Abstract
Alternative initiation, splicing, and polyadenylation are key mechanisms used by many organisms to generate diversity among mature mRNA transcripts originating from the same transcription unit. While previous computational analyses of alternative polyadenylation have focused on polyadenylation activities within or downstream of the normal 3′-terminal exons, we present the results of the first genome-wide analysis of patterns of alternative polyadenylation in the human, mouse, and rat genomes occurring over the entire transcribed regions of mRNAs using 3′-ESTs with poly(A) tails aligned to genomic sequences. Four distinct classes of patterns of alternative polyadenylation result from this analysis: tandem poly(A) sites, composite exons, hidden exons, and truncated exons. We estimate that at least 49% (human), 31% (mouse), and 28% (rat) of polyadenylated transcription units have alternative polyadenylation. A portion of these alternative polyadenylation events result in new protein isoforms.
It is well known that polyadenylation at different positions along pre-mRNAs leads to mature mRNAs with different 3′-untranslated regions (UTRs). 3′-UTRs have been shown to contain regulatory elements controlling mRNA stability (Touriol et al. 1999; Dreyfus and Regnier 2002), translational efficiency (Knirsch and Clerch 2000), and intracellular localization (Kislauskis et al. 1994). A recent review of the known molecular mechanisms involved in 3′-end formation in eukaryotes can be found in Zhao et al. (1999). Alternative initiation, splicing, and polyadenylation are key mechanisms used by organisms to generate diversity among mature mRNA transcripts originating from the same transcription unit. Recent studies have shown that mRNA processing, including capping, splicing, and polyadenylation, occur cotranscriptionally and that these are highly coupled and perhaps co-regulated reactions involving both cis- and trans-acting elements (Dye and Proudfoot 2001; Proudfoot et al. 2002; Levine and Tjian 2003; Kornblihtt et al. 2004).
Edwalds-Gilbert et al. (1997) first surveyed the recurring patterns of alternative polyadenylation based on the experimental literature. In this survey, three distinct patterns of alternative polyadenylation were noted: tandem poly(A) sites, composite exons, and skipped exons. Exons are generally categorized as 5′-terminal exons, internal exons, or 3′-terminal exons. In tandem poly(A) sites, multiple poly(A) sites are found within the same 3′-terminal exon. In composite exons, 5′-splice sites can some-time be silent, causing them to behave as 3′-terminal exons, or sometime be active, thereby causing them to behave as internal exons. In skipped exons, either the first alternative 3′-terminal exon is used, or that exon is skipped entirely and the second 3′-terminal exon is spliced into the transcript. Since this paper, there have been several computational studies on alternative polyadenylation, mostly using expressed sequence tags (ESTs). Gautheret et al. (1998) found alternative polyadenylation in 189 out of 1000 human EST clusters. Aligning ESTs to the 3′-UTRs from the UTRdb database (Pesole et al. 2000), Beaudoing et al. (2000) found that 29% of human genes were alternatively polyadenylated. Beaudoing and Gautheret (2001) detected the biases among the EST libraries in the alternative polyadenylation for 1450 human and 200 mouse mRNAs, suggesting that alternative polyadenylation is tissue- or disease-specific in each case. Iseli et al. (2002) found that at least half of the human genes were alternatively polyadenylated. They also noted that a significant portion of polyadenylation sites spread over distances in the kilobase range. However, all of the above computational analyses have focused on the polyadenylation activities within or downstream of the normal 3′-terminal exon. Therefore, they can only identify tandem poly(A) sites as described in Edwalds-Gilbert et al. (1997). In this paper, we carry out the first genome-wide analysis on the alternative polyadenylation over the entire transcribed regions of mRNAs in the human, mouse, and rat genomes.
Results
As described in detail in the Methods section, mRNA sequences from the RefSeq database (Pruitt and Maglott 2001) are aligned to the human, mouse, and rat genomes. The cluster of overlapping mRNAs on the genome is identified as a transcription unit. One mRNA sequence is chosen as the reference sequence to represent each transcription unit. The genomic sequence surrounding the reference sequence is extracted as the “extended genomic sequence.” ESTs with putative poly(A) tails from the dbEST database (Boguski et al. 1993) are aligned to these extended genomic sequences. Only the aligned ESTs satisfying stringent criteria are identified as poly(A) ESTs. Poly(A) sites are then identified from the 3′-ends of poly(A) ESTs.
