Abstract
Identification of the molecular machinery employed in cancer invasion, but not in normal adult cells, will greatly contribute to cancer therapeutics. Here we found that an ArfGAP, AMAP1/PAG2, is expressed at high levels in highly invasive breast cancer cells, but at very low levels in noninvasive breast cancer cells and normal mammary epithelial cells. siRNA-mediated silencing of AMAP1 effectively blocked the invasive activities. AMAP1 expression in human breast primary tumors also indicated its potential correlation with malignancy. Paxillin and cortactin have been shown to colocalize at invadopodia and play a pivotal role in breast cancer invasion. We found that AMAP1 is also localized at invadopodia, and acts to bridge paxillin and cortactin. This AMAP1-mediated trimeric protein complex was detected only in invasive cancer cells, and blocking this complex formation effectively inhibited their invasive activities in vitro and metastasis in mice. Our results indicate that AMAP1 is a component involved in invasive activities of different breast cancers, and provide new information regarding the possible therapeutic targets for prevention of breast cancer invasion and metastasis.
Keywords: AMAP1, breast cancer, cortactin, invasion and metastasis, paxillin
Introduction
The metastatic potential of carcinomas constitutes a major cause of the poor prognosis of patients and correlates well with the invasive phenotype. In the case of breast carcinomas, it has been widely believed that they may mostly develop via a sequence of pathologically discrete stages, initiating at the premalignant stage of atypical ductal hyperplasia (ADH) to progress into the preinvasive stage of ductal carcinoma in situ (DCIS) and finally into invasive ductal carcinoma (IDC) (Frykberg and Bland, 1994; Allred et al, 2001). However, most of the specific genetic alterations found in IDC are also found in DCIS, and DCIS has been characterized as a genetically far-advanced precursor of IDC, while some alterations, such as c-myc amplification, may be associated specifically with the invasive stage of a population of breast carcinomas (Robanus-Maandag et al, 2003). Analyses of gene expression profiles have also shown that the three pathologically discrete stages are highly similar at the transcriptome level, and there is no clear universal ‘invasive' tumor molecular signature (Ma et al, 2003; Porter et al, 2003). On the other hand, gene expression profiling analyses have shown that primary breast tumors can be subclassified into those with ‘good' or ‘poor' prognosis gene expression signatures (Perou et al, 2000; Sorlie et al, 2001; van't Veer et al, 2002; Kang et al, 2003; Ma et al, 2003). Thus, an adequate answer as to how the invasive phenotype arises in primary breast tumors has not yet been fully provided at the molecular level.
Tumor cells are highly diverse in their ways of invasion (Friedl and Wolf, 2003). Invasive characters are diverse even among the different cell lines of breast tumors (Bowden et al, 1999; Zajchowski et al, 2001). On the other hand, utilization of invasive activities is thought to be strictly restricted to normal epithelial cells in adults, and is only allowed during limited situations, such as epithelial–mesenchymal transdifferentiation in organogenesis and tissue remodeling (Grunert et al, 2003). Thus, identification of the molecular machinery involved in the invasive activities of carcinomas, which may not normally be active in normal epithelia in adults, will greatly contribute toward understanding the nature of cancer invasion and facilitate further development of cancer therapeutics. A direct correlation between in vivo invasive phenotypes and in vitro invasion activities has been demonstrated with a number of different breast cancer cell lines, including MDA-MB-231 (Thompson et al, 1992; Coopman et al, 1998; Bowden et al, 2001). Therefore, well-characterized breast cancer cell lines may provide excellent experimental model systems to study possible mechanisms of cancer invasion at the molecular level. Understanding the basic mechanisms of cancer invasion may also help to understand some aspects of cancer metastasis. Breast cancer is frequently associated with gene amplification at chromosome 11q13, resulting in overexpression of cortactin (Weed and Parsons, 2001; Ormandy et al, 2003). Cortactin was originally identified as a substrate of v-Src kinase, and has been shown to interact with several proteins such as the Arp2/3 complex, dynamin and F-actin, and play pivotal roles in coupling membrane dynamics to cortical actin assembly (Weed and Parsons, 2001). Cortactin has moreover been implicated in the migration, invasion and metastasis of a number of different types of tumor cells, and its amplified gene levels and overexpression also appear to be intimately associated with patients with poor prognosis or relapse (Weed and Parsons, 2001). Bowden et al (1999) have shown that cortactin localizes to invadopodia of MDA-MB-231 cells and microinjection of anti-cortactin antibodies blocks matrix degradation at the invadopodia. They also showed that other proteins, such as paxillin and protein kinase Cμ, also localize to invadopodia. Paxillin is a scaffold adaptor protein with multiple protein interaction modules, and has been shown to be involved in the regulation of Rho family GTPases and cell motility (Schaller, 2001; DeMali et al, 2003). By biochemical analyses, Bowden et al (1999) have moreover suggested that complex formation among cortactin, paxillin and protein kinase Cμ correlates with the invasive activities of different breast cancer cell lines, although these proteins do not directly bind to each other.
We have previously shown that Arf6 is localized at invadopodia and plays an essential role in invasive activities of different breast cancer cells, including MDA-MB-231 (Hashimoto et al, 2004b). We also previously isolated AMAP1/PAG2, a close isoform of another paxillin-binding ArfGAP, AMAP2/PAG3/Papα (Andreev et al, 1999; Kondo et al, 2000), as a paxillin-binding protein (clones #43 and #81; Kondo et al, 2000; Sabe, 2003). AMAP2 exhibits efficient GAP activity toward different Arf isoforms except Arf6 in vitro (Andreev et al, 1999), while AMAP2 overexpression antagonizes the cellular functions of Arf6 (Kondo et al, 2000; Uchida et al, 2001). We have recently shown that the mode of AMAP2 action toward Arf6 is different from those known of other GAPs toward their cognate small G proteins: AMAP2 binds stably to GTP-Arf6 and acts to recruit AMAP2-binding proteins to sites of Arf6 activation (Hashimoto et al, 2004a). We have confirmed that AMAP1 also binds stably to GTP-Arf6 and functions similarly (S Hashimoto and H Sabe, unpublished data, 2003).
In this report, we show that AMAP1 is involved in the invasive activities of different breast cancer cells. We show that high levels of AMAP1 expression and its association with paxillin and cortactin are crucial for the invasive phenotypes, the latter of which is consistent with a previous report by Bowden et al (1999). We use AMAP2 as a negative control when necessary. We performed model experiments in order to suggest that the interfaces involved in the AMAP1-mediated protein complex formation can be considered as molecular targets for drug design aiming at prevention of breast cancer invasion, and also metastasis. We also discuss possible reasons as to why the correlation between the upregulation of AMAP1 expression and the invasive phenotypes of breast carcinomas has not been identified in recent gene expression profiling analyses.
Results
AMAP1 binds to cortactin via its proline-rich sequence
AMAP1 has several protein interaction domains, including the proline-rich domain (PRD) and the src homology 3 (SH3) domain, and paxillin binds to the latter domain (Kondo et al, 2000). Comparison of the primary structures of AMAP1 and AMAP2 revealed that their PRDs are very different (Andreev et al, 1999; see Figure 1A). As a way to explore the function of AMAP1, we conducted yeast two-hybrid screening using the AMAP1 PRD as a bait. cDNAs encoding cortactin, Crk II, CIN85, PACSIN3 and intersectin1, each bearing single or multiple SH3 domains, were then identified. The PRDs of AMAP1 and AMAP2 contain 16 and eight repeats of proline-rich sequences, respectively (see Figure 1B). Using the yeast two-hybrid system, we next tested whether these molecules also bind to the AMAP2 PRD, and found that cortactin and Crk II bind only to the AMAP1 PRD, while others bind to both the AMAP1 and AMAP2 PRDs (data not shown). Binding of Crk II to AMAP1 has already been reported (Brown et al, 1998).
