Abstract
During spermatogenesis, male germ cells temporally synthesize many proteins as they differentiate through meiosis and become spermatozoa. The germ cell Y-box protein, MSY2, constituting ≈0.7% of total protein in male germ cells, binds to a consensus promoter element, and shows a general lack of RNA-binding specificity. Combining immunoprecipitation and suppressive subtractive hybridization, we identified populations of germ cell mRNAs that are not bound or bound by MSY2. The former population is enriched in cell growth and ubiquitously expressed mRNAs, whereas the latter population is enriched for stored or translationally delayed, male gamete-specific transcripts. Chromatin precipitation assays reveal that most of the MSY2 target mRNAs are transcribed from genes containing the Y-box DNA-binding motif in their promoters. In transgenic mice, mRNAs encoding exogenous GFP are directed or not directed into the MSY2-bound fraction by promoters containing or lacking the Y-box motif, respectively. We propose that MSY2 marks specific mRNAs in the nucleus for cytoplasmic storage, thereby linking transcription and mRNA storage/translational delay in meiotic and postmeiotic male germ cells of the mouse.
Keywords: mRNA storage, spermatogenesis, transcription and translation linkage, Y-box protein
During spermatogenesis, diploid spermatogonia differentiate into meiotic spermatocytes that then transform into haploid spermatids and species-specific shaped spermatozoa. This process requires a precise temporal regulation of gene expression with high levels of protein synthesis because meiotic cells increase many fold in volume at pachytene where DNA recombination and repair occurs. After two meiotic divisions, the haploid spermatid is also highly active in protein synthesis as it reorganizes its nucleus and synthesizes an axoneme and tail for the spermatozoon. Analyses of testes cDNA libraries have detected >18,000 sequence clusters with >2,000 sequence clusters being specifically from germ cells (1).
Y-box proteins, consisting of variable N and C termini and a highly conserved cold shock domain, are a family of evolutionarily conserved DNA- and RNA-binding proteins that function in both transcription and translation (2, 3). Y-box proteins recognize the DNA motif, CTGATTGGC/TC/TAA, sequences present in promoters of many germ cell-expressed genes. Although Y-box proteins are reported to recognize specific sequences in vitro (4, 5), in cells, they appear to repress translation by packaging mRNAs (4). The functions of Y-box proteins may vary, depending on protein:RNA ratios (6). In in vitro assays, high levels of Y-box proteins block translation (7, 8), whereas lower amounts stimulate translation (9).
A germ cell Y-box protein, FRGY2, was first identified in Xenopus laevis oocytes where it couples transcription and translation of maternal mRNAs derived from intron-less genes (2). This discovery has led to the proposal that specific intronless oocyte mRNAs are packaged as FRGY2-RNPs that allows a stable association of FRGY2 and the mRNA in the nucleus and cytoplasm (10). In contrast, mRNAs that require splicing of introns release FRGY2.
In mice and humans, a number of Y-box proteins, MSY1, MSY2a, MSY2b, and MSY4, have been identified in germ cells (11–13). Based on sequence similarities, the mouse protein, MSY2, and the human protein, Contrin (14), are orthologs of FRGY2. MSY2 constitutes ≈2% of total oocyte protein, is present in diplotene-stage oocytes and fully grown oocytes, and is totally degraded by the late two-cell stage, suggesting it stabilizes and/or regulates translation of maternal mRNAs (15). Consistent with this proposal is that reducing MSY2 levels in mouse oocytes results in reduced fertility (16). In the testis, MSY2 is highly abundant in both meiotic and postmeiotic germ cells (12). In in vitro assays, FRGY2 binds to Y-box sequences in the promoter of the mouse protamine (P)2 gene and stimulates transcription (17). The predominantly cytoplasmic location of MSY2 in male germ cells suggests it serves to stabilize and/or regulate the translation of paternal mRNAs.
The abundance of MSY2 in male germ cells and its general lack of RNA-binding specificity raise the question of how many meiotic or postmeiotic male germ cell mRNAs avoid random translational inactivation by MSY2, i.e., is there a mechanism to target MSY2 to a specific population of germ cell mRNAs? We report here that mRNAs transcribed from a Y-box promoter are preferentially bound by MSY2, thereby linking transcription and mRNA storage/translational delay in male germ cells.
Materials and Methods
Polysome Gradient Fractionation and Analysis. Polysomal gradients were prepared as described (18). RNA was purified from 20% of each fraction and protein extracts from the remainder. Northern blotting with 32P-labeled P2 cDNA was used for gradient calibration. Immunoblotting was performed as described (18).
Immunoprecipitation of MSY2-Bound mRNAs. Nonpolysomal fractions (tubes 2–8), containing 6–8 mg of adult CD-1 mouse testis cytoplasmic protein extract, were pooled, diluted 5-fold with TBS-T buffer (20 mM Tris·HCl, pH 7.4/137 mM NaCl/0.1% Tween 20/0.1% Empigen BB/protease inhibitor/RNasin), and incubated with Protein A agarose beads (100 μl of 50% slurry) for 1 h at 4°C. After centrifugation, the supernatants were incubated with 50 μgof anti-MSY2 (15) for 1 h and 200 μl of protein A agarose beads for 4 h at 4°C. After centrifugation (400 × g for 5 min), the supernatants were incubated with 20 μg of anti-MSY2 and 60 μl of Protein A agarose beads for 2 h at 4°C. After centrifugation, the pellets were combined, washed four times with 5 ml of TBS-T buffer, and RNA was purified (18). Polysomal RNA was purified directly from tubes 12–18. Poly(A) mRNAs were prepared by using the Qiagen (Valencia, CA) Oligotex mRNA minikit.
