Abstract
Retrotransposition of human LINE-1 (L1) element, a major representative non-LTR retrotransposon in the human genome, is known to be a source of insertional mutagenesis. However, nothing is known about effects of L1 retrotransposition on cell growth and differentiation. To investigate the potential for such biological effects and the impact that human L1 retrotransposition has upon cancer cell growth, we examined a panel of human L1 transformed cell lines following a complete retrotransposition process. The results demonstrated that transposition of L1 leads to the activation of the p53-mediated apoptotic pathway in human cancer cells that possess a wild-type p53. In addition, we found that inactivation of p53 in cells, where L1 was undergoing retrotransposition, inhibited the induction of apoptosis. This suggests an association between active retrotransposition and a competent p53 response in which induction of apoptosis is a major outcome. These data are consistent with a model in which human retrotransposition is sensed by the cell as a “genetic damaging event” and that massive retrotransposition triggers signaling pathways resulting in apoptosis.
INTRODUCTION
Retrotransposons are mobile retroelements that utilize reverse transcriptase and RNA intermediate to relocate to new locations within the cellular genome. Retrotransposons are subdivided into two subclasses: LTR-(long terminal repeats) and non-LTR-retrotransposons [1, 2]. Non-LTR-retrotransposons are typified by the LINE-1 (long interspersed nuclear element 1), or L1, in mammals [3, 4]. L1 is one of the repetitive sequences in the genome, with 500 000 copies comprising about 17% of the genome [5, 6, 7]. Most human L1s (> 99.8%) are unable to transpose as a result of 5′ truncations, rearrangements, or nonsense mutations [8, 9]. However, evidence exists that L1 transposition continues to occur.
Several examples of de novo transposition events have been identified largely as the result of germline and somatic mutations caused by the insertion of new L1 elements into functional genes [10, 11, 12, 13, 14, 15, 16, 17]. Constitutive methylation of CpG sites in an L1 promoter is considered to be one of the major mechanisms for repression of retrotransposition [18, 19, 20]. In these cases, the CpG sites in an L1 promoter are normally heavily methylated [21] and demethylation of core CpG sites in the promoter leads to increased levels of L1 transcription [18]. Interestingly, demethylation and subsequent activation of an L1 promoter have been observed in bladder cancer cells [22], suggesting that release of the methylation constraints and activation of L1 may be a common cancer-associated event. Indeed, DNA methylation is considered to be an important mechanism for silencing retroelements in the mammalian genome, and it has been demonstrated that loss of genomic methylation activates L1 elements and causes p53-dependent apoptosis [23]. Thus, there seem to be selective pressure for silencing of L1 in “normal” cells and contrary activation of L1 in cancer cells.
In this paper we show that L1 elements capable of high-frequency retrotransposition in cultured human cells have a differential impact upon cell growth depending on the cells p53 status. Human cancer cells that contained a functional p53 underwent apoptosis following L1 retrotransposition, while human cancer cells mutant for p53 did not. These results imply that increased retrotransposition is recognized as DNA damage and demonstrate that active retrotransposition by L1 induces p53-dependent cell killing.
MATERIALS AND METHODS
Cell culture, antibodies, and plasmids
HCT-116, SW480, and DLD-1 were maintained in Dulbecco's modified Eagle's medium (DMEM)/F-12 (Gibco-BRL, Calif) with 10% fetal bovine serum (FBS), 1% L-Glutamine, and 1% penicillin-streptomycin, in the presence of 5% CO2/95% air at 37°C. HT1080wt, HT1080mut, and MCF-7 were maintained in Iscove's modified Dulbecco's medium (Gibco-BRL) with 10% FBS, 1% L-Glutamine, and 1% penicillin-streptomycin. Normal human fibroblasts (NHF) were maintained in Dulbecco's modified Eagle's medium (DMEM)/H-21 with 10% FBS, 1% L-Glutamine, and 1% penicillin-streptomycin.
The antibodies anti-p53, anti-Bcl2, anti-Bax, and anti-HRP-conjugated secondary antibody were purchased from Santa Cruz Biotechnology, Calif. Anti-α-tubulin was purchased from Sigma (Saint Louis, Mo). The plasmid pJM101 containing human L1 tagged with an antisense NEO cassette was obtained from John V. Moran (University of Michigan Medical School, Mich). The control plasmid, pBRV1 containing only NEO gene under SV40 promoter, was a gift from Vladimir Larionov (NCI, NIH) and has been previously described [24]. The plasmids p1321 (E6), normal E6/E7 and a control E6/E7 deficient plasmid, p1318 (ΔE6) were obtained from Peter M. Howley (Harvard Medical School Mass). The HPV16 E6/E7 containing construct used, p1321, has been described previously [25]. To construct the plasmid, a SalI/HindIII fragment containing the E6/E7 ORFs of HPV16 was ligated to the HindIII/SalI site in the p1318 vector, which contained a human β-actin enhancer/promoter and a β-actin 3′ UTR polyadenylation signal sequence. A control E6/E7-deficient plasmid p1318 was produced by removing the SalI/HindIII fragment.