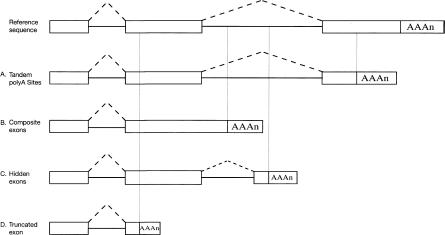
The poly(A) ESTs can be categorized into four classes as shown in Figure 1. In class I (Fig. 1A), the 3′-end of the EST falls within or downstream of the 3′-terminal exon of the reference sequence. In class II (Fig. 1B), the 3′-end of the EST falls in the intron of the reference sequence and the 3′-terminal exon of the EST overlaps with the internal exon of the reference sequence. In class III (Fig. 1C), the 3′-end of the EST falls in the intron of the reference sequence but the 3′-terminal exon of the EST does not overlap with any exon of the reference sequence. In class IV (Fig. 1D), the 3′-end of the EST falls in the internal exon of the reference sequence. The abundance of four classes of poly(A) ESTs is shown in Table 1.
Figure 1.
Four classes of patterns of alternative polyadenylation are shown together with the reference sequence. (A) Tandem poly(A) sites. (B) Composite exons. (C) Hidden exons. (D) Truncated exons.
Table 1.
The abundance of four classes of poly(A) ESTs in human, mouse, and rat
| Poly(A) ESTs | Class I | Class II | Class III | Class IV |
|---|---|---|---|---|
| Human | 329,966 | 3230 | 2257 | 1816 |
| Mouse | 86,598 | 394 | 359 | 656 |
| Rat | 27,999 | 148 | 138 | 109 |
For almost all poly(A) sites that we identified, the poly(A) ESTs using the same poly(A) site all belong to the same class as defined above. So we can also categorize most poly(A) sites into four classes according to their poly(A) ESTs. The abundance of four classes of poly(A) sites is shown in Table 2.
Table 2.
The abundance of four classes of poly(A) sites in human, mouse, and rat
| Poly(A) sites | Class I | Class II | Class III | Class IV |
|---|---|---|---|---|
| Human | 22,712 | 1221 | 730 | 907 |
| Mouse | 12,681 | 244 | 144 | 384 |
| Rat | 3379 | 88 | 59 | 62 |
When the transcription unit contains at least one poly(A) site, it is referred to as being polyadenylated. When the transcription unit contains more than one poly(A) site, it is referred to as being alternatively polyadenylated. When the transcription unit contains more than one class I poly(A) site, it is referred to as having tandem poly(A) sites, as described in Edwalds-Gilbert et al. (1997). When the transcription unit contains class II poly(A) sites, it is referred to as having composite exons, as described in Edwalds-Gilbert et al. (1997), because the internal exon in the reference sequence behaves as the 3′-terminal exon in class II ESTs. When the transcription unit contains class III poly(A) sites, it is referred to as having hidden exons because class III ESTs use the alternative 3′-terminal exon hidden inside the intron of the reference sequence. These are essentially the skipped exons as described in Edwalds-Gilbert et al. (1997) in that the reference sequence skips the 3′-terminal exon of class III ESTs. When the transcription unit contains class IV poly(A) sites, it is referred to as having truncated exons because the internal exon of the reference sequence is truncated by polyadenylation in class IV ESTs. The abundance of transcription units with polyadenylation, tandem poly(A) sites, composite exons, hidden exons, truncated exons, and alternative polyadenylation is summarized in Table 3.
Table 3.
The abundance of transcription units in human, mouse, and rat
| Transcription units | Polyadenylated | Tandem poly(A) sites | Composite exons | Hidden exons | Truncated exons | Alternatively polyadenylated |
|---|---|---|---|---|---|---|
| Human | 13,268 | 5521 | 1084 | 650 | 794 | 6535 (49%) |
| Mouse | 9227 | 2532 | 233 | 138 | 355 | 2898 (31%) |
| Rat | 2611 | 621 | 87 | 57 | 59 | 710 (28%) |
The percentages of alternative polyadenylation are calculated relative to the polyadenylated transcription units.
