Summary
The development of soluble envelope glycoprotein (Env) mimetics displaying ordered trimeric symmetry has ushered in a new era in HIV-1 vaccination. The recently reported native, flexibly linked (NFL) design allows the generation of native-like trimers from clinical isolates at high yields and homogeneity. As the majority of infections world-wide are of the clade C subtype, we examined responses in non-human primates to well-ordered subtype C 16055 trimers administered in soluble or high-density liposomal formats. We detected superior germinal center formation and enhanced autologous neutralizing antibodies against the neutralization-resistant (tier 2) 16055 virus following inoculation of liposome-arrayed trimers. Epitope-mapping of the neutralizing monoclonal antibodies (mAbs) indicated major contacts with the V2 apex and 3D electron microscopy reconstructions of Fab-trimer complexes revealed a horizontal binding angle to the Env spike. These vaccine-elicited mAbs target the V2 cap, demonstrating a means to accomplish tier 2 virus neutralization by penetrating the dense N-glycan shield.
Keywords: vaccine, HIV-1, clade C, envelope glycoproteins, trimers, liposome, monoclonal antibody, epitope, germinal centers, B cell responses
Graphical Abstract
Introduction
Selection for viral variants that successfully evade recognition by the host immune system is a hallmark of HIV-1 infection. Particularly, the functional HIV-1 envelope glycoprotein (Env) spike has evolved extreme resistance to antibody (Ab)-mediated neutralization. This is apparent from the spacing of N-linked glycans on solvent-exposed regions of the trimer spike that occlude Ab access to the underlying polypeptide surface (Ingale et al., 2014; Pritchard et al., 2015; Stewart-Jones et al., 2016). Constant Ab-mediated immune pressure during chronic infection results in a glycan shield that is continuously evolving within each infected individual, generating an enormous diversity of replication-fit HIV-1 variants that can mediate transmission (Wei et al., 2003). In addition to N-glycan shielding, highly variable (V) regions and conformational constraints of the Env trimer restrict Ab access to conserved neutralizing Ab determinants. Since Env is the sole neutralizing target on the viral surface, these immune evasion mechanisms pose major challenges for the development of an effective HIV-1 vaccine (Karlsson Hedestam et al., 2017). However, some HIV-1-infected individuals generate broadly neutralizing antibodies (bNAbs), indicating the capacity of the human immune system to infrequently generate Abs that penetrate these barriers (Burton and Mascola, 2015; Stamatatos et al., 2009). Such bNAbs often display unusual features such as uncommon precursor traits, N-glycan reactivity and high levels of somatic hypermutation (SHM). As a reflection of this challenge, the elicitation of cross-reactive neutralizing antibodies to primary isolate strains of HIV-1 (termed tier 2) has not been replicated in the context of vaccination, except in specialized systems such as in mice engineered to express various germline-reverted forms of human bNAbs (Dosenovic et al., 2015; Escolano et al., 2016).
In recent years, developments in Env trimer design have resulted in the generation of soluble spike mimics, such as the SOSIPs and NFLs that display well-ordered, native-like conformation faithfully mimicking the cleaved, native spike (Georgiev et al., 2015; Sanders et al., 2013; Sharma et al., 2015). Antigenic evaluation of the native-like trimers demonstrates that they are efficiently recognized by most bNAbs, but not by non-bNAbs. This favorable antigenic profile has raised expectations that these spike mimetics will elicit neutralizing antibodies that access the functional spike by angles of approach not accomplished by antibodies elicited by the previous generation of trimers, such as the foldon trimers (Navis et al., 2014; Sundling et al., 2012a; Tran et al., 2014). An accompanying paper describes the design and high-resolution structure of NFL trimers derived from the clade C isolate, 16055 (Guenaga et al.). This work emphasizes the concept that structure-guided modifications of Env can generate homogeneous trimers that display native-like conformation and antigenicity. Immunogenic evaluation of such trimers in animal models, particularly in primates, including identification of the elicited neutralizing and non-neutralizing specificities through the isolation of mAbs, is now a high priority in the HIV-1 vaccine field.
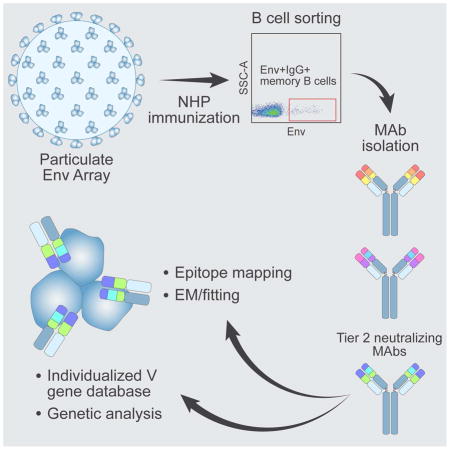
Accordingly, we analyzed in detail Ab responses induced by these well-ordered clade C-derived trimers in non-human primates (NHPs). We demonstrate that liposome-conjugated trimers generated superior germinal center (GC) responses and higher neutralizing Ab titers against the autologous tier 2 16055 virus compared to the same trimers administered in a soluble format. A panel of NHP mAbs was isolated, including mAbs that recapitulated the plasma 16055 neutralizing activity. We examined the genetic properties of these mAbs using an individualized V gene database generated from the donor animal using Next Generation Sequencing (NGS) and analysis by the computational tool IgDiscover (Corcoran et al., 2016). Knowledge of the specific allelic content of V genes in a given donor animal allows analysis of clonal relationships and precise calculations of SHM levels of mAbs isolated from that animal. By epitope-mapping and electron microscopy (EM) analyses, the 16055-neutralizing mAbs were shown to target the V2 apex region of the HIV-1 spike, using an angle of approach horizontal to the viral membrane. Our results provide encouraging impetus for ongoing and planned pre-clinical and clinical vaccine trials aimed to elicit neutralizing Abs against HIV-1 through Env-based immunization.
Results
16055 NFL trimers conjugated to liposomes display an intact antigenic profile
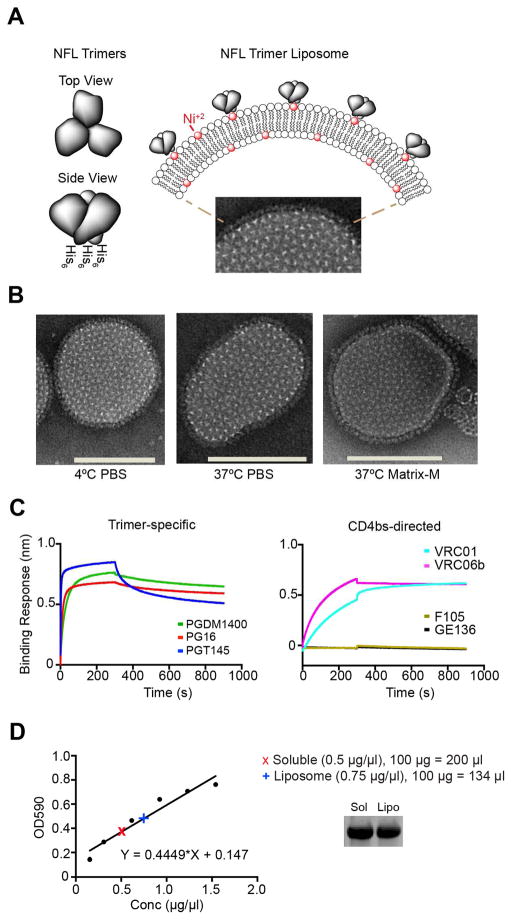
We generated single bilayer nickel-containing liposomes arrayed with well-ordered His-tagged subtype C-derived 16055 NFL TD CC trimers (Guenaga et al., 2015) as previously described for clade B trimer liposomes (Ingale et al., 2016) (Figure 1A). We assessed the resulting trimer-conjugated liposomes by negative stain electron microscopy (EM) and confirmed that the trimers were arrayed at high density on the Ni-liposome surface (Figure 1B). To verify that the trimer-liposomes remained intact at physiologic temperature, we incubated the trimer-liposomes at 37°C in buffer with or without Matrix-M™ adjuvant and assessed their integrity by EM. The trimer-conjugated liposomes were stable in Matrix-M up to 24 hours at 37°C (Figure 1B, right). We also confirmed that the trimers maintained a well-ordered conformation following conjugation by efficient recognition of bNAbs and inefficient recognition by non-bNAbs using biolayer light interferometry (BLI) with the trimer-conjugated liposomes captured on the wheat germ agglutinin sensor surface and the mAbs in solution (Figure 1C). Specifically, we confirmed that the trimer-specific bNAbs PG16, PGT145 and PGDM1400 efficiently recognized the 16055 NFL TD CC trimers (Figure 1C, left), consistent with maintenance of a well-ordered conformation (Guenaga et al., 2015). Similarly, efficient recognition by the CD4 binding site (CD4bs)-directed bNAbs VRC01 and VRC06b was maintained while binding by the non-neutralizing CD4bs-directed mAbs (F105 and GE136) was low to non-detectable (Figure 1C, right). To confirm that equal amounts of Env were used for the in vivo comparison of soluble trimers and liposome-conjugated trimers, we quantified the protein levels in each preparation using a colorimetric assay with a 16055 NFL TD CC protein standard curve (Figure 1D, left). The respective trimer quantifications were confirmed by subsequent SDS gel analysis using an aliquot from each immunogen stock solution (Figure 1D, right). Overall, these analyses show that the antigenic integrity of the 16055 NFL trimers was intact after liposome coupling and that a comparable amount of Env was used for immunization of the two groups.
Figure 1. The antigenicity of NFL TD CC Env trimers is intact after conjugation to liposomes.
(A) Schematic representation of soluble 16055 NFL TD CC trimers containing C-terminal His tags (left) and the trimers arrayed at high density on liposomes following nickel-NTA-lipid capture by the His tags (top right; DGPC lipid head group, white spheres; DGS-NTA(Ni) lipid, red spheres) with a representative negative stain EM of the liposomal trimer array shown below. (B) Negative stain EM images of representative liposomes, selected from hundreds in the field, after incubation for 24 hr at 4°C in PBS (left), at 37°C in PBS (middle), or at 37°C in Matrix-M adjuvant (right; 96,000x magnification; scale bars, 100 nm). (C) Confirmation of trimer-specific bNAb recognition (left; PGDM1400, PG16 and PGT145) of the trimers following liposome conjugation and recognition by CD4bs-directed bNAbs (right; VRC01 and VRC06) but not by CD4bs-directed non-bNAbs (F105 and GE136) by BLI. (D) Quantification of soluble trimer and trimer-conjugated liposome relative Env concentrations were calculated based on a standard curve equation generated from assays using the Advanced Protein Assay Reagent. Sample calculation for 100 μg is shown with SDS reducing gel of each to confirm equivalent Env amounts (right; 5 μg per lane). These experiments were performed at least two independent times.