Figure 1.
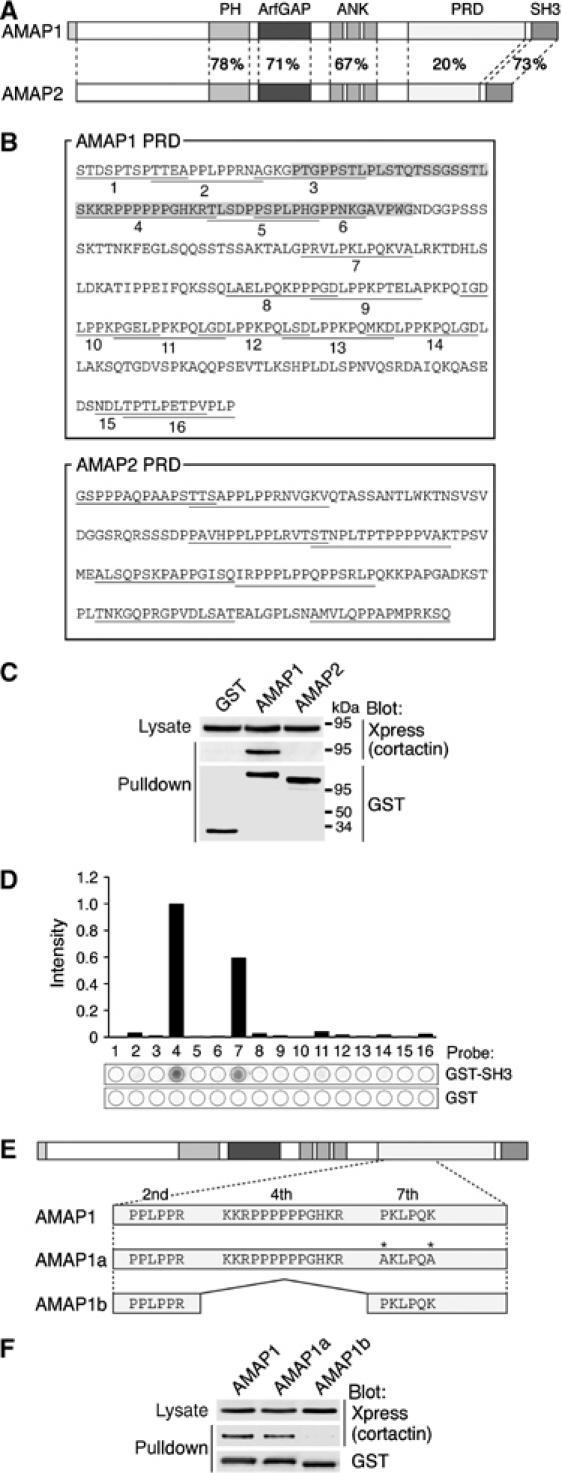
Binding of AMAP1 to cortactin. (A) Schematic representation of AMAP1 and AMAP2. Amino-acid sequence homology between each domain is indicated by percentages. (B) Amino-acid sequences of PRDs of AMAP1 and AMAP2. Each proline-rich sequence of the AMAP1 PRD is underlined and numbered (1st–16th). The shaded box indicates the alternate exon, which AMAP1b lacks. (C) In vivo binding of AMAP1 and cortactin. Xpress-cortactin was coexpressed with GST-AMAP1, GST-AMAP2 or GST alone in COS-7 cells, and GST proteins were pulled down using glutathione beads. After separation by SDS–PAGE, the precipitates were subjected to immunoblotting analysis, as indicated. (D) Biochemical identification of the AMAP1 proline-rich sequence(s) responsible for binding to the cortactin SH3 domain. Each proline-rich sequence of the AMAP1 PRD was synthesized on membranes, and probed with radiolabeled GST-cortactin SH3 or GST alone. Spots 1–16 correspond to the 1st–16th proline-rich sequences, respectively. Radioactivities retained on each spot were expressed as a value with spot 4 normalized as 1.0 (upper panel). (E) Schematic representation of a spliced isoform and a mutant of AMAP1. (F) Requirement of the fourth proline-rich sequence of AMAP1 for its binding to cortactin. Xpress-cortactin, coexpressed with GST-AMAP1, GST-AMAP1a or GST-AMAP1b in COS-7 cells, was pulled down and subjected to immunoblotting analysis, as above.
The intracellular binding of AMAP1 with cortactin was then confirmed by coexpressing GST-tagged AMAP1 with Xpress-tagged cortactin in COS-7 cells and pulling down GST-AMAP1 using glutathione beads (Figure 1C). GST-AMAP2 did not bind to Xpress-cortactin (Figure 1C). We next identified the proline-rich sequence within the AMAP1 PRD, which is primarily responsible for its binding to cortactin. Each proline-rich sequence was synthesized on a filter membrane, and incubated with radiolabeled GST-cortactin SH3 protein. We found that the fourth and seventh proline-rich sequences exhibited significant binding, of which the fourth sequence was more potent (Figure 1D). A natural splice variant of AMAP1, AMAP1b, exists, in which the fourth proline-rich sequence is deleted (Brown et al, 1998; see Figure 1B and E). We also made an AMAP1 mutant, AMAP1a, in which amino acids within and near the canonical SH3-binding Pro-x-x-Pro motif of the seventh proline-rich sequence were mutated (Figure 1E). AMAP1b and AMAP1a were each tagged with GST and coexpressed with Xpress-cortactin in COS-7 cells. As shown in Figure 1F, AMAP1b did not co-precipitate cortactin, while AMAP1a did. Therefore, the fourth proline-rich sequence of the AMAP1 PRD appears to be primarily responsible for the binding of AMAP1 to the cortactin SH3 domain. There are 25 different genes in the human genome found to encode proteins containing the ArfGAP domain (Sabe, 2003). A sequence similar to the fourth proline-rich sequence does not exist among them.
AMAP1 colocalizes with cortactin at the invadopodia of MDA-MB-231 cells
MDA-MB-231 cells produce invadopodia that extend into the matrix substratum, such as collagen and gelatin matrices (Coopman et al, 1996; Bowden et al, 1999, 2001). We next examined the subcellular localization of AMAP1 in MDA-MB-231 cells, cultured on a crosslinked gelatin matrix, by immunolabeling cells with an anti-AMAP1 antibody. We found that AMAP1 proteins were enriched in dot-like structures at the cell bottom, where they colocalized well with cortactin, F-actin and paxillin (Figure 2A, data not shown), markers for invadopodia (Bowden et al, 1999). To visualize the sites of matrix degradation, MDA-MB-231 cells were then cultured on a fluorescently labeled, crosslinked gelatin matrix. These cells actively reform invadopodia, leaving a cumulative record of matrix degradation. AMAP1, together with cortactin and paxillin, was found localized within sites of matrix degradation (Figure 2C and D). On the other hand, AMAP2 was diffusely distributed in the cytoplasm and not localized to invadopodia (Figure 2B and E).
Figure 2.