Cell transfection and L1 retrotransposition assay
5 × 105 cells were seeded in 60 mm dishes and grown to 70% confluence. Transfections were carried out using lipofectamine (Gibco-BRL). 2 μg of plasmid pJM101 DNA and 8 μL of lipofectamine reagent were used for each transfection. pJM101 contains a complete human L1 and is marked with two selectable genes, hygromycin and neomycin [26]. 16 hours after transfection the medium was changed, and 3 days later, HygR cells were selected by growth in the medium containing 250 μg/mL hygromycin for 14 days (Figure 1). HygR cells were trypsinized and plated in growth medium containing 300–400 μg/mL G418. After 14 days the resistant cells (G418R) were either fixed and stained to score the retrotransposition frequency or collected for DNA and protein extractions.
Figure 1.

Retrotransposition rationale. L1 plasmid is tagged with an antisense copy of the indicator gene NEO, which is disrupted by an intron, (I), in the sense orientation of L1. The NEO gene is flanked by a heterologous promoter (P2) and a polyadenylation signal (An). This ensures that G418R cells will only appear when an L1 transcript (P1: L1 promoter) is spliced (SD: splicing donor, SA: splicing acceptor), reverse transcribed, and reintegrated into chromosomal DNA, thus allowing expression of the uninterrupted NEO gene (NEO active). L1 tagged construct is subcloned into pCEP4 expression vector, which contains hygromycin selectable marker gene.
Retrotransposition frequency and colony forming ability
Retrotransposition frequency was scored as the number of G418R colonies per 106 HYGR cells plated. 106 HygR cells were plated in triplicate in the presence of G418 and maintained for 14 days under this selection. Growing clones were fixed, stained with Giemsa stain solution, and counted.
For the colony forming ability, one thousand cells (103) from HCT-116 and SW480 cell lines, either mock transfected or transfected with L1 and selected for either hygromycin or G418 resistance, were plated in triplicate. Colony forming ability is determined as the number of colonies generated from 103 plated cells and given as a percentage.
Polymerase chain reaction
Genomic DNA was extracted using DNAzol reagent (Molecular Research Center, Inc, Ohio). PCR was carried out in 50 μL reaction volume. Each PCR reaction contained 4 mM MgCl2, 5 μL 10 X buffer, 0.2 mM dNTPs, 200 ng of each primer, 10 U of Taq polymerase, and 200 ng of DNA template. The reactions were carried out under the following conditions: 94°C for 10 minutes, 30 cycles of 94°C for 1 minute, 64°C for 30 seconds, and 72°C for 30 seconds, then 72°C for 10 minutes. One fifth of the reaction volume was loaded and separated on 1% agarose gels containing ethidium bromide.
Immunoblotting
Cells, either mock-transfected or transfected with pJM101 or pBRV1 (as control), were lysed in the following buffer: 120 mM NaCl, 25 mM Tris, pH 7.5, 1% Triton x-100, and protease inhibitors cocktail (Complete Mini, Roche, Germany). The cells were centrifuged and the proteins collected and quantified using the Bio-Rad assay (Bio-Rad, Calif). The proteins were electrophoresed through 12% SDS-polyacrylamide gels, and transferred to nylon membranes (Immobilon-P). The membranes were first incubated in 5% blocking solution (nonfat dry milk in 1 X PBS) for two hours at room temperature, then incubated in blocking solution overnight at 4°C with a primary antibody: anti-p53 (1 : 500 dilution), anti-Bcl2 (1 : 500 dilution), anti-Bax (Santa Cruz Biotechnology) (1 : 250 dilution), or anti-α-tubulin (Sigma, Saint Louis, Mo) (1 : 1000 dilution) as a loading control. The membranes were washed first for 15 minutes and then 4 times for 5 minutes each in PBST (PBS 1 X + 0.1% Tween 20). The blots were incubated in blocking solution for 45 minutes at room temperature with the secondary HRP-conjugated antibody (Santa Cruz Biotechnology), and washed in PBST as described above. The detection was performed using the Western Blotting Luminol Reagent (Santa Cruz Biotechnology).
Apoptosis detection assay
Apoptosis was assayed using TiterTACS kit (Trevigen, Md) following the manufacturer's recommendations on cells plated and fixed in 96-well plates. Cells were either mock-transfected or transfected with L1 plasmid (pJM101) or with NEO plasmid (pBRV1) and selected for G418 resistance. As a control, cells were treated with nuclease allowing complete degradation of DNA. TiterTACS is a colorimetric assay to quantify apoptosis by detecting DNA fragmentation in cells.
Inactivation of p53 by human papillomavirus E6/E7 genes
HCT116 cells were seeded in 6-well dishes (2 × 105 cells/well) and grown to 70% confluence in DMEM-complete media. First, cells were transfected with pJM101 and selected for hygromycin resistance as described earlier. After 14 days, the HygR cells were trypsinized, pooled, and expanded in DMEM-Hyg until 70% confluence. HygR cells were then transfected with either no DNA (mock), 1 μg of complete E6 (E6), or 1 μg of ΔE6 (E6-deleted form) plasmid DNA and 8 μL of lipofectamine reagent. Three days later, the cells were placed in DMEM-complete containing 300–400 μg/mL G418 (DMEM-G418). After 10–14 days, the G418R cells were fixed and stained with 0.4% Giemsa solution for visualization and counting. The number of G418R colonies was scored and the retrotransposition frequency was determined.
