Abstract
ClpB and Hsp104 are conserved AAA+ protein disaggregases that promote survival during cellular stress. Hsp104 acts on amyloids, supporting prion propagation in yeast, and can solubilize toxic oligomers connected to neurodegenerative diseases. A definitive structural mechanism, however, has remained elusive. We have determined the cryo-EM structure of Hsp104 in the ATP state, revealing a near-helical hexamer architecture that coordinates the mechanical power of the twelve AAA+ domains for disaggregation. An unprecedented heteromeric AAA+ interaction defines an asymmetric seam in an apparent catalytic arrangement that aligns the domains in a two-turn spiral. N-terminal domains interact to form a broad channel entrance for substrate engagement and Hsp70 interaction. Middle-domain helices bridge adjacent protomers across the nucleotide pocket, explaining roles in hydrolysis and disaggregation. Remarkably, substrate-binding pore loops line the channel in a continuous spiral that appears optimized for substrate transfer across the AAA+ domains, establishing a directional path for polypeptide translocation.
INTRODUCTION
Heat shock protein (Hsp) 104, found in yeast, is a member of the Hsp100 class of molecular chaperones that contain highly conserved AAA+ (ATPases Associated with diverse cellular Activities) domains and serve essential roles in thermotolerance and protein quality control1–3. Hsp104 and its bacterial homolog, ClpB, form large hexameric-ring structures that cooperate with the Hsp70 system to unfold and rescue aggregated protein states by active translocation of polypeptide substrates through a central channel4–6. In addition to solubilizing stress-induced disordered aggregates5,7, Hsp104 recognizes and remodels cross-β structures of amyloid fibrils, such as those found in Sup35 prions, which enables Hsp104 to control prion inheritance in yeast8–10. This function in prion disaggregation is enhanced in Hsp104 compared to ClpB, and studies have identified potentiated Hsp104 variants that reduce the toxicity of proteins linked to neurodegenerative diseases including TDP-43, FUS and α-synuclein11–13. Despite fundamental roles in protein quality control and promising therapeutic activity in rescuing amyloidogenic states14, how Hsp104 and its family members function as powerful molecular motors to solubilize proteins is not fully understood.
Hsp104 and ClpB contain a mobile N-terminal domain (NTD) implicated in substrate engagement 15,16, two evolutionarily distinct AAA+ nucleotide binding domains (NBD1 and NBD2) that mediate oligomeric interactions and power polypeptide threading, and a middle domain (MD) that is required for disaggregation and interaction with Hsp70 (Figure 1a)17–21. Hsp104 also contains a 38-residue C-terminal domain (CTD) not found in ClpB that is implicated in substrate binding22, cochaperone interactions23, and is critical for hexamerization24. The crystal structure of ClpB25 from a thermophilic eubacterium (Thermus thermophilus) identified the conserved type II AAA+ domain architecture wherein the NBDs are comprised of large and small subdomains that form the ATP binding pocket and contain the respective Walker A and B motifs and ‘sensor’ residues required for hydrolysis. The MD, located within the small subdomain of NBD1, forms an ~85 Å-long flexible coiled-coil that is proposed to project outward from hexamer25,26, but alternatively has been modeled to intervene between the protomers27 as well as wrap tightly around the outside of the hexamer28,29.
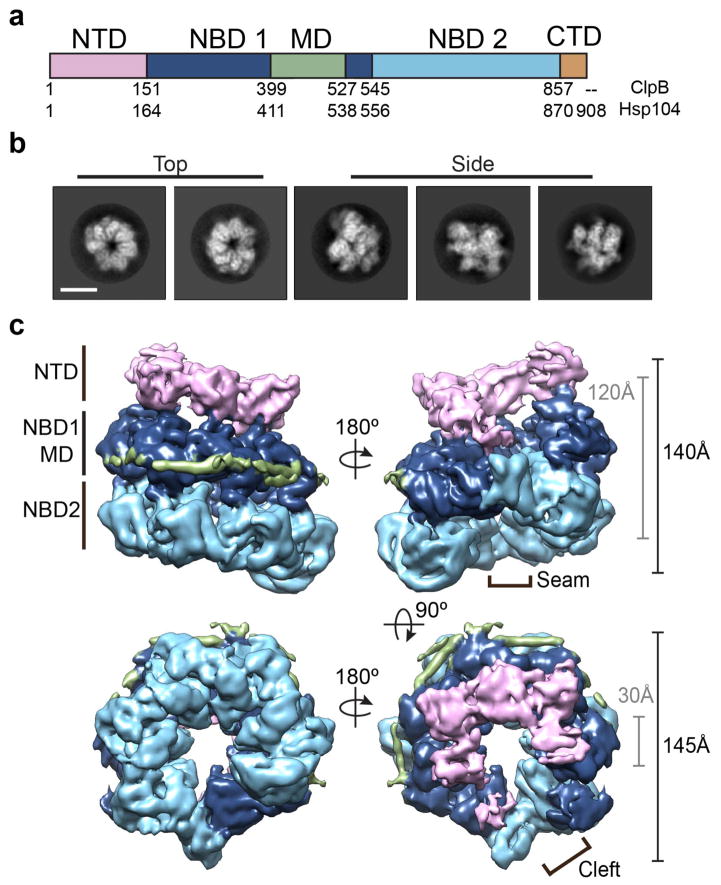
Figure 1.
Spiral architecture and three-tiered domain arrangement of the Hsp104 hexamer complex by cryo-EM. (a) Schematic showing individual domains and corresponding residue numbers for T. thermophilus ClpB and S. cerevisiae Hsp104. (b) Reference-free 2D cryo-EM class averages of Hsp104 showing representative top and side-views. Scale bar equals 100 Å. (c) Views of the final, sharpened 3D density map (4σ) of Hsp104 showing spiral, three-tier architecture. The domains are colored as in (a), based on the atomic model. Approximate dimensions of the channel (grey) and exterior (black) are shown, and the hexamer “seam” and channel “cleft” are indicated.
A number of cryo-EM models of Hsp104 mutants21,27,30,31 and ClpB25,26 have been described that propose different structural arrangements but generally show a symmetric hexamer with three distinct rings comprised of the NTD, NBD1, and NBD2. An asymmetric arrangement has been identified for Hsp104 bound to ATP or ATPyS31, supporting an asymmetric hydrolysis mechanism similar to ClpX32. Nucleotide-dependent conformations that involve changes in the pore diameter and position of the NTD, NBDs, and MD have been proposed based on small-angle x-ray scattering (SAXS)29 and cryo-EM models26,31. The oligomeric state, however, is dynamic, requiring presence of nucleotide for stability33 and the protomers exchange rapidly during the hydrolysis cycle34–36. These dynamics present significant challenges to achieving higher resolution views of the hexamer and cryo-EM structures of full-length, wild type Hsp104 have not been determined.