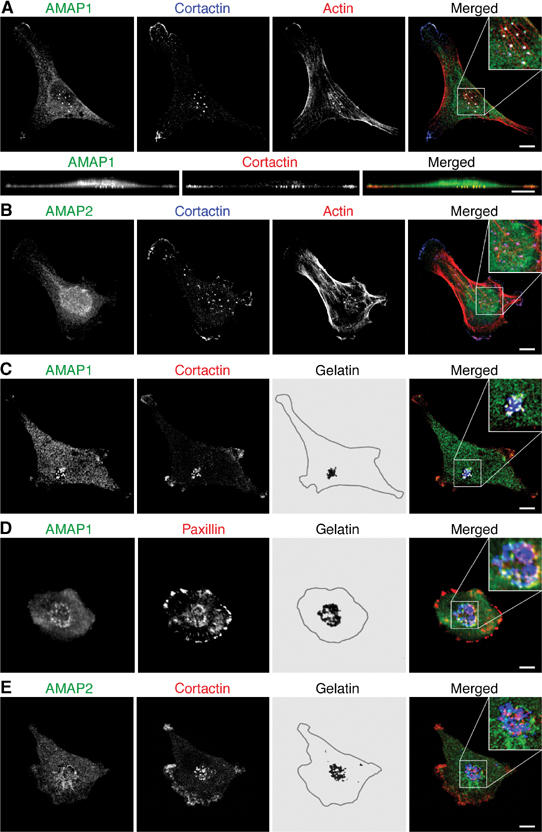
Localization of AMAP1 at invadopodia of MDA-MB-231 cells. Confocal projections of cells stained with anti-AMAP1 (A, C, D), anti-AMAP2 (B, E), anti-cortactin (A–C, E), anti-paxillin (D) and phalloidin (A, B). In (A), z-sections of the images are shown in the lower panels. Cells were cultured for 16 h on a crosslinked gelatin matrix (A, B) or a fluorescently labeled, crosslinked gelatin matrix, in which degraded gelatin zones are shown in black (C–E). In (A–E), representative images are shown from more than 50 cells analyzed. In the merged images, each protein is shown in the color as labeled in the left panels. Degraded gelatin zones in the merged images of (C–E) are indicated in blue. Bars, 10 μm.
AMAP1 is involved in the invasive activities of MDA-MB-231 cells
We next examined whether AMAP1 plays an essential role in invadopodia formation. Treatment of MDA-MB-231 cells by an AMAP1 siRNA duplex suppressed the AMAP1 protein level by about 70% (Figure 3A). In these cells, invadopodia formation and localized matrix degradation were both significantly inhibited as compared to those in control cells (Figure 3C, data not shown). AMAP1 siRNA treatment also effectively inhibited Matrigel chemoinvasion activity, in which cells migrate toward chemoattractants by breaking through a barrier of Matrigel (Figure 3B). In contrast, siRNA-mediated knockdown of AMAP2 did not affect these invasive activities (Figure 3A–C). The specificity of the effects of the AMAP1 siRNA was verified by expressing a rescue construct of wild-type AMAP1 (resWT) in the siRNA-treated cells (Figure 3B–D). We also expressed a deletion mutant lacking the ArfGAP domain (resΔGAP) to confirm the necessity of this domain for AMAP1 function (Figure 3B–D). Expression levels of these rescue and mutant proteins were adjusted to be similar to that of endogenous AMAP1 (data not shown).
Figure 3.
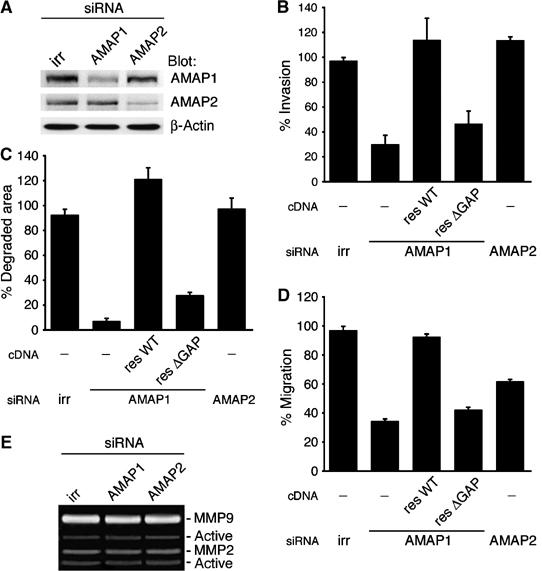
Silencing AMAP1 expression blocks the in vitro invasive activities of MDA-MB-231 cells. Cells were treated with siRNA duplexes against AMAP1or AMAP2, or with an irrelevant sequence (irr). (A) siRNA-mediated knockdown of AMAP1 and AMAP2 protein expression. Lysates prepared from siRNA-treated cells were analyzed by immunoblotting using antibodies as indicated. (B–D) Effects of siRNA-mediated knockdown of AMAP1 and AMAP2 on cellular activities. The percentages of siRNA-treated cells that transmigrated through a barrier of Matrigel toward chemoattractants (Matrigel chemoinvasion assay (B)), total area of gelatin degradation (C) and cells that transmigrated toward collagen in a modified Boyden chamber (haptotactic migration assay (D)) are shown. Data collection and presentation are described in Materials and methods. Results shown are the means±s.e.m. of three experiments. (E) Secretion of MMP2 and MMP9 was assayed in siRNA-treated cells using the gelatin zymography method.
We previously showed that overexpression of AMAP2 blocks haptotactic migration, a type of migration that does not require matrix degradation, of COS-7 cells (Kondo et al, 2000). Haptotactic migration of MDA-MB-231 cells was also significantly inhibited by AMAP1 knockdown and also by AMAP2 knockdown, although the degrees of inhibition differed notably between them (Figure 3D). On the other hand, no significant inhibition of cell adhesion activity onto collagen was observed by the knockdown of AMAP1 or AMAP2 (data not shown). MDA-MB-231 cells produce the matrix metalloproteinases MMP2 and MMP9, which are necessary for matrix degradation (Hashimoto et al, 2004b). The siRNA-mediated knockdown of neither AMAP1 nor AMAP2 blocked the secretion of active MMP2 and MMP9 into the culture media (Figure 3E). This result suggests that secretion of these proteases is not enough for invasion.
Correlation of AMAP1 protein levels with the invasiveness of different human breast cancer cells
We next examined the expression of AMAP1 in different human breast cancer cell lines. We found that other highly invasive cells, such as MDA-MB-435s and Hs578T, express the AMAP1 protein at high levels as in MDA-MB-231 cells, while cells with weakly invasive or noninvasive phenotypes express much lower amounts of the AMAP1 protein (Figure 4A and B). AMAP1 protein expression was also very low in cultured normal mammary epithelial cells (HMEC; Figure 4A and B), which do not exhibit invasive activity under our culture conditions. In contrast, such a correlation was not seen with AMAP2 expression (Figure 4A). On the other hand, the levels of AMAP1 mRNA in these cells do not correlate with the AMAP1 protein levels nor with invasiveness: these different cancer cell lines and normal cells express comparable amounts of AMAP1 mRNA (Figure 4B). Such disproportion between mRNA levels and protein levels was not observed with β-actin (Figure 4C). Thus, AMAP1 protein levels appear not to be simply regulated by its mRNA levels, and some mechanism may exist to suppress AMAP1 protein expression to very low levels in these noninvasive cells.
Figure 4.