RESULTS
Human L1 retrotransposition in cancer cells
In order to investigate the impact of human L1 transposition on cancer cell growth, we first established stable transformants using a retrotransposition assay which employed a selectable molecular marker in cultured cells (Figure 1). The assay is based on the excision of an inverted intron from a neomycin phosphotransferase (NEO) gene [26]. Before retrotransposition the intron prevents NEO expression. During retrotransposition the intron is spliced out of the RNA intermediate and allows NEO expression upon reinsertion of the L1. We analyzed two colorectal cancer cell lines with different p53 status: HCT-116 (p53wt) and SW480 (p53mut) for their ability to allow L1 retrotransposition by measuring their colony forming ability before and after transfection with L1 plasmid (Figure 2a). Both cell lines showed similar survivability, and both seem to equally support transfection with L1 and selection with hygromycin, as demonstrated in the colony formation assay. Interestingly, we found a dramatic difference between the two cell lines in the outcome of G418 selection, which allows for selection of cells harboring retrotransposition events. Indeed, while we were able to recover-large number of G418R clones from SW480 with mutant p53, only a few clones were recovered from HCT-116 cell line with wild-type p53 although the same number (106 cells) of HygR cells were plated for both cell lines to carry out G418 selection (Figure 2a). We concluded that it is the direct effect of retrotransposition in HCT-116 that results in reduced colony survival.
Figure 2.
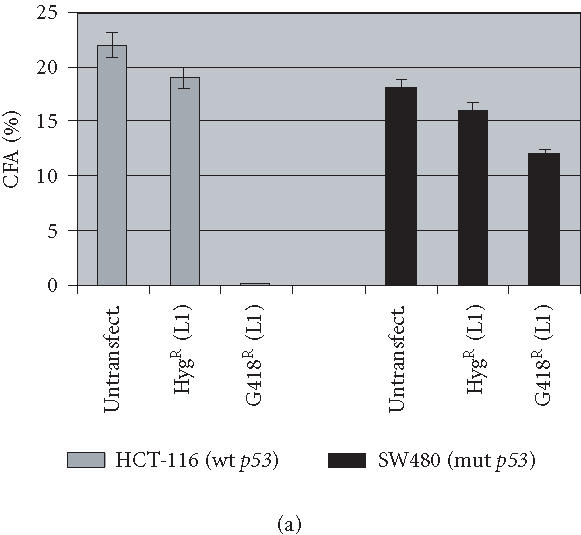
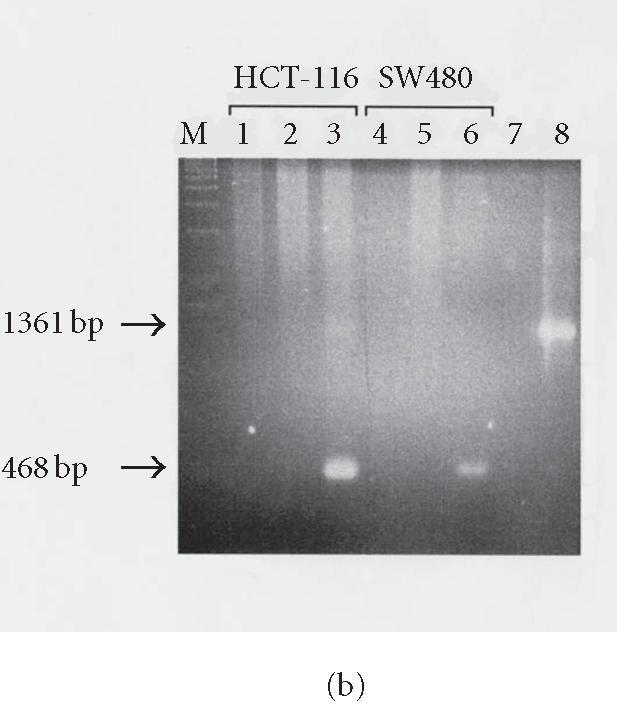
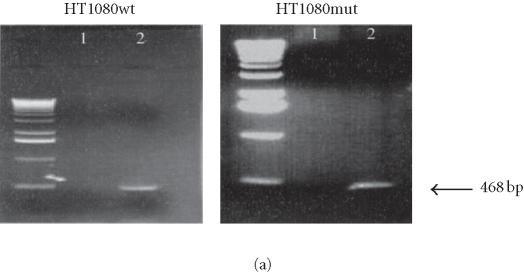
Human L1 retrotransposition in different cancer cell lines. (a) Colony forming ability of HCT-116 and SW480 cell lines. 103 cells either untransfected or transfected with L1 and selected for either hygromycin or G418 resistance were plated in triplicate until colonies were measurable, then stained and counted. Both cell lines exhibit similar colony forming ability when mock-treated or selected only for stable transformation with the plasmid (HygR). However, a dramatic difference in their survival ability was observed in G418-resistant clones bearing a retrotransposition-competent L1. (b) Confirmation of L1 retrotransposition by PCR in HCT-116 and SW480 cell lines. Lane M: DNA marker λ/HindIII; lane 1: HCT-116 mock-transfected cells; lane 2: HCT-116 HygR cells did not show the spliced form in either cell line; lane 3: HCT-116 G418R cells showing the correctly spliced form of NEO at 468 bp; lane 4: SW480 mock-transfected cells; lane 5: SW480 HygR cells; lane 6: SW480 G418R cells; lane 7: no DNA; and lane 8: the DNA plasmid alone with the unspliced form shown at 1361 bp.