Polypeptide threading between NBD1 and NBD2 rings is controlled via allosterically driven ATP hydrolysis events 31,37. Substrate binding is favored in the ATP-bound state38, and involves direct interaction with highly conserved tyrosine residues in flexible NBD “pore loops”5,6,39. A fully functional Hsp104 hexamer is required for the disaggregation of highly stable amyloids compared to disordered aggregates, indicating cooperativity between protomers is required to unfold structured fibril 35. The “pore loops” are proposed to line the axial channel and operate by hydrolysis-driven hand-off cycles to translocate polypeptides3, but how this occurs across the 100 Å-long channel and between the two AAA+ domains remains unknown.
Here we have determined the cryo-EM structure of wild-type Hsp104 from yeast (Saccharomyces cerevisiae) bound to AMP-PNP (a nonhydrolyzable ATP analog) at 5.6 Å resolution. Hsp104 adopts an asymmetric spiral architecture involving a substantial, 10-Å rise between protomers that results in a defined seam where the first protomer is offset by more than 40 Å from the sixth. This offset brings NBD1 from one protomer into contact with NBD2 in the adjacent protomer, forming an unusual heteromeric AAA+ interaction at the seam. The nucleotide pocket at the NBD1-NBD2 interaction site appears functional and contains a bound nucleotide. The NTD, CTD and coiled-coil MD are observed in functionally significant arrangements around the hexamer. Well-defined density corresponding to the conserved Tyr pore loops presents a helical spiral of substrate-binding surfaces along the axial channel. Remarkably, the protomer offset optimally positions the NBD1 and NBD2 pore loops at the hexamer seam for coordinated transfer of substrates around the two-turn spiral and across the AAA+ domains.
RESULTS
Hsp104 hexamers adopt an asymmetric, spiral architecture
We determined the cryo-EM structure of the intact, wild-type Hsp104 complex in the presence of AMP-PNP, a nonhydrolyzable ATP analog, to mimic the substrate-binding ATP state8. Purified Hsp104 was determined to be functionally active via established ATPase and disaggregase assays (Supplementary Fig. 1a, 1b). Cryo-EM images of Hsp104-AMP-PNP show homogeneous oligomeric complexes (Supplementary Fig 1c) and reference-free 2D classification reveals a variety of distinct views with well-resolved secondary structural features (Supplementary Fig 1d). In these 2D averages a well-defined three-tier architecture is observed and top-view and side-view projections show a clear asymmetry and a unique protomer arrangement compared to what has been described previously (Figure 1b)27,31. The final cryo-EM reconstruction following 3D refinement with no imposed symmetry was achieved at an indicated resolution of 6.5 Å for the unsharpened map and 5.6 Å for the sharpened map (Supplementary Fig. 1e). The final map is comprised of 65% of the total data, and 85% of the data following sorting by 2D classification. No additional conformations or oligomeric states were identified by 3D classification (data not shown), thus the final map represents the predominant form of Hsp104 in the dataset. The angular distribution of the particles that went into the final map showed multiple preferred orientations including distinct equatorial and axial views, while additional, less populated orientations were also present that aided the asymmetric refinement (Supplementary Fig 1f). 2D projections of the 3D map match the reference-free averages, confirming the overall asymmetric architecture (Supplementary Figure 1g).
The reconstruction identifies Hsp104-AMP-PNP as a ring-shaped hexamer with three distinct domain-layers that correspond to the NTD, NBD1-MD and NBD2, consistent with other studies (Figure 1c and Supplementary Video 1) 27,28. Surprisingly, the hexamer adopts an apparent helical spiral arrangement where the NBD rings connect together at a distinct asymmetric seam. The NBD1 and NBD2 show the highest resolution density, at approximately 5 Å, while the NTDs as well as regions at the hexamer seam are more flexible and at ~6–7 Å resolution (Supplementary Fig. 1h). Density for the MD coiled-coil is partly resolved on one face of the hexamer where it wraps around the outside of the hexamer adjacent to the NBD1s (Figure 1c). The overall dimensions are similar to those of previous studies with a 145 Å outer diameter and 140 Å length. The central channel is approximately 25–30 Å in diameter but, surprisingly, opens to form a wide cleft at the hexamer seam due to the spiral offset of the adjacent protomers.
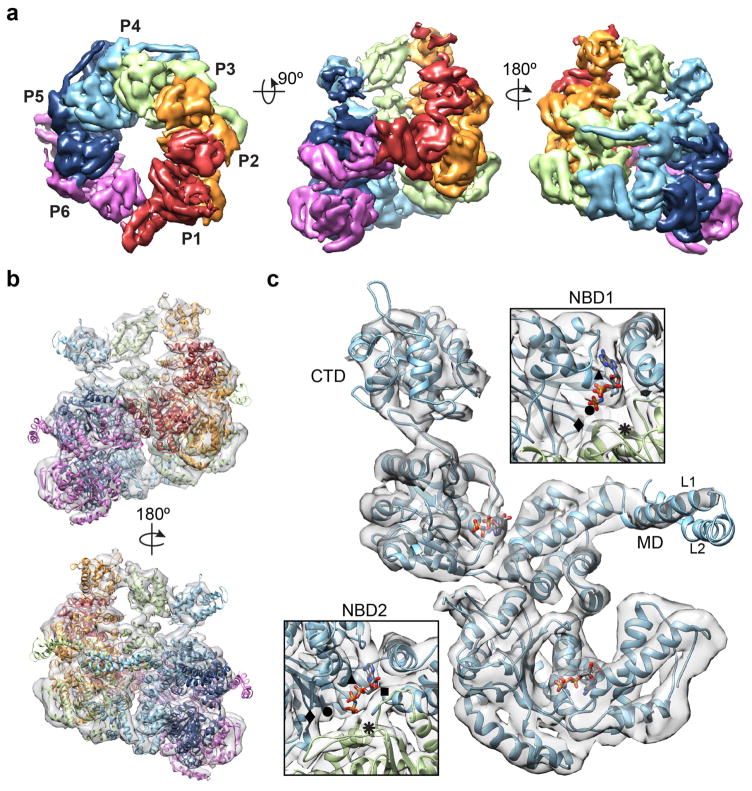
The crystal structure from bacterial (T. thermophilus) ClpB25, which has ~45% sequence identity, was used for initial fitting and to determine a homology model for Hsp104. Helices and β-strands are designated as they are for the ClpB structure (Supplementary Fig. 2a) The reconstruction was docked by rigid body fitting of the domains followed by flexible fitting to achieve an atomic model with the highest correlation to the map, at 0.92. The protomers are designated 1 through 6, with protomer 1 (P1) in the highest position in the side-view orientation, and proceeding counterclockwise when viewed from the NTD-face (Figure 2a, b). The NBDs fit well, with a cross-correlation value of 0.94 between the map and model, and identify the conserved AAA+ subdomain architecture with α-helical and β-sheet regions that are well resolved for each of the protomers (Figure 2c and Supplementary Video 2). For P2-P5, density that likely corresponds to a bound nucleotide is observed in all the NBDs. The NBD1s show a canonical AAA+ arrangement, including expected positions for the Walker A (K218), Walker B (E285), and sensor-1 (T317) residues. Putative Arg finger residues R334 and R33327 are within 7 and 10 Å, respectively, of the nucleotide pocket of the adjacent protomer, indicating a catalytically active arrangement. The NBD2s are similarly well defined showing conserved interprotomer interactions and positions for the Walker A (K620), Walker B (E687), sensor-1 (N728) and sensor-2 (R826) residues, and the proposed Arg finger (R765)40 is near the nucleotide pocket at less than 10 Å, based on our model. Protomers P1 and P6 that make up the unusual hexamer seam are in a different conformation and are discussed in further detail below.
