Abstract
Nearly 60 CCCH zinc finger proteins have been identified in humans and mice. These proteins are involved in the regulation of multiple steps of RNA metabolism, including mRNA splicing, polyadenylation, transportation, translation and decay. Several CCCH zinc finger proteins, such as tristetraprolin (TTP), roquin 1 and MCPIP1 (also known as regnase 1), are crucial for many aspects of immune regulation by targeting mRNAs for degradation and modulation of signalling pathways. In this Review, we focus on the emerging roles of CCCH zinc finger proteins in the regulation of immune responses through their effects on cytokine production, immune cell activation and immune homeostasis.
Graphical Abstract
Immune responses combating infectious microorganisms are under precise control, as unchecked or inappropriate responses can be detrimental to the host and lead to inflammatory or autoimmune diseases1. Although the transcriptional control of immune responses has been studied extensively2, the importance of post-transcriptional regulation of these processes is less well-defined. Post-transcriptional control can occur at each step of RNA metabolism, including splicing, capping, polyadenylation, export, localization, translation and decay. Recent studies have emphasized the importance of RNA metabolism, particularly mRNA degradation and translation, in the regulation of immune responses3.
Zinc finger proteins are generally thought of as DNA-binding transcription factors. However, certain classes of zinc finger proteins, such as CCCH zinc finger proteins, often function as RNA-binding proteins and regulate RNA metabolism4. CCCH zinc finger proteins are characterized by one or more CCCH zinc finger domains containing three cysteines and a histidine. Nearly 60 CCCH zinc finger proteins have been identified in humans and mice. Although the functions of most CCCH zinc finger proteins remain obscure, emerging evidence suggests that some CCCH zinc finger proteins are involved in a broad range of biological processes that are associated with immune responses, including cytokine production, immune cell activation, immune homeostasis and antiviral innate immunity, as well as in regulation of cell differentiation and cancer cell growth (TABLE 1; see Supplementary information S1 (table)).
Table 1.
Human CCCH zinc finger proteins
| Gene name* | Other names | CCCH zinc fingers (number of repeats) | Molecular functions | Biological functions |
|---|---|---|---|---|
| CNOT4 | CLONE243; NOT4 | CX8CX4CX3H (1) | Deadenylation | Megakaryocyte differentiation |
| CPSF4 | CPSF30; NAR; NEB1 | CX7–8CX4–5CX3H (5) | mRNA splicing | Antivirus |
| CPSF4L | – | CX7–8CX4–5CX3H (4) | ND | ND |
| DHX57 | DDX57 | CX7CX5CX3H (1) | ND | ND |
| DUS3L | DUS3 | CX10CX5CX3H (1) | ND | ND |
| HELZ | DHRC; HUMORF5 | CX8CX5CX3H (1) | RNA helicase | ND |
| HELZ2 | PDIP1; PR1C285 | CX5CX5CX3H (1) | RNA helicase | Adipocyte differentiation |
| LENG9 | – | CX7CX5CX3H (1) | ND | ND |
| MBNL1 | EXP; MBNL | CX7CX6CX3H (2) | mRNA splicing | Myoblast differentiation |
| MBNL2 | MBLL; MBLL39 | CX7CX4–6CX3H (4) | mRNA splicing | Myoblast differentiation |
| MBNL3 | CHCR; MBLX | CX7CX6CX3H (2) | mRNA splicing | Myoblast differentiation |
| MKRN1 | RNF61 | CX7CX5CX3H (3) | E3 ligase | Tumorigenesis |
| MKRN2 | RNF62; HSPC070 | CX7–9CX5CX3H (2) | Putative E3 ligase | Neurogenesis |
| MKRN3 | CPPB2; RNF63; ZNF127 | CX7–9CX5CX3H (3) | Putative E3 ligase | Central precocious puberty |
| NUPL2 | CG1; NLP1 | CX7CX5CX3H (1) | RNA export | ND |
| PAN3 | – | CX8CX5CX3H (1) | Deadenylation | ND |
| PPP1R10 | CAT53; FB19; PNUTS | CX8CX5CX3H (1) | Adaptor protein | Inhibition of HIV replication |
| PRR3 | CAT56 | CX8CX5CX3H (1) | ND | ND |
| RC3H1 | ROQUIN; ROQUIN 1; RNF198 | CX8CX5CX3H (1) | mRNA decay | Immune homeostasis |
| RC3H2 | ROQUIN 2; RNF164; MNAB | CX8CX5CX3H (1) | mRNA decay | Immune homeostasis |
| RNF113A | CWC24; TTD5; ZNF183 | CX8CX5CX3H (1) | ND | ND |
| RNF113B | RNF161; ZNF183L1 | CX8CX5CX3H (1) | ND | ND |
| TIPARP | PARP7; ARTD14 | CX7CX5CX3H (1) | mRNA splicing | Antivirus |
| TOE1 | – | CX8CX5CX3H (1) | Deadenylation | Inhibition of HIV replication |
| TRMT1 | TRM1 | CX7CX5CX3H (1) | tRNA methyltransferase | ND |
| U2AF1 | U2AF35; U2AFBP; FP793 | CX8CX5CX3H (2) | mRNA splicing | Blood cell differentiation |
| U2AF1L4 | U2AF26; U2AF-RS3 | CX8CX5CX3H (2) | mRNA splicing | T cell activation |
| ZC3HAV1L | C7ORF39 | CX7–12CX4–5CX3H (2) | ND | ND |
| ZC3H1 | PARP12; ARTD12; MST109 | CX7–12CX5CX3H (3) | mRNA translation | Inflammation and antivirus |
| ZC3H2 | ZC3HAV1; ZAP; PARP13 | CX7–11CX4–5CX3H (3) | mRNA decay | Antivirus |
| ZC3H3 | SMICL | CX7–8CX4–6CX3H (5) | Polyadenylation | ND |
| ZC3H4 | C19ORF1 | CX7–8CX5CX3H (3) | ND | ND |
| ZC3H5 | UNK; UNKEMPT | CX6–14CX5–6CX3H (5) | mRNA translation | Neuronal differentiation |
| ZC3H5L | UNKL | CX6–14CX5–6CX3H (5) | ND | ND |
| ZC3H6 | ZC3HDC6 | CX7–8CX5CX3H (3) | ND | ND |
| ZC3H7A | HSPC055 | CX7–8CX5CX3H (3) | ND | ND |
| ZC3H7B | ROXAN | CX7–8CX5CX3H (3) | ND | ND |
| ZC3H8 | FLIZ1 | CX7–8CX5CX3H (3) | Transcription repressor | Thymocyte homeostasis |
| ZC3H9 | ZGPAT; ZIP; GPATC6 | CX7CX5CX3H (1) | Transcription repressor | Tumour suppressor |
| ZC3H10 | ZC3HDC10 | CX7CX4–5CX3H (3) | ND | Putative tumour suppressor |
| ZC3H11A | ZC3HDC11A | CX7–8CX4–5CX3H (3) | mRNA export | ND |
| ZC3H12A | MCPIP1; REGNASE 1 | CX5CX5CX3H (1) | Ribonuclease | Inflammation and immunity |
| ZC3H12B | MCPIP2 | CX5CX5CX3H (1) | Putative ribonuclease | ND |
| ZC3H12C | MCPIP3 | CX5CX5CX3H (1) | Putative ribonuclease | Inflammation |
| ZC3H12D | MCPIP4; TFL; P34 | CX5CX5CX3H (1) | Putative ribonuclease | Inflammation and immunity |
| ZC3H13 | KIAA0853 | CX8CX5CX3H (1) | mRNA splicing | Inflammation |
| ZC3H14 | UKP68; MSUT2;SUT2 | CX5CX4–5CX3H (5) | Polyadenylation | Neuronal differentiation |
| ZC3H15 | LEREPO4; HTO10 | CX7CX5CX3H (1) | ND | HIV replication |
| ZC3H16 | RBM22; CWC2; FSAP47 | CX7CX5CX3H (1) | mRNA splicing | ND |
| ZC3H17 | RBM26; PPP1RB2; ARRS2 | CX8CX5CX3H (1) | ND | ND |
| ZC3H18 | NHN1 | CX7CX5CX3H (1) | mRNA export | Inflammation |
| ZC3H19 | ZMAT5; SNRNP20 | CX8CX5CX3H (1) | ND | ND |
| ZC3H20 | RBM27; ARSS1; PSC1 | CX8CX5CX3H (1) | ND | ND |
| ZC3H22 | ZRSR2; URP; U2AF1RS2 | CX7–8CX5CX3H (2) | mRNA splicing | Blood cell differentiation |
| ZFP36 | TTP; NUP475; TIS11; G0S24 | CX8CX5CX3H (2) | mRNA decay | Inflammation and immunity |
| ZFP36L1 | TIS11B; BRF1; BERG36 | CX8CX5CX3H (2) | mRNA decay | Immune cell maturation |
| ZFP36L2 | TIS11D; BRF2; ERF2 | CX8CX5CX3H (2) | mRNA decay | Immune cell maturation |
C, cysteine; H, histidine; ND, not determined; X, any amino acid.
57 human CCCH zinc finger proteins are identified based on updated information in GenBank.
Zc3h21 and Zfp36l3 are rodent-specific genes; therefore, the mouse genome encodes 59 CCCH zinc finger proteins. Additional information, such as the full description of gene names and references for biological function, is presented in Supplementary information S1 (table).
In this Review, we discuss the recent explosion of information on the role of CCCH zinc finger proteins in the regulation of immune responses. We focus on three protein families — tristetraprolin (TTP; encoded by ZFP36), roquin 1 and roquin 2 (encoded by RC3H1 and RC3H2, respectively), and monocyte chemotactic protein-induced protein 1 (MCPIP1; also known as regnase 1 and encoded by ZC3H12A) — which provide an important layer of regulation of both innate and adaptive immune responses by targeting mRNA for degradation and through the modulation of signalling pathways.