To test whether the G418R clones generated after transfection have acquired resistance because the tagged L1 elements have undergone retrotransposition, we performed a PCR analysis of these clones using primers flanking the neomycin cassette. The results show that the retrotransposition marker, the neomycin phosphotransferase gene, is correctly spliced as evidenced by the detection of a 468 bp DNA fragment (Figure 2b). This was also shown for HCT-116 and SW480 (Figure 2b, lane 3). As controls, we used genomic DNA from mock-transfected cells, no DNA, or the DNA plasmid alone. HygR cells did not show the spliced form in either cell line (lane 2). In most cases, only the spliced form (468 bp) was visualized on agarose gels, which indicates that only the spliced and active form of neomycin gene can be stably maintained in cells. However, the longer unspliced form (1368 bp) can also sometimes be seen as extrachromosomal molecules in early passage.
To test whether this association between low number of G418R clones and the p53 status is applicable to other cell lines, we decided to analyze a panel of cell lines from different tissues having either a wild-type or mutant p53. This group included two fibrosarcoma cell lines, HT1080wt (p53 wild type) and its derivative HT1080mut (p53 mutant); a breast adenocarcinoma cell line, MCF-7 (p53wt); and an NHF (p53wt). The retrotransposition frequency was scored as described. As before, cell lines harboring wild-type p53 were poorly supportive of retrotransposition (Table 1). In contrast, the cell line with mutant p53 exhibited higher frequencies of retrotransposition. At this stage, low-frequency retrotransposition can be explained either by the inability of cells expressing p53wt to allow high L1 retrotransposition, or the inability to detect clones undergoing retrotransposition in cells because of their immediate loss.
Table 1.
Analysis of L1 retrotransposition frequency in multiple cell lines. Cells with mutant p53 exhibit high retrotransposition frequency, while cell lines with wild-type p53 exhibit a lower frequency.
| Cell line | p53 status | Retrotransposition frequency (× 106) |
|---|---|---|
| HCT-116 | wt | 68 |
| SW480 | mut | 1000 |
| DLD-1 | mut | 760 |
| MCF-7 | wt | 1.5 |
| HT1080wt | wt | 6 |
| HT1080mut | mut | 793 |
| NHF | wt | < 0.1 |
L1 induction of apoptosis in p53 (wt) cell lines
Since induction of apoptosis is an early event in DNA damage-mediated p53-dependent response and since p53 wild-type cells exhibit a dramatic decrease in their survivability after L1 retrotransposition, we examined the role of apoptosis in the observed differences in retrotransposition frequency. We assayed for apoptosis using a colorimetric assay allowing quantification by detection of DNA fragmentation. A dramatic increase in apoptosis was detected in G418R HCT-116 cells, while no induction of apoptosis was detected in G418R SW480 cells (Figure 3a). A nuclease treatment was used as a positive control. To confirm that drug selection using G418 is not associated with the induction of apoptosis, we transfected these cells with a plasmid containing only the NEO gene (pBRV1) without the L1 element. No apoptosis was induced in these cells. Thus, these data demonstrate that the induction of apoptosis is associated with the presence of retrotransposition-competent L1.
Figure 3.
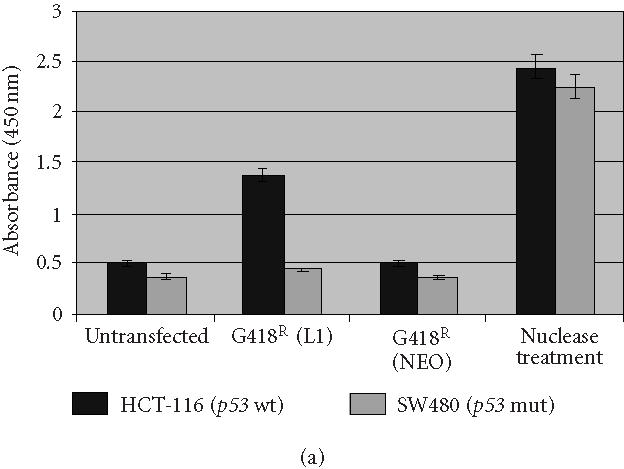
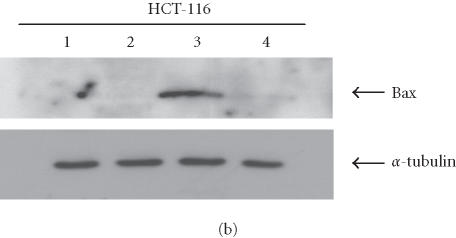
Analysis of apoptosis induction in the presence of RC-L1. (a) Apoptosis was highly induced in HCT-116 G418R cells with a wild-type p53 (G418R L1). No significant apoptosis induction was detected in SW480 cells with a mutant p53. Both HCT-116 and SW480 transfected with the vector alone (G418R NEO) did not show any apoptosis induction. Nuclease treatment was used as positive control. (b) Specific induction of Bax expression following L1 retrotransposition. HCT-116 cells transfected with either the L1 plasmid (JM101) or the vector alone (pBRV1). Bax induction was observed only in G418R L1-transfected cells (lane 3). No Bax expression was observed in mock-transfected cells (lane 1), HygR cells transfected with L1 (Lane 2), or G418R cells transfected with a plasmid containing a NEO gene alone (lane 4). α-tubulin is shown as loading control.