Figure 2.
Protomer arrangement and molecular model of Hsp104. (a) Top-view, showing NBD1 density and protomer labels, and side views of the sharpened map (5σ) with each protomer colored based on the molecular model. (b) Side views of the Hsp104 hexamer molecular model determined by flexibly fitting the homology model based on the ClpB structure25 (PDB: 1QVR). (c) Molecular model of a segmented protomer (P4) showing bound nucleotides and well-resolved electron density for the NBD1 and NBD2 AAA+ domains. Enlarged views of the nucleotide pockets are shown with indicated positions for Walker A (▲) and B (◆), Sensor 1 (●) and 2 (■) and Arginine finger (⁕) residues.
Density corresponding to the MD coiled-coil that is partly resolved for P3-P6 was docked with residues 409–467 that correspond to helix L1 and part of helix L2 that connect to the NBD1 large subdomain (Figure 2c). Density for the C-terminal-half of the extended, 85 Å helix L2 as well as helix L3 and helix L4, which includes Hsp70 interaction sites18, was unable to be modeled, indicating this portion of the MD remains flexible. The NTDs form globular lobes of density that interact and follow the helical arrangement of the NBDs, forming a broad entrance to the channel (Figure 1c and 2a). Despite the NTD flexibility, the NTD-NBD1 linkers are well defined, enabling unambiguous connection to the corresponding protomer. NTD structures were able to be modeled for protomers P2-P4 (Figure 2b), which exhibit the best-resolved features, and the cross correlation of the fit was determined to be 0.91. Further details of the MD and NTD conformations are discussed below.
A large protomer offset and an unusual NBD1-NBD2 interaction define the hexamer seam
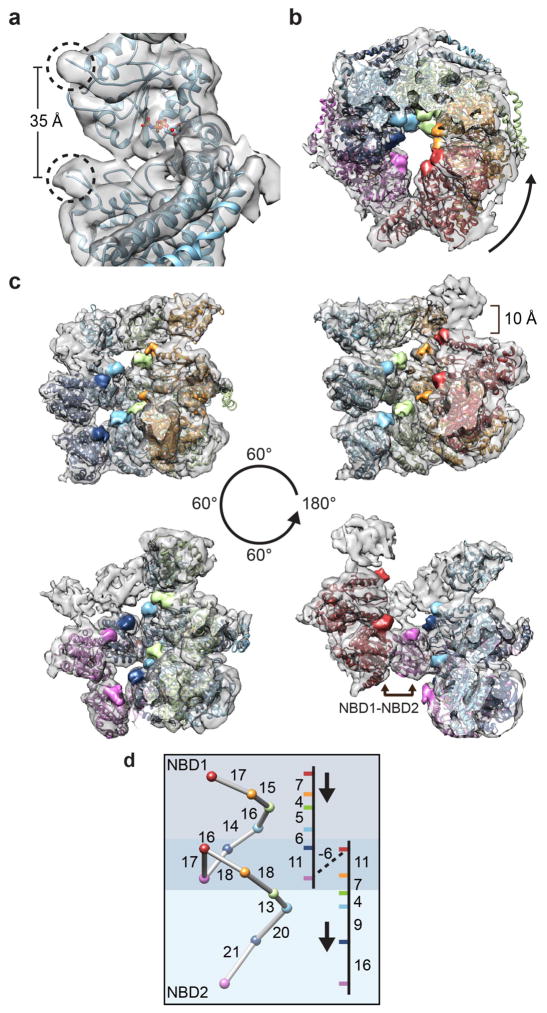
The most striking structural feature of the Hsp104 hexamer is the helical-like arrangement of the protomers. While canonical AAA+ interactions are generally maintained around the hexamer, each protomer is tilted slightly, resulting in an approximate 7-fold helical symmetry. Going from P1 to P6, the protomers each rise nearly 10 Å and rotate 53°, on average (Figure 3a). The individual protomers vary in conformation and helical change, indicating the complex is indeed asymmetric (Supplementary Fig. 3a). When the NBD1s are superimposed, continuous conformational changes are observed from P1-P6 (Supplementary Fig 3b), with the greatest protomer-protomer differences between P1 and P6 at the hexamer seam (Supplementary Fig. 3c). The conformational differences primarily involve rotations of the NBD1 and NBD2 small subdomains, which move inward relative to the channel axis by approximately 10° and 20°, respectively, resulting in a more compact P1 protomer compared to P6 (Supplementary Fig. 3c).
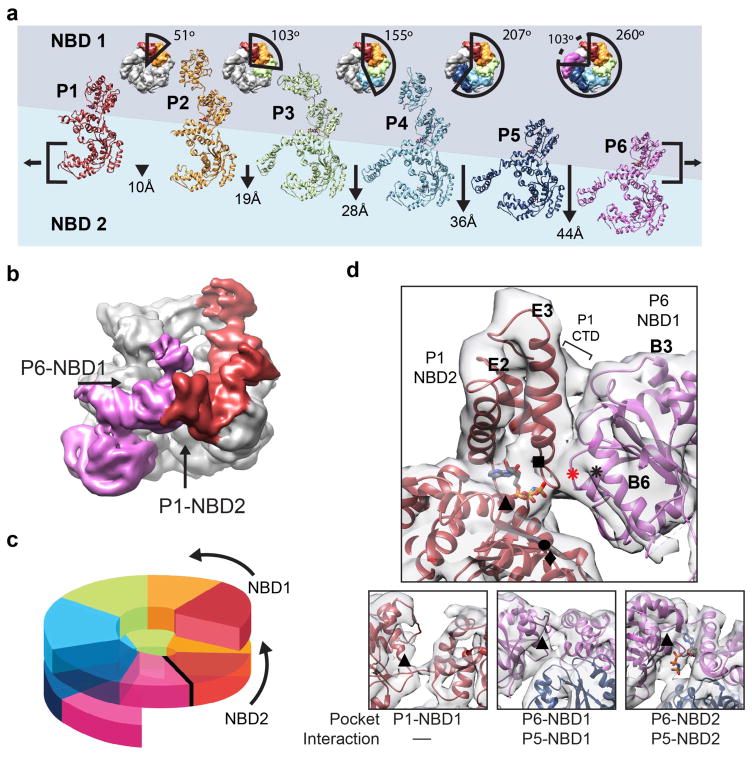
Figure 3.