CCCH zinc finger proteins
We previously identified 58 CCCH zinc finger proteins in mice and 56 CCCH zinc finger proteins in humans, through genome-wide surveys5. Based on updated information in GenBank, we identify here three new CCCH zinc finger proteins: CNOT4, HELZ2 and PAN3. In addition, two previously identified cDNAs that potentially encoded CCCH zinc finger proteins (BC003883 and BC019429) have now been deleted in the database. Thus, there are currently 59 and 57 CCCH zinc finger proteins in mice and humans, respectively (TABLE 1). Zfp36l3 is a rodent-specific gene6 and does not exist in humans. Human ZC3H21 is a pseudogene and not listed in TABLE 1. Furthermore, a recent genomic analysis revealed 68 and 67 CCCH zinc finger proteins in Arabidopsis thaliana and rice, respectively7, suggesting that CCCH zinc finger proteins are evolutionarily conserved.
Most CCCH zinc finger proteins with known functions act as regulators of RNA metabolism, including mRNA splicing, polyadenylation, export, translation and decay, and a number of CCCH zinc finger proteins also act as transcriptional repressors or signalling modulators (Supplementary information S2 (figure)). The molecular functions of 18 human CCCH zinc finger proteins have not yet been determined, and the biological functions of many CCCH zinc finger proteins remain unknown. However, most of the characterized CCCH zinc finger proteins have been shown to be crucial regulators of immune responses, through the regulation of cytokine production, immune cell activation, immune homeostasis and antiviral responses.
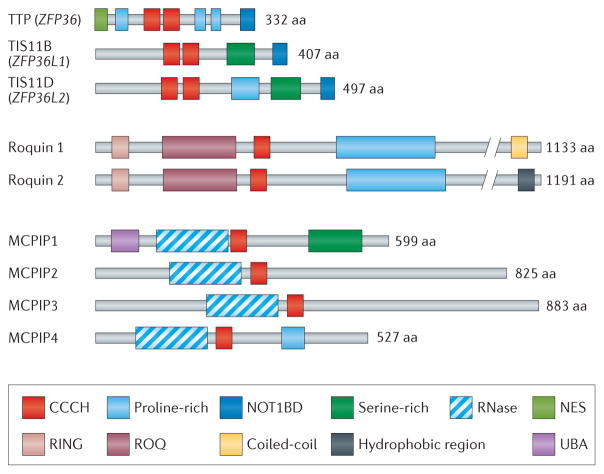
Emerging evidence suggests that several CCCH zinc finger proteins, such as TTP, roquin 1 and MCPIP1, constitute a novel regulatory network that promotes the resolution of inflammation, controls the magnitude and duration of adaptive immune responses, and maintains immune homeostasis by targeting mRNA decay and modulation of signalling pathways8–11. TTP is the best-studied member of a small family of three proteins in humans that is characterized by a specific tandem CCCH zinc finger domain (FIG. 1). TTP was initially discovered as a gene that could be rapidly and transiently induced by the stimulation of fibroblasts with growth factors and mitogens12. It is now known that this protein can bind to AU-rich elements (AREs) in mRNA, leading to the removal of the poly(A) tail from the mRNA and increased rates of mRNA decay13. In addition to the tandem CCCH zinc finger domains, TTP contains three proline-rich domains and a conserved carboxy-terminal sequence that can bind the NOT1 scaffolding protein. A nuclear export sequence (NES) is located at the amino terminus of TTP (FIG. 1). The other two human family members, encoded by ZFP36L1 and ZFP36L2, share very similar tandem CCCH zinc finger domains, as well as the putative C-terminal NOT1 binding sequences (FIG. 1).
Figure 1. Schematic structures of human tristetraprolin (TTP), roquin and monocyte chemotactic protein-induced protein (MCPIP) protein families.
The protein domains are presented as boxes as indicated; NOT1-binding domain (NOT1BD) has been identified by a recent study105. NES, nuclear export sequence; RNase, ribonuclease domain; UBA, ubiquitin-associated domain.
Roquin 1 was identified through an N-ethyl-N-nitrosourea (ENU)-induced mutagenesis screen in mice9. In addition to a single CCCH zinc finger domain, roquin 1 contains a RING finger domain, a ROQ domain and a proline-rich domain (FIG. 1). Roquin 1 recognizes stem–loop motifs in the 3′ untranslated region (UTR) of its target mRNAs, through its ROQ domain and adjacent CCCH zinc finger domain, and promotes mRNA decay by recruiting the helicase RCK (also known as DDX6) and enhancer of mRNA-decapping protein 4 (EDC4)14. A second family member, roquin 2, shares a similar structure and has some overlapping functions with roquin 1 (FIG. 1).
MCPIP1 was identified as a novel protein containing a CCCH zinc finger domain and a PIN domain-like RNase domain; it also contains an ubiquitin-associated domain at its N terminus and a serine-rich domain at its C terminus (FIG. 1). MCPIP1 targets mRNA degradation via its intrinsic RNase activity11. Three other related proteins (termed MCPIP2, MCPIP3 and MCPIP4) share similar domain structures with MCPIP1 (REF. 10) (FIG. 1). Interestingly, phylogenetic analysis5 showed that TTP, roquin 1 and MCPIP1 are evolutionarily closely related.
CCCH zinc finger proteins share many common features: the first is that each protein contains one or more CCCH zinc fingers, which defines this superfamily. Second, although other types of zinc fingers are well known as DNA-binding motifs, CCCH zinc fingers more commonly bind RNA by recognizing specific sequences or secondary structures in their mRNA targets4. For example, the tandem CCCH zinc finger motifs of the TTP protein family recognize specific ARE elements such as UUAUUUAUU in the 3′ UTR of their target mRNA13, which is further confirmed by structural analysis15. Of note, a single CCCH zinc finger can bind only weakly (albeit specifically) to AU-rich RNA16, suggesting that proteins that contain a single CCCH zinc finger domain may bind RNA as dimers. Biochemical analysis indicates that both the ROQ domain and the adjacent CCCH zinc finger of roquin 1 contribute to the recognition of RNA17. However, structural analysis suggests that the ROQ domain of roquin 1 is sufficient for binding to the conserved decay element (CDE) in the 3′ UTR of tumour necrosis factor (Tnf) mRNA18,19. Although the CCCH zinc finger domain of MCPIP1 has been shown to play a part in the control of inter-leukin-6 (Il6) mRNA decay11 (see later), the functional contributions of the CCCH zinc fingers in roquin 1 and MCPIP1 are not fully understood. Why CCCH zinc finger domains preferentially bind to RNA over DNA is also not well understood.
A third common feature of CCCH zinc finger proteins is that they commonly shuttle between different cellular compartments, such as from the nucleus to the cytoplasm, and between different RNA compartments, such as polysomes, stress granules and P-bodies, where they are thought to perform their various roles in RNA metabolism (BOX 1). A fourth common feature of this protein superfamily is that its members commonly interact with microRNA (miRNA) processing and effector pathways (BOX 2). Last, in addition to CCCH zinc finger domains, these family members contain other important functional domains. For example, MCPIP1 contains an RNase domain, and the roquin family proteins contain RING-type E3 ubiquitin ligase domains (FIG. 1). These domains greatly contribute to the functional variety within this protein superfamily.
Box 1. CCCH zinc finger proteins in RNA compartments.
Eukaryotic mRNAs are in dynamic equilibrium between different subcellular locations: actively translated mRNAs can be found in polysomes; mRNAs stalled in translation initiation can accumulate in stress granules; and mRNAs targeted for degradation or translational repression can accumulate in P-bodies97. During energy deprivation and oxidative stress, the CCCH zinc finger protein tristetraprolin (TTP) is recruited to stress granules82 — a process that is inhibited by phosphorylation of TTP by the mitogen- activated protein (MAP) kinase p38. TTP also localizes to P-bodies. Overexpression of TTP can increase the contacts of P-bodies with stress granules, which may allow for the transit of stalled mRNAs into the decay compartment that is P-bodies. Moreover, TTP and ZFP36L1 can nucleate P-body formation by moving the mRNAs with AU-rich elements (AREs) into P-bodies98,99.
Similar to TTP, roquin 1 can also localize in P-bodies. However, under stress conditions, roquin 1 can be completely re-localized to stress granules14. The ROQ domain of roquin 1 is necessary and sufficient for its localization to stress granules and for inducing the formation of these structures upon its overexpression, and is required to trigger Icos mRNA decay17.
Overexpression of monocyte chemotactic protein-induced protein 1 (MCPIP1) in cells usually results in the formation of granule-like structures in the cytoplasm. Most MCPIP1-containing granules are adjacent to or in contact with P-bodies; however, only small portions of MCPIP1-containing granules actually overlap with the P-bodies100. The identity of MCPIP1 containing granules is still controversial. We recently found that MCPIP1-containing granules overlapped with GW182 and argonaute 2 (AGO2), which are molecular markers of GW-bodies, suggesting that MCPIP1 may functionally interact with microRNA effector pathways38. However, a recent study indicated that MCPIP1 was localized on ribosomes on endoplasmic reticulum32. It is also possible that MCPIP1 protein is localized in both positions and exerts its specific roles. Nevertheless, further studies are required to define the identity of MCPIP1-containing granules, as this is important for understanding the molecular mechanisms of its action.
Interestingly, MCPIP1 expression completely blocks the formation of stress granules under different stress conditions100, through which it may protect macrophages from stress-induced apoptosis. These studies also indicate that MCPIP1 functions differently to TTP and roquin 1 in the formation of stress granules. However, the physiological implications of this difference remain unclear. Other CCCH zinc finger proteins, such as ZAP, have also been shown to transiently localize to stress granules and P bodies101. Taken together, as RNA granules serve as key modulators of post-transcriptional and epigenetic gene expression, the dynamic localization and nucleation of these CCCH zinc finger proteins in RNA granules may have important roles in the regulation of immune responses.
Box 2. Interplay of CCCH zinc finger proteins with miRNAs.