To further test whether the induction of apoptosis is directly associated with the presence of L1, proteins extracted from HCT-116 transfected either with the L1 plasmid or with a neomycin plasmid (vector alone) were analyzed by immunoblotting (Figure 3b). The induction of Bax expression was detected only in G418R cells derived from cells transfected with L1 plasmid (lane 3). No induction of Bax expression was observed in mock-transfected cells (lane 1), HygR cells transfected with L1 (lane 2), or G418R cells transfected with a plasmid containing only NEO gene (vector alone) (lane 4).
Suppression of p53 by expression of E6 ablates L1-dependent apoptosis
The E6 protein encoded by the oncogenic human papillomaviruses (HPVs) targets p53 for ubiquitin-dependent proteolysis [27]. To address the role of a competent p53 in the induction of apoptosis, we examined the effects of L1 activity in cells in which p53 is functionally inactivated by HPV E6 expression. We used the ability of the HPV E6 gene to inactivate p53 in order to examine the dependence of L1-mediated apoptosis on a functional p53 protein. When HCT-116 cells are transformed with L1 they undergo apoptosis and inhibition of cell growth. When these same cells were transfected to express a normal E6 gene (L1 + E6), the effect of L1 on cell growth is reduced and the number of G418 resistant colonies increased (Figure 4). When a parallel population of HCT-116 cells were transfected to express L1 and a deleted form of E6 (L1 + ΔE6), the L1 retrotransposition frequency was reduced and colony formation was similar to L1 alone. These results are consistent with the notion that L1-induced apoptosis requires a functional p53.
Figure 4.



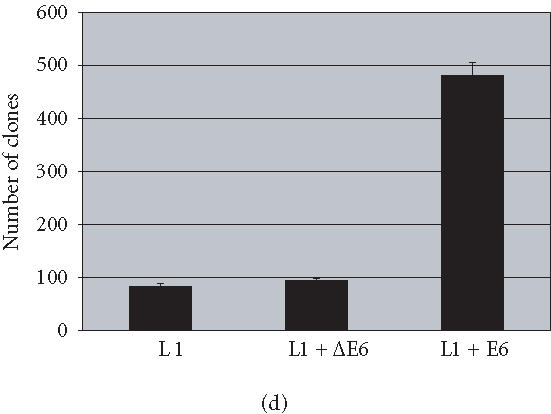
p53 inactivation by human papillomavirus E6 gene. HCT-116 cells transformed with L1 and selected for hygromycin resistance HygR were either (a) subjected to G418 selection directly (L1) or (b) exposed to either a deleted form of E6, ΔE6 (L1 + ΔE6), or (c) a complete form of E6 (L1 + E6). A histogram showing the effects of L1 retrotransposition on the number of clones in the presence and absence of E6 is shown in (d).
L1 retrotransposition in isogenic HT1080 cells
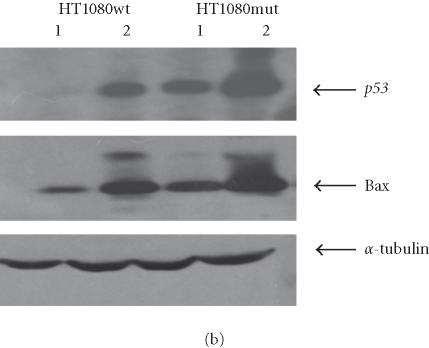
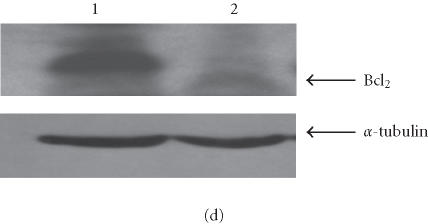
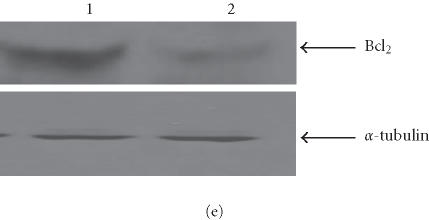
We have conducted most of our comparative studies in two colorectal cancer cell lines HCT-116 and SW480. These cells differ not only by their p53 status, but also by the status of some DNA repair genes, mainly mismatch repair genes hMLH1 and hMH6. Therefore, we cannot rule out the implication of other genes in the observed differences in these cells ability to undergo apoptosis after the addition of retrotransposition-competent L1 elements. Therefore, we further studied the association between L1 retrotransposition and p53 status in the isogenic cells HT1080wt, with wild-type p53, and its derivative HT1080mut, with mutant p53. HT1080 mutant cells (HT1080TG) have two independent mutations in each of the p53 alleles and can make p53 [28]. We showed that G418R clones have indeed undergone the retrotransposition for the marked L1 elements, using PCR analysis. The spliced form of 468 bp was detected in both HT1080wt and HT1080mut after transfection and selection for G418 resistance (Figure 5a). We analyzed protein expression levels in HT1080wt and its derivative HT1080mut by immunoblotting before and after transformation with human L1 (Figure 5b). We found that p53 is overexpressed after transfection and selection for G418R clones harboring retrotransposition-competent L1 elements in both HT1080wt and HT1080mut. We also found that Bax is overexpressed in HT1080wt in a much more pronounced manner in comparison with HT1080mut. We quantified the differences in Bax expression between HT1080wt and HT1080mut by densitometric analysis (Figure 5c). While we found that Bax expression was induced relatively about 8 times in HT1080wt, the difference was less than double in HT1080mut cells. In addition, we observed a decrease in Bcl2 expression level following L1 retrotransposition in HT1080wt cells (Figure 5d) and to a slightly lower extent in HT1080mut cells (Figure 5e). This is in agreement with p53 being involved in the induction of apoptosis following L1 retrotransposition, and in agreement with our finding in other cell lines where we have observed a more pronounced induction of apoptosis in cells with wild-type p53.