Liposome-arrayed trimers induce superior GC responses compared to soluble trimers
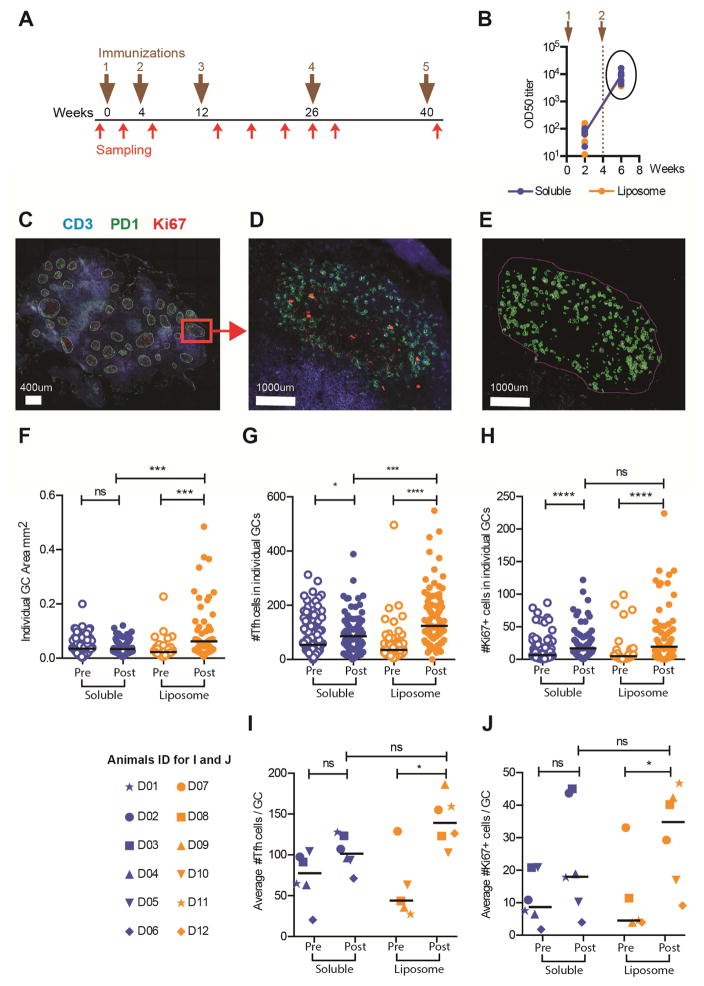
Previous studies in mice and rabbits demonstrated superior capacity of liposome-arrayed trimers over soluble trimers to activate Env-specific B cells ex vivo and to induce GC B cells and neutralizing Ab responses in vivo (Ingale et al., 2016). These studies also demonstrated that liposomes lacking Env do not induce GC responses in vivo and do not activate an Env-specific B cell line ex vivo, supporting the hypothesis that administration of liposomes conjugated with Env increases B cell receptor cross-linking, perhaps allowing low affinity B cell clonotypes with neutralizing potential to enter the GC reaction. Here, we performed an immunogenicity experiment in rhesus macaques to examine Ab responses elicited by the 16055 NFL TD CC trimers. Twelve rhesus macaques were divided in two groups of six, receiving either soluble trimers or liposome-arrayed trimers, both formulated in Matrix-M adjuvant. The animals were inoculated five times at weeks 0, 4, 12, 26 and 40; samples were collected prior to the first inoculation and two weeks after each inoculation. Additional blood samples were collected during the interval between the third and fourth immunization (Figure 2A). We first measured Env-specific Ab titers in plasma collected two weeks after the first boost (Figure 2A) and observed no difference in the magnitude of Env- specific IgG OD50 titers between the groups (Figure 2B). To investigate potential qualitative differences between the groups, we analyzed draining lymph nodes biopsied two weeks after the first boost. Pre-immunization lymph nodes were also analyzed for baseline quantification. Cryo-sections of lymph nodes were stained with antibodies against CD3, PD-1 (a marker of follicular T helper (Tfh) cells) and Ki67 (a marker of proliferation) and GC structures were defined as conglomerations of PD-1+ cells (green; mainly Tfh cells) and Ki67+ (red; mainly proliferating GC B cells) cells, surrounded by CD3+ T cells (blue) (Figure 2C and 2D). We manually identified GCs and applied the Cell profiler software (Carpenter et al., 2006) to calculate GC areas and numbers of Ki67+ and PD-1+ cells within each GC based on the fluorescence for each marker (Figure 2E). Data from pre- and post-immunization samples were collected and analyzed, revealing that animals inoculated with liposome-arrayed trimers displayed, on average, larger GCs compared to animals inoculated with the soluble trimers (p=0.0004). In the liposome group, the area of individual GCs was significantly greater post-immunization compared to pre-immunization (p=0.0001); while this was not observed in the animals inoculated with the soluble trimers (Figure 2F). We detected a greater content of PD-1+ Tfh cells within individual GCs in animals inoculated with the trimer-liposomes compared to animals inoculated with soluble trimers (p=0.0002). By this criterion, there was a significant difference between pre- and post-immunization samples in both groups, but the difference was greater in the trimer-liposome group (p<0.0001) (Figure 2G). Enumeration of Ki67+ cells revealed an increase in the number of proliferating cells between pre- and post-immunization in both groups (p<0.0001), but no difference post-immunization between the two groups (Figure 2H). We also calculated the average number of PD-1+ cells or Ki67+ cells per GC for each animal and found that animals inoculated with the liposome-arrayed trimers displayed higher average number of PD-1+ cells/GC than soluble trimer-immunized animals, but this difference was not statistically significant. When we compared the average number of Tfh+ cells/GC between pre- and post-immunization, there was a significant difference for the liposome group (p=0.03), but not for the soluble group (Figure 2I). Similarly, the average number of Ki67+ cells/GC was statistically different between pre- and post-immunization for animals inoculated with the liposome trimers (p=0.05), but not for those inoculated with soluble trimers. These analyses suggest that immunization with liposome-displayed trimers offers an advantage over soluble trimers as determined by analysis of the induced GC responses.
Figure 2. Immunization with liposome-displayed trimers elicits superior GC responses.
(A) The immunization scheme consisted of five injections on weeks 0, 4, 12, 26 and 40 (brown arrows), sampling was performed two weeks after each inoculation and four additional times as indicated (red arrows). The animals were divided in two groups: soluble trimers (Soluble, Blue) (n=6) and liposome arrayed-trimers (Liposome, Orange) (n=6), both formulated in Matrix-M. (B) Half maximum binding titers (OD50) of Env-specific IgG represented on a log10 scale. Lines represent group means and each dot represents an animal. (C) Cryosections of draining inguinal lymph nodes from rhesus macaques stained with antibodies against CD3 (blue), PD-1 (green) and Ki67 (red), where GCs structures were recognized. (D) GCs structures were identified by the conglomeration of PD-1+ cells and Ki67+ cells surrounded by CD3+ cells. (E) GCs were manually selected using cell profiler software, and in house pipelines allowed the automatic count of the fluorescent signal. The white bar represents 400μm in C and 1000μm in D and E. Pre- and post-immunization samples are shown in F–J. In F–H, each dot represents one GC parameter: (F) total individual GC area, (G) number of Tfh cells in each individual GC (H) number of Ki67+ in each individual GC. (I, J) Each dot represents the average number of Tfh or Ki67+ cells per animal. The average was calculated as the number of positive cells within a GC divided by the total number of GCs. (I) Average number of Tfh cells per GC. (J) Average number of Ki67+ cells per GC. Post-immunization samples were from two weeks after the second immunization. Statistical difference were evaluated by Mann-Whitney test; ns: non-significant; * for p≤0.05, ** for p≤0.01, *** for p≤0.001 and **** for p≤0.0001. These experiments were performed two independent times.
Liposome-arrayed trimers stimulate enhanced autologous tier 2 virus neutralizing Ab responses
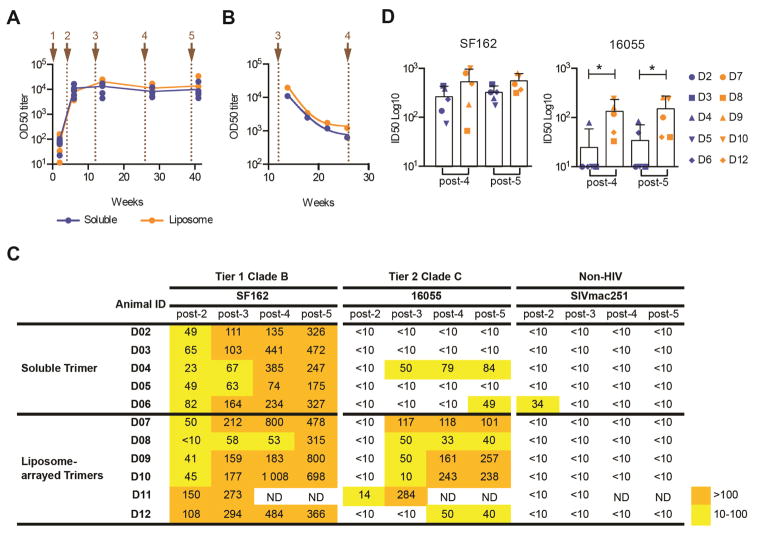
Next, we analyzed plasma binding and neutralizing Ab responses over the complete immunization regimen. We used an ELISA format where the Env trimers are captured via their His-tags to anti-His Ab coated wells to preserve the native-like trimer conformation, unlike the conventional ELISA format where Env is coated directly on the plate. We observed comparable Env-specific IgG OD50 titers in the two groups at all time points (Figure 3A), as well as similar decline kinetics of circulating Env-specific IgG response during the 14 week interval between the third and fourth vaccine inoculation (Figure 3B). Neutralizing Ab titers against tier 1 viruses were also similar as shown for SF162 (Figure 3C) and MW965 (Figure S1A). In contrast, we observed more consistent and higher ID50 neutralizing Ab titers against the autologous tier 2 16055 virus in animals immunized with the liposome-arrayed trimers compared to animals immunized with soluble trimers (Figure 3C). When post-4 and post-5 immunization samples were subjected to statistical analysis, we found no significant difference for the SF162 neutralizing Ab response between the groups, while there was significant difference (p=0.05) for the 16055 neutralizing response (Figure 3D). Since SF162 and MW965 are known to be sensitive to V3-directed antibodies induced in NHPs (Morner et al., 2009), we performed V3 peptide competition experiments, which confirmed that the neutralizing Ab response against both tier 1 viruses was predominantly V3-directed (Figure S1A). We detected weak neutralizing Ab activity against HXBc2 but no co-receptor-directed Ab neutralizing activity as determined by the HIV-2731A.V434M virus in the presence of soluble CD4 (sCD4), an assay that is diagnostic of such Abs (Decker et al., 2005) (Figure S1B). The latter result demonstrates that the CC bond, which was introduced to prevent the NFL TD CC trimers undergoing conformational changes upon binding to CD4 in vivo, was productive. We have previously shown that trimers that allow in vivo CD4 binding readily induce co-receptor-directed neutralizing Ab activity (Forsell et al., 2008).