MicroRNAs (miRNAs) are a class of non-coding RNAs that modulate gene expression at the post transcriptional level and are involved in regulating many aspects of the immune response. It is well established that AU rich elements (AREs) in the 3′ untranslated region (UTR) of tumour necrosis factor (Tnf) mRNA dictates its degradation by tristetraprolin (TTP) binding. Interestingly, dicer 1, argonaute 1 (AGO1) and AGO2, which are molecules involved in miRNA processing and effector pathways, are required for the rapid decay of TnfmRNA102, suggesting the potential interaction of TTP with miRNA pathways. Interestingly, miR-16, a human miRNA containing a UAAAUAUU sequence that is complementary to the ARE sequence, is required for the decay of ARE containing RNA, a process that also depends on the ARE binding protein TTP. Moreover, TTP does not directly bind to miR-16 but interacts with argonaute family members to form a complex with miR-16 and assists in the ARE-mediated mRNA decay102.
Roquin 1 was recently identified as a key factor to regulate miRNA homeostasis. Roquin 1 can decrease the half-life of mature miRNA by 50%, probably through increasing their mono-uridylation103. A loss of functional roquin 1 leads to accumulation of miR 146a in T cells, which also targets inducible T cell co stimulator (Icos) mRNA for degradation by binding to its complementary site that is different to roquin 1-binding site. These studies suggest that roquin 1 and miR-146a function in the same pathway. Loss of roquin 1 leads to increased levels of miR-146a, which may compensate for the role of roquin 1 in regulating Icos mRNA.
Moreover, roquin 1 interacts with AGO2, the core component of miRNA-induced silencing complex (miRISC), suggesting that roquin 1 may be involved in the miRNA effector pathway103. Similarly, monocyte chemotactic protein-induced protein 1 (MCPIP1) interacts with GW182, another core component of miRISC38. However, the functional significance of the interaction of these CCCH zinc finger proteins with miRISC is still an open question. One report also indicates that MCPIP1 counteracts dicer-mediated miRNA processing and suppresses miRNA biosynthesis via cleavage of the terminal loops of pre-miRNAs104. The implications of these studies in immune regulation need to be further explored.
Regulation of cytokine production
TNF
Cytokines are crucial mediators of inflammation and immune responses and their production is regulated at multiple levels. Although transcription is an essential first step, full regulation of cytokine production also involves many additional post-transcriptional checkpoints. These occur at the levels of mRNA splicing, polyadenylation, degradation and translation20. It is well established that many mRNAs that encode cytokines have short half-lives and are subject to ARE-mediated decay. For example, degradation of Tnf mRNA is crucial for restricting TNF production, and involves AREs in the 3′ UTR of Tnf mRNA. Indeed, specific deletion of AREs from endogenous Tnf transcripts results in more stable Tnf mRNA and hypersecretion of TNF21. The ARE-binding protein TTP is crucially involved in the regulation of TNF production by binding directly to AREs in the Tnf 3′ UTR and promoting TNF decay through recruitment of the CCR4–CAF1–NOT1 deadenylase complex and the 4EHP–GYF2 cap-binding complex22,23 (FIG. 2). Interestingly, TTP expression is induced by TNF signalling and by many of the same agents that stimulate TNF production. Thus, TTP acts as one component of a negative feedback loop that controls TNF production by destabilizing its mRNA8.
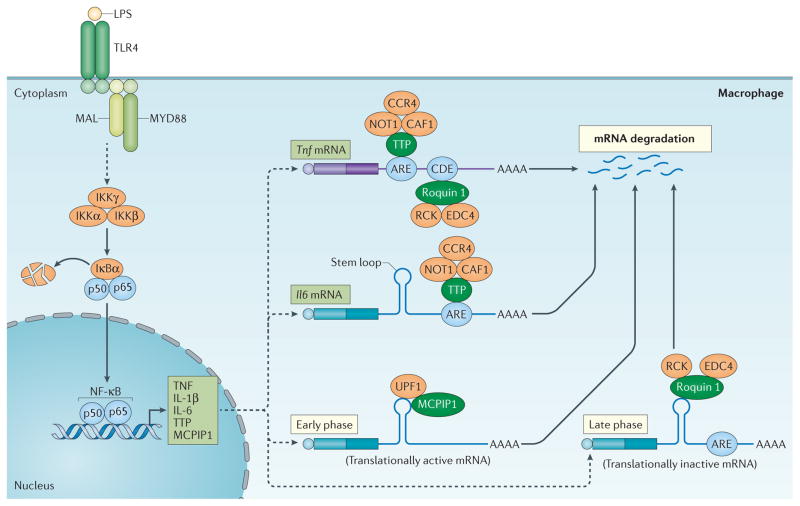
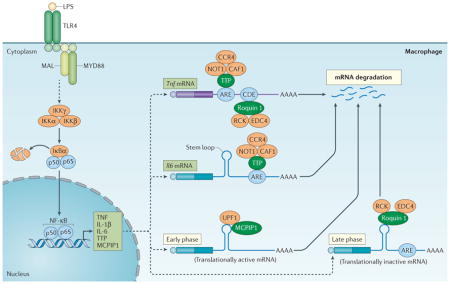
Figure 2. Regulation of cytokine production by CCCH zinc finger proteins.
Upon activation of Toll-like receptor (TLR4) by lipopolysaccharide (LPS), signalling results in activation of the inhibitor of nuclear factor-κB (NF-κB) kinase (IKK) complex (that is, IKKγ, IKKα and IKKβ) and the phosphorylation and degradation of IκBα. Upon release, NF-κB translocates into the nucleus and activates the expression of genes encoding tumour necrosis factor (TNF), interleukin-1β (IL-1β) and IL-6, as well as tristetraprolin (TTP) and monocyte chemotactic protein-induced protein 1 (MCPIP1). TTP can bind to the AU-rich elements (AREs) on the 3′ untranslated region (UTR) of Tnf and Il6 mRNAs and promote their decay, at least in part, by recruiting the CCR4–CAF1–NOT1 deadenylase complex. There is a second layer of regulation of Tnf mRNAs, involving the binding of roquin 1 to constitutive decay element (CDE), which promotes mRNA degradation through the recruitment of RCK and enhancer of mRNA-decapping protein 4 (EDC4). MCPIP1 and roquin 1 also bind to the stem–loop structures of the conserved elements in the 3′ UTR of Il6 mRNA and respectively promote its degradation in the early phase of the inflammatory response through the recruitment of UPF1 and in the late phase through the recruitment RCK and EDC4. MAL, MYD88 adaptor-like; MYD88, myeloid differentiation primary response gene 88; Ub, ubiquitylation.
TTP-deficient mice develop a complex syndrome of inflammatory arthritis, dermatitis, cachexia, autoimmunity and myeloid hyperplasia24. These phenotypes are very similar to those seen in Tnf-transgenic mice25 and mice lacking Tnf AREs21. In addition, treatment of young TTP-deficient mice with TNF blocking antibodies or crossing TTP-deficient mice with mice lacking TNF receptor 1 can prevent the development of most aspects of this syndrome24,26. Together, these studies suggest that TTP is essential for the normal control of TNF production.
Post-transcriptional control through AREs is often influenced by non-ARE sequences in the same transcripts. For example, the constitutive decay element (CDE) located downstream of the ARE in the 3′ UTR of Tnf mRNA has been proposed to prevent the pathological expression of TNF in conditions in which the ARE is inactive27. Indeed, roquin 1 has been shown to have a role in the regulation of Tnf mRNA decay via binding to the CDE28. Roquin 1 binds to the CDE through its unique ROQ domain; after doing so, it recruits the helicase RCK and decapping enzyme EDC4 for RNA degradation28. The coexistence of multiple regulatory RNA elements in a single mRNA ensures that several RNA-binding proteins can work together to regulate the expression levels of important transcripts, such as those encoding TNF. Both ARE-mediated control by TTP and CDE-mediated control by roquin 1 are of crucial importance for controlling the production of TNF. This conclusion is further supported by in vivo evidence that roquin 1san/san mice developed TNF-driven inflammation and arthritis that was comparable to the disease that develops in TTP-deficient mice29. Together, these studies reveal a complex interplay of two CCCH zinc finger proteins in the regulation of TNF production (FIG. 2). Given that TNF is the most potent pro-inflammatory cytokine in mammals, and that increased levels of TNF often have detrimental consequences, including the development of septic shock and chronic inflammatory diseases such as rheumatoid arthritis, these pathways may be novel targets for anti-inflammatory treatments for rheumatoid arthritis and other inflammatory conditions.
IL-6
IL-6 is a multifunctional pro-inflammatory cytokine that has important roles in a variety of diseases30, and its expression is tightly controlled at both the transcriptional and post-transcriptional levels. There are five ARE sites in the 3′ UTR of mouse Il6 mRNA. It has been reported that TTP can promote Il6 mRNA degradation by binding to ARE2, ARE3 and ARE4 (REF. 31) (FIG. 2). Moreover, IL-6 production was significantly elevated after injection of TTP-deficient mice with IL-1β, indicating that TTP directly regulated IL-6 production31.
Recent studies revealed that MCPIP1 is also a crucial regulator of IL-6 production by macrophages11. MCPIP1 binds to and cleaves a conserved stem–loop element in the 3′ UTR of Il6 mRNA via its endonuclease activity and the helicase activity of UPF1 (REFS 11,32). As a result, MCPIP1-deficent mice have significantly higher levels of IL-6 than normal mice, and Il6 mRNA is more stable in MCPIP1-deficient macrophages compared with wild-type macrophages11. Interestingly, a recent study showed that roquin 1 and MCPIP1 regulate an overlapping set of mRNAs, including Il6 mRNA, via their recognition of a common stem–loop structure32. However, MCPIP1 and roquin 1 promote the decay of inflammatory mRNAs at different phases of the inflammatory response: MCPIP1 controls the early phase of inflammation via the cleavage and degradation of translationally active mRNAs and requires the helicase activity of UPF1 (FIG. 2). By contrast, roquin 1 controls the later phase of inflammation by removing translationally inactive mRNAs via a mechanism that is independent of UPF1 and involving RCK and EDC4 (REF. 32) (FIG. 2). Taken together, these results suggest that three CCCH zinc finger proteins — MCPIP1, roquin 1 and TTP — act together to control IL-6 production through different elements in the 3′ UTR of its mRNA. Further studies are needed to understand how these various mechanisms are integrated and regulated to determine the level of cytokine production by innate immune cells.