More detailed information including complete lists of the four classes of alternative polyadenylation is available at http://physics.nyu.edu/~jy272/altA. One representative for each class of transcription unit is given below. As an explicit example of our alignment method, we show the mRNA and EST alignments of Lamin A/C (composite exons) in Figure 2.
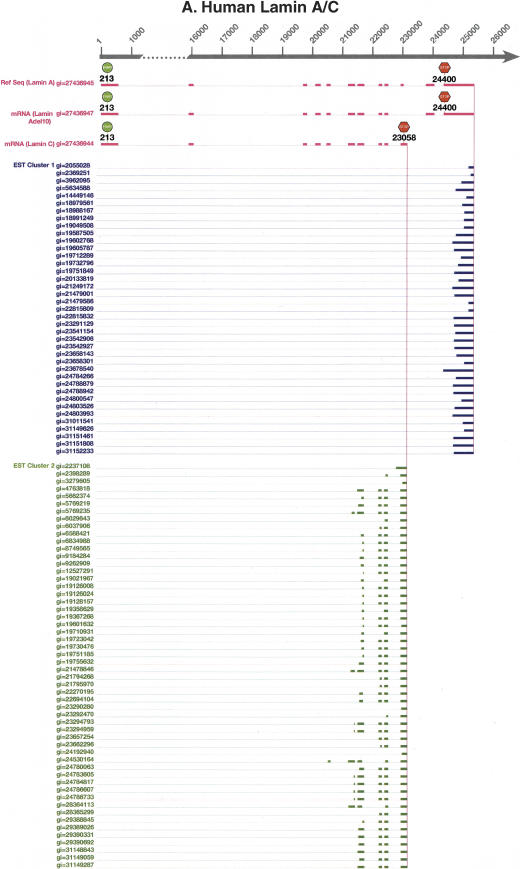
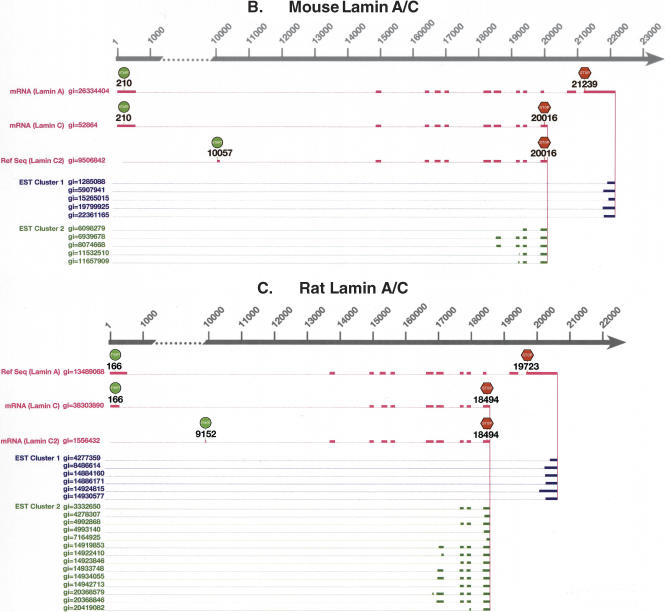
Figure 2.
Comparison between the full-length mRNA alignments and the poly(A) EST alignments of Lamin A/C in (A) human, (B) mouse, and (C) rat. In all three species, poly(A) ESTs are clustered into two clusters, each corresponding to a poly(A) site. Lamin A and C isoforms are present in all three species. In human, a third isoform, Lamin Adel10, skips exon 10 of Lamin A. In mouse and rat, a third isoform, Lamin C2, has alternative initiation. GenBank gi numbers are shown for all mRNAs and ESTs.
Rhodopsin has tandem poly(A) sites. We found five (human), six (mouse), and four (rat) class I poly(A) sites in rhodopsin. All of them are downstream of the stop codon of the reference sequence. Transcripts using these poly(A) sites have different 3′-UTRs but encode the same protein. Three of the poly(A) sites are highly conserved across human, mouse, and rat.