Basis for the protomer spiral and NBD1:NBD2 AAA+ interaction at the hexamer seam. (a) Protomer positions shown in top views and individually unrolled from the hexamer with the indicated rotation and translation shift relative to P1. Brackets indicate alignment of NBD1 and NBD2 AAA+ domains for P6 and P1, respectively. (b) Low-pass filtered map colored for P1 (red) and P6 (magenta), highlighting the P6-NBD1:P1-NBD2 interaction. (c) Cartoon model showing the NBD1 and NBD2 AAA+ domains connect and form a two-turn spiral. (d) Expanded view of the model and map showing the P6-NBD1:P1-NBD2 interaction from inside the channel. Interface regions are indicated: B3 and B6 helices (NBD1 large subdomain), E2 and E3 helices (NBD2 small subdomain) and the P1 CTD. Positions for the Walker A (▲) and B (u), Sensor 1 (●) and 2 (■) and putative Arginine finger R333/R334 (⁕) and R307 (⁕, red) residues are indicated. Sub-panels show nucleotide pockets for the P1-NBD1, P6-NBD1 and P6-NBD2 with Walker A residues (▲) indicated. Nucleotide is shown for sites where clear electron density is observed in the pocket.
The helical shift of the protomers results in a 44 Å-offset between P1 and P6, yet these protomers interact together, covering a 100° rotation along the channel axis to complete the hexamer ring (Figure 3b). Remarkably, this large offset positions the P1-NBD2 adjacent the P6-NBD1, resulting in an unusual interaction between the two different AAA+ domains. Thus, the AAA+ domains form a continuous two-turn spiral around the hexamer (Figure 3c). Although at a slightly lower resolution compared to other protomers, electron density is well defined in this region and reveals interactions between the P1-NBD2 and P6-NBD1 that are comprised of the small and large AAA+ subdomains, respectively (Figure 3d). While precise contacts were unable to be modeled, the interaction appears to be mediated by the B3-b2 and B6-b5 connecting loops from the P6-NBD1 and helices E2 and E3 from the P1-NBD2. Density corresponding to the P1-CTD also contacts the P6 NBD1, towards the outside of the channel, potentially stabilizing the interaction.
Despite the unusual NBD2-NBD1 interaction, density is present that corresponds to bound nucleotide for the P1-NBD2, indicating a potentially active catalytic site (Figure 3d). As indicated above, ATP hydrolysis by the AAA+ domains requires interactions with the adjacent protomer that includes contact by the Arg finger to stabilize the bound nucleotide. While the NBD2-NBD1 interface is different compared to the canonical AAA+ inter-protomer interface, putative Arg finger residues R334 and R333 in the P6-NBD1 are localized approximately 10–12 Å away from the P1-NBD2 nucleotide pocket. Interestingly, another highly conserved Arg residue, R307 in P6, is within 5 Å of the P1-NBD2 pocket. Thus, either a small conformational change that repositions R334/R333 or activation by R307 could trigger hydrolysis at this heteromeric NBD2-NBD1 site. In contrast, density is not observed in the P1-NBD1 pocket, indicating nucleotide is not present (Figure 3d). However, because of the protomer offset, P1-NBD1 is not supported by an adjacent AAA+ domain from P6, thus the nucleotide pocket is likely destabilized and inactive. Despite the adjacent P5-NBD1 in an apparent canonical orientation, density in the P6-NBD1 pocket appears weak relative to the other protomers, indicating partial or no nucleotide occupancy. Finally, density for nucleotide is observed for the P6-NBD2; this site is supported by the adjacent P5-NBD2, which is in a similar arrangement compared to what is observed for other protomers. Overall, comparison of the AAA+ domains for protomers 1 and 6 reveals a mix of nucleotide states and an asymmetric structural organization with distinct interactions relative to the rest of the hexamer that potentially support unique functions during disaggregation.
Arrangement of the NTD, CTD and MD highlight distinct functions in the hexamer
The conserved NTDs bind hydrophobic regions of substrates and are proposed to direct polypeptides to the NBD pore loops as well as enhance cooperative substrate binding required for amyloid disaggregation15,29. In our Hsp104 map the NTDs interact together, forming a broad, open-ended funnel that mirrors the helical arrangement of the NBD rings (Figure 4a). The P1-P5 NTDs surround the axial channel, forming a C-shaped entrance that is approximately 35 Å in diameter. The opening widens substantially to approximately 50 Å moving towards the P6 NTD at the hexamer seam, creating a large cleft that opens along the side of the hexamer. Although the P6-NTD is more flexible and less well resolved, interactions between the adjacent P5-NTD as well as an unusual interaction with P1-NBD2 are identified when viewed at a reduced threshold for the electron density (Figure 4a). Because of this NTD separation, the P1-NBD1 largely defines an equatorial channel entrance. Based on the arrangement in the other protomers, the P1-MD would extend across the cleft towards the P6-NTD, however density was not sufficient for localization due to its flexibility.
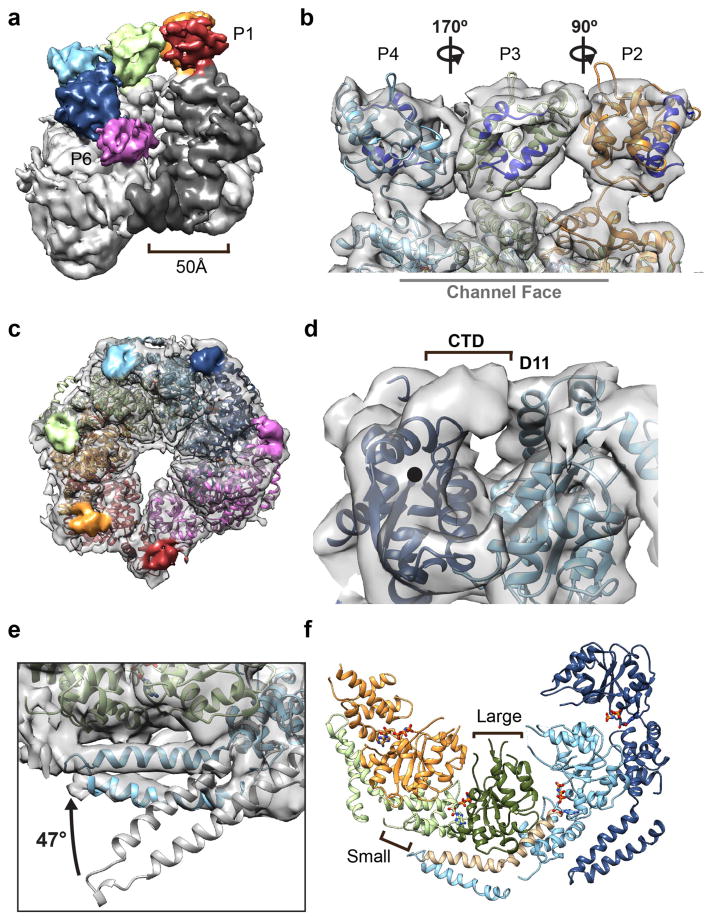
Figure 4.