IL-10 and other cytokines
The range of TTP-targeted mRNAs includes not only those encoding pro- inflammatory cytokines, but also the anti-inflammatory cytokine IL-10 (REF. 33), a cytokine important for the resolution of inflammation. IL-10 inhibits acute and chronic inflammation by suppressing pro-inflammatory cytokine production by activated macrophages. Il10 mRNA was identified as a target of TTP through genome-wide analysis, and TTP enhances Il10 mRNA degradation through binding to AREs in its 3′ UTR33. Furthermore, IL-10 was found to induce TTP expression in macrophages by activating signal transducer and activator of transcription 3 (STAT3), suggesting that IL-10-mediated TTP induction is part of a negative feedback loop that controls the production of this cytokine34. The discovery that TTP promotes the decay of Il10 mRNA suggests that post-transcriptional control mechanisms can regulate both the initiation and the resolution of inflammatory responses.
Intensive studies have demonstrated that the feedback control mediated by these CCCH zinc finger proteins can also be applied to other cytokines, chemokines and pro-inflammatory molecules, such as IL-16, IL-8, IL-22, IL-23, CCL3, IFNγ, CXCL1, GM-CSF, iNOS and VEGF35. Tiedje et al. recently identified numerous mRNA targets of TTP, such as Ler3, Dusp1 and Tnfaip3, which are feedback inhibitors of nuclear factor-κB (NF-κB) activation, suggesting a new role for TTP in the regulation of the NF-κB signalling pathway36. In addition, Sedlyarov et al.37 have recently shown an increased influence of TTP-dependent mRNA decay on the expression profile of inflammatory cytokines at the transition to the resolution phase of the inflammatory response, which suggests that TTP-mediated mRNA decay directly controls the switch from the inflammatory to the resolution phase of the immune response in macrophages37.
In addition to the three CCCH zinc finger proteins discussed above, other CCCH zinc finger proteins participate in the regulation of cytokine expression. For example, MCPIP4 (also known as TFL and p34 and encoded by ZC3H12D) is involved in the regulation of Il6, Il1b, Tnf and Il2 mRNA decay through the targeting of their 3′ UTR38–40. The consequences of this regulation could be seen in MCPIP4-deficient mice, which produced elevated levels of IL-2, IL-6, TNF and IL-17A40. In studies of experimental autoimmune encephalitis, Mcpip4−/− mice contained a higher proportion of T helper 17 (TH17) cells than did wild-type mice during the resolution phase. These results suggest that MCPIP4 may have an important role in attenuating local inflammation by suppressing the infiltration of TH17 cells and may contribute to recovery from T cell-mediated autoimmune diseases40.
Taken together, these studies suggest that mRNA stability is determined by the integration of multiple regulatory processes that have competing, additive or synergistic effects. These regulatory pathways are used to coordinate the expression of pro-inflammatory and anti-inflammatory cytokines to tune immune and inflammatory responses in a timely and efficient manner.
Regulation of immune cell activation
In addition to regulating immune responses through the control of cytokine production, CCCH zinc finger proteins can regulate immune cell activation via intrinsic pathways.
Macrophage activation
Macrophages are centrally involved in both innate and adaptive immunity. They express pattern recognition receptors (PRRs) that sense components of bacteria, viruses and parasites, as well as endogenous components. For example, lipopolyaccharide (LPS), a component of the outer membrane of Gram-negative bacteria, is recognized by Toll-like receptor 4 (TLR4) expressed on the surface of macrophages. Binding of LPS to TLR4 triggers signalling cascades leading to the activation of the transcription factors NF-κB and AP-1, which results in the expression of genes encoding cytokines and other molecules41 (FIG. 3).
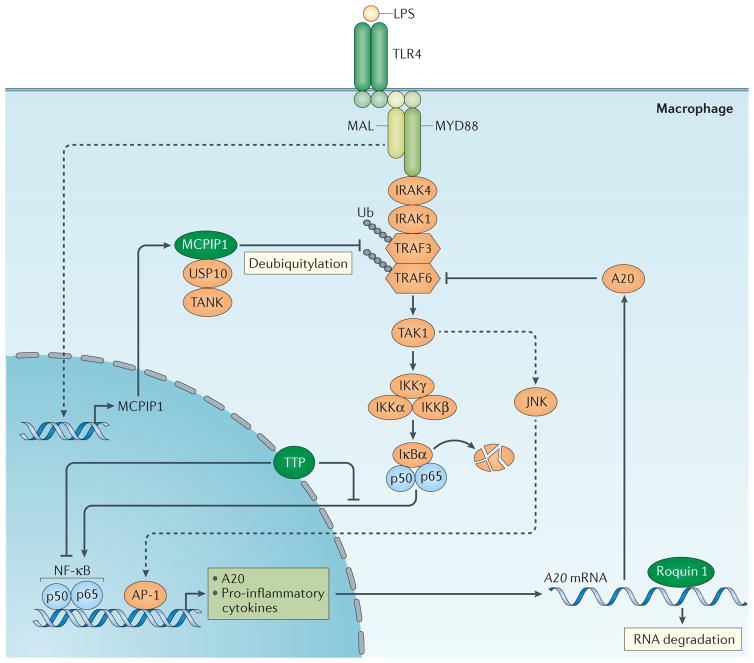
Figure 3. Regulation of macrophage activation by CCCH zinc finger proteins.
Macrophages are activated in response to environmental cues, and the activated cell surface receptors (exemplified by Toll-like receptor 4 (TLR4) in the figure) will initiate signalling, which ultimately transduces into the nucleus and activates gene transcription. The newly synthesized mRNAs are exported from the nucleus to the cytoplasm, where they can be translated into proteins or be subject to degradation. Tristetraprolin (TTP) may suppress signal transduction or transcription of pro-inflammatory cytokines by acting as a co-repressor of nuclear factor-κB (NF-κB) or by blocking NF-κB translocation into the nucleus. Monocyte chemotactic protein-induced protein 1 (MCPIP1) can act as an adaptor molecule to recruit the deubiquitinase USP10, which targets TNF-receptor-associated factor 6 (TRAF6) via TANK and promotes the deubiquitylation of TRAF6 to inhibit signal transduction. By contrast, roquin 1 can promote A20 mRNA degradation, which is a negative regulator of NF-κB signalling, thereby removing a negative feedback loop and facilitating macrophage activation. IKK, inhibitor of NF-κB kinase; IRAK, interleukin-1 receptor-associated kinase; LPS, lipopolysaccharide; MAL, MYD88 adaptor-like; MYD88, myeloid differentiation primary response gene 88.
During macrophage activation, MCPIP1 mRNA is induced by TLR ligands and pro-inflammatory cytokines such as TNF, IL-1β and CCL2 (REFS 10,11,42). Expression of MCPIP1, in turn, negatively regulates macrophage activation by multiple mechanisms (FIG. 3). As discussed above, MCPIP1 acts as an RNase to promote the degradation of a subset of mRNAs encoding pro-inflammatory cytokines, including IL-1β, IL-6 and IL-12p40, by directly targeting their 3′ UTRs11,43. In addition, several studies suggest that MCPIP1 may act as an adaptor molecule that recruits other proteins, such as TANK and USP10, to form a complex that inhibits NF-βB and the mitogen-activated protein kinase JNK by promoting the deubiquitylation of TNF-receptor-associated factor 3 (TRAF3) and TRAF6 (REFS 44–46).
There are some controversial reports on the regulation of NF-κB signalling by CCCH zinc finger proteins. An early study suggested that MCPIP1 did not regulate NF-κB signalling, as macrophages isolated from Mcpip1−/− mice did not show hyperactivation of NF-κB in response to LPS stimulation11. One possible explanation for this discrepancy is that some other CCCH zinc finger proteins may compensate for the effect of MCPIP1 deficiency on NF-κB activation. TTP was also reported to negatively regulate NF-κB signalling by acting as a co-repressor or by interfering with the nuclear translocation of p65 (REFS 47–49) (FIG. 3). Taken together, these studies suggest that these CCCH zinc finger proteins are multifunctional proteins. In controlling macrophage activation, they primarily act at a post-transcriptional level to promote mRNA decay of the inflammatory cytokines, but they may also attenuate the transcription of the inflammatory cytokines by inhibiting the signal transduction pathways that induce their expression (FIG. 3). The significance of these regulatory mechanisms can be seen from the phenotypes of the deficient mice. For example, mice specifically lacking TTP in myeloid cells are extremely sensitive to LPS-induced septic shock owing to a heightened production of TNF50. Mice lacking MCPIP1 spontaneously developed a systemic inflammatory syndrome with massive infiltration of inflammatory cells into many organs, especially the lungs and liver51. Furthermore, MCPIP1-deficient mice were extremely sensitive to LPS-induced septic shock due to overproduction of TNF52. It is noteworthy that TANK-deficient mice and MCPIP1-deficient mice share similar phenotypes, including splenomegaly, heightened production of inflammatory cytokines and early death53, which further supports the hypothesis that these factors may be mediating their effects through the same pathway.
Although TTP and MCPIP1 act as negative regulators of NF-κB signalling, a recent study suggests that roquin 1 promotes the activity of the inhibitor of NF-κB kinase (IKK)–NF-κB pathway by promoting mRNA degradation of A20 (REF. 54). A20 is an ubiquitin-editing enzyme that inhibits the NF-κB signalling pathway55. Roquin 1 uses its ROQ and CCCH zinc finger domains to contact a non-CDE-type stem–loop structure that is preceded by an ARE in the 3′ UTR of A20 mRNA and promotes its degradation54. These studies highlight the importance of post-transcriptional regulation of gene expression to control crucial cellular signal transduction pathways (FIG. 3).
In addition, the CCCH zinc finger proteins ZC3H13 and ZC3H18 were reported to positively regulate the NF-κB pathway. ZC3H13 was found to have an important role in the transcription of NF-κB-induced latent membrane protein 1 (LMP1), TNF and IL-1β, but not for IKK activation, whereas ZC3H18 was crucial for IKK activation56. However, the mechanisms and physiological significance of ZC3H13 and ZC3H18 in the regulation of NF-κB-mediating signalling pathways need to be further investigated.