Lamin A/C has a composite exon. Two poly(A) sites are present in human, mouse, and rat (Fig. 2). We searched LocusLink (Pruitt and Maglott 2001) for Lamin A/C in three species. We found all full-length mRNA transcripts using these two poly(A) sites. When the transcript uses the promoter-distal poly(A) site in exon 12, it gives rise to Lamin A mRNA. When the transcript uses the promoter-proximal poly(A) site hidden in the intron of Lamin A mRNA between exons 10 and 11, it gives rise to Lamin C mRNA. When translated into proteins, Lamin A has a different C-terminal domain from Lamin C (Alsheimer et al. 2000). Lamin A has a CaaX-box (C, cysteine; a, aliphatic; X, any amino acid) in the C-terminal domain. Isoprenylation of the cysteine residue of the CaaX-box is essential for lamin attachment to the inner nuclear membrane. Lamin C lacks the CaaX-box and requires the presence of other lamins for nuclear envelope attachment. In mouse and rat, there is a third isoform, Lamin C2 (Alsheimer et al. 2000). Like Lamin C, it uses the promoter-proximal poly(A) site but has an alternative 5′-terminal exon (alternative initiation). In human, there is also a third isoform, Lamin Adel10. Like Lamin A, it uses the promoter-distal poly(A) site but skips exon 10 (alternative splicing).
Cell Division Cycle 42 (CDC42) has a hidden exon. We found six (human), five (mouse), and five (rat) poly(A) sites in CDC42. With the exception that one poly(A) site appears to be human-specific, all of the other five poly(A) sites are conserved among human, mouse, and rat. Polyadenylation at three (four in human) promoter-distal poly(A) sites leads to the transcripts encoding protein isoform 1. Polyadenylation at the other two promoter-proximal poly(A) sites leads to the transcripts encoding protein isoform 2. These two protein isoforms have exactly the same lengths but differ in their last 10 C-terminal amino acids. Whether this leads to different functions of the two protein isoforms is worth further investigation.
WW domain binding protein 2 (Wbp2) has a truncated exon in rat but not in human or mouse. We found that 67 (human), 31 (mouse), and 14 (rat) poly(A) ESTs use a conserved class I poly(A) site. Only one rat poly(A) EST (UI-R-CA0-bgv-h-11-0-UI.s1; GenBank gi: 11378731) uses a class IV poly(A) site within the last internal exon of the reference sequence. This poly(A) EST truncates the coding sequence before it reaches the stop codon. No canonical poly(A) signals (AATAAA and ATTAAA) or their variants (Beaudoing et al. 2000) are found near the poly(A) site. This raises the suspicion that this poly(A) EST may be an experimental artifact.
Discussion
To study how alternative polyadenylation contributes to protein diversity, we must first identify the protein-coding regions on the alternative transcripts. This is a difficult task because ESTs are only partial sequences of the whole transcripts. So far, all the known examples (Edwalds-Gilbert et al. 1997; Zhao et al. 1999) suggest that alternative polyadenylation does not change the open reading frame. We therefore assume that all four classes of ESTs follow the same open reading frame as the reference sequence, even though open reading frame shift due to alternative initiation or splicing can occur in principle. Most class I poly(A) sites (99% in human) fall downstream of the stop codon of the reference sequence. They share the stop codon with the reference sequence and encode the same protein. Most class II poly(A) sites (93% in human) and class III poly(A) sites (90% in human) fall between the start and stop codons of the reference sequence on the genome. If they exist, the new stop codons of these class II and III ESTs can only lie in the region where they overlap with the introns of the reference sequence. We search for the new stop codons in these regions following the open reading frame of the reference sequence. The results are summarized in Table 4. The new stop codons exist for ∼80% of class II and ∼90% of class III poly(A) sites. The transcripts using these poly(A) sites are translated into new protein sequences with different C-terminal domains. Interestingly, we note that the percentage of the stop codon presence is about 10% higher in class III than in class II poly(A) sites in all three species. Most class IV poly(A) sites (87% in human) fall between the start and stop codons of the reference sequence on the genome. These class IV ESTs truncate the coding exon of the reference sequence and lack the stop codon. If not experimental artifacts, most class IV ESTs and a small portion of class II and III ESTs can only be noncoding transcripts. Noncoding transcripts have been shown to exist extensively in human (Cawley et al. 2004). They can simply be the mistakes made by the transcription machinery and undergo the degradation soon after the transcription, or they can have important regulatory functions (Cawley at al. 2004; Martens et al. 2004).
