Abstract
Retroviruses encounter dominant postentry restrictions in cells of particular species. Human immunodeficiency virus type 1 (HIV-1) is blocked in the cells of Old World monkeys by TRIM5α, a tripartite motif (TRIM) protein composed of RING, B-box 2, coiled-coil, and B30.2(SPRY) domains. Rhesus monkey TRIM5α (TRIM5αrh) more potently blocks HIV-1 infection than human TRIM5α (TRIM5αhu). Here, by studying chimeric TRIM5α proteins, we demonstrate that the major determinant of anti-HIV-1 potency is the B30.2(SPRY) domain. Analysis of species-specific variation in TRIM5α has identified three variable regions (v1, v2, and v3) within the B30.2 domain. The TRIM5α proteins of Old World primates exhibit expansion, duplication, and residue variation specifically in the v1 region. Replacement of three amino acids in the N terminus of the TRIM5αhu B30.2 v1 region with the corresponding TRIM5αrh residues resulted in a TRIM5α molecule that restricted HIV-1 nearly as efficiently as wild-type TRIM5αrh. Surprisingly, a single-amino-acid change in this region of TRIM5αhu allowed potent restriction of simian immunodeficiency virus, a phenotype not observed for either wild-type TRIM5αhu or TRIM5αrh. Some of the chimeric TRIM5α proteins that are >98% identical to the human protein yet mediate a strong restriction of HIV-1 infection may have therapeutic utility. These observations implicate the v1 variable region of the B30.2(SPRY) domain in TRIM5αrh antiviral potency.
The primate lentiviruses include human immunodeficiency virus type 1 (HIV-1) and HIV-2 and simian immunodeficiency viruses (SIVs) (2, 5, 7, 8, 11). HIV-1 and HIV-2 infect humans, HIV-1-like viruses infect chimpanzees, and SIV variants infect African monkeys. Humans infected by HIV-1 and HIV-2 and Asian macaques infected by certain SIV strains (SIVmac) often develop life-threatening immunodeficiency due to depletion of CD4-positive T lymphocytes (9, 19).
HIV and SIV tropism is determined by cell-type-specific and species-specific host factors. Following entry into the host cell, uncoating of the viral core, reverse transcription, nuclear access, and integration of the viral DNA into the host genome must occur to establish a permanent infection (1, 10, 33). Early postentry restrictions to retrovirus infection can determine tropism at the species level. HIV-1 encounters a postentry block in Old World monkeys, whereas SIVmac is blocked in most New World monkey cells (15, 16, 27). These species-specific restrictions occur prior to or concurrent with reverse transcription; at most, low levels of early reverse transcripts are made in restricted cells (6, 15, 20, 27). The viral determinant of susceptibility to these blocks is the capsid protein (6, 13, 18, 22, 23, 30). The early restriction of HIV-1 and SIV is mediated by dominant host factors, the activities of which can be titrated by the introduction of virus-like particles containing proteolytically processed capsid proteins of the restricted viruses (3, 4, 6, 12, 22, 31, 32). Thus, in the cells of specific monkey species, host restriction factors apparently interact, directly or indirectly, with the HIV-1 or SIV capsid and prevent its progression along the infectious pathway. Recently, a genetic screen has identified a major restriction factor in monkey cells that acts on HIV-1 and, to a lesser extent, on SIVmac (29). The factor, TRIM5α, was selected from a cDNA library prepared from primary rhesus monkey lung fibroblasts (PRL cells). TRIM5α was shown to be sufficient to confer potent resistance to HIV-1 infection in otherwise susceptible cells. Moreover, TRIM5α was necessary for the maintenance of the block to the early phase of HIV-1 infection in Old World monkey cells, as demonstrated by interference with TRIM5α expression in these cells. HIV-1 infection in cells expressing rhesus monkey TRIM5α (TRIM5αrh) was blocked at the earliest stage of reverse transcription. Cells expressing rhesus monkey TRIM5α exhibited partial inhibition of SIVmac infection but were as susceptible as control cells to infection by Moloney murine leukemia virus (MLV) vectors. The human ortholog, TRIM5αhu, is 87% identical in amino acid sequence to TRIM5αrh. Even when expressed at comparable levels, TRIM5αhu was less potent at suppressing HIV-1 and SIVmac infection than TRIM5αrh (29).
