the modified DNA base 5-methylcytosine (5mC) to 5-hydroxymethyl-cytosine (5hmC)
- Topologically associated domains
3D spatial organization units of mammalian genomes, within which most enhancer–promoter interactions occur
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
ENCODE: http://encodeproject.org
ENCSR000EBQ | ENCSR000EBV | ENCSR000BSI | ENCSR000ECC | ENCSR000ECF | ENCSR000BJW | ENCSR000BHO
FURTHER INFORMATION
Irreproducible Discovery Rate: https://www.encodeproject.org/software/idr
References
- 1.Bestor TH. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Phil. Trans. R. Soc. Lond. B. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- 2.Bird AP, Wolffe AP. Methylation-induced repression — belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 4.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 5.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 6.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 7.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 9.Krauss V, Reuter G. DNA methylation in Drosophila — a critical evaluation. Prog. Mol. Biol. Transl. Sci. 2011;101:177–191. doi: 10.1016/B978-0-12-387685-0.00003-2. [DOI] [PubMed] [Google Scholar]
- 10.Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 11.Takayama S, et al. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014;24:821–830. doi: 10.1101/gr.162412.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selker EU. Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- 14.Jeon J, et al. Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus Magnaporthe oryzae. Sci. Rep. 2015;5:8567. doi: 10.1038/srep08567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 17.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 18.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 20.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 22.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 23.Li M, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neurosci. Biobehav. Rev. 2010;34:882–888. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang YH, et al. A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am. J. Med. Genet. A. 2004;131A:1–10. doi: 10.1002/ajmg.a.30297. [DOI] [PubMed] [Google Scholar]
- 26.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–e11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001;20:3166–3173. doi: 10.1038/sj.onc.1204340. [DOI] [PubMed] [Google Scholar]
- 34.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang XY, et al. Binding sites in mammalian genes and viral gene regulatory regions recognized by methylated DNA-binding protein. Nucleic Acids Res. 1990;18:6253–6260. doi: 10.1093/nar/18.21.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J. Biol. Chem. 2002;277:35434–35439. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Springer NM, Kaeppler SM. Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiol. 2005;138:92–104. doi: 10.1104/pp.105.060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2 encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 40.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat. Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 41.Prokhortchouk A, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rishi V, et al. CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc. Natl Acad. Sci. USA. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. This paper demonstrates that ZFP57 and its cofactor KAP1 affect chromatin by interacting with methylated ICRs in embryonic stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsson QH, Schelcher C, Verrall E, Petosa C, Sinclair AJ. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog. 2008;4:e1000005. doi: 10.1371/journal.ppat.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He X, Futterer J, Hohn T. Sequence-specific and methylation-dependent and -independent binding of rice nuclear proteins to a rice tungro bacilliform virus vascular bundle expression element. J. Biol. Chem. 2001;276:2644–2651. doi: 10.1074/jbc.M006653200. [DOI] [PubMed] [Google Scholar]
- 47.Bahar Halpern K, Vana T, Walker MD. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J. Biol. Chem. 2014;289:23882–23892. doi: 10.1074/jbc.M114.573469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hantusch B, Kalt R, Krieger S, Puri C, Kerjaschki D. Sp1/Sp3 and DNA-methylation contribute to basal transcriptional activation of human podoplanin in MG63 versus Saos-2 osteoblastic cells. BMC Mol. Biol. 2007;8:20. doi: 10.1186/1471-2199-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niesen MI, et al. Activation of a methylated promoter mediated by a sequence-specific DNA-binding protein, RFX. J. Biol. Chem. 2005;280:38914–38922. doi: 10.1074/jbc.M504633200. [DOI] [PubMed] [Google Scholar]
- 50.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spruijt CG, et al. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. This paper describes the identification of proteins that interact with mCpG sites, 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in ES cells and neuronal progenitor cells using a MS/MS-based approach. [DOI] [PubMed] [Google Scholar]
- 52.Hu S, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu S, et al. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. doi: 10.7554/eLife.00726. This study identifies the transcription factors that preferentially bind to methylated DNA using a protein microarray-based approach and verified that endogenous KLF4 binds to methylated DNA in human ES cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger MF, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mann IK, et al. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013;23:988–997. doi: 10.1101/gr.146654.112. This paper describes the use of DNA microarrays to identify proteins that interact with methylated DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brinkman AB, et al. Sequential ChIP–bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Statham AL, et al. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP–seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012;22:1120–1127. doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao F, et al. Direct ChIP–bisulfite sequencing reveals a role of H3K27me3 mediating aberrant hypermethylation of promoter CpG islands in cancer cells. Genomics. 2014;103:204–210. doi: 10.1016/j.ygeno.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Strogantsev R, et al. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015;16:112. doi: 10.1186/s13059-015-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Lopes EC, et al. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res. 2008;68:7258–7263. doi: 10.1158/0008-5472.CAN-08-0344. [DOI] [PubMed] [Google Scholar]
- 63.Qin S, et al. Kaiso mainly locates in the nucleus in vivo and binds to methylated, but not hydroxymethylated DNA. Chin. J. Cancer Res. 2015;27:148–155. doi: 10.3978/j.issn.1000-9604.2015.04.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blattler A, et al. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin. 2013;6:13. doi: 10.1186/1756-8935-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li JJ, Jiang CR, Brown JB, Huang H, Bickel PJ. Sparse linear modeling of next-generation mRNA sequencing (RNA-Seq) data for isoform discovery and abundance estimation. Proc. Natl Acad. Sci. USA. 2011;108:19867–19872. doi: 10.1073/pnas.1113972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y, et al. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014;42:4859–4867. doi: 10.1093/nar/gku134. This study determined the crystal structure of the KLF4-methylated DNA complex and provided the structural basis for mCpG-TF interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dantas Machado AC, et al. Evolving insights on how cytosine methylation affects protein-DNA binding. Brief. Funct. Genom. 2015;14:61–73. doi: 10.1093/bfgp/elu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He HH, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lazarovici A, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl Acad. Sci. USA. 2013;110:6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buck-Koehntop BA, et al. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc. Natl Acad. Sci. USA. 2012;109:15229–15234. doi: 10.1073/pnas.1213726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tippin DB, Sundaralingam M. Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA. J. Mol. Biol. 1997;267:1171–1185. doi: 10.1006/jmbi.1997.0945. [DOI] [PubMed] [Google Scholar]