T cell activation
The failure of mechanisms that control T cell activation and function results in autoimmune diseases, which are characterized by the generation of autoantibodies and systemic inflammatory injury. As MCPIP1, TTP and roquin 1 are highly expressed by T cells, their function in T cell activation is of interest. MCPIP1-deficient mice showed severe systemic inflammation characterized by T cell and B cell hyper-activation, hyperimmunoglobulinaemia and production of autoantibodies11.
Mice with a T cell-specific deletion of Mcpip1 have a similar phenotype to that of Mcpip1-global knockout mice, suggesting that MCPIP1 is crucial for preventing autoimmunity in a T cell-intrinsic manner57. The molecular mechanisms by which MCPIP1 negatively regulates T cell activation may involve the decay of a set of mRNAs encoding immunoregulatory molecules, such as inducible T cell co-stimulator (ICOS), REL, OX40, IL-2 and IL-2Rβ57. REL is a major component of the NF-κB pathway and has been shown to have an essential role in TH1 cell activation associated with autoimmune disease58. The development of autoimmune disease in Mcpip1−/− mice was partially inhibited by a loss of REL, suggesting that increased REL expression, due to defective mRNA degradation, contributes to the activation of T cells and subsequently B cells in Mcpip1−/− mice57.
As antigen recognition is required for T cell activation, the source (or sources) of antigens responsible for activating T cells in MCPIP1-deficient mice remain to be determined. The indigenous microbiota of mucosal surfaces, particularly the intestinal micro-flora, is thought to account for a substantial proportion of the antigenic stimuli for innate and adaptive immune cells59. Treatment of MCPIP1-deficient mice with broad-spectrum antibiotics significantly decreased the inflammatory phenotype and increased the lifespan of these mice, suggesting that microflora antigens contribute to T cell activation in the absence of MCPIP1 (REF. 51). In addition, adoptive transfer of activated T cells from MCPIP1-deficient mice into normal animals results in the generation of autoantibodies, suggesting that B cell activation in MCPIP1-deficient mice might be secondary to T cell activation57. Nevertheless, the intrinsic role of MCPIP1 in B cells warrants further investigation. Together, these studies suggest that MCPIP1 not only regulates the activation of macrophages but also controls T cell activation by targeting mRNA degradation.
In addition to mRNA decay, mRNA splicing is also crucial for suppressing aberrant T cell activation. For example, CD45 is a transmembrane tyrosine phosphatase that is expressed by all nucleated haematopoietic cells. CD45 is essential for T cell activation as it removes an inhibitory phosphate group from kinases, such as SRC and LCK, and thereby allows T cell receptor (TCR) signalling60. However, expression of the spliced isoform of CD45 that lacks exon 4, 5 and 6 (CD45RO) has been shown to limit T cell activation61. During T cell activation, there is a shift in expression of CD45 isoforms from the full protein to the CD45RO isoform. This activation-induced alternative splicing may be part of a negative feedback regulatory loop to prevent aberrant TCR signalling. The CCCH zinc finger protein U2AF1L4 (also known as U2AF26) functions as a regulator of mRNA splicing and has been shown to cooperate with the zinc finger protein GFI1 in determining the splicing of CD45. U2AF1L4 expression facilitates the formation of the less-active CD45RO isoform, and by which it negatively regulates T cell activation61.
In response to different antigens, activated T cells differentiate into different effector T cells such as TH1, TH2 and TH17 cells. A recent study suggests that roquin 1 and MCPIP1 are crucial factors for controlling TH17 cell differentiation by cooperatively repressing a set of common target genes encoding TH17 cell-promoting factors such as IL-6, ICOS, REL, IRF4, IκBNS and IκBζ62. Mice with a combined deficiency of roquin 1 and roquin 2 specifically in T cells and their precursors developed severe TH17 cell-mediated lung inflammation and gastritis62. Similar hyperinflammatory phenotypes were also observed in MCPIP1-deficient mice11,51,63. Furthermore, a recent study indicates that MCPIP1 negatively regulates IL-17 signalling and inflammation via suppressing the expression of IL-17 target genes and inducing decay of mRNA transcripts encoding IL-17 receptor subunits64. Although several studies reported that MCPIP1 was induced by IL-17 (REFS 65, 66), these studies suggest that these CCCH zinc finger proteins act as feedback inhibitors of IL-17 signalling.
In summary, these studies reveal that the regulation of mRNA metabolism by CCCH zinc finger proteins is crucial for suppressing aberrant activation of both innate and adaptive immune cells. The CCCH zinc finger proteins may control the immune cell activation by multiple mechanisms: inducing the decay of mRNA transcripts encoding immunoregulatory molecules, signalling transducers and inflammatory targets; recruiting other inhibitors to suppress crucial signalling pathways; and/or facilitating the formation of specific splicing isoforms that limit T cell activation. There are still many other questions waiting for answers. For example, the roles of these CCCH zinc finger proteins in the activation of other immune cells such as dendritic cells, natural killer cells and B cells remains to be determined. Further understanding of these post-transcriptional pathways may provide novel therapeutic targets for treatment of autoimmune diseases and inflammatory conditions.
Regulation of immune homeostasis
The regulation of costimulatory signalling in CD4+ T cells is crucial for maintaining peripheral T cell tolerance and immune homeostasis. Failure to control costimulatory signalling may break immunological tolerance and contribute to the development of autoimmune diseases such as systemic lupus erythematosus (SLE). The expression of ICOS, which is a member of the CD28 superfamily of costimulatory molecules, is upregulated by T cells upon activation. ICOS signalling is required for the development and proliferation of T follicular helper (TFH) cells67. TFH cells promote the maturation of B cells into memory B cells or antibody-secreting plasma cells in germinal centres68. Dysfunctional TFH cells can lead to the aberrant selection of autoreactive germinal centre B cells and the production of autoantibodies that cause diseases such as SLE69. Roquin 1 is a crucial factor that controls Icos mRNA levels and maintains activation-induced expression of ICOS by CD4+ T cells70. Recognition of the 3′ UTR of this mRNA by the RNA-binding domain of roquin 1 facilitates the degradation of the transcript through interactions with the decapping enzyme EDC4 and the helicase RCK14.
Roquin 1san/san (also called sanroque), a single point mutation in the ROQ domain of the gene encoding roquin 1, leads to a lupus-like autoimmune phenotype in mice, which is marked by enhanced numbers of TFH cells and spontaneous germinal centre formation9. These mice had high-level ICOS expression. Partial correction of ICOS overexpression in roquin 1san/sanIcos+/− mice was accompanied by a reduction in lymphadenopathy, splenomegaly, TFH cell population expansion and germinal centre B cell numbers, suggesting that ICOS overexpression is an essential contributor to the lupus phenotype in roquin 1san/san mice70. Interestingly, unlike roquin 1san/san mice, the cell-specific ablation of roquin 1 in T cells or B cells did not cause autoimmunity71, and similarly to roquin 1, T cell-specific deletion of roquin 2 does not affect immune cell homeostasis72. However, mice deficient in both roquin 1 and roquin 2 developed an autoimmune phenotype with increased TFH cell numbers and germinal centre expansion, which phenotypically resembles roquin 1san/san mice72. These results suggest redundant functions of roquin 1 and roquin 2 in regulating ICOS expression in TFH cells (FIG. 4).
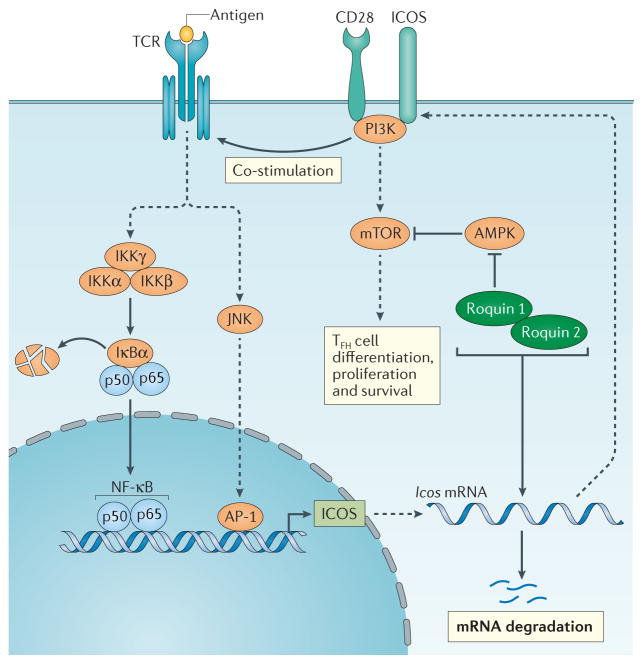
Figure 4. Roquin 1 and roquin 2 regulate TFH cell differentiation.
Inducible T cell co stimulator (ICOS) is upregulated by T cells upon activation. ICOS signaling is required for T follicular helper (TFH) cell differentiation and proliferation. Roquin 1 and roquin 2 redundantly promote Icos mRNA degradation by recruiting the RNA decay proteins enhancer of mRNA-decapping protein 4 (EDC4) and RCK through their ROQ domain and CCCH zinc finger domains (not shown), thereby negatively regulating TFH cell differentiation. However, the RING domain of roquin 1 protein promotes TFH cell differentiation by directly interacting with the catalytic unit of adenosine monophosphate-activated kinase (AMPK) and suppressing AMPK activity, by which it enhances mechanistic target of rapamycin (mTOR) signalling and increases TFH cell differentiation, proliferation and survival. IKK, inhibitor of NF-κB kinase; NF-κB, nuclear factor-κB; PI3K, phosphatidylinositol 3 kinase; TCR, T cell receptor.
The paradoxical observations between the autoimmune-prone roquin 1san/san mice and the healthy roquin 1-deficient mice are not fully understood. A recent elegant study showed that a RING-less roquin 1-mutant protein failed to localize to stress granules and resulted in a compensatory role for roquin 2 in the repression of ICOS and TFH cells29, thus roquin 1RINGless mice did not show any phenotypes of autoimmunity29. By contrast, in roquin 1san/san mice, the ROQ-mutant roquin 1 still localized to mRNA-regulating stress granules and prevented a compensationary response by roquin 2; thus roquin 1san/san mice showed severe phenotypes of autoimmunity29. Nevertheless, more investigation into this paradoxical observation is required for a full understanding.