Figure 3. Liposome-arrayed trimers stimulate enhanced autologous tier 2 neutralizing Ab responses compared to soluble trimers.
(A) Binding titers (OD50) of Env-specific IgG responses two weeks after each immunization represented as log10. Lines represent group means and dots represent each animal, Soluble (blue) and Liposome-arrayed (orange). (B) Half-life of Env-specific IgG between the third and fourth immunization. Lines represent group means, dots represent each time point mean. Brown arrows and dotted lines represent each immunization point in A and B (C) Plasma neutralization activity against SF162, 16055 and SIV pseudoviruses was calculated for each animal two weeks after the 3rd, 4th and 5th immunization as the reciprocal dilution giving 50% of neutralization (ID50 titer). The background neutralization ID50 titer threshold was <10. ND: not determined. (D) Comparison of the plasma neutralization activity (ID50 titer) between soluble (blue) and liposome-arrayed groups (orange) against SF162 and 16055 pseudoviruses two weeks after the 4th and 5th immunization. Bars indicate group means, symbols represents an animal. Statistical difference were evaluated by Two-way ANOVA followed by Sidak’s multiple comparison test; ns: non-significant; *(p<0.05); **(p<0.01); ***(p<0.001); ****(p<0.0001). These experiments were performed two independent times. See also Figure S1.
Because we observed both increased titers of 16055-neutralizing Abs and superior GC responses after immunization with liposome-displayed Env compared to soluble Env, we wondered whether these two results were linked. Havenar-Daughton et al. recently reported that induction of autologous BG505 neutralizing antibodies correlates with GC B cell frequencies in draining lymph node cells of vaccinated rhesus macaques (Havenar-Daughton et al., 2016). Their report demonstrates that the frequencies of GC B cells in “top”-BG505 neutralizers are statistically higher than in the “low/non”-neutralizers. Using a similar means of analysis, we examined cells from draining lymph nodes from animals inoculated with soluble or liposome-conjugated Env trimers by flow cytometry and detected a significant difference in GC B cell frequencies (p=0.04) between “top” and “low/non”-neutralizers (Figure S1C). While the trend was clear, additional studies are necessary to support a causal relationship between GC B cell frequencies and autologous neutralizing antibodies. The results reported here demonstrate that autologous tier 2 neutralizing Ab responses were consistently elicited in NHPs injected with liposome-arrayed trimers.
mAb isolation identifies diverse Ab binding specificities and genetic properties
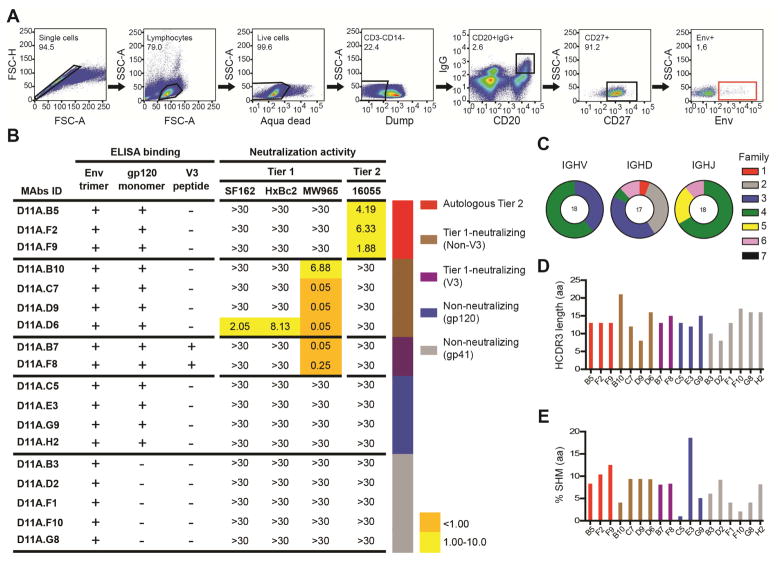
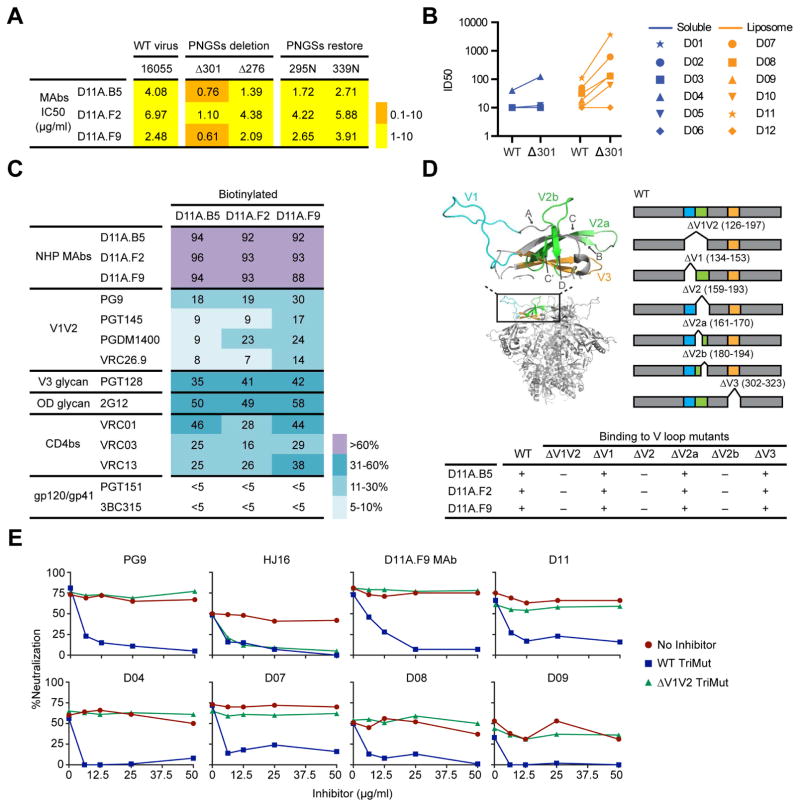
To define immunogenic determinants on the well-ordered 16055 NFL TD CC trimers and identify specificities mediating neutralization of the autologous tier 2 virus, we selected animal D11, which displayed the most potent 16055 neutralizing Ab response after the third immunization and scored among the highest in the GC analysis. We sorted single Env-specific IgG-switched memory B cells by fluorescence-activated cell sorting (FACS) using a panel of Abs specific for macaque memory B cells and a C-terminally biotinylated probe matched to the 16055-trimer immunogen (Figure 4A). We sorted 94 Env-specific CD20+IgG+CD27+ B cells, from which 20 mAbs were successfully cloned and 18 were confirmed to be Env-specific (Figure S2). We performed ELISA of the Env-specific mAbs to assess their binding to monomeric 16055 gp120 and to a linear 16055 V3 peptide. In parallel, we investigated the neutralizing activity of the mAbs using a panel of tier 1 viruses (SF162, HXBc2 and MW965) and the autologous tier 2 16055 virus. Based on these analyses, we categorized the Env-specific mAbs in five groups as follows: those that neutralized the 16055 virus (Autologous Tier 2; n=3); those that neutralized tier 1 viruses but did not bind the V3 peptide (Tier 1-neutralizing (non-V3); n=4); those that neutralized tier 1 viruses and bound the V3 peptide (Tier 1-neutralizing (V3); n=2); those that did not display detectable neutralizing activity and bound both the trimers and the monomers (Non-neutralizing (gp120); n=4) and those that did not display detectable neutralizing activity and bound the trimers but not the monomers (Non-neutralizing (gp41); n=5) (Figure 4B and S3).
Figure 4. Isolated mAbs display diverse binding and neutralization properties.
(A) FACS plots showing the gating strategy used for single cell sorting Env+ memory B cells from animal D11. PBMCs were sampled and stained two weeks after the third immunization. (B) Categorization of the isolated mAbs (n=18) based on Env specificities and neutralization capacities. Neutralization activity was reported as the reciprocal dilution giving 50% of neutralization (ID50 titer). The background neutralization ID50 titer threshold was >30. Genetic features of the mAbs are shown: (C) Distribution of the VDJ family usage for the mAb HC. (D) HCDR3 amino acid length. (E) SHM of VH genes at the amino acid level calculated as the percentage divergence from the assigned germline gene. The sorting and mAb isolation was performed once, the ELISA and neutralization assays twice. See also Figures S2–S5 and Table S1.
The Env-specific mAbs were characterized for their genetic properties including V(D)J gene usage, complementarity determining region 3 (CDR3) lengths and levels of SHM. Most of the vaccine-induced Env-specific mAbs were clonally unrelated, consistent with previous reports (Phad et al., 2015). We applied the recently developed IgDiscover tool (Corcoran et al., 2016) to generate individualized VH, VK and VL databases of animal D11 for accurate gene assignment and SHM calculations. The individualized database resulted in a total of 93, 73 and 44 sequences for VH, VK and VL, respectively, of which 47 VH, 52 VK and 28 VL were not previously described (submitted to Genbank under accession numbers KY196572 – KY196664). From the 47 new VH alleles discovered in D11, the Env-specific mAbs isolated here used seven. About half of the mAbs used kappa light chains and half used lambda light chains. The mAbs used V gene segments belonging to the VH3 and VH4 families with a variety of D and J segments (Figure 4C). The HCDR3 lengths varied between 8 and 21 amino acids (aa) (Figure 4D) and the average SHM at the aa level for the VH region was 7.7% (Figure 4E). The three 16055-neutralizing mAbs, D11A.B5, D11A.F2 and D11A.F9, which were assigned to a new D11 allele, VH4_3T-S3452 (Figure S4A), displayed SHM levels at the aa level ranging from 8.3–12.5% and highly similar HCDR3 regions of 13 residues. These features, together with the fact that they used the same D, JH, VL and JL segments were strong indications that they were clonally related. Of note, initial use of a public VH rhesus macaque database assigned these mAbs to two different VH segments but when the IgDiscover-generated donor-derived database was used the three mAbs were assigned to the same germline V gene confirming clonal relationship (Figure S4B). For complete information of the mAb genetic features (see Figure S5). Thus, from the polyclonal B cell response to Env, we identified one class of clonally related antibodies that mediated autologous tier 2 neutralizing activity.