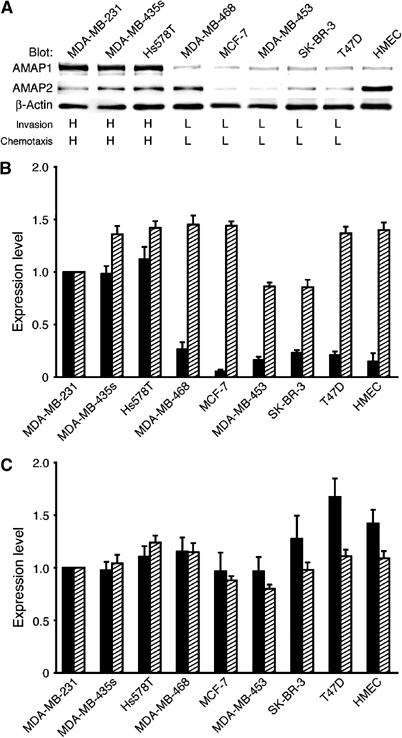
Protein and mRNA expression of AMAP1 in different breast cancer cell lines. (A) Cell lysates were subjected to immunoblotting analysis using antibodies against AMAP1, AMAP2 and β-actin. Invasive activities from the literature (Thompson et al, 1992; Bowden et al, 1999; Zajchowski et al, 2001) are shown in the bottom rows, graded as % MDA-MB-231 cells: L, 0–40%; M, 40–60%; H, >60%. (B, C) Amounts of mRNAs for AMAP1 (B) and β-actin (C), quantified by a real-time PCR amplification method (hatched bars), and their protein levels (closed bars), measured by densitometric scanning of the immunoblots, are shown. Results shown are the means±s.e.m. of three experiments, by normalizing the values obtained for MDA-MB-231 cells as 1.0.
AMAP1 is involved in the invasive activities of different breast cancer cell lines
We then tested whether AMAP1 siRNA treatment can also block the invasive activities of these other highly invasive cancer cell lines. The AMAP1 siRNA duplex effectively suppressed AMAP1 protein expression in these cells, by 60–80% (Figure 5A). In all of these cell lines, effective inhibition of Matrigel chemoinvasion activity was observed upon AMAP1-siRNA treatment, although the degrees of inhibition somewhat differed among them (Figure 5B). Haptotactic migration, but not the adhesion activity onto collagen, was also inhibited, with large differences in the degrees of inhibition among the cell lines (Figure 5C, data not shown). Such differences may be due to different mechanisms employed by these cancer cell lines for their invasion and migration, as mentioned earlier. These results nevertheless suggest that AMAP1 is used in various, if not all, human breast cancers for their invasion. We also found that AMAP1 is highly expressed in a mouse mammary tumor cell line, 4T1/luc, which is also highly invasive. siRNA-mediated suppression of AMAP1 in 4T1/luc cells also effectively inhibited invasion and migration, but not adhesion (Figure 5, data not shown).
Figure 5.
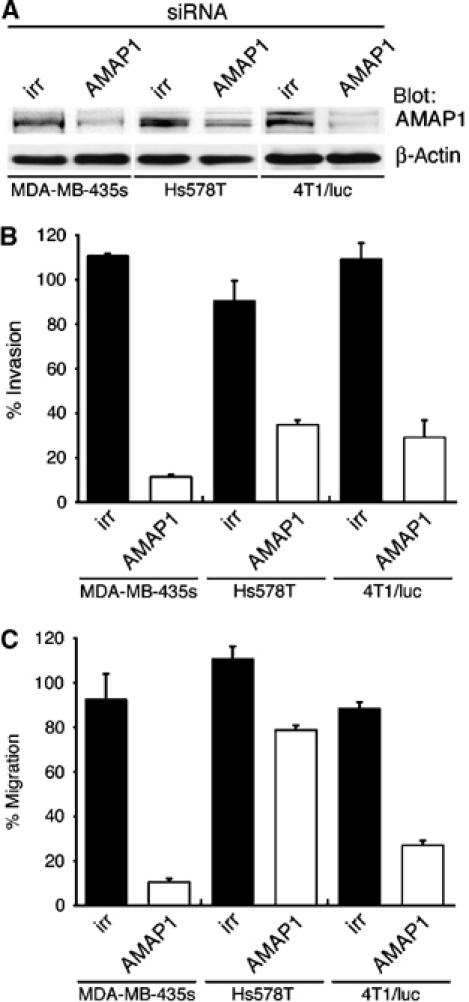
Silencing of AMAP1 expression blocks the in vitro invasive activities of different breast cancer cell lines. Effects of AMAP1 siRNA treatment on AMAP1 protein expression (A), Matrigel chemoinvasion activity (B) and haptotactic migration toward collagen (C) are measured as in Figure 3, with the indicated cancer cell lines. irr, siRNA duplex with an irrelevant sequence. Results shown are the means±s.e.m. of three experiments.
Expression of AMAP1 in primary human breast tumors
We then examined AMAP1 protein expression in primary human breast tumors, by immunohistochemical staining of clinical specimens. We found high levels of AMAP1 expression in specimens pathologically diagnosed as IDC, although the adjacent noncancerous mammary ductal epithelia and tissue components gave rise to very low or background levels of staining (Figure 6A and D). We examined 10 independent cases of IDC, in which patients apparently only had IDC at the time of diagnosis, and found that all of them express AMAP1 at significantly high levels (IDC only in Figure 6E). In contrast, in specimens of patients diagnosed as DCIS (n=7), who did not seem to also have IDC coincidentally, expression of AMAP1 was very low and almost comparable to that in noncancerous cells, although there was one case in our samples that expressed AMAP1 at a high level (Figure 6B and E). We moreover examined specimens of patients who had both IDC and DCIS at the time of the diagnosis (n=9, DCIS+IDC in Figure 6E), and found that in these specimens, expression of AMAP1 is already significantly augmented even at the sites of DCIS (Figure 6C and E).
Figure 6.
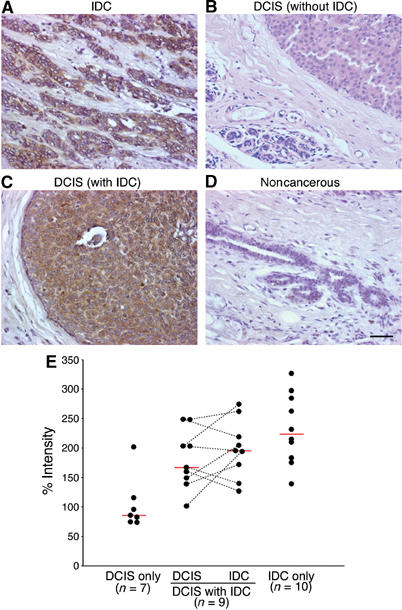
Immunohistochemical staining of AMAP1 in primary tumors of the human breast. Tissue sections were immunostained with an affinity-purified anti-AMAP1 polyclonal antibody. Positive staining for the AMAP1 protein is shown in a reddish-brown color. (A) Breast carcinoma diagnosed as IDC. (B) Breast carcinoma diagnosed as DCIS without the co-occurrence of IDC. (C) Site of a breast carcinoma diagnosed as DCIS, in which the patient simultaneously has IDC. (D) Noncancerous breast components from the same patient as (A). Bar, 50 μm in (A–D). (E) Relative levels of AMAP1 staining in primary tumors plotted into four groups by their tumor types. The percentage staining intensity of cancerous cells relative to the staining of noncancerous cells on the same slides set as 100% is shown. In samples of DCIS+IDC, values from the same patient were connected with dotted lines. Median values were calculated and are presented as red lines.
AMAP1 bridges cortactin and paxillin in highly invasive cells
Since AMAP1 binds to paxillin via its SH3 domain and to cortactin via its PRD, we examined whether AMAP1 acts to bridge these proteins. As shown in Figure 7A, both paxillin and AMAP1 could be co-precipitated with an anti-cortactin antibody. Moreover, consistent with a previous report (Bowden et al, 1999), this co-precipitation was only detected in highly invasive cells, such as MDA-MB-231 and 4T1/luc, but was undetectable in weak or noninvasive cells, such as MDA-MB-468 and MCF7, as well as in normal mammary epithelial cells (Figure 7A). AMAP1 siRNA treatment, and also the predepletion of AMAP1 using AMAP1 antibodies, abolished the co-precipitation of paxillin with cortactin in MDA-MB-231 and 4T1/luc cells (Figure 7B). We also confirmed that AMAP2 was not co-precipitated with cortactin (Figure 7A). Judging from Figure 7A, the percentage of AMAP1 associated with cortactin and paxillin seemed to be no less than 0.3% in MDA-MB-231 and 1.6% in 4T1/luc cells.