In view of the important role of RING domains in driving protein ubiquitylation, determining whether roquin 1 and roquin 2 contain E3 ubiquitin ligase activity is crucial for understanding their complicated roles. Roquin 2 was identified as an E3 ubiquitin ligase required for reactive oxygen species (ROS)-induced ubiquitylation and degradation of apoptosis signal- regulating kinase 1 (ASK1; also known as MAP3K5)73. As evident from research using Caenorhabditis elegans, mutation of the gene encoding the roquin 2 orthologue RLE1 results in the accumulation of the activated form of the ASK1 orthologue NSY1, which conferred resistance to Pseudomonas aeruginosa infection73. Moreover, Zhang et al. recently demonstrated that both roquin 1 and roquin 2 are functional E3 ubiquitin ligases that drive the assembly of polyubiquitin chains of different linkages74. Interestingly, a recent study showed that the RING domain of roquin 1 directly binds to the catalytic a1 subunit of adenosine monophosphate-activated kinase (AMPK) and represses its enzymatic activity75. As AMPK is a central regulator of cellular metabolism in response to cellular stress and an inhibitor of mechanistic target of rapamycin (mTOR) signalling, roquin 1 may positively regulate TFH cell formation by inhibition of AMPK activity (FIG. 4). Although this surprising observation75 on the role of roquin 1 on TFH cell formation needs more investigation, these results suggest that roquin 1 may fine-tune the regulation of TFH cell differentiation through two different domains (RING domain and ROQ domain) and two different mechanisms (inhibition of AMPK and degradation of ICOS) (FIG. 4).
Taken together, these studies revealed the importance of roquin 1- and roquin 2-mediated post-transcriptional networks in TFH cell differentiation and the maintenance of immune homeostasis. Investigations into the mechanisms through which roquin proteins and MCPIP1 repress their target mRNA molecules have highlighted the complex and intricate feedback loops these RNA-binding proteins use to augment or control adaptive immune responses.
Regulation of the regulators
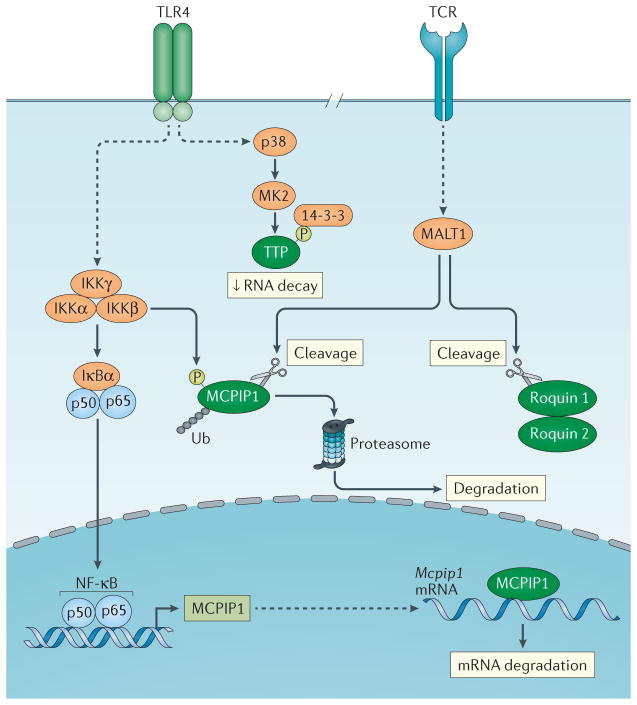
The expression and function of CCCH zinc finger proteins are precisely regulated by multiple mechanisms, which add another layer of control on the regulation of innate and adaptive immune responses (FIG. 5). For example, Mcpip1 mRNA is induced by TLR ligands and inflammatory cytokines through the NF-κB signalling pathway76. However, during LPS- and IL-1β-induced macrophage activation, MCPIP1 protein is rapidly degraded by the ubiquitin–proteasome system, which is dependent on IKK-mediated phosphorylation77 (FIG. 5). Consistent with this observation, MG132, which is a potent proteasome inhibitor, has been shown to significantly increase MCPIP1 protein levels in HepG2 or HeLa cells78. In addition, MCPIP1 protein can target the 3′ UTR of its own mRNA and promote its degradation77. In activated T cells, MCPIP1 is regulated through TCR-induced MALT1 (mucosa-associated lymphoid tissue lymphoma translocation 1)-mediated cleavage. MALT1 is an arginine-specific protease that cleaves MCPIP1 at arginine 111; the resulting fragments are rapidly degraded by other proteases57. Similarly to MCPIP1, both roquin 1 and roquin 2 are also cleaved by MALT1 upon T cell activation62 (FIG. 5).
Figure 5. Expression and function of CCCH zinc finger proteins are regulated by multiple mechanisms.
Toll-like receptor 4 (TLR4) signalling results in the activation of the inhibitor of nuclear factor-κB (NF-κB) kinase (IKK) complex and NF-κB. Then NF-κB translocates into the nucleus and activates the expression of monocyte chemotactic protein-induced protein 1 (Mcpip1) mRNA. MCPIP1 can bind its own mRNA and promote its own mRNA degradation. Moreover, the activated IKK complex can phosphorylate MCPIP1, which is followed by ubiquitylation and degradation by proteasomes. T cell receptor (TCR) activation can activate mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), a paracaspase. MCPIP1, roquin 1 and roquin 2 can be cleaved by MALT1, and the fragments are rapidly degraded by the proteasome. Tristetraprolin (TTP) can be phosphorylated by p38–MK2 and be inactivated by the recruitment of 14-3-3 protein.
Importantly, the function of MCPIP1 on mRNA decay is regulated by other RNA-binding proteins. For instance, AT-rich interactive domain-containing protein 5α (ARID5α) can compete with MCPIP1 for binding to the 3′ UTR of Il6 mRNA and protect it from MCPIP1-mediated degradation. Furthermore, ARID5a deficiency in mice attenuates acute inflammation and autoimmunity79. Together, these results suggest that both the expression and function of MCPIP1 are dynamically regulated during the course of an immune response: MCPIP1 expression is suppressed by TLR signalling during the early phase of an infection, allowing the immune system to be activated and fully respond to the microbial challenge, whereas, in the later phases of the response, MCPIP1 expression is restored by increased transcription76 and helps to promote the resolution of inflammation by promoting the degradation of inflammatory cytokines and immune molecules.
Similarly to MCPIP1, the transcription of Zfp36 is also regulated by NF-κB signalling in LPS-stimulated macrophages80. TTP can also promote the degradation of its own mRNA by binding to the ARE on its 3′ UTR81. Phosphorylation of TTP by p38–MK2 has been reported to promote its sequestration by 14-3-3 proteins, resulting in diminished mRNA decay activity82 (FIG. 5). Additionally, phosphorylated TTP may be impaired in its ability to recruit mRNA decay enzymes, resulting in decreased deadenylation of target transcripts82–84. In addition, the function of TTP in the regulation of cytokine production may be antagonized by another RNA-binding protein. For example, as an ARE-binding protein, HuR can compete with TTP for ARE-binding sites on the Il6 3′ UTR, thereby stabilizing Il6 mRNAs85. TTP can also be ubiquitylated by the adaptor molecule TRAF2 in a phosphorylation-dependent manner86,87. Ubiquitylated TTP no longer affects NF-κB activity but promotes the activation of JNK86. A further study suggests that TTP degradation by proteasomes is ubiquitin independent but instead is dependent on intrinsically disordered N- and C-termini of the protein88. These studies suggest that the expression and function of TTP are regulated by multiple mechanisms.
Taken together, these studies suggest that the expression and function of TTP, roquin 1 and MCPIP1 can be governed by the same signalling pathways that control immune cell activation in response to TCR, TLR and cytokine receptor signalling. It is likely that some of CCCH zinc finger proteins may act as a point of convergence for the activity of numerous kinases, including IKK, ERK1, ERK2, JNK and p38 MAP kinase. For example, TTP can be modified by different kinases in response to different environmental cues, and the different modified states of TTP have different roles in the regulation of target decay or inhibition of signalling pathways. Precise regulation of these regulators provides further layers of control to fine tune immune responses (FIG. 5).
Associated diseases and therapeutic potential
Although emerging evidence from studies of genetically engineered animals indicates that TTP, roquin 1 and MCPIP1 are crucial regulators of inflammation and immune homeostasis, mutations of these genes in humans have not been clearly implicated in disease development. Suzuki et al.89 reported that a single nucleotide polymorphism (SNP) within the human ZFP36 promoter resulted in twofold greater promoter activity. These authors reported that patients with rheumatoid arthritis with the GG genotype might be prone to higher disease activity than those with the AG or AA genotypes. This suggests that this SNP mildly affects ZFP36 promoter activity, and may thus influence the activity of rheumatoid arthritis and perhaps other inflammatory diseases89. Another polymorphism in the protein coding domain of ZFP36 has also been shown to be significantly associated with rheumatoid arthritis in African–Americans90. Associations between human disease and mutations of RC3H1 (encoding roquin 1) and MCPIP1 have not been the subject of published investigations but certainly warrant future study.
Given their importance in immune regulation and RNA metabolism, CCCH zinc finger proteins are possible targets for the treatment of autoimmunity, viral infections and cancer, although the effects of targeting CCCH zinc finger proteins on an organismal level are currently unknown. Based on their complex function, both increasing and decreasing CCCH zinc finger protein expression and activity could have therapeutic benefits for specific indications. For instance, increasing MCPIP1 expression, by the selective suppression of degradative enzymatic activity within the cells, such as that exhibited by MALT1 and proteasomes, could potentially improve outcomes in diseases in which excessive inflammatory responses are considered detrimental, such as septic shock, atherosclerosis and certain viral infections. MI-2, a specific inhibitor of MALT1, was recently reported to selectively enhance MCPIP1 expression in CD4+ T cells and consequently promote apoptosis of T cell lines that were latently infected with HIV-1 in the presence of cell stimuli in vitro91. Given that MI-2 is a highly specific and non-toxic small molecular inhibitor of MALT1 (REF. 92), it would be interesting to explore its therapeutic potential for inflammatory diseases and viral infections in vivo. By contrast, suppressing MCPIP1 and roquin 1 activity may be a novel strategy for enhancing cancer immunotherapy or responses to vaccines. Recent studies with a mouse model of regulated TTP overexpression demonstrated a protective effect against several models of immune and inflammatory disease, supporting the potential role of TTP as a therapeutic target93,94.