Epitope mapping of the 16055 tier 2 neutralizing mAbs reveals binding to the V2 region
We next focused on the three 16055-neutralizing mAbs, D11A.B5, D11A.F2, and D11A.F9. Since N-glycans often influence neutralization sensitivity of HIV-1, we began by performing genetic modifications at potential N-linked glycosylation sites (PNGS) of Env. We eliminated PNGS at residues N301 and N276, two sites previously shown to moderate sensitivity to Ab-mediated neutralization. In addition, following inspection of 16055 N-linked glycosylation in the context of the available high-resolution SOSIP structures, we observed a potential hole in the N-glycan shield. Accordingly, we genetically restored PNGS at residues 295 and 339 in full-length 16055 Env. The N301 glycan deletion increased neutralization sensitivity to all three 16055-neutralizing mAbs, resulting in IC50 values of less than 1 μg/ml, compared to wildtype (wt) (Figure 5A). No major changes in neutralization sensitivity were detected for the other N-glycan modified viruses tested in this initial screen. Based on these results, we inspected the plasma neutralizing activity of the delta N301 virus, confirming that increased neutralization of this virus relative to wt was detectable in the polyclonal plasma from all animals displaying 16055 neutralizing activity, including animal D11 (Figure 5B).
Figure 5. The vaccine-induced autologous neutralizing mAbs target a V2 epitope.
(A) Neutralization potency (IC50, μg/ml) of the NHP mAbs with selected N-glycan modifications of 16055 Env compared to wt. (B) Plasma neutralization titers (ID50) from each animal against wt 16055 compared to N301 N-glycan deleted virus. (C) Binding of biotinylated NHP mAbs to 16055 NFL TD CC trimers (His-captured) in the presence of non-biotinylated mAb competitors (left column) as assessed by ELISA, where 0% competition was the absorbance measured with no competitor present. (D) Structure of the V1 (cyan) V2 (green) V3 (orange) region from the clade G X1193.c1 trimer crystal structure (PDB 5FYJ, top left) with strands A, B, C/C′, D and loops V2a and V2b indicated; orientation within one protomer of the trimer shown below. Bar diagram schematic indicating locations of the variable region deletions in 16055 gp120 to map binding (top right) with relative binding to each deletion mutant compared to wt gp120 shown below; +, binding; –, no binding. (E) Differential adsorptions of 16055 virus entry. Control bNAbs PG9 (V2-directed) and HJ16 (CD4bs-directed) along with D11A.F9 and the post 3 NHP plasma were tested at a fixed concentration and pre-incubated with culture medium (no inhibitor) or titrating amounts of wt or ΔV1V2 TriMut gp120 to assess depletion of 16055 neutralizing activity in the TZM-bl assay. The experiments were performed at least two independent times. See also Figure S6.
To broaden the binding specificity screen, we performed cross-competition analysis on the 16055 NFL TD CC trimers using a panel of bNAbs directed to different regions of Env. To validate the cross-competition ELISA, we pre-incubated the His-captured trimers with unlabeled excess Ab and detected nearly complete cross-competition with biotinylated versions of all three 16055-neutralizing mAbs (Figure 5C and S6A), confirming that they formed a distinct cross-competition group. We then analyzed the ability of bNAbs directed toward the trimer apex to reduce binding of the biotinylated NHP mAbs using the V2-directed bNAbs (VRC26, PGT145, PG9 and PGDM1400) and detected weak cross-competition ranging from 7 to 30%. Moving downward from the trimer apex, we analyzed cross-competition generated by the N332-glycan-dependent bNAbs, PGT128 and 2G12. The 16055 NFL TD CC trimers have the glycan at N332 restored at the C-terminal base of V3 as part of the immunogen design process, whereas the virus naturally lacks this N-glycan. With these bNAbs, we detected increased cross-competition (35–58%), consistent with the effects observed following deletion of the 301 N-glycan PNGS at the base of V3. We generated N332-glycan restored 16055 pseudovirus to determine any effects on neutralization, but did not detect any change in neutralizing potency compared to wt 16055 (Figure S6B), indicating no direct interaction with this N-glycan or region of Env. Next, we assessed cross-competition by the CD4bs-directed bNAbs VRC01, VRC03 and VRC13, detecting binding inhibition at 25–46% (Figure 5C and S6A). Finally, we did not detect any cross-competition of the NHP mAbs with bNAbs directed toward the more membrane-proximal regions of the spike (PGT151 and 3BC315) or for other 16055 non-neutralizing mAbs (Figure 5C and S6A). Because of the partial cross-competition with CD4bs-directed bNAbs, we examined binding of the vaccine-induced mAbs to wt 16055 gp120 and 16055 gp120 possessing a D368R substitution that eliminates binding by most CD4bs-directed mAbs. We did not detect any difference in binding of the vaccine-induced mAbs to the D368R-substituted gp120 compared to wt, but did so with previously described CD4bs-specific NHP and human mAbs (Figure S6C) suggesting that the 16055-neutralizing Abs were not CD4bs-directed
To better define the region(s) of mAb recognition, we performed targeted variable (V) region deletions of V1, V2 and V3 of 16055 gp120 and analyzed binding of the mAbs by ELISA (Figure 5D; curves are shown in Figure S6D). There was no detectable effect of mAb binding with the V3 deletion (residues 302-323); however, full deletion of V1V2 (residues 126-197) eliminated nearly all Env recognition by the three mAbs. Targeted deletion of V1 (residues 134-153) revealed detectable but limited effects on mAb recognition, whereas the V2 deletion (residues 159-193) eliminated binding comparable to the V1V2 deletion, indicating that most of the critical Ab contacts are located within V2. We then performed partial deletions of V2 to better define the mAb critical contacts. Based upon current SOSIP structures (Stewart-Jones et al., 2016) we deleted two loops in V2. The first loop (V2a, residues 161-170) overlaps with the binding site of V2-directed apex bNAbs (e.g. PG9, PGT145, CH04); however, no effect on recognition by the 16055-neutralzing mAbs was detected. In contrast, deletion of a second V2 loop (V2b, residues 180-194) resulted in a substantial loss of mAb binding, indicating that this region is a major protein contact of the three mAbs. Inspection of this region in the context of the current high-resolution BG505 SOSIP structures revealed a V2 variable “mini-loop” amidst a cluster of N-linked glycans (Garces et al., 2015; Stewart-Jones et al., 2016). We therefore performed a focused alanine scan within the V2b mini-loop of Env. Using pseudovirus possessing these substitutions, we confirmed that changes in residues 184-186c resulted in a complete loss of neutralizing activity, consistent with the interpretation that this region is a major determinant of the 16055-neutralizing mAbs (Figure S6E).
To determine if the autologous neutralizing Ab activity demonstrated in all liposome-immunized animals from Group 2 and the animal that displayed such activity in Group 1 (D04), we performed differential adsorptions of the plasma as follows. We generated gp120 proteins possessing a set of three mutations in the bridging sheet that eliminate CD4 binding but retain recognition by most CD4bs-directed mAbs (TriMut) (Feng et al., 2012). The lack of CD4 binding allows these proteins to be added directly to the HIV neutralization assay without interfering with viral entry. Additionally, to focus the differential analysis on V1V2, we deleted this domain of TriMut (ΔV1V2 TriMut) and confirmed that deletion did not affect recognition by conformationally sensitive antibodies that bind outside of V1V2, and that binding by the known V1V2-specific bNAb, PG9, was completely eliminated (Figure S6F).
We next assessed the impact of selected neutralizing Abs and plasma on the ability of 16055 pseudovirus to enter the TZM-bl target cells in the presence of each TriMut pair (Figure 5E). To validate the assay, we demonstrate that while the V2-directed bNAb, PG9, can inhibit 16055 virus entry, pre-incubation of PG9 with WT TriMut depleted PG9-mediated neutralization, whereas pre-incubation with ΔV1V2 TriMut had no effect. As another control, neutralization by the CD4bs-directed bNAb, HJ16, could be depleted by both WT and ΔV1V2 TriMut, confirming the integrity of both TriMut variants. Similar to PG9, when the 16055 neutralizing NHP mAb, D11A.F9, was assayed with the TriMut pair, WT but not ΔV1V2 TriMut completely depleted D11A.F9 neutralizing activity, further supporting its targeting of the V1V2 Env region. The same was observed with the corresponding post-3 NHP D11 anti-Env trimer hyper-immune plasma. Similar results were detected for all plasma samples displaying 16055 autologous neutralizing Ab activities, indicating that the major tier 2 autologous neutralization specificity was directed to the 16055 Env V1V2 region.
EM analysis of Fabs from vaccine-induced mAbs reveals their mode of binding to the Env spike complex
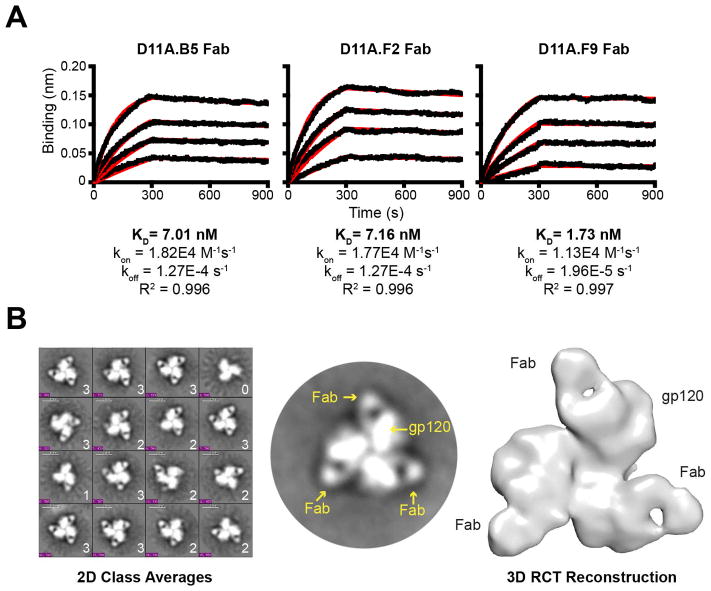
To define further the properties of mAb recognition, we generated Fabs from each of the 16055-neutralizing mAbs to determine affinity to the 16055 NFL TD CC trimers by BLI analysis. With the trimers arrayed on the sensor surface and the monomeric Fabs in solution, we detected high affinity binding ranging from 2 to 7 nM (Figure 6A). Detecting a relatively slow off-rate for the Fabs, we anticipated that negative stain EM might be feasible to determine the angle of access used by these neutralizing mAbs to recognize the trimeric functional spike. Accordingly, EM analysis of the D11A.F9 Fab in complex with the 16055 NFL TD CC soluble trimers was performed. We incubated the Fab in excess with the trimer and following negative stain EM, a variety of Fab:trimer complexes and unliganded single particle proteins were detected (Figure S7A, left). Following classification, trimers bound by zero, one, two or three Fabs were identified and the relative ratios were determined (Figure 6B, left). Of the 8,950 total particles analyzed, the most dominant species was composed of trimers bound with three Fabs per single particle, accounting for approximately 55% of the Fab:trimer mixture (two Fab bound, 33%; one Fab bound, 6%; no Fab bound, 6%). The end-to-end distance of one Fab bound protomer to the next Fab bound protomer within the trimer was estimated to be 17nm. Each protomer of the trimer was estimated to be 6 nm in length along its long axis emanating from the center of the propeller like structure (Figure S7A, right). Using the 2D classifications containing three D11A.F9 Fabs bound per trimer, a 3D reconstruction of the Fab:trimer complex density was generated at 32 Å resolution (Figure 6B, right).