TRIM5 is a member of a family of proteins that contain a tripartite motif, leading to the designation TRIM (24). The tripartite motif includes a RING domain, a B-box 2 domain, and a coiled-coil domain; hence, TRIM proteins have also been called RBCC proteins. Some TRIM proteins contain a C-terminal B30.2, or SPRY, domain. Differential splicing of the TRIM5 primary transcript gives rise to the expression of several isoforms of the protein product. The TRIM5α isoform is the largest product (∼493 amino acid residues) and contains the B30.2(SPRY) domain. The other TRIM5 isoforms (γ and δ are the best substantiated of these) lack an intact B30.2(SPRY) domain. The rhesus monkey TRIM5γ isoform lacks anti-HIV-1 and anti-SIV activities, indicating the importance of the B30.2(SPRY) domain to the antiviral function (29). In fact, TRIM5γrh has been shown to exhibit weak dominant-negative activity in repressing the ability of wild-type TRIM5αrh to inhibit HIV-1 infection (29). An intact RING domain also contributes, either directly or indirectly, to the anti-HIV-1 activity of TRIM5αrh. Alteration of two cysteine residues that are highly conserved in RING domains markedly diminished, but did not abolish, the antiviral potency of TRIM5αrh (29).
Here, we investigate the basis for the differences in the potencies of HIV-1 inhibition observed for TRIM5αrh and TRIM5αhu.
MATERIALS AND METHODS
TRIM5α chimerae.
The trim5 cDNAs from humans and rhesus monkeys were obtained from a kidney cDNA library (Clontech) and from a primary rhesus monkey lung cDNA library, respectively (29). The nomenclature for the chimerae is TRIM5α A(Bx-y), in which the encoded TRIM5α amino acids from x to y from species B are inserted into the TRIM5α protein of species A (H, human; R, rhesus monkey). The numbering scheme is based on the human TRIM5α residue numbers; the same numbers are used for the rhesus monkey TRIM5α residues, after the TRIM5αrh sequence is aligned to that of TRIM5αhu.
Some of the chimeric trim5 constructs were created by exchanging fragments generated by digestion with the restriction enzymes BsmII [TRIM5α R(H286-493) and TRIM5α H(R286-493)] or BsmII and BamHI [TRIM5α R(H286-371) and TRIM5α H(R286-371)]. The following chimeric proteins were expressed by plasmids created by QuikChange mutagenesis (Stratagene): TRIM5α H(R305-314), R(H305-314), H(R323-332), R(H323-332), R(H335-340), and R(H337-338a). The predicted amino acid sequence of the TRIM5α R(H337-338a) protein in the region affected by the mutation is GTLFQSLTNF. The mutated plasmids were sequenced in the regions surrounding the introduced changes.
TRIM5αhu mutants.
Plasmids expressing TRIM5αhu with single-amino-acid changes were created by QuikChange mutagenesis. The nomenclature for these mutants is TRIM5αhu followed by the wild-type amino acid residue in single-letter code, the amino acid position, and the amino acid residue to which the change was made.
Creation of cells stably expressing TRIM5α variants.
Retroviral vectors encoding TRIM5αhu, TRIM5αrh, or chimeric TRIM5α proteins were created using the pLPCX vector (29). The pLPCX vectors contain only the amino acid coding sequence of the TRIM5α cDNA. In all constructs, the TRIM5α proteins possess C-terminal epitope tags derived from influenza hemagglutinin (HA). Recombinant viruses were produced in 293T cells by cotransfecting these pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. The resulting virus particles were used to transduce ∼106 HeLa cells in the presence of 5 μg of Polybrene/ml. Cells were selected in 1 μg of puromycin (Sigma)/ml.
Immunoblotting.