Conclusions and perspectives
Regulation of RNA metabolism is an important component of gene expression that facilitates the fine-tuning of transcript levels during physiological conditions and during the rapid and profound switch in global gene expression associated with inflammation and immune responses. Long-term dysregulation of RNA metabolism can often result in disease states, including inflammatory and autoimmune diseases95. CCCH zinc finger proteins have emerged as important regulators of multiple facets of RNA metabolism and immune responses, with promising therapeutic potential. During the innate immune response, several CCCH zinc finger proteins, such as TTP, roquin 1 and MCPIP1, promote the resolution of inflammation by helping to eliminate the mRNAs of certain pro-inflammatory cytokines. During the activation of B cells and T cells, these CCCH zinc finger proteins intrinsically control the magnitude and duration of adaptive immune responses and maintain immune homeostasis via multiple mechanisms. Failure of these important regulatory controls (for example, in mice that are genetically deficient for TTP, roquin 1 or MCPIP1) often leads to the development of systemic inflammatory responses and autoimmune syndromes. In addition to the regulation of immune responses, other CCCH zinc finger proteins such as ZAP (encoded by ZC3HAV1), PARP12 (encoded by ZC3HDC1) and TOE1 are important regulators of viral replication (Supplementary information S3 (box)).
In the future, searching for associations between mutations in the genes for CCCH zinc finger proteins and various inflammatory disorders and cancer should help to assess the biological roles of members of this protein superfamily in humans. Furthermore, elucidating how these CCCH zinc finger proteins are regulated and exploiting this knowledge for therapeutic application, by enhancing or repressing their expression and/or function, is a crucial next step in understanding these multifaceted proteins. At a more basic level, it would be extremely interesting to determine how cross-regulation of two different domains occurs in a single protein. For example, does the E3 ubiquitin ligase activity of roquin 1 RING domain influence the RNA decay process mediated by its ROQ domain and CCCH zinc finger? Alternatively, does ROQ and CCCH zinc finger-mediated RNA binding affect its RING domain- mediated ubiquitylation function? Based on structural studies, it seems that the RING domain of roquin 1 can interact with its CCCH zinc finger domain and affect its RNA-binding activity74. A recent study also showed that the N-terminal domain of MCPIP1 was associated with its PIN domain and significantly enhanced its RNase activity96. Lastly, as most CCCH zinc finger proteins have not been well-characterized, further studies on the function and mechanisms of these less- characterized proteins will provide valuable information for understanding this fascinating group of proteins.
Supplementary Material
Acknowledgments
The authors thank C. J. Papasian and V. Heissmeyer for critical reading and comments on the manuscript. This work was supported by a US National Institutes of Health Grant (AI103618) and a University of Missouri Research Board Award (to M.F.) and by the Intramural Research Program of the National Institute of Environmental Health Sciences, US National Institutes of Health (to P.J.B.).
Glossary
- RNA metabolism
Refers to any events in the life cycle of RNA molecules, including their synthesis, folding and unfolding, modification, processing and degradation.
- Zinc finger
A finger-shaped fold in a protein that permits it to interact with DNA and RNA. The fold is created by the binding of specific amino acids in the protein to a zinc atom.
- AU-rich elements
(AREs). Found in the 3′ untranslated region (3′ UTR) of many mRNAs that encode proto-oncogenes, nuclear transcription factors and cytokines. AREs are one of the most common determinants of RNA stability in mammalian cells.
- RING finger domain
RING (really interesting new gene) finger domain is a protein structural domain of zinc finger type that contains a C3HC4 amino acid motif and binds two zinc cations. Many proteins containing a RING finger domain have a key role in the ubiquitylation pathway.
- Polysomes
Polysomes (or polyribosomes) are a cluster of ribosomes that are attached along the length of a single molecule of mRNA. Polysomes read this mRNA simultaneously, helping to synthesize the same protein at different spots on the mRNA.
- Stress granules
Dense aggregations in the cytosol composed of proteins and RNA molecules that appear when the cell is under stress. The RNA molecules stored in these granules are stalled translation pre-initiation complexes.
- P-bodies
Cytoplasmic domains that contain proteins involved in diverse post-transcriptional processes, such as mRNA degradation, nonsense- mediated mRNA decay, translational repression and RNA-mediated gene silencing.
- MicroRNA
(miRNA). A small, RNA molecule that regulates the expression of genes by binding to the 3′ untranslated region of specific mRNAs.
- Roquin 1san/san mice
Mice with a single point mutation (M199R) in the ROQ domain of the gene encoding roquin 1. These mice develop a lupus-like autoimmune phenotype, marked by enhanced numbers of T follicular helper cells and spontaneous germinal centre formation.
- MALT1
(Mucosa-associated lymphoid tissue lymphoma translocation protein 1). A protein of the paracaspase family that shows proteolytic activity. Since many of its substrates are involved in the regulation of inflammatory responses, the protease activity of MALT1 has emerged as an interesting therapeutic target.
- 14-3-3
A family of proteins that functions as adaptor molecules in protein interactions and can regulate protein localization and enzyme activity.
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
GenBank: https://www.ncbi.nlm.nih.gov/genbank/
See online article: S1 (table) | S2 (figure) | S3 (box)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol. 2014;15:492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- 2.Man K, Kallies A. Synchronizing transcriptional control of T cell metabolism and function. Nat Rev Immunol. 2015;15:574–584. doi: 10.1038/nri3874. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 4.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. This study showed that there are 56 and 58 CCCH zinc finger proteins in humans and mice, respectively, through genome-wide surveys. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingerich TJ, et al. Emergence and evolution of Zfp36l3. Mol Phylogenet Evol. 2016;94:518–530. doi: 10.1016/j.ympev.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, et al. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics. 2008;9:44. doi: 10.1186/1471-2164-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. This report is the first to identify TTP as a key component of a negative feedback loop that controls TNF production through a post-transcriptional mechanism. [DOI] [PubMed] [Google Scholar]
- 9.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. This report is the first to identify roquin 1 as a crucial regulator of ICOS expression, TFH cell differentiation and autoimmunity. [DOI] [PubMed] [Google Scholar]
- 10.Liang J, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. This report is the first to identify MCPIP1 as a negative regulator of macrophage inflammatory activation. [DOI] [PubMed] [Google Scholar]
- 11.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. These authors are the first to report that Zc3h12a acts as an endonuclease to selectively control the expression of a set of genes by promoting their mRNA decay. [DOI] [PubMed] [Google Scholar]
- 12.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 13.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 14.Glasmacher E, et al. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- 15.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 16.Michel SL, Guerrerio AL, Berg JM. Selective RNA binding by a single CCCH zinc-binding domain from Nup457 (Tristetraprolin) Biochemistry. 2003;42:4626–4630. doi: 10.1021/bi034073h. [DOI] [PubMed] [Google Scholar]
- 17.Athanasopoulos V, et al. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 2010;277:2019–2127. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
- 18.Schlundt A, et al. Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation. Nat Struct Mol Biol. 2014;21:671–678. doi: 10.1038/nsmb.2855. [DOI] [PubMed] [Google Scholar]
- 19.Tan D, Zhou M, Kiledjian M, Tong L. The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat Struct Mol Biol. 2014;21:679–685. doi: 10.1038/nsmb.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 21.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 22.Lai WS, et al. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. This study shows that CCCH zinc finger motifs of TTP are RNA-binding domains, through which TTP promotes mRNA degradation by facilitating deadenylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu R, Olsen MT, Webb K, Bennett EJ, Lykke-Andersen J. Recruitment of the 4EHP–GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016;22:373–382. doi: 10.1261/rna.054833.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor GA, et al. A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. This study is the first to establish that TTP is a key regulator of Tnf mRNA degradation through the characterization of TTP-deficient mice. [DOI] [PubMed] [Google Scholar]
- 25.Probert L, et al. Dissection of the pathologies induced by transmembrane and wild-type tumor necrosis factor in transgenic mice. J Leukoc Biol. 1996;59:518–525. doi: 10.1002/jlb.59.4.518. [DOI] [PubMed] [Google Scholar]
- 26.Carballo E, Blackshear PJ. Roles of tumor necrosis factor-α receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98:2389–2395. doi: 10.1182/blood.v98.8.2389. [DOI] [PubMed] [Google Scholar]
- 27.Stoecklin G, Lu M, Rattenbacher B, Moroni C. Aconstitutive decay element promotes tumor necrosis factor α mRNA degradation via an AU-rich element-independent pathway. Mol Cell Biol. 2003;23:3506–3515. doi: 10.1128/MCB.23.10.3506-3515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leppek K, et al. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881. doi: 10.1016/j.cell.2013.04.016. The paper shows that roquin 1 binds a conserved stem–loop structure in the 3′ UTR of Tnf mRNA and promotes mRNA decay. [DOI] [PubMed] [Google Scholar]
- 29.Pratama A, et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Nishimoto N. Interleukin-6 as a therapeutic target in candidate inflammatory diseases. Clin Pharmacol Ther. 2010;87:483–487. doi: 10.1038/clpt.2009.313. [DOI] [PubMed] [Google Scholar]
- 31.Zhao W, Liu M, D’Silva NJ, Kirkwood KL. Tristetraprolin regulates interleukin-6 expression through p38 MAPK-dependent affinity changes with mRNA 3′ untranslated region. J Interferon Cytokine Res. 2011;31:629–637. doi: 10.1089/jir.2010.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mino T, et al. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015;161:1058–1073. doi: 10.1016/j.cell.2015.04.029. This paper shows that MCPIP1 and roquin 1 regulate an overlapping set of mRNAs via a common stem–loop structure, but they target the mRNAs at different times during an immune response and in different locations within the cell. [DOI] [PubMed] [Google Scholar]