Figure 6. The autologous neutralizing mAbs bind Env with high affinity and target the V2 cap.
(A) Binding curves of vaccine-induced Fabs at selected concentrations (250 nM, top, serial 2-fold reductions) to the 16055 NFL TD CC trimers for affinity measurements by BLI. (B) Representative EM 2D class averages of Fab:trimer complexes with 1, 2 and 3 Fabs per particle as indicated (left); selected 2D image of a single trimer in complex with three Fabs (middle); 3D reconstruction of the 16055 NFL TD CC trimer in complex with three D11A.F9 Fabs, top view (right). See also Figure S7.
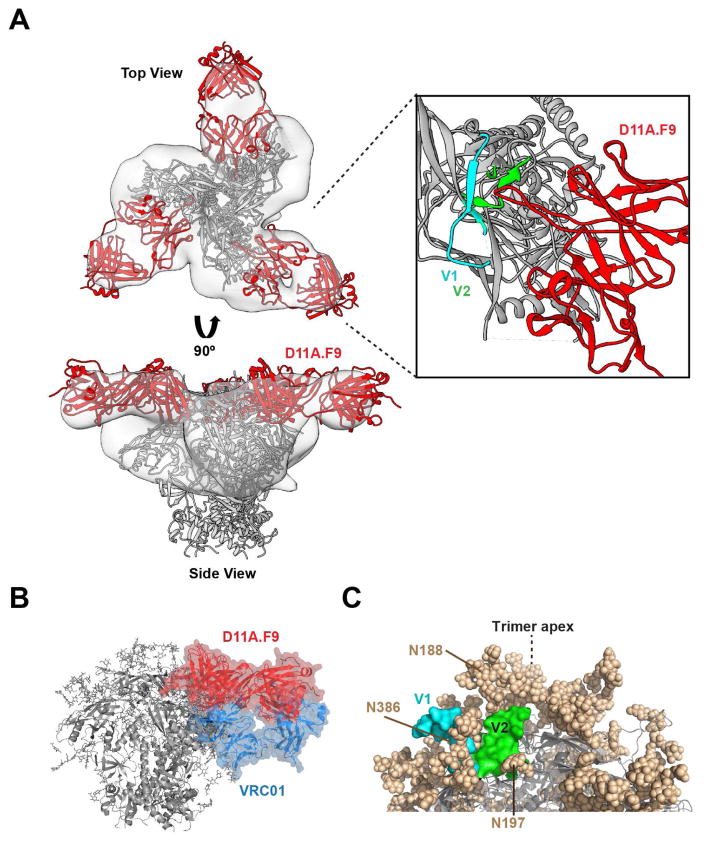
To better approximate the mode of binding of D11A.F9 Fab to the trimer, we fitted in the high-resolution BG505 SOSIP.664 crystal structure (PDB 5T3Z) into the trimer density. To fit the remaining density, we first generated a D11A.F9 Fab model using LYRA; other programs resulted in very similar structural overlays (Figure S7B). Inspection of the HCDR3 sequence revealed a characteristic “signature” of two cysteine residues separated by four residues present in all three vaccine-induced mAbs (Figure S4C). Although most programs predicted the known Fab disulfide bonds, none predicted formation of a disulfide in the relatively short HCDR3. The modeled D11A.F9 Fab structure fit well into the remaining density emanating from each protomer (Figure 7A, top). When viewed from the side of the complex, D11A.F9 appears to approach the Env trimer with a horizontal route of access (Figure 7A, bottom). The docking indicated that the modeled D11A.F9 would be positioned proximal to the V1V2 region (Figure 7A, right). This fitting was consistent with the deletion mapping and Ala scan analysis, indicating that the major contacts of the mAbs were located in V2. Docking of the structurally defined bNAb VRC23 (PDB 4J6R) into the density was also consistent with this horizontal mode of access and the proximity of the Fab-defined density to V2 (Figure S7C). This angle of access to the V2 region is considerably different the trimer-specific V2-directed bNAbs (Figure S7D), perhaps related to the limited cross-competition detected for these antibodies.
Figure 7. Docking of the EM 3D reconstruction of the Fab:trimer density suggests a horizontal binding approach to the Env trimer apex.
(A) Top and side views of the D11A.F9-liganded trimer EM density (light gray) with the high-resolution BG505 SOSIP.664 crystal structure (PDB 5T3Z; gray ribbon) and the D11A.F9 Fab model (red) fit within the 3D reconstruction (left); V1 (cyan) and V2 (green) are highlighted on each protomer. Expanded view of the D11A.F9 Fab in proximity of the V2 (green) and V1 (cyan) region; dashed lines indicate the predicted structure of the loops which are incomplete in the crystal structure. (B) Side view of the VRC01 crystal structure (PDB 5KZC, blue) and the D11.A.F9 model (red) docked into the natively glycosylated BG505 SOSIP.664 crystal structure (PDB 5T3Z). (C) The molecular surfaces of V1 (cyan) and V2 (green) regions of one protomer in the clade G X1193.c1 crystal structure (PDB 5FYJ) are shown along with proximal N-glycans (tan spheres). See also Figure S7.
The horizontal angle of approach displayed by D11A.F9 is close to that used by the prototypic CD4bs-directed bNAb, VRC01, but on a plane closer to the trimer apex (or more distal to the viral membrane; see Figure 7B). Docking of VRC01 (PDB 5KZC) and D11A.F9 Fabs into the natively glycosylated BG505 trimer crystal structure (PDB 5T3Z) revealed considerable overlap of the densities for these two mAbs (Figure 7B), consistent with the cross-competition displayed by VRC01 and the other CD4bs-directed bNAbs. In addition, the fitting revealed that D11A.F9 would be proximal to the N-glycan at residue 301, consistent with increased neutralization potency following disruption of this PNGS (Figure S7E). Inspection of the high-resolution crystal structure of the clade G trimer (PDB 5FYJ), which possesses a V1V2 loop length and N-glycan distribution similar to 16055, reveals the region of V2 that we identified to contain the major D11A.F9 contacts is surrounded by N-glycans. Elements of V2 would likely be most accessible to Ab from a route of access perpendicular to the orientation shown in (Figure 7C). These analyses reveal that the vaccine-induced 16055-neutralizing mAbs display a horizontal mode of binding to the trimer apex.
Discussion
Here, we inoculated NHPs with well-ordered, structurally defined clade C 16055 NFL TD CC trimers either administered as soluble protein or displayed on synthetic liposomes, and analyzed the elicited B cell response. We found that the high-density liposome format induced superior GC responses and higher tier 2 virus autologous neutralizing Ab titers compared to the same trimers in the soluble format. Thus, the advantage of particulate-array observed in the preceding small animal studies (Ingale et al., 2016) was supported by the current NHP study, demonstrating that this effect translates to primates. We further found that the animals displaying autologous tier 2 neutralizing activity generally displayed more potent GC responses, a potential linkage that will be important to investigate in subsequent studies.
To define the specificities mediating autologous tier 2 neutralizing activity, we isolated a panel of functional Env-specific mAbs from a trimer-liposome immunized NHP. Of these, three clonally related mAbs were capable of neutralizing the autologous tier 2 16055 isolate and thus recapitulated the plasma activity against this virus. By mutagenesis studies, cross-competition analysis and targeted Env deletion, we mapped the critical contacts of these mAbs to the V2 region of Env. Negative stain EM of mAb D11A.F9 in complex with the 16055 NFL TD CC trimers revealed a horizontal approach to the V2 region. This binding mode is unique for the antibodies described here, but reminiscent of the VRC01-class CD4bs-directed bNAbs to the trimeric spike, albeit on a higher plane proximal to the V2 apex. This binding mode differs from the more vertical angle toward V2 used by bNAbs PG9, PGT145 and VRC26 that target another epitope region with the V2 (Andrabi et al., 2015; Doria-Rose et al., 2014; McLellan et al., 2011). Although the V region “cap” is likely to be one of the most accessible sites for Ab recognition on Env, it is heavily shrouded with N-linked glycans. This may prevent vertical access by most Abs, requiring the infrequent very long HCDR3s present in the apex-targeting bNAbs isolated from chronic HIV-1 infection, highlighting the capacity of these vaccine-elicited antibodies to penetrate the evolved N-glycan shroud to mediate tier 2 virus neutralizing activity.
It is therefore encouraging that Ab responses against this glycan-shielded region were generated after just three vaccine inoculations even if neutralization breadth was not, as expected, achieved with this single immunogen. Autologous neutralizing Ab titers were boosted by the fourth vaccine inoculation in four animals but only in one animal by the fifth inoculation, possibly reflecting that the autologous neutralizing Ab response had reached an “affinity ceiling” at this point of the regimen (Batista and Neuberger, 1998; Poulsen et al., 2011), perhaps because performed with the homologous immunogen. In this regard, we note that neutralizing mAbs isolated from animal D11 after the third immunization, D11A.B5, D11A.F2 and D11A.F9, already displayed high affinity binding (nM levels) to the immunogen. In addition, differential adsorptions indicated that V1V2 antibodies appear to be the major specificity mediating tier 2 autologous neutralization in all animals displaying such capacity following 3 inoculations of the well-ordered trimers. This is in contrast to the neutralizing specificity reported for neutralizing antibodies to the BG505 SOSIP trimers, which target a relatively large gap in the glycan shield located near the base of the trimer (McCoy et al., 2016). That the V2 surface recognized by the 16055-induced mAbs is variable likely arises from Ab pressure at this site, highlighting that this epitope region may be relatively immunogenic and may present opportunities to expand neutralization breadth by immunogen modification.