HA-tagged proteins were expressed in HeLa cells by transduction with pLPCX vectors as described above. The HA-tagged TRIM5α proteins were detected in whole-cell lysates [100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 7.5), 10% glycerol, 1% Nonidet P40] by Western blotting with horseradish peroxidase-conjugated 3F10 antibody (Roche). β-Actin was detected with A5441 antibody (Sigma).
Infection with viruses expressing GFP.
Recombinant HIV-1 expressing green fluorescent protein (GFP) was prepared as described previously (16, 22, 23, 29), and MLV-GFP was prepared by cotransfecting 293T cells with 15 μg of pFB-hrGFP, 15 μg of pVPack-GP, and 4 μg of pVPack-VSV-G (all from Stratagene). HIV-1 stocks were quantified by measuring reverse transcriptase activity as described previously (25). MLV reverse transcriptase activity was determined by the same procedure, except that 20 mM MnCl2 was used instead of MgCl2. For infections, 3 × 104 HeLa cells seeded in 24-well plates were incubated in the presence of virus for 24 h. The cells were washed and returned to culture for 48 h and then subjected to fluorescence-activated cell sorter (FACS) analysis with a FACScan (Becton Dickinson).
RESULTS
A carboxy-terminal TRIM5α determinant of anti-HIV-1 potency.
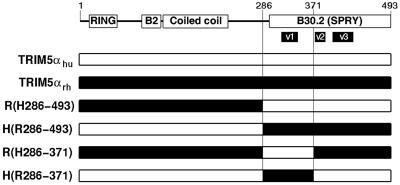
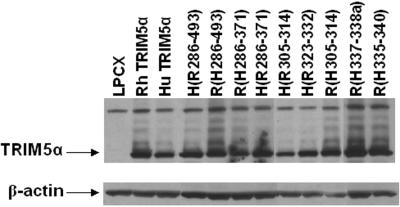
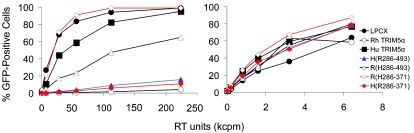
TRIM5αrh exhibits significantly greater inhibitory activity against HIV-1 than TRIM5αhu (29). To map the determinants of this potency, chimeric proteins between TRIM5αrh and TRIM5αhu were created (Fig. 1). HeLa cells were transduced by pLPCX retroviral vectors expressing the wild-type and chimeric TRIM5α proteins, as described previously (29). All of the TRIM5α proteins have a carboxy-terminal HA tag, allowing documentation of expression levels in the transduced cells. We first studied TRIM5α chimerae containing the RING, B-box 2, and coiled-coil domains of one parent protein and the B30.2 domain of another. These chimerae, designated TRIM5α R(H286-493) and TRIM5α H(R286-493), were both expressed in HeLa cells at levels comparable to those of the parental TRIM5α proteins (Fig. 2). The HeLa cells were incubated with a recombinant HIV-1 vector (HIV-1-GFP) expressing GFP or, as a control, with an MLV vector (MLV-GFP). Figure 3 shows that, compared with HeLa cells transduced with the empty pLPCX vector, HeLa cells expressing TRIM5αrh were strongly resistant to HIV-1-GFP infection. A modest decrease in HIV-1-GFP infection was observed in cells expressing TRIM5αhu. HeLa cells expressing the TRIM5α H(R286-493) protein were almost as resistant to infection by HIV-1-GFP as cells expressing TRIM5αrh. An intermediate level of HIV-1-GFP inhibition was observed in the cells expressing TRIM5α R(H286-493). MLV-GFP infection of the cells expressing the various TRIM5α chimerae was similar to that of the control cells transduced with the empty pLPCX vector. We conclude that the B30.2 domain of TRIM5αrh is sufficient to confer potent anti-HIV-1 activity on TRIM5αhu.
FIG. 1.