Figure 7.
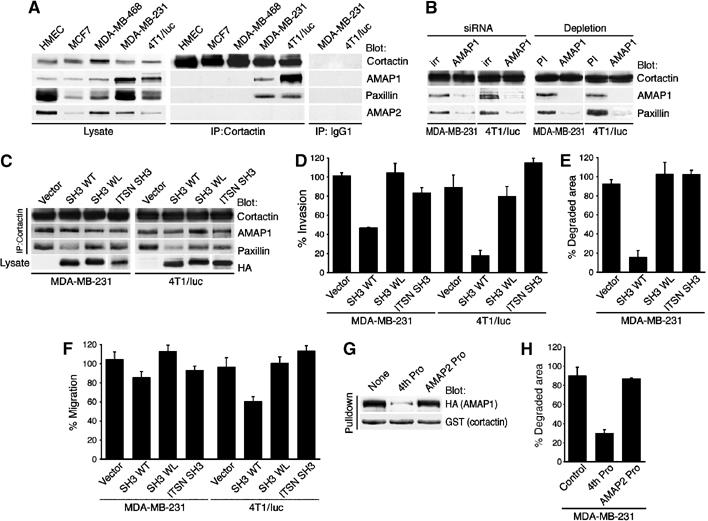
Roles of the AMAP1-mediated protein complex formation with paxillin and cortactin in in vitro invasion activities of MDA-MB-231 and 4T1/luc cells. (A) Assessment of AMAP1-mediated trimeric protein complex formation in breast cancer cell lines and normal mammary epithelial cells (HMEC). Anti-cortactin immunoprecipitates (middle panel) or control nonimmune mouse IgG1 precipitants (right panel) were analyzed for co-precipitation of AMAP1, AMAP2 and paxillin by immunoblotting using antibodies as indicated (middle panel). Protein levels in total cell lysates are shown in the left panel. (B) Requirement of AMAP1 for the co-precipitation of paxillin and cortactin. AMAP1 was knocked down by its siRNA (left panel) or predepleted from the lysates using anti-AMAP1 antibodies (right panel). irr, siRNA with an irrelevant sequence; PI, control preimmune rabbit IgG. Lysates were then analyzed as above. (C–F) Effects of overexpression of the AMAP1 SH3 domain on AMAP1-mediated trimeric protein complex formation (C), Matrigel chemoinvasion activity (D), total area of gelatin degradation (E) and haptotactic migration activity toward collagen (F). MDA-MB-231 cells and 4T1/luc cells were transfected with pcDNA3HA/AMAP1SH3 encoding the wild-type AMAP1 SH3 domain (SH3 WT), pcDNA3HA/AMAP1SH3WL encoding its WL mutant (SH3 WL), pcDNA3HA/ITSN SH3 encoding the intersectin SH3 domains (ITSN SH3) or the empty vector (vector). Overexpression of the AMAP1 SH3 domain reduced the amount of paxillin co-precipitated with anti-cortactin by 68.6±6.2% in MDA-MB-231 cells and by 77.5±2.8% in 4T1/luc cells as compared to that in vector control cells, calculated from three independent experiments. (G, H) Effects of the fourth proline-rich peptide of the AMAP1 PRD. GST-cortactin purified on glutathione beads was incubated with COS-7 cell lysates expressing HA-AMAP1 in the absence (none) or presence of the fourth proline-rich peptide (4th pro) or the control AMAP2 proline-rich peptide (AMAP2 pro), and the amount of HA-AMAP1 co-precipitated with GST-cortactin was analyzed by anti-HA immunoblotting (G). MDA-MB-231 cells microinjected with these peptides were analyzed for the total area of degradation (H). In (D–F, H), results shown are the means±s.e.m. of three independent experiments.
Inhibition of breast cancer cell invasion by the inhibition of AMAP1-mediated protein complex formation
We next validated the role of AMAP1-mediated trimeric protein complex formation in breast cancer invasion. The AMAP1 SH3 domain is primarily responsible for the binding of AMAP1 to paxillin. We transiently overexpressed this domain in both MDA-MB-231 and 4T1/luc cells by cDNA transfection. This overexpression reduced the amount of paxillin co-precipitated with cortactin by about 70% in both cell types, without affecting the amounts of AMAP1 co-precipitated with cortactin (Figure 7C). It also effectively blocked Matrigel chemoinvasion activity, invadopodia formation and localized matrix degradation in both cell lines, while it did not affect their adhesion to collagen (Figure 7D and E, data not shown). Slight inhibition of haptotactic cell migration toward collagen was also observed (Figure 7F). We used intersectin SH3 domains (ITSN SH3) and a mutant of the AMAP1 SH3 domain (SH3WL), whose Trp1107 was mutated into leucine in order to diminish its binding to paxillin, as negative controls (Figure 7C–F).
To inhibit protein complex formation between AMAP1 and cortactin, we used the fourth proline-rich sequence of the AMAP1 PRD (4th Pro). In vitro analysis using GST-cortactin and HA-AMAP1 showed that the presence of 0.1 mM of this peptide blocked their binding by about 90% (Figure 7G). As a control, we used the peptide AMVLQPPAPMPRKSQ, derived from the AMAP2 PRD (AMAP2 Pro), which did not block the binding of cortactin and AMAP1 (Figure 7G). MDA-MB-231 cells form robust invadopodia in which the area of matrix degradation can be easily quantified (Bowden et al, 2001). The ability of the AMAP1 peptide to inhibit invasive activity was then assessed in MDA-MB-231 cells by peptide microinjection. About 70% inhibition of matrix degradation was observed, while no significant blockage was seen with the control peptide (Figure 7H).
Trails to inhibit breast cancer metastases
Most human breast carcinomas arise from the mammary ductal epithelia, and it is believed that they mostly initiate metastasis by invading the extracellular matrices. We finally examined whether inhibition of AMAP1-mediated protein complex formation can also inhibit metastatic activities. In this regard, 4T1/luc cells have the experimental advantage of being able to be injected into syngeneic mice of the wild-type Balb/c strain, while human cells have to be injected into immunodeficient mice. We established 4T1/luc cell clones stably overexpressing the AMAP1 SH3 domain (SH3-1, SH3-2 and SH3-3; Figure 8A), and confirmed that complex formation between AMAP1 and paxillin, and Matrigel chemoinvasion activity are largely inhibited in these cell clones (Figure 8A and B). These cells were then injected into the mammary fat pad of adult female Balb/c mice, and assessed for their metastases into the lung by measuring luciferase activity. We found that metastatic activities were severely suppressed in these SH3-overexpressing cells as compared to that of the vector control cells (Figure 8C). Similar overexpression of the WL mutant of the AMAP1 SH3 domain did not exert such blockage (Figure 8B and C). Tumor growth at the original sites of injection was much the same among these SH3-expressing cells and control cells (Figure 8D).
Figure 8.