Figure 5.
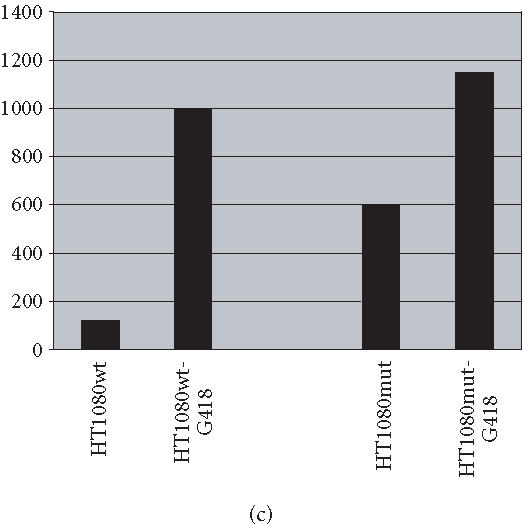
L1 retrotransposition in isogenic HT1080 cells. (a) Confirmation of L1 retrotransposition in HT1080 wt and HT1080 mut cells: a 468 bp DNA fragment was generated by PCR in HT1080 wt and HT1080 mut G418R clones demonstrating a correct splicing of the NEO marker gene. (b) Western blot analysis: whole cell extracts from HT1080 wt and HT1080 mut cells showing an increase of Bax expression following transfection with human L1 (1) and selection for G418R clones (2) harboring L1-retrotransposition elements predominantly in HT1080 wt. (c) Densitometric analysis of Bax expression levels in HT1080 wt and HT1080 mut before and after generation of G418R clones. More pronounced increase of Bax expression in HT1080 wt in comparison with HT1080 mut. Shown are relative arbitrary numbers. (d) Western blot analysis of HT10807thinsp;wt cells: Bcl2 expression before transfection (1) and after selection of G418R clones following L1 retrotransposition (2). (e) Western blot analysis of HT1080 mut cells: Bcl2 expression before transfection (1) and after selection of G418R (2). α-tubulin is shown as loading control.
DISCUSSION
Our understanding of the biology of L1 element, its retrotransposition mechanism, and its ability to retrotranspose in human cultured cells has increased over the last several years [29, 30]. However, the cellular consequences of the presence of L1 sequences and the support of L1 retrotransposition on the stability of the genome are still poorly understood. Indeed, it is becoming increasingly apparent that human L1 retrotransposition impacts the stability of the human genome and evidence is mounting that human L1 is more than just an insertional mutagen [31, 32, 33]. Interestingly, Morrish et al reported that L1 is capable of mediating DNA repair. Specifically, the authors showed that the L1 element is capable of retrotransposition using a pathway independent of endonuclease but dependent on reverse transcriptase, suggesting that L1 can integrate into preexisting DNA lesions resulting in retrotransposition-mediated DNA repair [33]. Conversely, it has also been shown that human L1 retrotransposition induces genetic instability in vivo through numerous L1 element inversions, extra nucleotide insertions, exon deletions, chromosomal inversion, and 5′ transduction [30]. Thus, L1 has been associated with both stabilizing and destabilizing molecular activities. The vast majority of these studies have not addressed the consequences of L1 transposition on genetic instability and cell growth.
In this study we analyzed the impact of L1 retrotransposition on human cancer cell growth and showed that L1 retrotransposition can elicit a DNA damage response leading to apoptosis in p53 competent cells. Thus, the overriding cellular consequences of L1 retrotransposition would appear to derive from genome destabilizing activities. We suggest two possible mechanisms through which L1 elements might induce apoptosis. First, since L1 creates DNA strand breaks using its own endonuclease to create nicks in the insertion sites, these events might be recognized by the cell as unrepaired DNA damage. This would be especially true if the transposition were massively induced thus triggering pathways for apoptosis. Despite the fact that the proteins encoded by L1 elements work preferentially in cis, members of the Alu class of retroelements are believed to misappropriate L1 proteins in order to proliferate [6, 34, 35]. Therefore, the introduction of exogenous retrotransposition-competent L1 elements into cells by transfection may activate not only endogenous L1 but probably also Alu sequences by providing in trans encoded proteins such as reverse transcriptase, thus contributing to a burst of transposition. Pertinent to this discussion it was recently reported that L1 reverse transcriptase can transactivate other endogenous L1 sequences and even other L1-unrelated sequences [36, 37, 38]. Thus, the delivery of retrotransposition-competent L1 elements into cancer cells can transactivate other endogenous repressed or “dormant” L1 and Alu sequences leading to a release of “jumping” L1s and Alus, that are ultimately sensed by the DNA repair machinery as a stress signal. A second possible mechanism is that a class of ribonucleoprotein particles (RNPs), consisting of transposition complexes that contain L1 RNA and the L1-encoded proteins, might separately elicit apoptosis. The heteronuclear ribonucleoproteins (hnRNPs) C1 and C2 are targets of destruction within the cell that has been induced to undergo apoptosis by a variety of stimuli [39]. L1-RNPs might be inducers in addition to targets of apoptosis. While we have yet to uncover the detailed mechanism through which human L1 is capable of inducing apoptosis in cancer cells with wild-type p53, our findings demonstrate an interplay between L1 retrotransposition and p53-mediated-induction of apoptosis in cells harboring retrotransposition-competent L1s.