Table 4.
The percentage of the presence of stop codons in class II and III poly(A) sites
| Presence of stop codons | Class II poly(A) sites | Class III poly(A) sites |
|---|---|---|
| Human | 81% | 88% |
| Mouse | 73% | 84% |
| Rat | 79% | 89% |
We observe that different poly(A) sites in the same transcription unit can have different polyadenylation efficiencies. Some poly(A) sites are used more often and are associated with a large number of poly(A) ESTs. The others are used more rarely and associated with only one or a few poly(A) ESTs. We define the polyadenylation efficiency of the poly(A) site as the number of poly(A) ESTs using the poly(A) site. Within each class of poly(A) site, the average polyadenylation efficiency is simply the total number of poly(A) ESTs (shown in Table 1) divided by the total number of poly(A) sites (shown in Table 2). In human, the average polyadenylation efficiencies are 14.53 (class I), 2.65 (class II), 3.09 (class III), and 2.00 (class IV). In all three species, we found that the average polyadenylation efficiencies follow the same order: class I > class III > class II ≈ class IV. As pointed out in Edwalds-Gilbert et al. (1997) and Zhao et al. (1999), there is competition between splicing and polyadenylation during the mRNA post-transcriptional processing. Recently, Qiu and Pintel (2004) showed an interesting distance effect on the polyadenylation efficiency. They found that the alternative polyadenylation of AAV5 RNA within an intron is inhibited by U1 snRNP binding to the 5′-splice site immediately upstream to the poly(A) site. When the distance between the 5′-splice site and the poly(A) site was increased by inserting a heterologous DNA sequence, the inhibition was reduced. A similar inhibition and distance effect was observed in HIV-1, where the alternative polyadenylation at the promoter-proximal poly(A) site was inhibited by U1 snRNP binding to the 5′-splice site immediately downstream of the poly(A) site. In our study, class II and class III poly(A) sites are immediately downstream of the 5′-splice site, whereas class IV poly(A) sites are immediately upstream of the 5′-splice site. Generally, the distance between the poly(A) site and the 5′-splice site is longer for class III (9708 bp in rat CDC42), but shorter for class II and IV (110 bp in rat Lamin A/C and 40 bp in rat Wbp2). This may explain why class III sites are more efficient than class II and IV sites. Qiu and Pintel (2004) also pointed out that the polyadenylation efficiency is proportional to the distance between promoter and poly(A) site. This is consistent with the observation that the most efficient poly(A) site is often the most distal one from the promoter. Class I poly(A) sites are most distal to the promoter among four classes of poly(A) sites. This may explain why class I poly(A) sites are most efficient at polyadenylation.
We compared the transcription units containing class II, III, and IV poly(A) sites across human, mouse, and rat. We found that alternative polyadenylation in Lamin A/C (class II), CDC42 (class III), Nucleophosmin (class III), and Olfactomedin 1 (class III) are conserved in all three species. They all result in new protein isoforms with different C-terminal domains. Since there are only a relatively small number of transcription units in common among three species in our study and human, mouse, and rat have different redundancies of ESTs, the above list is by no means complete.
Four classes of alternative polyadenylation are distinct in their splicing-polyadenylation patterns, protein-coding capacities, and polyadenylation efficiencies. Strong evidence including full-length mRNA sequences and protein sequences exists for class I, II, and III transcripts. But the role of class IV transcripts remains elusive. To tell whether they are experimental artifacts or noncoding transcripts, future experimental verifications will be very necessary.