Chimeric TRIM5αhu/TRIM5αrh proteins. A diagram of the TRIM5α protein is shown, with the known domains indicated (B2, B-box 2). The numbers of the residues at the N and C termini of the protein and at the chimeric junctions are indicated. The species-specific variable regions (v1, v2, and v3) within the B30.2 domain (28) are shown. The numbering scheme and nomenclature used for the chimeric proteins is based upon the TRIM5αhu sequence and is described in Materials and Methods.
FIG. 2.
Expression of chimeric TRIM5α proteins. Lysates from HeLa cells expressing the parental and chimeric TRIM5α proteins, which contain C-terminal HA epitope tags, were subjected to Western blotting with an antibody against HA. Control lysates from HeLa cells transduced with the empty pLPCX vector were analyzed in parallel. The lysates were also Western blotted with an antibody directed against β-actin.
FIG. 3.
Effects of expression of the chimeric TRIM5α proteins on HIV-1 infection. HeLa cells expressing the parental and chimeric TRIM5α proteins, or control HeLa cells transduced with the empty pLPCX vector, were incubated with various amounts of HIV-1-GFP or MLV-GFP. Infected GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. RT, reverse transcriptase.
Functional importance of variation in the v1 region of the TRIM5α B30.2 domain.
Analysis of the sequences of rodent and primate TRIM5α proteins has revealed the existence of three regions within the B30.2 domain that exhibit dramatic species-specific length and amino acid variations (28). These regions have been designated v1, v2, and v3 (Fig. 1). The v1 region of the TRIM5α B30.2 domain in Old World primates is longer than those of other primates (28). Thirteen of the 33 differences in amino acid sequence between the B30.2 domains of TRIM5αhu and TRIM5αrh occur within the v1 region. We hypothesized that variation in the B30.2 v1 region contributes to the differences between the HIV-1-restricting activities of human and rhesus monkey TRIM5α proteins. To test this hypothesis, we created additional chimeric proteins that would allow an assessment of the contribution of the v1 region independently of the other B30.2 variable regions. These chimeric proteins, TRIM5α R(H286-371) and TRIM5α H(R286-371) (Fig. 1), were expressed stably in HeLa cells transduced with pLPCX vectors. The levels of expression of these chimeric proteins were equivalent to those of the wild-type TRIM5αhu and TRIM5αrh proteins (Fig. 2). The abilities of the wild-type and chimeric proteins to inhibit HIV-1-GFP infection were examined (Fig. 3). The TRIM5α H(R286-371) chimera, which contains the B30.2 v1 region of TRIM5αrh in a TRIM5αhu background, suppressed HIV-1-GFP infection nearly as efficiently as TRIM5αrh. Conversely, the reciprocal chimera, TRIM5α R(H286-371), exhibited no inhibitory activity against HIV-1-GFP infection. Expression of these chimeric TRIM5α proteins did not significantly affect the susceptibility of the HeLa cells to infection by MLV-GFP. These results indicate that the major determinant of anti-HIV-1 potency in TRIM5α is located between residues 286 and 371 in a segment including the B30.2 v1 region.
Mapping of the potency determinant within the B30.2 v1 region.
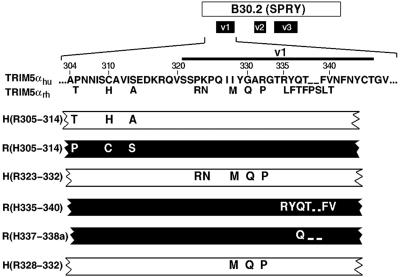
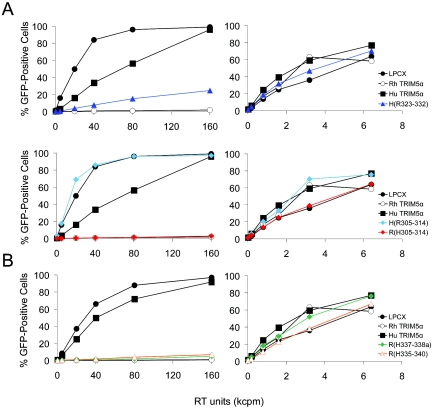
To investigate whether the determinant of anti-HIV-1 potency could be defined more precisely, specific changes within the B30.2 v1 region were made. The v1 region was arbitrarily divided into two segments, and the appropriate chimerae were constructed (Fig. 4). We also tested the importance to the TRIM5α phenotype of amino acid differences in a region N-terminal to the B30.2 v1 region (residues 305 to 314) (Fig. 4). After verifying that approximately equivalent levels of the chimeric proteins were made in transduced HeLa cells (Fig. 2), the ability of the cells to support HIV-1-GFP infection was tested. The TRIM5α H(R323-332) chimera, which contains the amino-terminal segment of the B30.2 v1 region, strongly inhibited HIV-1 infection (Fig. 5A). The inhibition observed for the TRIM5α H(R323-332) chimera was slightly less than that seen for the TRIM5αrh protein. The levels of inhibition of HIV-1-GFP infection by the TRIM5α H(R323-332) and H(R286-371) chimerae were comparable (data not shown). We conclude that the amino-terminal segment of the B30.2 v1 region is the major determinant of anti-HIV-1 potency.