Distinct arrangments of the NTD, MD and CTD in the hexamer. (a) 3D map in a tilted orientation showing the NTDs, colored by protomer, interacting around the channel entrance and the 50 Å-wide cleft defined by the P1 and P6 NTDs and P1 NBD1. (b) Enlarged view of the modeled P2-P4 NTDs showing different rotations around the NBD1 connecting linker. Rotations are relative to P3, and hydrophobic substrate binding sites15 are indicated (blue). (c) View of the NBD2 channel exit face of the hexamer with the density corresponding to the CTDs colored by protomer. (d) Enlarged view of the P5-CTD density extending from the NBD2 C-terminus (●) and interacting with P4-NBD2, adjacent helix D11. (e) Comparison between the MD L1-L2 orientation based on the docked ClpB structure25 (grey) and Hsp104 model showing 47° rotation around C3-L1 connecting residues (409–410) to fit the electron density. (f) View of the NBD1-MD interactions for P2-P5, showing helices C3-L1 (brown) bridging across the large (dark green) and small (light green) subdomains of the adjacent protomer.
Modeling the P2-P4 NTDs was facilitated by localization of the well-defined NTD-NBD1 connecting density as well as regions that could be attributed to the larger A1 and A6 helices that make up the hydrophobic substrate-binding cleft15 (Figure 4b). Based on our model only the P3 NTD appears to be positioned with its substrate-binding surface facing towards the channel, while the P2 and P4 NTDs each appear to be in different orientations. Thus, while the NTDs bind one another to form a defined ring at the channel entrance, the interaction interfaces are variable and multiple conformations are adopted around the hexamer.
Conversely, the channel exit on the opposite face of the hexamer is largely defined by the NBD2, however additional density is observed, which we predict corresponds to the 38-residue CTD (Figure 4c). While the CTD is considered to be disordered based on sequence41, this distinct lobe of density is clearly observed in all the protomers, indicating the CTD is partially structured and interacts with the NBD2 domains around the hexamer. The density extends from the NBD2 C-terminal residue that was modeled based on ClpB and extends across, forming a defined bridge connection to the adjacent protomer, contacting helix D11 in the NBD2 (Figure 4d). The interaction likely stabilizes the hexamer, supporting previous studies 24 and the CTD position around the channel exit also rationalizes previously proposed functions in cochaperone binding23, and thus could serve as a docking point for substrate hand-off to downstream chaperone systems.
The fit of the MD L1-L2 helices for P3-P6 revealed a substantial, 47° rotation of the MD compared to the crystal structure that enables direct connection to the adjacent protomer (Figure 4e). In this conformation, the MD helix L1 and L2 are positioned alongside the NBD1 of the clockwise neighboring protomer, and together with helix C3, form a 60 Å-long strap across the small and large subdomains (Figure 4f). This interaction is with the opposite protomer that makes up the canonical AAA+ interface, such that each NBD1 nucleotide pocket is supported by the MD of one neighbor and the NBD1 of the other neighbor. Density in this region is well defined for helix L1, which contacts helix C1, the C1-C2 connecting loop and part of helix B3 in the neighboring protomer. Specifically, we identify that conserved charged residues, R419, E427, and D434, that have previously identified functional roles27,42,43, are positioned adjacent the conserved residues E190, R353 and R366 in the NBD1, respectively, potentially making stabilizing salt-bridge interactions (Supplementary Fig. 4).
Spiral arrangement of the pore loops reveals a directional NBD1-NBD2 substrate translocation mechanism
Each NBD contains flexible loop regions that harbor the conserved Tyr residues (Tyr 257 for NBD1 and Tyr 662 in NBD2) that bind polypeptide substrates and are required for disaggregation functions5,6,39,44. The pore loops are proposed to face the axial channel in cryo-EM models26 and coordinate substrate threading based on asymmetric structures of the related ClpX single-ring AAA+ complex32. However, the pore loops for both NBD1 and NBD2 are not resolved in the ClpB structure25 and how polypeptides could be actively transferred either across the two distinct AAA+ domains or between protomers has been a mystery. Based on our flexible fitting we are able to model pore loop residues up to 252 and 259 for NBD1 and 659 and 667 for NBD2 within the large lobes of density that extend from these positions (Figure 5a). The NBD1 and NBD2 pore loop densities are separated by 35–40 Å, depending on the protomer, and indeed project toward the channel axis in a staggered, counter clockwise arrangement looking down the N-terminal channel entrance (Figure 5b).
Figure 5.
Two-turn spiral arrangement of the substrate-binding Tyr pore loops around the channel. (a) Electron density corresponding to the Tyr pore loops (circled) for NBD1 and NBD2 is shown for P1. (b) View down the channel from the NBD1 face with the pore loop densities colored according to protomer. Arrow indicates counter-clockwise shift between adjacent protomers moving down the channel. (c) Rotating views from inside channel showing two-turn spiral organization of the pore loops and putative P6-NBD1 to P1-NBD2 substrate transfer site. (d) Distances between the pore loops and corresponding movement down the channel axis are shown in Å.
Seen from inside the channel, the NBD1 and NBD2 pore loops form a striking two-turn spiral staircase (Figure 5c and Supplementary Video 3). The P1-NBD1 pore loop is nearest to the NTD channel cleft and each subsequent loop is 15–20 Å away, moving further into the channel towards the P6-NBD2 (Figure 5d). The distances between the pore loops vary, with greater separation near the hexamer seam, particularly between P5 and P6, which are separated by 20 Å, reflecting conformational differences in the protomers. Notably, we observe additional lobes of density located beneath the NBD1 pore loops, corresponding to residues 288–298, which could serve as additional substrate binding surfaces via electrostatic interactions (Supplementary Fig 5). Indeed, this corresponding region in ClpA, a related Hsp100, has been shown to interact directly with substrate45. The position of NBD1 loops are approximately 10 Å away from the respective NTDs for each protomer, thus substrates could be transferred to the NBD at multiple sites around the channel. Remarkably, the heteromeric NBD1-NBD2 interaction at the hexamer seam positions the respective pore loops 17 Å apart, similar to the other distances, revealing a continuous substrate transfer pathway between the AAA+ domains.
DISCUSSION
Hsp104 catalyzes protein disaggregation reactions that protect cells during thermal stress and facilitate prion propagation and dissolution in yeast. Despite these fundamental roles and a growing interest in harnessing disaggregase functions in neurodegenerative disease pathways, a definitive structure of the Hsp104 hexamer has remained elusive. Previous cryo-EM structures were critical in revealing the overall three-tier architecture and nucleotide-dependent conformational changes26,27,30,31 but were limited in resolution, likely due to available cryo-EM technology, heterogeneity, and imposed symmetry. By utilizing the latest cryo-EM technical advances and acquiring a substantial single particle dataset for exhaustive classification and alignment, we have achieved a structure of Hsp104 at 5.6 Å resolution that reveals, for the first time, that the hexamer adopts an asymmetric, near-helical architecture that connects the AAA+ domains in a two-turn spiral.