Methods
We extracted 20,860 (human), 16,743 (mouse), and 4843 (rat) mRNA sequences from the NCBI RefSeq database release 4 (Pruitt and Maglott 2001). Only the mRNAs with accession numbers starting with NM (mRNA sequences that are experimentally verified) were selected. The accession numbers, start codon positions, and stop codon positions were also extracted. These sequences were masked by the RepeatMasker (http://repeatmasker.org; setting -w) program using the WU-BLAST program (http://blast.wustl.edu). The repeat databases were obtained from the RepBase Update (http://www.girinst.org). Human (build 34), mouse (build 32), and rat (build 2) genomes were downloaded from NCBI. We aligned the masked sequences onto their genomes using the megablast program (Altschul et al. 1990; settings -p 98, -D 3). Only the alignments with percent identities >98% were kept. The minimum and maximum positions of each mRNA sequence alignment on each genomic contig were calculated, and the genomic sequence starting from 40 kb upstream of the minimum position to 40 kb downstream of the maximum position was extracted from the genomic contig. The unmasked mRNA sequences were realigned to their corresponding extracted genomic sequences one by one using the sim4 program (Florea et al. 1998; settings A = 0, P = 1). Only the sim4 alignments satisfying the following high-quality controls were kept: (1) All splice sites were in the same orientations. (2) The entire sequence except the 50 bp at both ends was aligned. (3) The average percent identity of alignments was >95%. The average percent identity given by sim4 was defined as the alignment score. Overall, we aligned 20,095 (96%) human mRNAs, 14,861 (89%) mouse mRNAs, and 3803 (79%) rat mRNAs to their genomes. The disparity in the percentages of mRNAs aligned was largely due to the differences in the completeness of three genomes.
We clustered all splicing variants based on their common exon boundaries. Two mRNA sequences were clustered together if they shared at least one 5′- or 3′-splice site. The mRNA sequence with the highest alignment score in the cluster was chosen as the reference sequence to represent that cluster. If the same reference sequence was aligned to multiple positions on the genome, the position with the highest alignment score was chosen. We identified the resulting clusters as transcription units. We obtained 16,018 (human), 14,406 (mouse), and 3771 (rat) transcription units. We defined the “extended genomic sequence” as the genomic sequence spanning between 10 kb upstream and 10 kb downstream of the reference sequence alignments. The extended genomic sequences were extracted and converted into the same orientation as the reference sequences (5′ to 3′). The new alignments of the reference sequences on their extended genomic sequences were calculated.
We downloaded and parsed the NCBI dbEST database (08 April 2004 release; Boguski et al. 1993) for ESTs of human, mouse, and rat. The ESTs shorter than 100 bp were discarded. The EST sequences were searched for poly(A) tracks in the forward orientation and poly(T) tracks in the reverse orientation. The poly(A) or poly(T) tracks must have more than 10 contiguous As or Ts. One end of the poly(A) or poly(T) track must be within 10 bp of the EST end. The other end of the poly(A) or poly(T) track was identified as the putative start of the poly(A) tail. These ESTs were identified as the putative poly(A) ESTs and were converted into the forward orientation such that they all have poly(A) tracks at the 3′-ends. Overall, we obtained 691,293 ESTs with putative poly(A) tails for human, 227,576 for mouse, and 223,367 for rat. They were then masked by the RepeatMasker program (setting -w). Masked EST sequences were aligned to the extended genomic sequences by the megablast program only in the forward orientation (settings -p 95, -D 3, -S 1). Only the alignments with total coverage higher than 50% of the EST sequence lengths and percent identities higher than 95% were kept. The minimum and maximum positions of each EST sequence alignment on each extended genomic sequence were calculated, and the sequence spanning between 4 kb upstream of the minimum position and 4 kb downstream of the maximum position was extracted from the extended genomic sequences. The unmasked EST sequences were realigned to their corresponding extracted sequences one by one using the sim4 program (settings A = 0, P = 1). Only the sim4 alignments satisfying the following quality controls were kept: (1) All splice sites were in the same orientation. (2) The entire sequence was aligned except the 50 bp at the 5′-end and the 3′-end of alignment must occur within 5 bp of the putative start of the poly(A) tail. (3) The average percent identities of alignment must be higher than 90%.