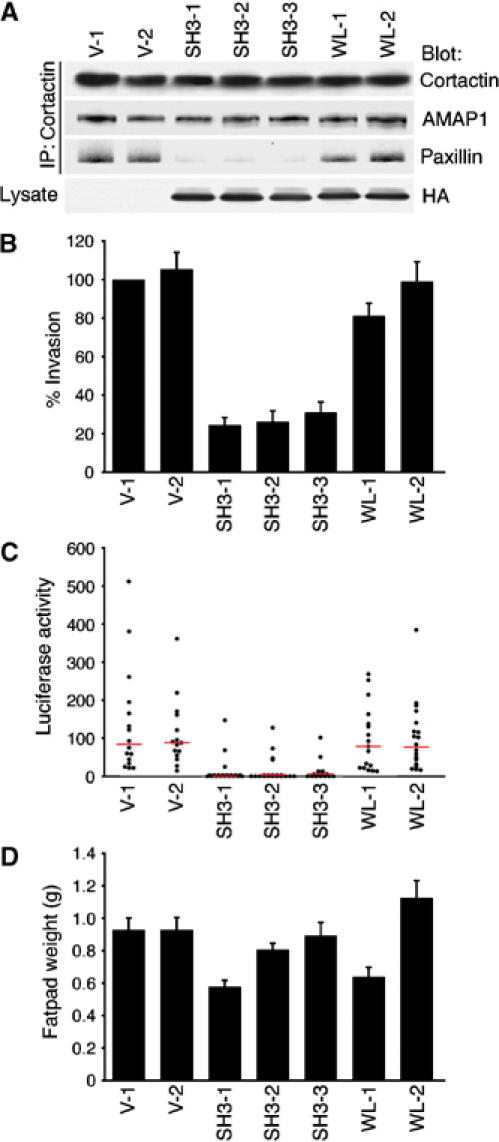
Effect of the inhibition of AMAP1-mediated protein complex formation on metastasis in vivo. 4T1/luc cell clones, stably overexpressing the HA-tagged AMAP1 SH3 domain (SH3-1, SH3-2 and SH3-3) or its WL mutant (WL-1 and WL-2), or stably transfected with the empty vector (V-1 and V-2), were established. (A) Levels of the AMAP1 SH3 domain or the WL mutant in each cell clone (upper panel), and co-precipitation of paxillin with AMAP1 in these cell clones (lower panel) were examined, as described in Figure 7B by anti-HA immunoblotting. (B) In vitro Matrigel invasion activity of cell clones. (C, D) Tumor metastasis in the left lung via injection of these clones into the right inguinal mammary fat pad of Balb/c mice. The luciferase activity in the left lungs isolated from these mice 19 days after the injection was measured, and plotted as activity per mg of the total protein after subtracting the background values. Median values were calculated and are presented as red lines (C). The weight of the tumors at sites of the originally injected mammary fat pad was measured simultaneously and is also plotted (D). A total of 20 mice were used for each cell clone. In (B, D), results shown are the means±s.e.m.
Discussion
Most human breast carcinomas arise from the mammary ductal epithelia. They are nevertheless highly diverse in their natural history (Tavassoli and Schnitt, 1992). We have previously shown that Arf6 plays an essential role in the invasive activities of different breast cancer cells (Hashimoto et al, 2004b). In this study, we found that AMAP1, an ArfGAP whose function is related to Arf6 activity, plays pivotal roles in the invasive activities of MDA-MB-231 cells. Before analyzing the detailed role of AMAP1 in invasion, we set our major aims in this paper to investigate (1) whether AMAP1 participates in and plays a crucial part in the invasion activities of the majority, if not all, breast cancers, and if so, then (2) whether AMAP1-mediated complex formation with certain proteins, such as paxillin and cortactin, is specifically utilized for the invasion of breast cancer cells, rather than being generally used even in normal breast epithelial cells, and if it is also the case, then finally (3) whether we can provide possible methods to inhibit breast cancer cell invasion and metastases by use of our knowledge of AMAP1.
Our results provide several lines of evidence supporting that AMAP1 may satisfy the above criteria. We found that AMAP1 protein levels show a good correlation with the invasive phenotype of not only MDA-MB-231 cells but also of other breast cancer cells, and AMAP1 protein suppression by an siRNA duplex effectively inhibits their invasion activities. Primary cultures of normal mammary epithelial cells expressed the AMAP1 protein only at a marginal level. We found that AMAP1 binds both to cortactin and paxillin and acts to bridge these proteins. Our results also indicate that this trimeric protein complex is detected only in highly invasive cell lines and not in weakly invasive and noninvasive cells, which is consistent with a previous report by Bowden et al (1999). Both paxillin and cortactin are expressed ubiquitously in a variety of cells and tissues (Miglarese et al, 1994; Mazaki et al, 1998). Their expression is also seen in different breast cancer cell lines, although their levels of expression do not immediately correlate with the invasive phenotype (Bowden et al, 1999; also see Figure 7A). Rather than the levels of cortactin and paxillin, our results indicate that expression of the AMAP1 protein at high levels appears to be crucial for the association of paxillin and cortactin, and for the invasive phenotypes of cancer cells.
To demonstrate possible methods to inhibit breast cancer cell invasion and metastasis, we showed that inhibition of AMAP1-mediated protein complex formation, such as by overexpression of the AMAP1 SH3 domain and by microinjection of an AMAP1 PRD peptide, can effectively inhibit the in vitro invasion activities of both MDA-MB-231 and 4T1/luc cells, in addition to the siRNA-mediated suppression of AMAP1 expression. To show that such inhibition of AMAP1-mediated protein complex formation is also effective in vivo, we generated 4T1/luc cell clones overexpressing the AMAP1 SH3 domain and demonstrated that their metastasis from mammary fat pads to the lungs can be significantly suppressed. We have, on the other hand, so far tried in vain to suppress AMAP1 expression by a plasmid construct encoding the siRNA duplex, which is necessary to examine the AMAP1 siRNA effect on metastases in mice.
We have previously shown that high levels of Arf6 protein expression correlate well with the invasive phenotypes of different breast cancer cell lines, while its mRNAs are already expressed at high levels in noninvasive breast cancer cells and normal mammary epithelial cells (Hashimoto et al, 2004b). The expression patterns of AMAP1 at both the protein and mRNA levels were found to be similar to those of Arf6. Consistent with our results, gene expression profile analyses have not identified mRNA expression of Arf6 and AMAP1 as being upregulated in correlation with the invasive phenotypes of primary breast tumors. A number of studies have shown that individual mRNAs, whose products are involved in cell–cell interaction, signal transduction, growth control and transformation, are translationally regulated (Rajasekhar and Holland, 2004). Protein degradation also plays pivotal roles in regulating cellular protein levels, without immediately affecting mRNA levels. Some transcription-independent mechanism might function in noninvasive mammary epithelial cells to suppress the expression of certain proteins, such as Arf6 and AMAP1, to basal levels, and dysfunction of such putative mechanisms in primary breast tumors may participate in the acquisition of the invasive phenotype and the progression to malignancy.
Immunohistochemical staining indicates high levels of AMAP1 protein expression in invasive primary tumors of the human breast, while AMAP1 protein expression is very low in most of our DCIS samples, in which the patients were diagnosed as being free of IDC. However, we also found relatively high levels of AMAP1 protein expression in most sites of DCIS if the patients simultaneously have lesions of IDC. These observations suggest that upregulation of AMAP1 protein expression may be correlated with the potential malignancy of breast cancers rather than being simply correlated only with the invasive phenotypes. To further assess the significance of AMAP1 protein expression with regard to its relationship to invasiveness and potential malignancy, we are preparing to analyze AMAP1 protein levels with larger numbers of clinical specimens, together with analysis of its mRNA levels.