FIG. 4.
Chimeric TRIM5α proteins containing heterologous elements of the B30.2 domain v1 region. A diagram of the TRIM5α B30.2 domain, with the variable regions (v1, v2, and v3) indicated is provided at the top of the figure. The region of interest is expanded, with the primary amino acid sequence of TRIM5αhu shown. Amino acid residues in TRIM5αrh that differ from those in TRIM5αhu are shown. The numbers refer to the human TRIM5α residue. The relevant segments of the chimeric TRIM5α proteins are shown. The white segments indicate that the protein is identical to TRIM5αhu except for the amino acid residues shown. The black segments indicate that the protein is identical to TRIM5αrh except for the indicated residues.
FIG. 5.
The N-terminal half of the B30.2 domain v1 region as a determinant of anti-HIV-1 potency. HeLa cells expressing the parental and chimeric TRIM5α proteins, or control HeLa cells transduced with the empty pLPCX vector, were incubated with various amounts of HIV-1-GFP or MLV-GFP. Infected, GFP-positive cells were counted by FACS. In the top row of panel A, a human TRIM5α variant [TRIM5α H(R323-332)] containing the N-terminal half of the rhesus monkey TRIM5α B30.2 domain v1 region was tested. In the bottom row of panel A and in panel B, TRIM5α chimerae containing heterologous segments other than the N-terminal portion of the B30.2 v1 region were tested. The results of typical experiments are shown. In panel A, the results of one experiment are separated into upper and lower rows for ease of viewing. Similar results were obtained in two independent experiments. RT, reverse transcriptase.
Insertion of carboxy-terminal segments of the B30.2 v1 region from the TRIM5αhu protein had no significant effect on the ability of the rhesus monkey TRIM5α molecule to restrict HIV-1 infection [Fig. 4 and 5B, TRIM5α R(H335-340) and TRIM5α R(H337-338a)]. Similarly, substitution of the human TRIM5α sequences amino-terminal to the B30.2 v1 region into the TRIM5αrh protein did not affect the anti-HIV-1 potency of the latter molecule [Fig. 4 and 5A, TRIM5α R(H305-314)]. The reciprocal chimera, TRIM5α H(R305-314), did not restrict HIV-1 infection (Fig. 4 and 5A). These results indicate that, of the species-specific amino acid differences in the TRIM5α sequences extending from residues 304 to 340, only those in the segment comprised of residues 323 to 332 appreciably affect the anti-HIV-1 potency of the molecule.
Contributions of individual amino acids in the v1 region to TRIM5α antiviral potency.