The RV144 vaccine clinical trial demonstrated a correlation between V1V2-directed Abs and protection (Haynes et al., 2012), increasing interest in Ab responses to this region. The immunogenic properties of the V2 region were also shown by the isolation of glycan-dependent V2-targeting bNAbs from chronic infected individuals (McLellan et al., 2011; Walker et al., 2009) and isolation of monoclonal antibodies with ADCC activity from RV144 clinical trial vaccine recipients (CH58-59) (Liao et al., 2013), encouraging the design of V2-based immunogens (Jiang et al., 2016; Mayr et al., 2013). Further, the V2 region was shown to interact with the integrin α4β7 (Arthos et al., 2008) proximal to the region where the vaccine-induced mAbs reported here bind. The mAbs isolated and described in the current study confirm that the V2 cap region of a well-ordered trimer can elicit functional neutralizing antibodies after vaccination. The current study also demonstrates the value of generating individualized databases for Ab germline V segments and to use these for gene assignment and SHM analysis. We recently reported the extensive allelic diversity in rhesus macaque germline V genes, highlighting that current public NHP V gene databases, such as those hosted by IMGT (Lefranc et al., 1999), are far from complete. The use of IgDiscover as shown here allows rapid and reliable identification of germline alleles in each studied subject giving high confidence in subsequent analysis of SHM.
In conclusion, we report here Env trimer vaccine-induced neutralizing Ab responses and the isolation of autologous tier 2 neutralizing mAbs induced by a clade C HIV-1 clinical isolate, of relevance as the majority of infections world-wide are of this subtype. The clade C focus, the use of structurally homogeneous trimers, the high-density trimer array and the multi-faceted analysis presented here are important steps toward defining the range of immunogenic neutralizing determinants on well-ordered Env trimers targeted by the adaptive humoral response following immunization.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gunilla Karlsson Hedestam (Gunilla.Karlsson.Hedestam@ki.se).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics statement
The animal work was conducted with the approval of the regional Ethical Committee on Animal Experiments (Stockholms Norra Djurförsöksetiska Nämnd). All methods were carried out in accordance with the approved guidelines.
Animals
Twelve female rhesus macaques (Macaca mulatta) of Chinese origin, 4–10 years old, were housed at the Astrid Fagraeus Laboratory at Karolinska Institutet. Housing and care procedures were in compliance with the provisions and general guidelines of the Swedish Board of Agriculture, and the facility has been assigned an Animal Welfare Assurance number by the Office of Laboratory Animal Welfare (OLAW) at the National Institutes of Health (NIH).
The macaques were housed in pairs in 4 m3 cages, enriched to give them possibility to express their physiological and behavioral needs. They were habituated to the housing conditions for more than 6 weeks before the start of the experiment, and subjected to positive reinforcement training in order to reduce the stress associated with experimental procedures. All immunizations and blood samplings were performed under sedation with ketamine 10–15 mg/kg intramuscularly (i.m.) (Ketaminol 100 mg/ml, Intervet, Sweden). The macaques were weighed at each sampling. All animals were confirmed negative for simian immunodeficiency virus (SIV), simian T-cell lymphotropic virus, simian retrovirus type D and simian Herpes B virus.
METHOD DETAILS
Generation of clade C 16055 NFL trimers
The design, expression, and purification of 16055 NFL TD CC trimers, from transient expression in FreeStyle 293F cells using 293fectin (Invitrogen) have been previously described (Guenaga et al., 2015). In brief, the 16055 Env coding sequence was modified as follows: the native signal sequence was replaced by the CD5 leader sequence, the furin cleavage motif at the C terminus of gp120 was replaced with two copies of the G4S flexible linker, K334S was introduced to restore the 332N-glycan supersite, eight TD modifications to stabilize trimer formation along with I559P, I201C, and A433C substitutions. The sequence was terminated at D664 followed by a G4S linker and His6 tag. Cell culture supernatants containing the expressed Env proteins were collected 5–6 days post transfection and purified over a Galanthus nivalis lectin-agarose (Vector Laboratories) column followed by size exclusion chromatography to isolate the trimer.
Generation of 16055 NFL trimer-conjugated liposomes
Liposome preparation and protein conjugation has been described elsewhere (Ingale et al., 2016). In brief, liposomes were composed of a molar ratio of 60:36:4 of DGPC, cholesterol, and DGS-NTA(Ni) and extruded through a series of filters to obtain 0.1 μm in diameter. To conjugate the His-tagged trimers to liposomes, 2.2 mg of trimer was added to 500 ul of liposomes and incubated for 2 hr at room temperature (RT). The mixture was then passed through a Superdex 200 column to remove unbound protein. Liposome fractions were pooled and stored at 4°C. The 16055 NFL TD CC-conjugated liposomes were quantitated using a standard curve generated by soluble 16055 NFL TD CC with the Advanced Protein Assay Reagent (Cytoskeleton). To ensure proper and uniform conjugation of the trimer to liposome and that the trimers remained well-ordered, the samples were further visualized by negative stain EM and antigenicity assessed by biolayer light interferometry (BLI) using an Octet Red system (ForteBio, Pall) as previously described (Ingale et al., 2016). In brief, samples were stained with 2% phosphotungstic acid solution (pH 6.9) and grids were examined on a Philips CM100 electron microscope (FEI) at 80 kV and images were acquired with a Megaview III charge-coupled device camera (Olympus Soft Imaging Solutions). For BLI, biotinylated wheat germ agglutinin (WGA) (Vector Laboratories) on streptavidin biosensors (ForteBio) was used to capture the trimer-conjugated liposomes. Association of the mAbs to the immobilized trimers was measured for 5 min followed by dissociation in buffer for 10 min. Assayed mAbs included the trimer-specific bNAbs (PGDM1400, PG16, and PGT145), the CD4bs-directed bNAbs (VRC01 and VRC06b) and non-bNAbs (F105 and GE136). Reference sensors were included to ensure minimal non-specific binding of mAbs to the WGA sensors. The stability of the trimer-liposomes were further assessed in PBS or Matrix M (Novavax AB, Uppsala) adjuvant at 37°C for 24 hr. Samples taken over the time course were visualized by negative stain EM as described above.
Immunization and sampling
The macaques were divided into two experimental groups according to the delivery platform of the trimers: soluble trimers (n=6) and liposome arrayed-trimers (n=6). Both the soluble and liposome-arrayed 16055 NFL TD CC trimers (100 μg per inoculation) were formulated in Matrix-M adjuvant (Novavax) at 75 μg per inoculation. All inoculations were given i.m. in a total volume of 1 ml, divided equally between the left and right quadriceps and performed at weeks 0, 4, 12, 26 and 40. Peripheral blood samples were taken at nine different times, while lymph node (LN) biopsies were taken twice. Animal D11 was terminated 2 weeks after the 3rd immunization for comprehensive B cell and tissue archiving. Therefore, post-4 and post-5 samples were not and could not be collected.
PBMC isolation
Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation from EDTA blood by Ficoll-Paque™ PLUS (GE Healthcare), washed extensively in PBS treated with red-blood cell lysis buffer and frozen in 90% heat-inactivated FBS and 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich).
Lymph node biopsies
Samples of external inguinal lymph nodes (LNs) were obtained from the macaques 2 weeks after vaccine inoculations. The animals were anaesthetized by intramuscular injection of 10–15 mg/kg of ketamine and 0.05 mg/kg of medetomidine. Carprofen (4 mg/kg) was given i.m. as analgesia. A strict aseptic technique was used throughout the procedure. One or two LNs were localized and collected by blunt dissection through a 10–15 mm incision. After suturing, the anesthesia was reversed with atipamezole, 0.25 mg/kg i.m.
Immunofluorescence analysis of lymph node (LN) sections
LNs were snap frozen in optimal cutting temperature (OCT) with dry ice and stored at −80°C until use. OCT blocks were transferred to −20C freezer to thaw for approximately 60 min. Cryostat settings: Knife temperature −17°C and tissue −14°C. Sections (6 μm) were obtained using C35 knives and mounted on Superfrost plus glass. Slides were dried for 20 min before fixing with 2% PFA (Merck) at RT for 20 min. Cryosections of LN biopsies (6 μm) were stained to detect CD3, Ki-67 and PD-1 positive cells, with a combination of donkey anti-rabbit CD3 (Dako, Carpinteria, CA), rabbit anti mouse Ki67 (BD) and donkey anti-goat PD-1 (R&D systems, Minneapolis, MN, USA) antibodies. Briefly, tissue sections were blocked 30 min at RT with perm wash (Sigma) and 1% fetal calf serum followed by 15 min with Avidin/Biotin blocking kit (Bionordika). Each antibody was diluted in perm wash, added one at a time and incubated overnight (o/n) at 4°C. After incubation of each antibody, the tissue slides were washed three times with perm wash and blocked with 1% serum from the same species the secondary antibody was from. Secondary antibodies were: biotinylated anti-mouse (Dako), biotinylated anti-goat or biotinylated anti-rabbit (Jackson ImmunoResearch). Subsequently, streptavidin-conjugated AlexaFluor-405, -555, -647 were added and incubated for 30 min at RT (Invitrogen). After the stain, tissue slides were washed with water, air-dried in the dark (5–10 min) and mounted with Prolong Gold (without DAPI; ThermoFisher Scientific) and 24×50mm cover slip. Tiled images of entire LNs were acquired using a Nikon Eclipse Ti-E, Nikon A1 confocal microscope, 20x objective and analyzed in an automated fashion with Cell Profiler software (Carpenter et al., 2006). In brief, GCs were identified manually with Cell Profiler software as collections of CD3+, PD1+ and Ki67+ cells forming clear sub-follicular structures. PD1+ cells were CD3+ (Tfh cells) while Ki67+ cells were mostly CD3- (GC B cells). Statistical analysis was performed using GraphPad Prism.
Flow cytometry of LN cells
Frozen single cell suspensions from inguinal LNs were thawed at 37°C and washed twice in pre-warmed supplemented media. Cells were washed twice with PBS and counted using 0.4% Trypan Blue exclusion by a Countess cell counter (Life Technologies); cell viability was consistently >80%. Cells were suspended in 100μL of PBS on a 96 well-plate (1–4×106 cells/well) and incubated for 10 minutes at 4°C with Fc receptor (FcR) blocking (eBiosciences) and Aqua Dead Cell Stain Kit (Life Technologies). Cells were washed with FACS buffer (PBS+2% FBS) and resuspended in Brilliant Violet buffer (BD Biosciences) following the manufacturer’s instructions. Surface stain included antibodies (BD Biosciences) against CD3 clone SP34-2 FITC, CD20 clone 2H7 BV711, CD4 clone L200 FITC, CD14 clone M5E2 FITC, staining was performed at 4°C during 15 min incubation. Cells were washed twice with FACS buffer, prior to permeabilization with the transcription factor kit (BD Biosciences) for 30 min at 4°C. Cells were washed twice with perm wash, stained with antibodies (BD Biosciences) against Bcl-6 clone K112-91 PECy7 and Ki-67 clone B56 PE, for 15 min at 4°C. and washed twice again with perm wash. All Abs were previously titrated for optimal staining of rhesus macaque LN cells. Samples were collected on a FACS LSRII (BD Biosciences) and analyzed using FlowJo software.