- 74.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 75.Liu S, et al. Characterization of monoclonal antibody’s binding kinetics using oblique-incidence reflectivity difference approach. MAbs. 2015;7:110–119. doi: 10.4161/19420862.2014.985919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 77.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. This paper demonstrates that some proteins, such as CTCF and REST, can reduce DNA methylation levels at the genomic regions near their binding regions. [DOI] [PubMed] [Google Scholar]
- 79.Charlet J, et al. Bivalent regions of cytosine methylation and H3K27 acetylation suggest an active role for DNA methylation at enhancers. Mol. Cell. 2016;62:422–431. doi: 10.1016/j.molcel.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 81.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 82.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 83.Krebs AR, Dessus-Babus S, Burger L, Schubeler D. High-throughput engineering of a mammalian genome reveals building principles of methylation states at CG rich regions. eLife. 2014;3:e04094. doi: 10.7554/eLife.04094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han L, Lin IG, Hsieh CL. Protein binding protects sites on stable episomes and in the chromosome fromde novo methylation. Mol. Cell. Biol. 2001;21:3416–3424. doi: 10.1128/MCB.21.10.3416-3424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fujiki K, et al. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- 86.Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem. Biophys. Res. Commun. 2006;344:845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 87.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 88.Bednarik DP, et al. DNA CpG methylation inhibits binding of NF-kappa B proteins to the HIV-1 long terminal repeat cognate DNA motifs. New Biol. 1991;3:969–976. [PubMed] [Google Scholar]
- 89.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ehrlich KC, Cary JW, Ehrlich M. A broad bean cDNA clone encoding a DNA-binding protein resembling mammalian CREB in its sequence specificity and DNA methylation sensitivity. Gene. 1992;117:169–178. doi: 10.1016/0378-1119(92)90726-6. [DOI] [PubMed] [Google Scholar]
- 91.Falzon M, Kuff EL. Binding of the transcription factor EBP-80 mediates the methylation response of an intracisternal A-particle long terminal repeat promoter. Mol. Cell. Biol. 1991;11:117–125. doi: 10.1128/mcb.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 93.Inamdar NM, Ehrlich KC, Ehrlich M. CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol. Biol. 1991;17:111–123. doi: 10.1007/BF00036811. [DOI] [PubMed] [Google Scholar]
- 94.Kovesdi I, Reichel R, Nevins JR. Role of an adenovirus E2 promoter binding factor in E1A–mediated coordinate gene control. Proc. Natl Acad. Sci. USA. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 96.Maurano MT, et al. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 2015;12:1184–1195. doi: 10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 97.Domcke S, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. This paper shows that the removal of DNA methylation would create novel binding sites for NRF1 and thus affect the NRF1-DNA interactions in vivo, whereas other studies showed that DNA methylation could affect TF-DNA interactions in vitro. [DOI] [PubMed] [Google Scholar]
- 98.Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J. Biol. Chem. 2013;288:34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baylin SB. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2:S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 100.Yu DH, et al. Developmentally programmed 3’ CpG island methylation confers tissue- and cell-type-specific transcriptional activation. Mol. Cell. Biol. 2013;33:1845–1858. doi: 10.1128/MCB.01124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wan J, et al. Characterization of tissue-specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genomics. 2015;16:49. doi: 10.1186/s12864-015-1271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 103.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Soufi A, et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 109.Laverriere AC, et al. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 110.Liu JK, DiPersio CM, Zaret KS. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol. Cell. Biol. 1991;11:773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. This study demonstrates that MeCP2 affects splicing events through its interaction with methylated DNA in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lyko F, et al. The honey bee epigenomes: differentia methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wan J, et al. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013;41:8503–8514. doi: 10.1093/nar/gkt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2015 doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, an clinical opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 121.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo JU, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. This paper describes genome-wide methylation profiling in adult mammalian brain and the discovery of MeCP2 as a reader of non-CpG methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gabel HW, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl Acad. Sci. USA. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pfaffeneder T, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 132.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun Z, et al. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell. 2015;57:750–761. doi: 10.1016/j.molcel.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 136.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Song CX, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 139.Neri F, et al. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 140.Wu H, Wu X, Shen L, Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xia B, et al. Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat. Methods. 2015;12:1047–1050. doi: 10.1038/nmeth.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Iurlaro M, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. This paper identifies proteins that interact with 5hmC and 5fC using promoter sequences as bait in an MS/MS-based screens. Numerous 5fC interaction partners were discovered, including transcriptional regulators, DNA repair factors and chromatin regulators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khrapunov S, et al. Unusual characteristics of the DNA binding domain of epigenetic regulatory protein MeCP2 determine its binding specificity. Biochemistry. 2014;53:3379–3391. doi: 10.1021/bi500424z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.