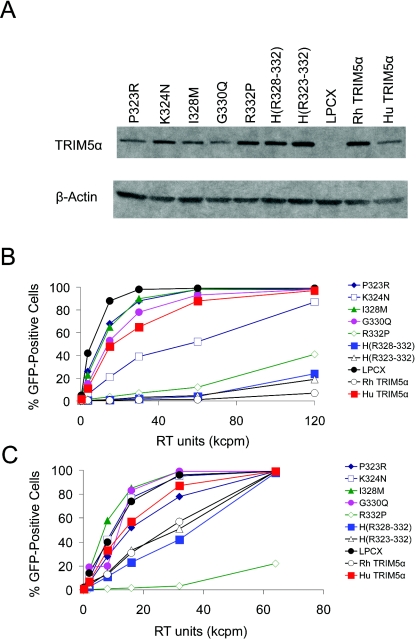
To dissect the species-specific determinants of TRIM5α antiviral potency further, additional TRIM5αhu mutants were created in which individual amino acid residues in the 323 to 332 segment were changed to those found in TRIM5αrh. In addition, the TRIM5αhu sequence from residues 328 to 332 was changed to that found in TRIM5αrh [Fig. 4, TRIM5α H(R328-332)]. HeLa cells were transduced with vectors expressing these TRIM5αhu variants. All of the TRIM5α variants were expressed in these HeLa cells, although a slight variation in the level of expression of the TRIM5α proteins was seen (Fig. 6A). The abilities of these TRIM5αhu mutants to inhibit HIV-1-GFP infection were examined and compared with those of TRIM5αrh, TRIM5αhu, and TRIM5α H(R323-332) (Fig. 6B). The levels of inhibition of HIV-1-GFP infection by the TRIM5α H(R328-332) and TRIM5α H(R323-332) proteins were comparable and only slightly less than that seen in cells expressing the wild-type TRIM5αrh protein. Of the TRIM5αhu mutants with single-amino-acid changes, TRIM5αrh R332P was most potent at restricting HIV-1 infection. The TRIM5αhu K324N mutant was reproducibly more effective than the wild-type TRIM5αhu protein at inhibiting HIV-1. The other single-amino-acid changes did not potentiate the anti-HIV-1 activity of TRIM5αhu. We conclude that the major determinant of anti-HIV-1 potency of TRIM5αrh resides in the sequence 328 to 332, in which three amino acids differ between TRIM5αrh and TRIM5αhu. Of the B30.2 domain v1 residues that differ between these two TRIM5α orthologs, differences in residues 332 and, to a lesser extent, 324 contribute to potency in restricting HIV-1 infection.
FIG. 6.
Expression and antiviral activities of mutant TRIM5α proteins. (A) Lysates from HeLa cells expressing the parental and mutant TRIM5α proteins, which contain C-terminal HA epitope tags, were subjected to Western blotting with an antibody against HA. Control lysates from HeLa cells transduced with the empty pLPCX vector were analyzed in parallel. The lysates were also Western blotted with an antibody directed against β-actin. (B and C) HeLa cells expressing TRIM5αrh, TRIM5αhu, or TRIM5αhu mutants or control HeLa cells transduced with the empty pLPCX vector were incubated with various amounts of HIV-1-GFP (B) or SIV-GFP (C). Infected GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in two independent experiments. RT, reverse transcriptase.
The abilities of the mutant TRIM5αhu proteins to restrict SIVmac infection were also studied. As has been previously reported (29), TRIM5αhu exerted little effect on the efficiency of SIV-GFP infection, whereas TRIM5αrh partially inhibited SIV-GFP infection (Fig. 6C). Most of the TRIM5αhu mutants were no better than the wild-type TRIM5αhu in restricting SIVmac infection. Two variants, TRIM5α H(R323-332) and TRIM5α H(R328-332), inhibited SIV-GFP infection to the same degree as TRIM5αrh. Surprisingly, the TRIM5αhu R332P mutant very potently inhibited SIV-GFP infection. This inhibition was specific, as these cells were infectible by MLV-GFP to the same degree as cells transduced with the empty control vector (data not shown). We conclude that a single-amino-acid change in the TRIM5αhu B30.2 v1 region results in a gain in SIVmac inhibitory activity beyond that exhibited by either TRIM5αhu or TRIM5αrh.