ELISA analysis of plasma samples
HIV-1 Env-specific IgG titers in plasma were measured by ELISA as previously described (Ingale et al., 2016). MaxiSorp 96-well plates (Nalgene Nunc International) were coated overnight at 4°C with a mouse anti-His tag mAb (1.5 μg/ml; R&D Systems). After blocking the plates with PBS containing 2% of bovine serum albumin for 2hr at RT, the plates were incubated with 16055 NFL TD CC at 3 μg/ml for 2 hr at RT. Next, the plates were incubated with 5-fold serial dilutions of plasma starting at 1:50 for 1hr at RT. Env-specific IgG was detected by adding a secondary horseradish peroxidase (HRP) conjugated anti-monkey IgG antibody (1:10000) (Nordic Immunology, Tillburg, The Netherlands) and the signal developed by addition of tetramethylbenzidine (TMB) substrate (Invitrogen). Adding equal volume of 1M H2SO4 stopped the reaction and the optical density (OD) was read at 450 nm. Between each incubation step, the plates were washed 6 times with PBS supplemented with 0.05% Tween 20. The half-max binding titers (OD50) for each sample was calculated by interpolation from mean OD50 values calculated from an immunized plasma using the formula (ODmax-ODmin)/2).
HIV-1 neutralization assays
Neutralization assays were performed using a single round infectious HIV-1 Env pseudovirus assay with TZM-bl target cells (Li et al., 2005). To determine the Ab concentration and the plasma dilution that resulted in a 50% reduction in relative luciferace units (RLU), serial dilutions of the mAbs and the plasma were performed and the neutralization dose-response curves were fit by non-linear regression using a 5-parameter hill slope equation using the R statistical software package. Diverse HIV-1 virus isolates were used in the neutralization assays. The clade C MW965 Env plasmid was obtained from the AIDS Research and Reagent Repository and the sources of other Env-encoding plasmids were described previously (Li et al., 2005). Site-directed mutagenesis using QuikChange (Agilent Technologies) or Q5 (NEB) mutagenesis kits per the manufacturer’s protocol generated Env mutants. Peptide competitions were done in the same assay format as the neutralization assay, except that the peptide was added to the plasma 30 min prior to the addition of virus. The V3 peptide (TRPNNNTRKSIRIGPGQTFYATGDIIGNIRQAY) based on the 16055 V3 sequence was synthesized by Genscript. Neutralization capacities of mAbs were reported as the Ab concentration resulting in 50% virus neutralization (IC50) whereas the result for plasma were reported as the plasma neutralization ID50, which is the reciprocal of the plasma dilution producing 50% virus neutralization.
Env-specific memory B cell sorting by flow cytometry
Env-specific memory B cells were single cell sorted from fresh isolated PBMCs from animal D11 sampled two weeks after the third immunization. Single cells were sorted with a three-laser FACSAria cell sorter (BD Bioscience) by gating Aqua Blue−, CD3−,CD14−, CD20+, IgG+, CD27+, Env+ cells into 96-well PCR plates containing 4 μl of cell lysis buffer (Tiller et al., 2008). Plates were sealed and immediately frozen on dry ice prior to storage at −80°C. All fluorescently labeled antibodies and the Env-trimer conjugated to streptavidin-allophycocyanin (Invitrogen) were carefully titrated to stain rhesus macaque PBMCs.
Single B cell RT-PCR
Cell lysates from single sorted HIV-1 Env-specific memory B cells from animal D11 were used as a source of RNA for reverse transcription and IgG V(D)J sequences amplification as described previously (Sundling et al., 2012a). Briefly, the 96-well plates, containing single B cell, were thawed at 4°C and reverse transcribed to cDNA by addition of random hexamers (Roche), dNTPs (Invitrogen), RNAsin (Promega) and SuperScript III reverse transcriptase (Invitrogen). Heavy and light chain V(D)J segments were amplified by nested PCR: 3μl of product (both rounds), HotStar Taq Plus Kit (Qiagen, CA) and 5′ leader sequence–specific and 3′ IgG-specific primers were used (Table S1) (Sundling et al., 2012b). Positive PCR products from heavy and light chain plates were purified and sequenced (GATC biotech). Restriction sites were added to productive heavy chain (HC) and light chain (LC) sequences, by high fidelity cloning PCR containing a total volume of 25 μl, 1 μl nested PCR product (1:100 dilution), Kapa mix (Kapa Biosystems) and 5′ and 3′ custom cloning primers containing restriction sites previously described (Sundling et al., 2012a; Sundling et al., 2012b).
Cloning and expression of monoclonal antibodies
Cloning PCR products were evaluated on 1% agarose gel (~450 bp for HC and ~350 bp for κ or λ LC), and then PCR purified (Thermo Scientific kit). Cloning of the Ab sequences into expression vectors containing human Igγ1 H, Igκ1 L, or Igλ2 L constant regions was performed with FastDigest restriction enzymes (Thermo Scientific) according to the manufacturer’s instructions. Digested and purified PCR products ligated into linearized, shrimp alkaline phosphatase-treated vectors using T4 DNA ligase (Thermo Scientific) were used to transform XL10-Gold ultracompetent cells by heat shock at 42 °C for 45 sec according to manufacturer’s protocol (Agilent Technologies). Bacterial colonies containing plasmids with inserts of the correct size were verified by PCR and expanded followed by plasmid purification (Qiagen, CA) and Sanger sequencing (GATC biotech). For expression, equal amounts of each HC and LC vector DNA (15 μg of each) were transfected together with 30 μl of Freestyle Max reagent (Invitrogen) in the presence of Opti-MEM medium (Gibco) into FreeStyle 293F cells, cultured in 30 ml of FreeStyle 293 expression medium (Life Technologies) at cell density 1 × 106 cells/ml and ≥ 90% viability. On day 4 or 5 after transfection cell culture supernatants were tested for total Ab production and binding to different HIV-1 Env ligands by ELISA. Cultures containing functional Env-specific Abs were then harvested and purified 7 days after transfection using Protein G Sepharose columns (GE Healthcare). All purified recombinant mAbs were further analyzed by SDS-PAGE under reducing condition using NuPAGE Novex 4–12% Bis-Tris polyacrylamide gels and NuPAGE reducing agent (Life Technologies) according to the manufacturer’s instructions.
Generation of individualized germline V gene databases and Ig gene sequence analysis
To identify the germline VH, VK and VL sequences utilized in each antibody rearrangement the IgDiscover germline program was used. IgM, IgK and IgL libraries were produced from a pre-immunization PBMC sample from animal D11 and sequenced by NGS. In brief, total RNA was isolated from a sample of 5 million cells, of which 400 ng was reverse transcribed with Superscript IV (Invitrogen) using gene-specific primers for the IgM, IgK and IgL constant regions. In addition to a unique molecular identifier (UMI), each primer contained a universal reverse primer sequence that enabled amplification when used in a PCR reaction containing multiplex forward primer sets for VH, VK and VL genes, respectively. The amplified libraries of IgM were sequenced in forward and reverse directions using the Illumina MiSeq 2 × 300 bp kit and paired, resulting in library sizes of 737,111 (IgM), 893,975 (IgK) and 1,116,529 (IgL). These were analyzed using IgDiscover to produce individualized germline databases for the heavy and light chain variable alleles.
To assign V(D)J segments of vaccine-elicited mAbs, single cell RT-PCR-generated HC and LC sequences were analyzed by using IgBLAST to identify their V(D)J germline gene segments. Assignments of VH genes were performed using the D11 individualized database, VK and VL genes were assigned using a combination of the individualized and the IMGT databases, while J and D segments were assigned using the IMGT database.
Characterization mAb binding analysis by ELISA
The mAbs were tested for binding against 16055 NFL TD CC trimer, gp120, gp120 V region deletion mutants, or V3 peptide directly coated on the ELISA plate as previously described (Navis et al., 2014). The gp120 deletion mutants included: ΔV1V2 (126-197), ΔV1 (134-153), ΔV2 (159-193), ΔV2a (161-170), ΔV2b (180-194), and ΔV3 (302-323) with residues replaced with GAG or GGSGG for ΔV2. The mAbs were tested for binding using MaxiSorp 96-well plates (Nalgene Nunc International) coated at 2 μg/ml with wt gp120, gp120 V region deletion mutants, V3 peptide or an anti-His tag mAb (to capture the trimer) in PBS at 4 C overnight. After incubation with blocking buffer (PBS containing 2% non-fat milk), the mAbs were added and incubated for 1 hour at 37 C. Binding was detected by secondary HRP-conjugated anti-human Fcγ (Jackson ImmunoResearch) at 1:10,000 for 1 hour. The signal was developed by addition of TMB substrate (Invitrogen) for 5 min, reactions were terminated with 1 N sulfuric acid, and the optical density (OD) was read at 450 nm. Between each incubation step, the plates were washed six times with PBS containing 0.05% Tween.
Cross-competition ELISA
D11A.B5, D11A.F2, and D11A.F9 were biotinylated using EZ-Link NHS-Biotin (Pierce Biotechnology, Thermo Scientific) per the manufacturer s protocol. For cross-competition ELISA assays, 16055 NFL TD CC trimers were captured on the ELISA plate by a mouse anti-His tag mAb (R&D Systems) coated at 2 μg/ml in PBS at 4°C overnight. Five-fold serial dilutions of various bNAbs and non-bNAbs were pre-incubated with the captured trimer at RT for 30 min prior to addition of the biotinylated mAbs at a concentration previously determined to give ~75% of the maximum binding signal (i.e. binding to trimer with no competitor present) for 60 min at RT. The bound biotinylated mAbs were detected using HRP-conjugated streptavidin (Sigma). The signal was detected with TMB substrate and reaction stopped with 1 N sulfuric acid as above. Wells were blocked with 5% non-fat milk in PBS-T (0.2% Tween-20) after coating with the anti-His mAb and washed with PBS-T after each incubation step. Samples were diluted in 10% blocking buffer. Competition is expressed as percent inhibition where 0% was the absorbance measured with no inhibitor present.
Differential adsorptions
Post 3 NHP plasma samples, D11A.F9, PG9, and HJ16, at a fixed concentration corresponding roughly to their IC70 values, were pre-incubated with culture medium (no inhibitor) or serially diluted amounts of wt or ΔV1V2 TriMut gp120 starting at 50 μg/ml for 1 hr at 37°C prior to addition of pseudovirus in the TZM-bl neutralization assay, as previously described. TriMut refers to three mutations (I423M, N425K, and G431E) that inhibit binding to CD4 on the TZM-bl target cells so that the proteins can be left in during assessment of viral entry.