Our finding supports a hypothesis in which the Bax gene plays a role in the L1-induced apoptosis in p53 wild-type cells. In addition to Bax gene, there is still a growing list of genes stimulated by p53 which are associated with apoptosis induction, such as Fas, CD95, DR5/killer, p85, IGF-BP3, PAG608, and so forth. It should be noted that several of these proapoptotic genes, as is also the case with Bax, appear to be cell-type-specific and signal-dependent. In our study, the induction of Bax expression and repression of Bcl2 expression following L1 retrotransposition in both HCT-116 and HT1080 cells indicate that induction of apoptosis by L1 involves activation via pathway regulated by the Bax gene.
The apoptotic gene Fas which is a cell surface “death receptor” mediates apoptosis upon engagement by its ligand, FasL. Many studies of transformed colon cell lines, such as these used in this study as well as freshly isolated tumor cells of diverse origin have revealed that the majority of cancer cells are resistant to Fas-mediated apoptosis [40, 41, 42, 43]. For these reasons we did not analyze Fas and FasL-mediated apoptosis in the colon cancer cell lines after L1 transposition.
In any case, the induction of retrotransposition would facilitate the apoptotic response in p53+/+ cells and the molecular events involved in sensing L1-induced stress need to be confirmed in order to shed some light on an important mechanism of inducing apoptosis in p53 competent cancer cells.
Human L1 as a potential novel cancer therapeutic strategy based on the induction of apoptosis: perspectives
Both chemotherapeutic agents and irradiation can induce tumor cell death primarily by causing DNA damage. However, it has been very difficult to limit damage to normal tissue to an acceptable level using these therapies. Although a number of methods to induce target cells to undergo apoptosis exist and are being evaluated, any approach which can be selectively targeted to the cancer cell offers an advantage to existing methods. The activation of L1 retrotransposition induces genomic instability, thereby triggering a DNA damage response, which results in the delay of cell cycle progression and/or induction of apoptosis. Using an endogenous component of the genome for a preferential induction of apoptosis within tumor cells could provide a novel mode of attack for cancer therapeutics. The employment of sophisticated cell-specific vectors as gene therapy tools will enable targeting of L1 only to tumor cells.
ACKNOWLEDGMENT
We thank Drs K. Tindall and A. Umar (NIEHS, NC) for the gift of the human colon cancer cell lines HCT-116, SW480, and DLD-1 and Ms Annab Lois (NIEHS, NC) for the gift of the NHF, MCF-7, and HT1080 cells. We thank Dr J. Moran (University of Michigan Medical School, Mich) for the gift of pJM101, and Dr Peter M. Howley (Harvard Medical School) for the gift of the HPV 16 E6/E7 plasmids. We are grateful to Annie Eisinger, Daphne Kantz, and Rodney C. Daniels for their technical assistance, and to Dr Kenneth Ramos for critical reading of the manuscript. This work was supported by the Fogarty International Center Award (AH), HHS CA76595 (OJS), and by a grant from the Commonwealth Health Research Board of Virginia (AH).