DISCUSSION
Here, we show that the B30.2 domain is the major determinant of the differences in anti-HIV-1 potency between human and rhesus monkey TRIM5α. The B30.2 domain of TRIM5αrh has been shown to be essential for the ability to restrict HIV-1 infection (29). The B30.2 domain is a component of a number of proteins, including some other TRIM proteins, butyrophilin-like proteins, and stonustoxin (14). Although B30.2 domain-containing proteins have proliferated in chordate lineages, the functions of these domains are not well understood (14, 26). In a few instances, the B30.2 domain has been implicated in binding to intracellular ligands. For example, the B30.2 domain of butyrophilin has been reported to bind xanthine oxidase (17). The B30.2 domain of TRIM11 has been shown to be important for interaction with humanin (21). Our results are consistent with the B30.2 domain of TRIM5α playing a role in interaction with the viral capsid. Interestingly, in an HIV-1-restricting factor, TRIMCyp, found in owl monkeys, the TRIM5α B30.2 domain is replaced by cyclophilin A, which is known to bind the HIV-1 capsid (21a, 25a).
The amino acid sequences of the TRIM5α proteins of a number of primate and rodent species have been determined (28). Although species-specific variation is observed in all of the TRIM5α domains, the type of variation in the B30.2 domain is noteworthy. Significant length polymorphism, as well as individual amino acid variation, is found in three regions within the B30.2 domains of TRIM5α proteins from different species (28). These variable regions (designated v1, v2, and v3) probably represent surface-exposed loops, as such length variation would not be readily tolerated within the core fold of the protein. The v1 region is particularly long in Old World monkeys, apes, and humans, whereas the v3 region exhibits additional length in New World monkeys (28). Thus, among the Old World primates, it appears that the B30.2 v1 region has evolved special features. Our results demonstrate that the major determinant of the anti-HIV-1 potency of the TRIM5αrh protein resides within the amino-terminal half of the B30.2 v1 segment. The sequence of African green monkey TRIM5α, which also restricts HIV-1 infection (13a, 17a, 28a, 34), is nearly identical to that of TRIM5αrh in this part of the v1 region. By contrast, within a stretch of 10 residues in this region, five amino acid differences between TRIM5αrh and TRIM5αhu exist. Our studies indicate that three differences, in residues 328, 330, and 332, largely explain the different anti-HIV-1 potencies of TRIM5αrh and TRIM5αhu. Particularly important is residue 332, in which a change can increase the potency of HIV-1 restriction and result in a gain in anti-SIVmac activity beyond that of either parental TRIM5α protein. The amino acid composition of the TRIM5α 320 to 333 region is suggestive of that of a surface-exposed loop, consistent with a role in interaction with the HIV-1 capsid and/or TRIM5α cofactors.
Additional studies will be required to address the mechanism of the enhanced antiviral potency of TRIM5αhu variants with alterations in the v1 region of the B30.2 domain. Previous studies (29) and some of the experiments in this study indicate that differences in the expression levels of TRIM5αhu and TRIM5αrh cannot account for the observed differences in anti-HIV-1 potency. Nonetheless, for the panel of TRIM5α variants presented in Fig. 6, a correlation between anti-HIV-1 potency and the steady-state level of expression is apparent. This correlation, however, was not evident with respect to anti-SIVmac activity. Nevertheless, the observed differences in steady-state levels may reflect properties of the TRIM5α variants relevant to antiviral activity, such as subcellular compartmentalization or association with cofactors.
The TRIM5α H(R328-332) and H(R323-332) chimerae created in this study exhibit >98% sequence identity to human TRIM5α yet inhibit HIV-1 with potencies that approach that of the rhesus monkey TRIM5α protein. Expression of these chimeric proteins in human cells would be expected to be tolerated better and to be less immunogenic than the expression of TRIM5αrh. Therefore, the TRIM5α H(R328-332) and H(R323-332) proteins, or similarly designed chimerae, might be useful in gene therapy approaches designed to protect human cells from HIV-1 infection.
Acknowledgments
We thank Yvette McLaughlin and Sheri Farnum for manuscript preparation.
This work was supported by a grant from the National Institutes of Health (HL54785) and by a Center for AIDS Research award (P30 AI28691). We also acknowledge the support of the Bristol-Myers Squibb Foundation, the International AIDS Vaccine Initiative, and the late William F. McCarty Cooper.