AMAP1-mediated protein complex formation with paxillin and cortactin was undetectable in cultured normal mammary epithelial cells. If AMAP1-mediated protein complex formation with paxillin and cortactin is indeed an event not employed in most types of normal cells in adults at their steady state, this protein complex will provide excellent targets for drug development. The binding of both AMAP1/paxillin and AMAP1/cortactin is mediated by SH3 domains. The three-dimensional structure of several SH3 domains has already been well established, including the structure of the interfaces involved in their binding to proline-rich ligands (Mayer, 2001). Thus, detailed studies on the interface structures involved in the formation of the AMAP1/paxillin complex and the AMAP1/cortactin complex will provide logical bases to design, or to screen, chemical compounds effective for breast cancer therapy, which may exhibit only minimal side effects on normal tissues and organs in adults. Further studies to determine whether normal cells utilize the AMAP1-mediated protein complex with paxillin and cortactin under some situations, such as epithelial–mesenchymal transdifferentiation and tissue remodeling, and if so, then to clarify at what stages of embryonic development and where in the adult body this complex is used might help to further assess their probability to be drug targets. It is also worthy to examine what other types of tumors also use this protein complex for their invasion.
Materials and methods
Cells
Human breast cancer cell lines were obtained from the American Type Culture Collection (ATCC). MDA-MB-231 cells were cultured as described previously (Bowden et al, 1999), and other cells were cultured according to the ATCC instructions. A primary preparation of human normal mammary epithelial cells (Cambrex, East Rutherford, NJ) was cultured according to the manufacturer's instructions. 4T1/luc cells expressing firefly luciferase were from T Yoneda (Osaka University) and were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FCS (HyClone). COS-7 cells were cultured in DMEM with 10% FCS.
cDNAs and transfection
cDNAs for AMAP1 and its splice variant, AMAP1b, were amplified by PCR from the first-strand cDNAs prepared from mRNAs of human fetal human brain (Clontech). Mutations were introduced into the seventh proline-rich sequence of AMAP1, in which Pro898 and Lys903 were changed to alanines, to generate AMAP1a. The rescue cDNA for AMAP1 was constructed by substituting the nucleotides within the siRNA target to 5′-AGGATTTAACTAAGGCGATAA-3′, without changing the coding amino acids. The ΔGAP mutant of this cDNA was made by deletion of the ArfGAP domain and the first ankyrin repeat (aa 457–668). cDNAs for cortactin and intersectin-I were isolated by yeast two-hybrid screening. cDNAs for the AMAP1 SH3 domain (aa 989–1132) and the intersectin-I SH3 domains (SH3(D) and SH3(E), aa 1068–1220) were amplified by PCR using appropriate oligonucleotides. The WL mutant of the AMAP1 SH3 domain was generated by PCR-based mutagenesis, in which Trp1107 was changed into leucine. For mammalian expression, these cDNAs were ligated in-frame to the COOH terminus of tag sequences of pEBG (for the GST-tag; Mayer et al, 1995), pcDNA3 HA (for the HA-tag, Invitrogen), pcDNA3.1 His (for the Express-His-tag, Invitrogen), as indicated in the text. cDNAs for cortactin and the AMAP1 SH3 domain were each ligated into pGEX2TK to be in-frame to the COOH terminus of the GST-TK-tag, to be expressed in Escherichia coli.
For the in vivo protein binding assay, 5 × 105 COS-7 cells were cotransfected with 1 μg of pcDNA3.1His C/cortactin and 3 μg of pEBG/AMAP1 or pEBG/AMAP2 using Polyfect (Qiagen) according to the manufacturer's instructions. For the in vitro binding assay, 5 × 105 COS-7 cells were transfected with 3 μg of pcDNA3HA/AMAP1. For invasion, migration and adhesion assays, 1 × 106 cells were cotransfected with 5 μg of pcDNA3HA/AMAP1 SH3 and 0.2 μg of pEGFP-C1 using Trans IT-LT1 (Mirus), and incubated for 24 h in growth medium before being subjected to analyses. Under these conditions, we observed that more than 95% of EGFP-positive cells also expressed the HA-tagged AMAP1 SH3 protein, by detecting the autofluorescence from EGFP and immunostaining cells with an anti-HA antibody.
siRNA-mediated silencing of protein expression
Protein knockdown was performed by use of the RNA interference (RNAi) technique (Elbashir et al, 2001). Cells cultured in the growth medium were transfected with 25 nM of oligonucleotide duplexes using LipofectAMINE 2000 (Invitrogen) and incubated for 48 h before being subjected to analyses. Duplex oligonucleotides were chemically synthesized and purified by Japan BioServices (Saitama, Japan). siRNA duplexes used were as follows: human AMAP1, 5′-GACCUGACAAAAGCCAUUAdTdT-3′ and 5′-UAAUGGCUUUUGUCAGGUCdTdT-3′; mouse AMAP1, 5′-GCUACCCAGUGUGAAGAUCdTdT-3′ and 5′-GAUCUUCACACUGGGUAGCdTdT-3′; and human AMAP2, 5′-AGAGGACUCCCAAAUUCGUdTdT-3′ and 5′-ACGAAUUUGGGAGUCCUCUdTdT-3′.
Immunohistochemical staining
Immunohistochemistry was performed on formalin-fixed paraffin-embedded tissues. Sections (4 μm) were deparaffinized in xylene and dehydrated in a graded series of ethanol, and processed for antigen retrieval by heating in 10 mM sodium citrate (pH 6.0) at 120°C for 15 min. Endogenous peroxidase was then blocked by incubating in 0.3% H2O2–methanol at room temperature for 30 min. After being rinsed in PBS and blocked with 10% BSA in PBS at room temperature for 15 min, the sections were incubated with an affinity-purified AMAP1 rabbit polyclonal antibody in 1% BSA in PBS overnight at 4°C. Immunocomplexes were visualized using a biotinylated anti-rabbit IgG antibody and peroxidase-conjugated streptavidin (DakoCytomation), and incubated with 3,3′-diaminobenzidine. Each section was counterstained with hematoxylin.
Quantitative analysis of immunohistochemical staining
Quantitative analysis of the immunohistochemical staining for AMAP1 protein expression in human breast tumor tissue was performed as follows. RGB color images of the stained slides were captured by a microscope (Axiovert 200, Carl Zeiss) equipped with a color CCD camera (AxioCam, Carl Zeiss). The staining intensity of noncancerous cells and that of cancerous cells were measured in inverted gray level values between 0 and 255 (representing pure white and pure black, respectively) using a computerized image analysis system (MacScope version 2.56, Mitani, Fukui, Japan). After background subtraction, the average staining intensity of cancerous cells was calculated as a percentage of that of noncancerous cells on the same slides, to estimate the amplification of AMAP1 protein expression. The data were classified into four groups—the DCIS-only stage, the DCIS in mixed stage (both DCIS and IDC were observed in the same patient), the IDC in the mixed stage and the IDC-only stage—according to the pathological determination.
In vitro invasive activities
The number of cells forming invadopodia and the degraded areas were determined as described previously (Bowden et al, 1999). Briefly, porcine skin gelatin (Type A with 300 Bloom, Sigma-Aldrich) was conjugated with Alexa Fluor 594 (Molecular Probes), and glass-bottomed dishes (MatTek) were coated with gelatin and subsequently crosslinked with 0.5% glutaraldehyde. Cells in the growth medium were then replated onto these dishes, incubated for 16 h and then fixed in 4% paraformaldehyde for 10 min at room temperature. Data were collected from three independent experiments, each carried out in duplicate, in which more than 100 cells were analyzed using a confocal laser scanning microscope (LSM 510, Carl Zeiss) at a × 63 magnification.
Matrigel chemoinvasion assay was performed using Biocoat Matrigel chambers (Becton Dickinson), as described previously (Thompson et al, 1992). Briefly, 1 × 105 cells were seeded on the upper wells of 24-well chambers in the absence of serum, in which the lower wells were filled with conditioned medium of NIH3T3 cells cultured for 24 h in the absence of serum. After incubation for 12 h, the number of cells that migrated out onto the lower surface of membranes was scored by their staining with 1% crystal violet (for siRNA-treated cells) or by identifying the EGFP-positive cells using a laser scanning microscope (for AMAP1 SH3 cDNA-transfected cells), after fixing the cells in 4% paraformaldehyde. Data were collected from three independent experiments each carried out in duplicate (more than 100 cells were scored in each experiment).