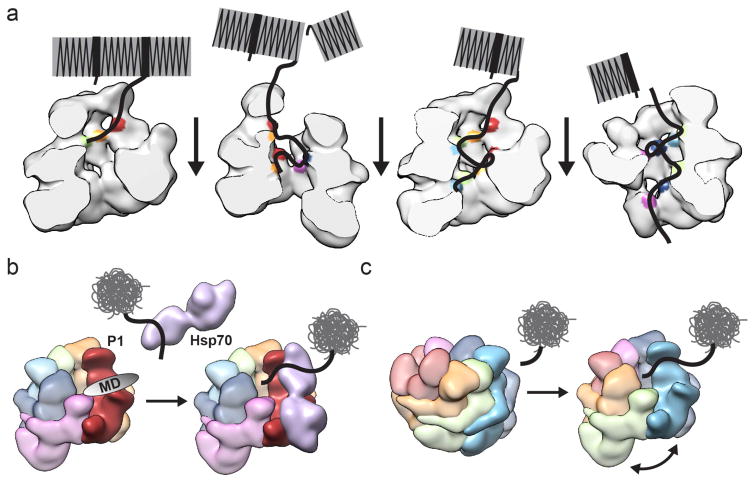
From this work a model emerges whereby the left-handed helical organization, which involves an 8–10 Å rise per subunit, establishes a substrate-binding path that appears optimally organized for unidirectional movement of polypeptides as they are transferred from protomer to protomer (Figure 6a). The Tyr pore loops are positioned in a continuous array that allows for discrete stepwise movement down the channel, suggesting how a cooperative mechanism of disaggregation might be achieved. In this scenario, polypeptides move in a counter clockwise direction around the channel, from the NTD entrance to the NBD2/CTD exit. The unusual NBD1-NBD2 interaction at the hexamer seam would therefore be critical for facilitating substrate transfer between the AAA+ domains, thus enabling two cycles of interaction per protomer during translocation. This mechanism is likely necessary for substrates such as structured amyloids that are dependent on high protomer cooperativity and action from both AAA+ domains 35. Additionally, polypeptide threading could occur more stochastically or involve a subset of protomers, such as for disordered, unstructured aggregates, as proposed in previous studies16,35. Nonetheless, the staggered arrangement of binding surfaces explains how unidirectional threading can confer more powerful unfolding. While the translocation step size is unknown for Hsp104, it is expected to be just a few amino acids for ClpB46, and has been measured to be 5–8 amino acids for ClpX47,48 and even smaller and more regular for ClpA49. Thus, with the 15–20 Å distance between pore loops we identify, interaction sites spanning a small number of amino acids along a polypeptide can be transferred stepwise from one protomer to the next during ATP hydrolysis, facilitating processive translocation through the channel.
Figure 6.
Models for cooperative disaggregation and substrate engagement by Hsp104. (a) Disaggregation of structured amyloids is depicted, showing cooperativity between protomers facilitated by the spiral architecture, which enables ordered polypeptide transfer across short distances and between the two AAA+ domains. (b) The hexamer seam and cleft between P1 and P6 presents a channel entrance and an optimal configuration for substrate binding and Hsp70 interaction via the P1-MD. (c) Depiction of protomer rearrangements triggered by substrate binding that could result in formation of the hexamer seam at the site of engagement to facilitate polypeptide translocation into the channel.
Structures of ClpX reveal an asymmetric hexamer and a staggered arrangement of the pore loops in its single AAA+ ring32 that have led to models for how Hsp100 family members ensure unidirectional threading3. The spiral architecture of Hsp104 is a remarkable extension of this mechanism that would also facilitate processive substrate transfer across its two AAA+ domains. Based on sequence conservation, we expect ClpB and other type II Hsp100 family members could adopt a similar asymmetric spiral architecture as Hsp104 in solution. Indeed, the ClpA50 and ClpB25 protomers form a continuous right-handed spiral in the crystal. However the conformations are overall different compared to our Hsp104 cryo-EM structure and both display mechanistic differences; ClpA functions in conjunction with a bound ClpP protease51, while ClpB is proposed to operate via non-processive translocation events35,46, thus these complexes may adopt different hexamer arrangements in solution.
In the Hsp104 cryo-EM structure, the P1 NTD and NBD1 substrate binding surfaces are at the topmost axial position and accessible due to the 50 Å-wide cleft formed by separation of P1 and P6 NTDs (Figure 4a). Thus, initial substrate engagement could involve preferential interactions with this protomer at the hexamer seam, followed by sequential downstream transfer to P2-P6. Furthermore, the cleft at the hexamer seam opens directly to the P1-NTD and MD, thereby presenting an optimal site for Hsp70 to interact and transfer substrates directly to the channel (Figure 6b). The interface at the hexamer seam may also play critical roles in the functional plasticity of Hsp10429, facilitating recognition and engagement of different aggregate structures. Finally, because the hexamer organization is known to be dynamic and coupled to the ATP hydrolysis cycle36, a compelling mechanism is that the protomers reorganize at the site of substrate binding and Hsp70 interaction, potentially forming the hexamer seam and NBD1-NBD2 configuration to facilitate transfer down the channel (Figure 6c). Indeed, a key question that emerges from these results is how the NBD1-NBD2 interaction and arrangement of the hexamer seam responds to substrate binding and ATP hydrolysis events during disaggregation.
A number of different MD conformations have been proposed based on previous low-resolution models25–28,30, thus the arrangement has remained a question. Our structure reveals the clearest view to date of the MD arrangement in the hexamer, resolving the L1-L2 portion of the MD where well-defined α-helical density enables precise localization along the NBD1 of the neighboring protomer (Figure 4e). We identify conserved MD residues, including R419, E427, and D434, which make specific contacts with the NBD1, and bridge the small and large subdomains, potentially stabilizing the nucleotide pocket (Figure 4f and Supplementary Fig 4). This observation supports previous fluorescence resonance energy transfer experiments in ClpB identifying contact between L1 and the adjacent NBD143. Given that Hsp104 is bound to AMP-PNP in our structure, these bridging interactions by the MD likely play key roles in activating ATP hydrolysis in the hexamer. Indeed, R419M exhibits impaired ATPase activity in vitro and is defective in thermotolerance in vivo27. DnaK/Hsp70 is known to bind the opposite end of the MD coil-coil at helix L2-helix L3 18,20,52, which is flexible and not observed in our reconstruction. Nonetheless, the L1-NBD1 interactions may serve as control elements for allosteric activation by DnaK/Hsp70 during substrate delivery19,43. Indeed, mutations in this L1 region, specifically A430W and V426L, eliminate collaboration with Hsp70 21 and enable disaggregation to occur independently of Hsp70 and Hsp4012, respectively.