REFERENCES
- 1.Arts, E. J., and M. A. Wainberg. 1996. Human immunodeficiency type 1 reverse transcriptase and early events in reverse transcription. Adv. Virus Res. 46:97-163. [DOI] [PubMed] [Google Scholar]
- 2.Barre-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rosenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immunodeficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 5.Clavel, F. 1987. HIV-2, the West African AIDS virus. AIDS 1:135-140. [PubMed] [Google Scholar]
- 6.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, M. D., N. L. Letvin, N. W. King, M. Kannagi, P. K. Sehgal, R. D. Hunt, P. J. Kanki, M. Essex, and R. C. Desrosiers. 1985. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228:1201-1204. [DOI] [PubMed] [Google Scholar]
- 8.Desrosiers, R. C. 1990. The simian immunodeficiency viruses. Annu. Rev. Immunol. 8:557-578. [DOI] [PubMed] [Google Scholar]
- 9.Fauci, A., A. M. Macher, D. L. Longo, H. C. Lane, A. H. Rook, H. Masur, and E. P. Gelman. 1984. NIH Conference. Acquired immunodeficiency syndrome: epidemiologic, clinical, immunologic, and therapeutic considerations. Ann. Intern. Med. 100:91-106. [DOI] [PubMed] [Google Scholar]
- 10.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai, G. White, P. Foster, and P. D. Markham. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref 1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatziioannou, T., S. Cowan, U. K. von Schwedler, W. I. Sundquist, and P. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry, J., I. H. Mather, M. F. McDermott, and P. Pontarotti. 1998. B30.2-like domain proteins: update and new insights into a rapidly expanding family of proteins. Mol. Biol. Evol. 15:1696-1705. [DOI] [PubMed] [Google Scholar]
- 15.Himathongkham, S., and P. A. Luciw. 1996. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219:485-488. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii, T., N. Aoki, A. Noda, T. Adachi, R. Nakamura, and T. Matsuda. 1995. Carboxy-terminal cytoplasmic domain of mouse butyrophilin specifically associates with a 150-kDa protein of mammary epithelial cells and milk fat globule membrane. Biochim. Biophys. Acta 1245:285-292. [DOI] [PubMed] [Google Scholar]
- 17a.Keckesova, Z., L. M. J. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 200:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letvin, N. L., M. D. Daniel, P. K. Sehgal, R. C. Desrosiers, R. D. Hunt, L. M. Waldron, J. J. MacKey, D. K. Schmidt, L. V. Chalifoux, and N. W. King. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71-73. [DOI] [PubMed] [Google Scholar]
- 20.Munk, C., S. M. Brandt, G. Luccero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niikura, T., Y. Hashimoto, H. Tajima, M. Ishizaka, Y. Yamagishi, M. Kawasumi, M. Nawa, K. Terashita, S. Aiso, and I. Nishimoto. 2003. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer's disease-relevant insults. Eur. J. Neurosci. 17:1150-1158. [DOI] [PubMed] [Google Scholar]
- 21a.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to post-entry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rho, H. M., B. Poiesz, W. Ruscetti, and R. C. Gallo. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355-360. [DOI] [PubMed] [Google Scholar]
- 25a.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 26.Seto, M. H., H.-L. C. Liu, D. Zajchowski, and M. Whitlow. 1999. Protein fold analysis of the B30.2-like domain. Proteins 35:235-249. [PubMed] [Google Scholar]
- 27.Shibata, R., H. Sakai, M. Kawamura, K. Tokunaga, and A. Adachi. 1995. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76:2723-2730. [DOI] [PubMed] [Google Scholar]
- 28.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean and J. Sodroski. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 28a.Song, B., H. Javanbakht, M. Perron, D. Park, M. Stremlau and J. Sodroski. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 29.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts human immunodeficiency virus (HIV-1) infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 30.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref 1 retrovirus restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 33.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 34.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5 protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]