The 3′-end of alignment was identified as a putative poly(A) cleavage site, and 50-bp downstream sequences of the putative poly(A) cleavage sites were extracted and searched for the poly(A) tracks. The poly(A) track was identified if there were more than five contiguous As or more than eight As out of any 10-bp sliding window. If the poly(A) track was found in the downstream sequence, the poly(A) cleavage site was identified as the putative internal priming site. The aligned ESTs without the putative internal priming sites were identified as poly(A) ESTs. To further filter out the poly(A) ESTs that may belong to the intronic genes or overlapping genes, the poly(A) ESTs must satisfy the following criteria: if the EST was spliced, it must share at least one 3′- or 5′-splice site with the reference sequence; if the EST was not spliced, it must overlap with the 3′-terminal exon of the reference sequence. The 3′-ends of the poly(A) ESTs on the same extended genomic sequence were clustered if they were <50 bp apart. This was because the 3′-ends of the polyadenylated transcripts were known to fluctuate within a small distance after the poly(A) signals. The clusters of the 3′-ends of poly(A) ESTs were identified as the poly(A) sites.
Acknowledgments
This project was supported by NIH grant RR-16466-01, NSF grant EPS-0092040, the Alaska BRIN program, and the University of Alaska Foundation. Leone Thierman in the Arctic Region Supercomputing Center kindly produced the figures. We thank anonymous referees for helpful comments.
Footnotes
[Supplemental material is available online at www.genome.org and at http://physics.nyu.edu/~jy272/altA.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3109605.
References
- Alsheimer, M., von Glasenapp, E., Schnolzer, M., Heid, H., and Benavente, R. 2000. Meiotic lamin C2: The unique amino-terminal hexapeptide GNAEGR is essential for nuclear envelope association. Proc. Natl. Acad. Sci. 97: 13120-13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Beaudoing, E. and Gautheret, D. 2001. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST Data. Genome Res. 11: 1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing, E., Freier, S., Wyatt, J.R., Claverie, J.M., and Gautheret, D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10: 1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. 1993. dbEST—Database for expressed sequence tags. Nat. Genet. 4: 332-333. [DOI] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499-509. [DOI] [PubMed] [Google Scholar]
- Dreyfus, M. and Regnier, P. 2002. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell 111: 611-613. [DOI] [PubMed] [Google Scholar]
- Dye, M.J. and Proudfoot, N.J. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105: 669-681. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert, G., Veraldi, K.L., and Milcarek, C. 1997. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res. 25: 2547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret, D., Poirot, O., Lopez, F., Audic, S., and Claverie, J.M. 1998. Alternate polyadenylation in human mRNAs: A large-scale analysis by EST clustering. Genome Res. 8: 524-530. [DOI] [PubMed] [Google Scholar]
- Iseli, C., Stevenson, B.J., de Souza, S.J., Samaia, H.B., Camargo, A.A., Buetow, K.H., Strausberg, R.L., Simpson, A.J., Bucher, P., and Jongeneel, C.V. 2002. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 12: 1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis, E.H., Zhu, X., and Singer, R.H. 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127: 441-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirsch, L. and Clerch, L.B. 2000. A region in the 3′ UTR of MnSOD RNA enhances translation of a heterologous RNA. Biochem. Biophys. Res. Commun. 272: 164-168. [DOI] [PubMed] [Google Scholar]
- Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., and Nogues, G. 2004. Multiple links between transcription and splicing. RNA 10: 1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M. and Tjian, R. 2003. Transcription regulation and animal diversity. Nature 424: 147-151. [DOI] [PubMed] [Google Scholar]
- Martens, J.A., Laprade, L., and Winston, F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571-574. [DOI] [PubMed] [Google Scholar]
- Pesole, G., Liuni, G., Grillo, G., Licciulli, F., Larizza, A., Makalowski, M., and Saccone, C. 2000. UTRdb and URTsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of the eukaryotic mRNA's. Nucleic Acids Res. 28: 193-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J. and Pintel, D.J. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by the distance between the promoter and the intron and is inhibited by U1 small nuclear RNP binding to the intervening donor. J. Biol. Chem. 279: 14889-14898. [DOI] [PubMed] [Google Scholar]
- Touriol, C., Morillon, A., Gensac, M.C., Prats, H., and Prats, A.C. 1999. Expression of human fibroblast growth factor 2 mRNA is post-transcriptionally controlled by a unique destabilizing element present in the 3′-untranslated region between alternative polyadenylation sites. J. Biol. Chem. 274: 21402-21408. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://blast.wustl.edu; BLAST.
- http://physics.nyu.edu/~jy272/altA; authors' Web site.
- http://repeatmasker.org; RepeatMasker.
- http://www.girinst.org; RepBase Update.