In addition to our results here, a number of recent cryo-EM structures of AAA+ complexes, including, in particular, the Pex1-Pex653 and NSF54 double AAA+ ring complexes, have identified asymmetric states. Together this work is beginning to reveal that AAA+ proteins operate via large asymmetries in the protomer organization, agreeing with known asymmetric hydrolysis mechanisms32. The Hsp104 structure we have determined here is the first AAA+ complex, to our knowledge, identified to adopt a two-turn spiral architecture that is supported by an interaction between different AAA+ (NBD1-NBD2) domains. Hsp104/ClpB arose from a gene fusion event and NBD1 and NBD2 are members of different AAA+ clades (clade 3 and clade 5, respectively)41, making the interaction identified here more surprising. Both Pex1-Pex6 and NSF complexes exhibit a planar, symmetric D1 ring that is flexibly connected to an asymmetric D2 ring (compared to NBD1 and NBD2 rings for Hsp104, respectively). In Hsp104, the orientation between the two AAA+ domains within a protomer is relatively consistent around the ring except for the hexamer seam. At this position, large conformational differences between P6 and P1, including a 20° rotation of NBD2 relative to NBD1, enable the NBD1-NBD2 interaction that maintains a closed hexamer ring despite the substantial, 100° rotation around the channel axis (compared to ~50° for the other protomers) (Figure 3a). In addition we are able to identify density corresponding to a bound nucleotide for 10 out of 12 ATP binding sites. This falls in between previous estimates for nucleotide occupancy of 8–12 nucleotides for ClpB hexamers28,55, but is consistent with an overall asymmetric hydrolysis mechanism. With this high nucleotide occupancy and spiral architecture, the power of the two AAA+ domains appears to be uniquely coupled for Hsp104 compared to other AAA+ complexes and is perhaps reflective of the mechanical force required for disaggregation. Additional studies will be critical in elucidating conformational changes associated with ATP hydrolysis-dependent substrate binding and translocation events to understand the mechanical changes that drive disaggregation.
METHODS
Protein Purification, ATPase and Luciferase Reactivation Assay
Wild type Hsp104 was purified as described56. Hexameric Hsp104 (0.042 μM) was incubated with ATP (1 mM) at 25°C before monitoring the release of inorganic phosphate over 5 minutes using a malachite green kit (Innova). ATPase turnover was measured at a maximum rate of ~14/min, which corresponded to a functional, ATPase-active complex. Disaggregase function was measured by a luciferase reactivation assay as previously described35. Aggregated luciferase (50nM) was incubated with Hsp104 (0.167μM hexamer) in the presence of equimolar Hsc70 and Hdj2 (Enzo Life Sciences) plus ATP (5.1 mM) and an ATP regeneration system (1 mM creatine phosphate, 0.25μM creatine kinase) for 90 min at 25°C.
Cryo Sample Preparation and Data Collection
WT Hsp104 (8.5 mg/mL ) was incubated with AMP-PNP (5 mM ) for 20 minutes at 25°C. Samples were diluted to 0.7 mg/ml in 40 mM HEPES pH=7.5, 40 mM NaCl, 10 mM MgCl2, 1 mM DTT, 5mM AMP-PNP and 3.5 μl was applied to plasma cleaned C-Flat 2/2 holey carbon grids (Protochips). Vitrification was performed using a Vitrobot (FEI Company) and samples were blotted for 1.5–2 seconds prior to plunge freezing in liquid ethane. Of note, during initial attempts at imaging Hsp104 routinely dissociated into monomers and significant optimization of buffer conditions, sample concentration and freezing conditions was required to achieve proper ice thickness and a homogeneous spread of hexameric particles. Furthermore, the presence of DDM detergent was tested to increase the angular distribution but resulted in dissociation of the hexamer at a variety of concentrations (data not shown).
Samples were imaged using a Titan Krios TEM (FEI Inc.) operated at 300 kV. Images were recorded on a Gatan K2 Summit direct electron detector operated in counted mode at 50,000X nominal magnification corresponding to a calibrated to 1.00 Å/pixel. Dose fractionated imaging was performed by semi-automated collection methods using UCSF Image 457 with a defocus range of 1.5–3 um. Total exposure time was 8 seconds with 0.2 second frames with a cumulative dose of ~45 e− per Å2 for 40 frames. Motion corrected frames were summed with the first two frames excluded, and the FFT was visually inspected for sufficient Thon rings prior to additional processing 58.
Image Processing and 3D Refinement
All micrographs were CTF corrected using CTFFIND459 and poorly corrected micrographs were removed following visual inspection of the FFT and CTF estimation. An initial single particle dataset was achieved by manual particle picking using e2boxer (EMAN2) 60, which yielded ~50,000 particles from 1731 micrographs. Well-populated reference-free 2D class averages, determined with Relion61, were used for templated automated particle picking with the Template Picker in Appion62 to achieve a ~200,000 single particles dataset from 1930 micrographs. The total dataset of ~250,000 particles was initially sorted following 2D classification by removing particles images from poorly resolved class averages, resulting in a total dataset of ~190,000 particles. All subsequent 3D processing was performed using Relion61 with no symmetry imposed based on the asymmetric arrangement identified in the 2D class averages (Figure 1b). A previously determined cryo-EM 3D reconstruction of ATP-Hsp104 (EMDB: 1600) 31, low pass filtered to 50Å, served as an initial model for 3D classification and refinement. The 8 models generated from 3D classification appeared homogenous (data not shown), therefore, the full dataset was used for initial gold-standard 3D refinement. The resulting 3D model refined to an estimated resolution of 7.5 Å (data not shown). Z-score parameters based on this refinement, defined in Relion, were written for the data set and the dataset was trimmed to the highest Z-score for 160,000 particles. Additional 3D refinement was performed with this trimmed data set and resulted in a final model with an estimated resolution of 6.54 Å using the FSC = 0.143 FSC criterion (Supplementary Fig 1e). Additional trimming based on the Z-score showed no improvement and extensive 3D classification and refinement of individual classes as well as local refinement using 3D masking was tested and did not did not yield improvements in the map. Thus, the resolution is likely limited by flexibility of multiple regions in the complex (Supplementary 1h) and moderate preferred orientation (Supplementary Fig. 1f). For the final sharpened map, the “Post-processing” procedure was used to generate a soft mask for the two half maps prior to FSC estimation, which was determined to be 5.64 Å (Supplementary Fig 1e). Automated B-factor sharpening was carried on the combined map with an estimated -188 B-factor. The local resolution was estimated using ResMap on the unsharpened map63.