References
- 1.Hutchison C.A, Hardies S.C, Loeb D.D, Shehee W.R, Edgell M.H. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome. In: Berg D.E, Howe M.M, editors. Mobile DNA. Washington, DC, Wash: ASM Press; 1989. pp. 593–617. [Google Scholar]
- 2.Boeke J.D, Stoye J.P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J.M, Hughes S.H, Varmus H.E, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 3.Fanning T.G, Singer M.F. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987;910(3):203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- 4.Moran J.V, Gilbert N. Mammalian LINE-1 retrotransposons and related elements. In: Craig N, Craigie R, Gellert M, et al., editors. Mobile DNA II. Washington, DC, Wash: ASM Press; 2002. pp. 836–869. [Google Scholar]
- 5.Smit A.F, Toth G, Riggs A.D, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246(3):401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 6.Smit A.F. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6(6):743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 7.Lander E.S, Linton L.M, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Sassaman D.M, Dombroski B.A, Moran J.V, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16(1):37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 9.Brouha B, Schustak J, Badge R.M, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100(9):5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazazian H.H. Jr, Wong C, Youssoufian H, Scott A.F, Phillips D.G, Antonarakis S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 11.Morse B, Rotherg P.G, South V.J, Spandorfer J.M, Astrin S.M. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333(6168):87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- 12.Miki Y, Nishisho I, Horii A, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–645. [PubMed] [Google Scholar]
- 13.Narita N, Nishio H, Kitoh Y, et al. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest. 1993;91(5):1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bratthauer G.L, Cardiff R.D, Fanning T.G. Expression of LINE-1 retrotransposons in human breast cancer. Cancer. 1994;73(9):2333–2336. doi: 10.1002/1097-0142(19940501)73:9<2333::aid-cncr2820730915>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Dombroski B.A, Mathias S.L, Nanthakumar E, Scott A.F, Kazazian H.H. Jr. Isolation of an active human transposable element. Science. 1991;254(5039):1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 16.Holmes S.E, Dombroski B.A, Krebs C.M, Boehm C.D, Kazazian H.H. Jr. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7(2):143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 17.Kazazian H.H. Jr. Mobile elements and disease. Curr Opin Genet Dev. 1998;8(3):343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 18.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189(2):227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 19.Woodcock D.M, Lawler C.B, Linsenmeyer M.E, Doherty J.P, Warren W.D. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272(12):7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 20.Florl A.R, Lower R, Schmitz-Drager B.J, Schulz W.A. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80(9):1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender J. Cytosine methylation of repeated sequences in eukaryotes: the role of DNA pairing. Trends Biochem Sci. 1998;23(7):252–256. doi: 10.1016/s0968-0004(98)01225-0. [DOI] [PubMed] [Google Scholar]
- 22.Steinhoff C, Prior A, Reichmann G, Seifert H.H, Schulz W.A. Activity of E2F-dependent promoters in bladder carcinoma cells and their use for tumour-specific targeting of p53-induced apoptosis. Int J Oncol. 2002;21(5):1033–1040. [PubMed] [Google Scholar]
- 23.Jackson-Grusby L, Beard C, Possemato R, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27(1):31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 24.Kouprina N, Larionov V. Selective isolation of mammalian genes by TAR cloning. In: Dracopoli N.C, Haines J.L, Korf B.R, et al., editors. Current Protocols in Human Genetics. Vol. I. New York, NY: John Wiley and Sons; 1999. pp. 5.17.1–5.17.21. [DOI] [PubMed] [Google Scholar]
- 25.Arbeit J.M, Munger K, Howley P.M, Hanahan D. Neuroepithelial carcinomas in mice transgenic with human papillomavirus type 16 E6/E7 ORFs. Am J Pathol. 1993;142(4):1187–1197. [PMC free article] [PubMed] [Google Scholar]
- 26.Moran J.V, Holmes S.E, Naas T.P, DeBerardinis R.J, Boeke J.D, Kazazian H.H. Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 27.Talis A.L, Huibregtse J.M, Howley P.M. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273(11):6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 28.Anderson M.J, Casey G, Fasching C.L, Stanbridge E.J. Evidence that wild-type TP53, and not genes on either chromosome 1 or 11, controls the tumorigenic phenotype of the human fibrosarcoma HT1080. Genes Chromosomes Cancer. 1994;9(4):266–281. doi: 10.1002/gcc.2870090407. [DOI] [PubMed] [Google Scholar]
- 29.Ostertag E.M, Kazazian H.H. Jr. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 30.Kazazian H.H. Jr, Goodier J.L. LINE drive: retrotransposition and genome instability. Cell. 2002;110(3):277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert N, Lutz-Prigge S, Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 32.Symer D.E, Connelly C, Szak S.T, et al. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 33.Morrish T.A, Gilbert N, Myers J.S, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 34.Boeke J.D. LINEs and Alus—the polyA connection. Nat Genet. 1997;16(1):6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 35.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24(4):363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 36.Dhellin O, Maestre J, Heidmann T. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo mRNA reverse transcription. EMBO J. 1997;16(21):6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haoudi A, Mason J.M. Reverse transcriptase can stabilize or destabilize the genome. Genome. 2000;43(6):949–956. [PubMed] [Google Scholar]
- 38.Cost G.J, Feng Q, Jacquier A, Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterhouse N, Kumar S, Song Q, et al. Heteronuclear ribonucleoproteins C1 and C2, components of the spliceosome, are specific targets of interleukin β-converting enzyme-like proteases in apoptosis. J Biol Chem. 1996;271(46):29335–29341. doi: 10.1074/jbc.271.46.29335. [DOI] [PubMed] [Google Scholar]
- 40.O'Connell J, O'Sullivan G.C, Collins J.K, Shanahan F. The Fas counterattack: Fas-mediated T cell killing by colon cancer cells expressing Fas ligand. J Exp Med. 1996;184(3):1075–1082. doi: 10.1084/jem.184.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shima Y, Nishimoto N, Ogata A, Fujii Y, Yoshizaki K, Kishimoto T. Myeloma cells express Fas antigen/APO-1 (CD95) but only some are sensitive to anti-Fas antibody resulting in apoptosis. Blood. 1995;85(3):757–764. [PubMed] [Google Scholar]
- 42.Owen-Schaub L.B, Zhang W, Cusack J.C, et al. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15(6):3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connell J, Bennett M.W, O'Sullivan G.C, Collins J.K, Shanahan F. The Fas counterattack: a molecular mechanism of tumor immune privilege. Mol Med. 1997;3(5):294–300. [PMC free article] [PubMed] [Google Scholar]