- 33.Stoecklin G, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaba A, et al. Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. J Immunol. 2012;189:2089–2093. doi: 10.4049/jimmunol.1201126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiedjie C, et al. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 2016;44:7418–7440. doi: 10.1093/nar/gkw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedlyarov V, et al. Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol Syst Biol. 2016;12:868. doi: 10.15252/msb.20156628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, et al. Monocyte chemotactic protein-induced protein 1 and 4 form a complex but act independently in regulation of interleukin-6 mRNA degradation. J Biol Chem. 2015;290:20782–20792. doi: 10.1074/jbc.M114.635870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, et al. ZC3H12D attenuated inflammation responses by reducing mRNA stability of proinflammatory genes. Mol Immunol. 2015;67:206–212. doi: 10.1016/j.molimm.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Minagawa K, et al. Posttranscriptional modulation of cytokine production in T cells for the regulation of excessive inflammation by TFL. J Immunol. 2014;192:1512–1524. doi: 10.4049/jimmunol.1301619. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 42.Zhou L, et al. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res. 2006;98:1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizgalska D, et al. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1β mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 44.Liang J, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J Exp Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu J, et al. USP10 inhibits genotoxic NF-κB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013;32:3206–3219. doi: 10.1038/emboj.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, et al. TRAF family member-associated NF-κB activator (TANK) inhibits genotoxic nuclear factor κB activation by facilitating deubiquitinase USP10-dependent deubiquitination of TRAF6 ligase. J Biol Chem. 2015;290:13372–13385. doi: 10.1074/jbc.M115.643767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang J, et al. RNA-destabilizing factor tristetraprolin negatively regulates NF-κB signaling. J Biol Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-κB/p65 nuclear translocation. J Biol Chem. 2009;284:29571–28581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu L, et al. Suppression of IL-12 production by tristetraprolin through blocking NF-κB nuclear translocation. J Immunol. 2013;191:3922–3930. doi: 10.4049/jimmunol.1300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu LQ, Stumpo DJ, Blackshear PJ. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol. 2012;188:5150–5159. doi: 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao R, et al. Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol. 2013;91:368–376. doi: 10.1038/icb.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang S, et al. MCPIP1 negatively regulates Toll-like receptor 4 signaling and protects mice from LPS-induced septic shock. Cell Signal. 2013;25:1228–1234. doi: 10.1016/j.cellsig.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawagoe T, et al. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat Immunol. 2009;10:965–972. doi: 10.1038/ni.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murakawa Y, et al. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat Commun. 2015;6:7367. doi: 10.1038/ncomms8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gewurz BE, et al. Genome-wide siRNA screen for mediators of NF-κB activation. Proc Natl Acad Sci USA. 2012;109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uehata T, et al. Malt1-induced cleavage of regnase-1 in CD4+ helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. This study shows that MCPIP1 is essential for preventing aberrant T cell activation in a cell-autonomous manner and that the protease activity of MALT1 is crucial for controlling the mRNA stability of T cell effector genes by cleaving MCPIP1. [DOI] [PubMed] [Google Scholar]
- 58.Hilliard BA, et al. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J Clin Invest. 2002;110:843–850. doi: 10.1172/JCI15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saleh M, Elson CO. Experimental inflammatory bowel disease: insights into the host microbiota dialog. Immunity. 2011;34:293–302. doi: 10.1016/j.immuni.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mustelin T, Tasken K. Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem J. 2003;371:15–27. doi: 10.1042/BJ20021637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heyd F, ten Dam G, Möröy T. Auxiliary splice factor U2AF26 and transcription factor Gfi1 cooperate directly in regulating CD45 alternative splicing. Nat Immunol. 2006;7:859–867. doi: 10.1038/ni1361. This study shows that the CCCH zinc finger protein U2AF26 regulates T cell activation by controlling mRNA splicing of the transmembrane tyrosine phosphatase CD45. [DOI] [PubMed] [Google Scholar]
- 62.Jeltsch KM, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote TH17 differentiation. Nat Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. This study shows that roquin 1 and MCPIP1 work together to repress target mRNAs and enhance TH17 cell differentiation. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Z, et al. MCPIP1 deficiency in mice results in severe anemia related to autoimmune mechanisms. PLoS ONE. 2013;8:e82542. doi: 10.1371/journal.pone.0082542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garg AV, et al. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity. 2015;43:475–487. doi: 10.1016/j.immuni.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhamija S, et al. Interleukin-17 (IL-17) and IL-1 activate translation of overlapping sets of mRNAs, including that of the negative regulator of inflammation, MCPIP1. J Biol Chem. 2013;288:19250–19259. doi: 10.1074/jbc.M113.452649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Somma D, et al. CIKS/DDX3X interaction controls the stability of the Zc3h12a mRNA induced by IL-17. J Immunol. 2015;194:3286–3294. doi: 10.4049/jimmunol.1401589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linterman MA, et al. Roquin differentiates the specialized functions of duplicated T cell costimulatory receptor genes CD28 and ICOS. Immunity. 2009;30:228–241. doi: 10.1016/j.immuni.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 68.Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nat Rev Rheumatol. 2012;8:337–347. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol. 2009;9:845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- 70.Yu D, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. The authors show that roquin 1 is essential for the prevention of autoimmunity by limiting Icos mRNA levels. [DOI] [PubMed] [Google Scholar]
- 71.Bertossi A, et al. Loss of Roquin induces early death and immune deregulation but not autoimmunity. J Exp Med. 2011;208:1749–1756. doi: 10.1084/jem.20110578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogel KU, et al. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity. 2013;38:655–668. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Maruyama T, et al. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci Signal. 2014;7:ra8. doi: 10.1126/scisignal.2004822. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Q, et al. New insights into the RNA-binding and E3 ubiquitin ligase activities of Roquins. Sci Rep. 2015;5:15660. doi: 10.1038/srep15660. This study shows that roquin 1 and roquin 2 contain E3 ubiquitin ligase activity and bind with overlapping, but not identical, E2 enzymes to drive the assembly of polyubiquitin chains of different linkages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramiscal RR, et al. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. eLife. 2015;4:e08698. doi: 10.7554/eLife.08698. This study shows that the RING domain of roquin 1 paradoxically promotes TFH cell differentiation by attenuating AMPK signalling, suggesting that roquin 1 may fine-tune the regulation of TFH cell differentiation through two different domains and two different mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skalniak L, et al. Regulatory feedback loop between NF-κB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 77.Iwasaki H, et al. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12:1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 78.Skalniak L, Koj A, Jura J. Proteasome inhibitor MG-132 induces MCPIP1 expression. FEBS J. 2013;280:2665–2674. doi: 10.1111/febs.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masuda K, et al. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci USA. 2013;110:9409–9414. doi: 10.1073/pnas.1307419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen YL, et al. Transcriptional regulation of tristetraprolin by NF-κB signaling in LPS-stimulated macrophages. Mol Biol Rep. 2013;40:2867–2877. doi: 10.1007/s11033-012-2302-8. [DOI] [PubMed] [Google Scholar]
- 81.Brooks SA, Connolly JE, Rigby WF. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J Immunol. 2004;172:7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- 82.Stoecklin G, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marchese FP, et al. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem. 2010;285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi JX, Su X, Xu J, Zhang WY, Shi Y. HuR post-transcriptionally regulates TNF-α-induced IL-6 expression in human pulmonary microvascular endothelial cells mainly via tristetraprolin. Respir Physiol Neurobiol. 2012;181:154–161. doi: 10.1016/j.resp.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 86.Schichl YM, Resch U, Lemberger CE, Stichlberger D, de Martin R. Novel phosphorylation-dependent ubiquitination of tristetraprolin by mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 (MEKK1) and tumor necrosis factor receptor-associated factor 2 (TRAF2) J Biol Chem. 2011;286:38466–38477. doi: 10.1074/jbc.M111.254888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Resch U, et al. Polyubiquitinated tristetraprolin protects from TNF-induced, caspase-mediated apoptosis. J Biol Chem. 2014;289:25088–25100. doi: 10.1074/jbc.M114.563312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ngoc LV, et al. Rapid proteasomal degradation of posttranscriptional regulators of the TIS11/tristetraprolin family is induced by an intrinsically unstructured region independently of ubiquitination. Mol Cell Biol. 2014;34:4315–4328. doi: 10.1128/MCB.00643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suzuki T, et al. Tristetraprolin (TTP) gene polymorphisms in patients with rheumatoid arthritis and healthy individuals. Mod Rheumatol. 2008;18:472–479. doi: 10.1007/s10165-008-0085-5. [DOI] [PubMed] [Google Scholar]
- 90.Carrick DM, et al. Genetic variations in ZFP36 and their possible relationship to autoimmune diseases. J Autoimmun. 2006;26:182–196. doi: 10.1016/j.jaut.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 91.Li H, He H, Gong L, Fu M, Wang TT. Short communication: preferential killing of HIV latently infected CD4+ T cells by MALT1 inhibitor. AIDS Res Hum Retroviruses. 2016;32:174–177. doi: 10.1089/aid.2015.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fontan L, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22:812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Patial S, et al. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc Natl Acad Sci USA. 2016;113:1865–1870. doi: 10.1073/pnas.1519906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patial S, Blackshear PJ. Tristetraprolin (TTP) as a therapeutic target in inflammatory disease. Trends Pharmacol Sci. 2016;37:811–821. doi: 10.1016/j.tips.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rigby RE, Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015;36:179–188. doi: 10.1016/j.it.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yokogawa M, et al. Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions. Sci Rep. 2016;6:22324. doi: 10.1038/srep22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 98.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 100.Qi D, et al. Monocyte chemotactic protein-induced protein 1 (MCPIP1) suppresses stress granule formation and determines apoptosis under stress. J Biol Chem. 2011;286:41692–41700. doi: 10.1074/jbc.M111.276006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goodier JL, et al. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. 2015;11:e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 103.Srivastava M, et al. Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis. Nat Commun. 2015;6:6253. doi: 10.1038/ncomms7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki HI, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 105.Fabian MR, et al. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.