Modeling
This final sharpened map was used for all rigid body docking and flexible fitting of atomic structures. Rigid body fitting was performed with UCSF Chimera64. Initial comparisons were performed by fitting individual protomers with the protomers in T. thermophilus ClpB crystal structure (PBD_ID=1qvr), but resulted in a low cross correlation value and clear conformational differences were apparent. Individual subdomains (NTD, NBD1 large, NBD1 small, MD (residues 409–467), NBD2 large, NBD2 small) from the crystal structure were then docked as rigid bodies and resulted in an improved fit, however a number of AAA+ domain helices did not align with the density. Therefore, to more accurately interrupt the map, a homology model of Hsp104 (residues 6–857) was determined from the ClpB structure (residues 4–850) using SWISS_MODEL65 followed by rigid body docking the subdomains. Flexible fitting of the protomer sub volumes and the entire hexamer map was then performed using phenix. real_space_refine66 using backbone carbons of our homology model. The real space refinement was implemented at 6σ, resolution 6Å and the cross correlation improved from 0.72 to 0.9 for the complete map after refinement. Nucleotide density was seen for 10 of the nucleotide binding pockets, therefore this molecule was included in the model and docked based on the ClpB structure. Manual modeling of the tyrosine loop regions was performed in COOT67. Loop regions within the NBD1 domain (248–260,287–300) were modeled by backbone positioning within the electron density; residues 251–259, 292–297 were unable to be modeled in the density. Of note, a major shift was required to model the NBD2 tyrosine loop (656–669), which showed a distinct curve away from the flanking helices. Similarly, the most central portion of the loop lacked density and was not modeled (659–666). All images were generated using UCSF Chimera64.
Accession codes
The cryo-EM electron density map and molecular model have been deposited in the Electron Microscopy Data Bank (EMDB-3416).
Supplementary Material
Acknowledgments
The authors thank J. Smith, Z. March, and J. Lin for reading the manuscript as well as Y. Skiniotis for many helpful discussions and advice. We thank A. Tariq for technical assistance with the Hsp104 purification. This work was supported by the National Institutes of Health (NIH) grant: R01GM099836 (J.S.). A.Y. is supported by an American Heart Association Predoctoral fellowship; M.J. is supported by a Target ALS Springboard Fellowship. K.M. is supported by an NSF Graduate Research Fellowship (DGE-1321851). J.S. is supported by a Muscular Dystrophy Association Research Award (MDA277268), the Life Extension Foundation, the Packard Center for ALS Research at Johns Hopkins University, and Target ALS.
Footnotes
AUTHOR CONTRIBUTIONS
A.Y. designed experiments, performed cryo-EM sample preparation, data collection and analysis, and wrote the manuscript; S.G. performed cryo-EM data collection and analysis; M.J. purified proteins, performed biochemical analysis and edited the manuscript; K.M. performed biochemical analysis; M.S. supervised cryo-EM data collection; J.S. designed experiments and edited the manuscript; D.S. designed and supervised the study and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 2.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 3.Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 5.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 7.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci U S A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 9.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 10.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo Bianco C, et al. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackrel ME, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cushman-Nick M, Bonini NM, Shorter J. Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet. 2013;9:e1003781. doi: 10.1371/journal.pgen.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackrel ME, Shorter J. Reversing deleterious protein aggregation with re-engineered protein disaggregases. Cell Cycle. 2014;13:1379–1383. doi: 10.4161/cc.28709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenzweig R, et al. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc Natl Acad Sci U S A. 2015;112:E6872–6881. doi: 10.1073/pnas.1512783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle SM, Hoskins JR, Wickner S. DnaK chaperone-dependent disaggregation by caseinolytic peptidase B (ClpB) mutants reveals functional overlap in the N-terminal domain and nucleotide-binding domain-1 pore tyrosine. J Biol Chem. 2012;287:28470–28479. doi: 10.1074/jbc.M112.383091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedzierska S, Akoev V, Barnett ME, Zolkiewski M. Structure and function of the middle domain of ClpB from Escherichia coli. Biochemistry. 2003;42:14242–14248. doi: 10.1021/bi035573d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 19.Seyffer F, et al. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol. 2012;19:1347–1355. doi: 10.1038/nsmb.2442. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, et al. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc Natl Acad Sci U S A. 2013;110:8513–8518. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desantis ME, et al. Conserved distal loop residues in the Hsp104 and ClpB middle domain contact nucleotide-binding domain 2 and enable Hsp70-dependent protein disaggregation. J Biol Chem. 2014;289:848–867. doi: 10.1074/jbc.M113.520759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cashikar AG, et al. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–760. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 23.Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay RG, Helsen CW, Tkach JM, Glover JR. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry. 2008;47:1918–1927. doi: 10.1021/bi701714s. [DOI] [PubMed] [Google Scholar]
- 25.Lee S, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wendler P, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroni M, et al. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife. 2014;3:e02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweeny EA, et al. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol Cell. 2015;57:836–849. doi: 10.1016/j.molcel.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Sielaff B, Lee J, Tsai FT. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci U S A. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wendler P, et al. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- 34.Aguado A, Fernandez-Higuero JA, Cabrera Y, Moro F, Muga A. ClpB dynamics is driven by its ATPase cycle and regulated by the DnaK system and substrate proteins. Biochem J. 2015;466:561–570. doi: 10.1042/BJ20141390. [DOI] [PubMed] [Google Scholar]
- 35.DeSantis ME, et al. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J Mol Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 37.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J Biol Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 39.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 40.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 41.Sweeny EA, Shorter J. Mechanistic and Structural Insights into the Prion-Disaggregase Activity of Hsp104. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dulle JE, Stein KC, True HL. Regulation of the Hsp104 middle domain activity is critical for yeast prion propagation. PLoS One. 2014;9:e87521. doi: 10.1371/journal.pone.0087521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oguchi Y, et al. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat Struct Mol Biol. 2012;19:1338–1346. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- 44.Schlieker C, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 45.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Li T, et al. Escherichia coli ClpB is a non-processive polypeptide translocase. Biochem J. 2015;470:39–52. doi: 10.1042/BJ20141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olivares AO, Nager AR, Iosefson O, Sauer RT, Baker TA. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat Struct Mol Biol. 2014;21:871–875. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 51.Alexopoulos JA, Guarne A, Ortega J. ClpP: a structurally dynamic protease regulated by AAA+ proteins. J Struct Biol. 2012;179:202–210. doi: 10.1016/j.jsb.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Haslberger T, et al. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell. 2007;25:247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Blok NB, et al. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci U S A. 2015;112:E4017–4025. doi: 10.1073/pnas.1500257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao M, et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin J, Lucius AL. Examination of ClpB Quaternary Structure and Linkage to Nucleotide Binding. Biochemistry. 2016;55:1758–1771. doi: 10.1021/acs.biochem.6b00122. [DOI] [PubMed] [Google Scholar]
- 56.Jackrel ME, Shorter J. Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Model Mech. 2014;7:1175–1184. doi: 10.1242/dmm.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Zheng S, Agard DA, Cheng Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol. 2015;192:174–178. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 61.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.