In each assay, data are presented as percentages calculated by normalizing the values obtained for the untreated cells as 100%.
Haptotactic migration assay, cell adhesion assay and gelatin zymography
The haptotactic cell migration assay was performed in the absence of serum using modified Boyden chambers (Transwell with 8 μm pores, Costar) in which the lower surface of the membrane was coated with collagen type I (10 μg/ml, Upstate), cell adhesion assay was performed using collagen type I-coated dishes (10 μg/ml) and gelatin zymography was performed as described previously (Hashimoto et al, 2004b). Data collection and presentation are the same as above.
In vivo metastasis studies
4T1/luc cells (5 × 105) were transfected with 5 μg of pcDNA3/AMAP1SH3, pcDNA3/AMAP1SH3WL or the empty vector, together with 0.5 μg of pBabePuro, using LipofectAMINE 2000, and independent clones were isolated after selection for 2 weeks with 1 μg/ml puromycin. Under anesthesia with pentobarbital (0.05 mg/g body weight), a 2 mm skin incision was made in the right inguinal mammary fat pad of female Balb/c mice (6–8 weeks old, SLC, Japan), and 1 × 106 4T1/luc cell clones in a volume of 0.1 ml in PBS were injected into the tissue through a 27-gauge needle. After 19 days, under deep anesthesia with ether the left lungs were excised, homogenized and determined for luciferase activity using a luciferase assay system (Promega), as described previously (Michigami et al, 2002). Tumor weights at the originally injected fat pads were also measured.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Manami Hiraishi and Yumiko Shibata for their technical assistance and Mayumi Iwahara for her secretarial work. We also thank Toshiyuki Yoneda for 4T1/luc cells and Helena Akiko Popiel for her critical reading of the manuscript. This work was supported in part by Grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (MESSC) and by grants from Takeda Pharmaceutical Co. YO is a recipient of an MESSC studentship of the 21st Century COE Program.
References
- Allred DC, Mohsin SK, Fuqua SA (2001) Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer 8: 47–61 [DOI] [PubMed] [Google Scholar]
- Andreev J, Simon JP, Sabatini DD, Kam J, Plowman G, Randazzo PA, Schlessinger J (1999) Identification of a new Pyk2 target protein with Arf-GAP activity. Mol Cell Biol 19: 2338–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC (1999) An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene 18: 4440–4449 [DOI] [PubMed] [Google Scholar]
- Bowden ET, Coopman PJ, Mueller SC (2001) Invadopodia: unique methods for measurement of extracellular matrix degradation in vitro. Methods Cell Biol 63: 613–627 [DOI] [PubMed] [Google Scholar]
- Brown MT, Andrade J, Radhakrishna H, Donaldson JG, Cooper JA, Randazzo PA (1998) ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol Cell Biol 18: 7038–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coopman PJ, Do MT, Thompson EW, Mueller SC (1998) Phagocytosis of cross-linked gelatin matrix by human breast carcinoma cells correlates with their invasive capacity. Clin Cancer Res 4: 507–515 [PubMed] [Google Scholar]
- Coopman PJ, Thomas DM, Gehlsen KR, Mueller SC (1996) Integrin alpha 3 beta 1 participates in the phagocytosis of extracellular matrix molecules by human breast cancer cells. Mol Biol Cell 7: 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMali KA, Wennerberg K, Burridge K (2003) Integrin signaling to the actin cytoskeleton. Curr Opin Cell Biol 15: 572–582 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K (2003) Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 3: 362–374 [DOI] [PubMed] [Google Scholar]
- Frykberg EB, Bland KI (1994) Management of in situ and minimally invasive breast carcinoma. World J Surg 18: 45–57 [DOI] [PubMed] [Google Scholar]
- Grunert S, Jechlinger M, Beug H (2003) Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol 4: 657–665 [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Hashimoto A, Yamada A, Kojima C, Yamamoto H, Tsutsumi T, Higashi M, Mizoguchi A, Yagi R, Sabe H (2004a) A novel mode of action of an ArfGAP, AMAP2/PAG3/Papalpha, in Arf6 function. J Biol Chem 279: 37677–37684 [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, Sabe H (2004b) Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci USA 101: 6647–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549 [DOI] [PubMed] [Google Scholar]
- Kondo A, Hashimoto S, Yano H, Nagayama K, Mazaki Y, Sabe H (2000) A new paxillin-binding protein, PAG3/Papalpha/KIAA0400, bearing an ADP-ribosylation factor GTPase-activating protein activity, is involved in paxillin recruitment to focal adhesions and cell migration. Mol Biol Cell 11: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC (2003) Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA 100: 5974–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ (2001) SH3 domains: complexity in moderation. J Cell Sci 114: 1253–1263 [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Hirai H, Sakai R (1995) Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol 5: 296–305 [DOI] [PubMed] [Google Scholar]
- Mazaki Y, Uchida H, Hino O, Hashimoto S, Sabe H (1998) Paxillin isoforms in mouse. Lack of the gamma isoform and developmentally specific beta isoform expression. J Biol Chem 273: 22435–22441 [DOI] [PubMed] [Google Scholar]
- Michigami T, Hiraga T, Williams PJ, Niewolna M, Nishimura R, Mundy GR, Yoneda T (2002) The effect of the bisphosphonate ibandronate on breast cancer metastasis to visceral organs. Breast Cancer Res Treat 75: 249–258 [DOI] [PubMed] [Google Scholar]
- Miglarese MR, Mannion-Henderson J, Wu H, Parsons JT, Bender TP (1994) The protein tyrosine kinase substrate cortactin is differentially expressed in murine B lymphoid tumors. Oncogene 9: 1989–1997 [PubMed] [Google Scholar]
- Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL (2003) Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat 78: 323–335 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, Schnitt S, Gabrielson E, Gelman R, Polyak K (2003) Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res 1: 362–375 [PubMed] [Google Scholar]
- Rajasekhar VK, Holland EC (2004) Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene 23: 3248–3264 [DOI] [PubMed] [Google Scholar]
- Robanus-Maandag EC, Bosch CA, Kristel PM, Hart AA, Faneyte IF, Nederlof PM, Peterse JL, van de Vijver MJ (2003) Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J Pathol 201: 75–82 [DOI] [PubMed] [Google Scholar]
- Sabe H (2003) Paxillin-associated ArfGAPs—their isoform specificities and roles in coordination. In ARF Family GTPases, Kahn R (ed) pp 185–207. Dordrecht, Netherlands: Kluwer Academic Publishers [Google Scholar]
- Schaller MD (2001) Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20: 6459–6472 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli FA, Schnitt SJ (1992) Pathology of the Breast. New York, USA: Elsevier [Google Scholar]
- Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME, Martin GR, Dickson RB (1992) Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol 150: 534–544 [DOI] [PubMed] [Google Scholar]
- Uchida H, Kondo A, Yoshimura Y, Mazaki Y, Sabe H (2001) PAG3/Papalpha/KIAA0400, a GTPase-activating protein for ADP-ribosylation factor (ARF), regulates ARF6 in Fcgamma receptor-mediated phagocytosis of macrophages. J Exp Med 193: 955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- Weed SA, Parsons JT (2001) Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene 20: 6418–6434 [DOI] [PubMed] [Google Scholar]
- Zajchowski DA, Bartholdi MF, Gong Y, Webster L, Liu HL, Munishkin A, Beauheim C, Harvey S, Ethier SP, Johnson PH (2001) Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res 61: 5168–5178 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