Binding kinetics of the NHP Fabs
D11A.B5, D11A.F2, and D11A.F9 Fab binding to 16055 NFL TD CC were assessed by BLI. The trimers (10 μg/ml) were captured onto HIS2 biosensors (ForteBio, Pall Life Sciences), followed by a wash in kinetics buffer (0.1% BSA, 0.02% Tween-20 in PBS) to remove unbound trimers and establish a baseline. The immobilized trimers were then immersed in wells containing 2-fold serially diluted Fabs (starting at 250 nM) for 300 s to assess binding and subsequently in kinetics buffer for 900 s to obtain the dissociation phase curve. The assays were performed at 30°C at a shaking speed of 1,000 rpm. Data Analysis 7.0 evaluation software (ForteBio) was used to assess the response curves and to calculate kinetic parameters using a global fit 1:1 model.
EM negative staining and modeling
The 16055 NFL TD CC trimers were incubated with ten molar excess of D11A.F9 Fab at RT for 1 hr. A 3 μl sample (~0.01 mg/ml) was applied onto a thin layer of continuous carbon placed over a C-flat holey carbon grid and then stained with uranyl formate. Data were obtained using an FEI Tecnai T12 electron microscope, operating at 120 eV and equipped with an FEI Eagle 4k x 4k CCD camera. Images were acquired at multiple scales to assess overall distribution. High magnification (67,000x, 0.16 nm/pixel) were acquired at a nominal underfocus of −2 μm to −1 μm and electron doses of ~25 e/A2. Automated picking protocols were used to select individual tilted and untilted images taken at 67,000x. Particles were matched across tilted image pairs by auto alignment (Voss et al., 2009) and separated into classes by a reference-free alignment strategy based on the XMIPP processing package. Random Conical Tilt (RCT) geometry was used for 3D reconstructions based on 817 particles. The nominal resolution was determined to be ~32 Å using a Fourier Shell Correlation cut-off of 0.5. Surface renderings of the density maps were produced with UCSF Chimera.
A structural model of the D11A.F9 variable fragment (Fv) was generated from the corresponding aa sequence using three different molecular modeling servers: LYRA (Lymphocyte Receptor Automated Modeling) (Klausen et al., 2015), RaptorX (Kallberg et al., 2012), and Rosetta Antibody Protocol (Lyskov et al., 2013). The Fvs were superimposed using Chimera. Structures of BG505 SOSIP.664 (PDB 5T3Z) with the D11A.F9 Fab model generated by LYRA were manually fitted into the EM densities and refined using the Chimera “fit in map” function (Goddard et al., 2007). The ligands, IOMA and 10-1074 Fabs, in the 5T3Z structure were removed prior to fitting the BG505 SOSIP.664 trimer into the D11A.F9:16055 NFL TD CC EM density. The D11A.F9 model or the HIV bNAb VRC23 (PDB 4J6R) was then fitted into the unoccupied EM density. To determine the relative angle of approach and overlap of D11A.F9 with other bNAbs, we used VRC01 (PDB 5KZC) and PG9 (PDB 3U4E) from crystal structures. We aligned the gp120 subunit of the 5KZC structure to the gp120 subunit of the BG505 SOSIP.664 (PDB 5T3Z) to visualize VRC01 relative to D11A.F9. Similarly, we aligned the Env V1V2 of the PG9 scaffold of the 3U4E structure to visualize PG9 relative to D11A.F9.
QUANTIFICATION AND STATISTICAL ANALYSIS
Comparison of < 3 groups was done by Mann-Whitney to perform simultaneous evaluation of statistical differences between samplings and groups over the course of the study. Two-way ANOVA was used followed by Sidak’s post-test for multiple comparisons. All statistical analysis was done with GraphPad Prism software version 5 or 6 and considered significant at * for p≤0.05, ** for p≤0.01, *** for p≤0.001 and **** for p≤0.0001.
DATA AND SOFTWARE AVAILABILITY
The Env-specific mAb sequences have been deposited in GenBank, HC sequences accession numbers KY196532 – KY196551, LC sequences accession numbers KY196552 – KY196571.
Supplementary Material
Acknowledgments
We thank Dr. Bengt Eriksson and all personnel at Astrid Fagraeus laboratory for expert assistance with rhesus macaques and Richard Wilson and Barbara Lemaire for assistance with protein production. We also thank Novavax, AB, Uppsala, Sweden, for making the Matrix-M adjuvant available and Christina Corbaci for help with the graphical abstract. EM analysis of the trimer-conjugated liposomes was performed at the TSRI Microscopy Core with assistance from Malcom Wood and Theresa Fassel. EM imaging of Fab:trimer complexes and 3D reconstructions were performed by NanoImaging Services, Inc, La Jolla, CA. This work was funded by an NIH P01 grants AI104722 (HIVRAD), the intramural research program of the Vaccine Research Center, NIAID, NIH, the Scripps CHAVI-ID grant AI100663 (RTW), grants for doctoral education to PM, GL and GP and from the IAVI Neutralizing Antibody Consortium (RTW and GKH) and its generous donors including the Bill and Melinda Gates Foundation CAVD and USAID. The full list of IAVI donors can be found at www.iavi.org. The authors have no conflict of interest.
Footnotes
Author Contribution:
P.M.M., G.K.H and R.T.W. planned the immunization scheme. J.G. designed and produced the 16055 NFL TD CC trimers. J.I. and S.B. produced trimer-conjugated liposomes. M.S. performed immunizations and collected samples. P.M.M., M.A., M.C., L.P., N.V., G.P. and V.D. prepared vaccine formulations and processed the samples. G.L. and K.L. analyzed lymph node sections. S.O. and J.M. evaluated Ab neutralizing activity. P.M.M., M.A., G.P. and N.V. sorted B cells, cloned mAbs and performed binding analysis. M.C. generated the individualized D11 germline V gene database. P.M.M, G.P. and M.C., analyzed the genetic properties of the mAbs. K.T. generated Fabs for EM analysis, performed cross-competition studies and analyzed Env point mutants. Y.F. generated and evaluated binding to Env deletion mutants. J.G. performed Fab modeling and EM density fitting analysis. P.M.M., K.T., J.G., M.A. F.Y., G.P., N.V., M.C., R.T.W. and G.K.H. analyzed the data and prepared the figures. P.M.M., K.T., R.T.W. and G.K.H. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrabi R, Voss JE, Liang CH, Briney B, McCoy LE, Wu CY, Wong CH, Poignard P, Burton DR. Identification of Common Features in Prototype Broadly Neutralizing Antibodies to HIV Envelope V2 Apex to Facilitate Vaccine Design. Immunity. 2015;43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nature immunology. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nature immunology. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran MM, Phad GE, Nestor VB, Stahl-Hennig C, Sumida N, Persson MA, Martin M, Karlsson Hedestam GB. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun. 2016;7:13642. doi: 10.1038/ncomms13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166:1445–1458. e1412. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, McKee K, Tran K, O’Dell S, Schmidt SD, Phogat A, Forsell MN, Hedestam GB, Mascola JR, Wyatt RT. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. Journal of Biological Chemistry. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell MN, Dey B, Morner A, Svehla K, O’Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, et al. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 2008;4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, et al. Single-Chain Soluble BG505.SOSIP gp140 Trimers as Structural and Antigenic Mimics of Mature Closed HIV-1 Env. Journal of virology. 2015;89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers. Journal of virology. 2015;90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Carnathan DG, Torrents de la Peña A, Pauthner M, Briney B, Reiss SM, Wood JS, Kaushik K, van Gils MJ, Rosales SL, et al. Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell reports. 2016;17:2195–2209. doi: 10.1016/j.celrep.2016.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, Wyatt RT. High-Density Array of Well-Ordered HIV-1 Spikes on Synthetic Liposomal Nanoparticles Efficiently Activate B Cells. Cell reports. 2016;15:1986–1999. doi: 10.1016/j.celrep.2016.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingale J, Tran K, Kong L, Dey B, McKee K, Schief W, Kwong PD, Mascola JR, Wyatt RT. Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies. Journal of virology. 2014;88:14002–14016. doi: 10.1128/JVI.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Totrov M, Li W, Sampson JM, Williams C, Lu H, Wu X, Lu S, Wang S, Zolla-Pazner S, Kong XP. Rationally Designed Immunogens Targeting HIV-1 gp120 V1V2 Induce Distinct Conformation-Specific Antibody Responses in Rabbits. Journal of virology. 2016;90:11007–11019. doi: 10.1128/JVI.01409-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Hedestam GB, Guenaga J, Corcoran M, Wyatt RT. Evolution of B cell analysis and Env trimer redesign. Immunological reviews. 2017;275:183–202. doi: 10.1111/imr.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen MS, Anderson MV, Jespersen MC, Nielsen M, Marcatili P. LYRA, a webserver for lymphocyte receptor structural modeling. Nucleic Acids Res. 2015;43:W349–355. doi: 10.1093/nar/gkv535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Muller W, Bontrop R, Lemaitre M, Malik A, Barbie V, Chaume D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of virology. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HXX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KKK, Chen X, Tsao CYY, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Chou FC, Conchuir SO, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, et al. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE) PLoS One. 2013;8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr LM, Cohen S, Spurrier B, Kong XP, Zolla-Pazner S. Epitope mapping of conformational V2-specific anti-HIV human monoclonal antibodies reveals an immunodominant site in V2. PLoS One. 2013;8:e70859. doi: 10.1371/journal.pone.0070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morner A, Douagi I, Forsell MN, Sundling C, Dosenovic P, O’Dell S, Dey B, Kwong PD, Voss G, Thorstensson R, et al. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. Journal of virology. 2009;83:540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis M, Tran K, Bale S, Phad GE, Guenaga J, Wilson R, Soldemo M, McKee K, Sundling C, Mascola J, et al. HIV-1 receptor binding site-directed antibodies using a VH1-2 gene segment orthologue are activated by Env trimer immunization. PLoS Pathog. 2014;10:e1004337. doi: 10.1371/journal.ppat.1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phad GE, Vazquez Bernat N, Feng Y, Ingale J, Martinez Murillo PA, O’Dell S, Li Y, Mascola JR, Sundling C, Wyatt RT, Karlsson Hedestam GB. Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization. J Immunol. 2015;194:5903–5914. doi: 10.4049/jimmunol.1500122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. 2011;187:4229–4235. doi: 10.4049/jimmunol.1000928. [DOI] [PubMed] [Google Scholar]
- Pritchard LK, Spencer DI, Royle L, Bonomelli C, Seabright GE, Behrens AJ, Kulp DW, Menis S, Krumm SA, Dunlop DC, et al. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Science translational medicine. 2012a;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Phad G, Douagi I, Navis M, Karlsson Hedestam GB. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J Immunol Methods. 2012b;386:85–93. doi: 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, et al. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.