Suppressive Subtractive Hybridization (SSH). One microgram of MSY2 bound and polysomal (hereafter, this fraction will be called nonbound because it lacks MSY2) mRNAs was used for SSH with the Clontech PCR-select cDNA subtraction kit. After two rounds of subtractive hybridization and selective amplification of differentially expressed genes, MSY2-bound and -nonbound subtracted cDNAs were generated by forward and reverse SSH. Two hundred clones of each cDNA library were randomly chosen for PCR amplification.
Differential screening was carried out by using the Clontech PCR-Select differential screening kit. PCR products were hybridized with radiolabeled probes generated from the MSY2-bound and -nonbound subtracted cDNAs. The relative differential expression of each clone was quantified by densitometry and expressed as the ratio of the signal of the homologous to the heterologous subtracted probes. Clones with a differential expression ratio of >5 were sequenced and subjected to a blast search.
Quantitative RT-PCR Analysis of Selected Genes. The specificity of MSY2-bound and -nonbound mRNAs identified above was confirmed by real-time RT-PCR. All primers (see Table 3, which is published as supporting information on the PNAS web site) were checked by PCR to ensure they generated single bands of the predicted size. PCR was performed by using the SYBR green PCR master mix and the ABI 7900 HT thermal cycler at typical amplification parameters (at 50°C for 2 min and at 95°C for 10 min, followed by 40 cycles of 95°C, for 15 s and at 60°C for 1 min) and differences were displayed as the cycle of threshold value for each gene.
Chromatin Immunoprecipitation (ChIP). The ChIP assay was carried out by using a ChIP kit (Upstate Biotechnology, Lake Placid, NY) following the manufacturer's protocol. Testes were incubated with collagenase to separate seminiferous tubules (19). After washing twice, formaldehyde (1%) was added, and the preparation was incubated at 37°C for 20 min. After adding 0.125 M glycine to stop the cross-linking, male germ cells were dissociated, centrifuged, and rinsed in cold PBS containing protease inhibitor mixture (Roche Molecular Biochemicals). The cells were then incubated at 4°C for 10 min in swelling buffer (10 mM potassium acetate/15 mM magnesium acetate/0.1 M Tris·HCl, pH 7.6/protease inhibitor), and disrupted in a Dounce homogenizer. Germ cell nuclei were resuspended in sonication buffer (1% SDS/10 mM EDTA/50 mM Tris·HCl, pH 8.0) and incubated on ice for 10 min. The samples were sonicated on ice to generate DNA fragments of 200–800 bp. After centrifugation, the chromatin solution was incubated with salmon sperm DNA/Protein A agarose beads for 1 h at 4°C, and anti-MSY2 (2 μg) and ssDNA/agarose beads were added to precipitate the immune complexes. After reversing the crosslinking by incubating at 65°C for 4 h in a final concentration of 0.2 M NaCl, the eluted immune complexes were extracted with phenolchloroform, and the purified DNA was analyzed by quantitative real-time RT-PCR. The time and conditions of cross-linking are important as variable results were obtained when cells were dissociated before cross-linking, perhaps because of changes in their physiological states.
To analyze each promoter sequence, genome browser gateway software was used (which can be accessed at http://genome.ucsc.edu). Primers were designed to investigate from -800 bp to their transcription start site (see Table 4, which is published as supporting information on the PNAS web site), and sequences around the Y-box sequence were preferentially chosen. Equal amounts of input DNA were added before immunoprecipitation and amplified with each sample as controls for differences in amplification efficiencies and DNA quantities. Fold differences were determined by comparing the ΔΔ cycle of the threshold of target genes from the DNA immunoprecipitated with anti-MSY2 to controls where no antibody was added after normalization for amplification efficiency (determined from the input DNA amplification).
Analysis of mRNA Distributions in Polysomal Gradients from Transgenic Mice. Adult testes extracts were fractionated and immunoprecipitated as above. Poly(A) mRNAs from the MSY2-bound fraction and polysomes were isolated, and, after addition of rabbit α-globin mRNA (1 pg) for normalization, aliquots (1 μg) were reverse-transcribed and quantified by real-time RT-PCR.
Results
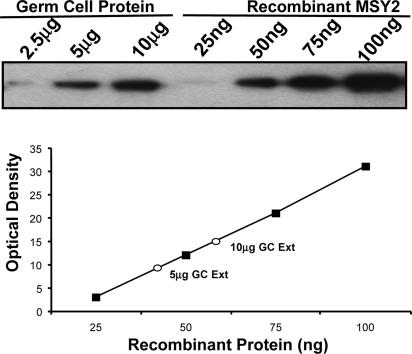
MSY2 Is Highly Abundant in Male Germ Cells and Present in Nonpolysomal Ribonucleoproteins (RNPs). To quantify the amount of MSY2 in testis, an enriched population of dissociated spermatogenic cells was extracted with RIPA buffer (0.15 mM NaCl/0.05 mM Tris· HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). By using recombinant MSY2 to create a calibration curve, ≈0.7% of total soluble protein was MSY2 (Fig. 1). Because MSY2 is solely expressed in the meiotic and postmeiotic male germ cells (20), this result represents a minimal estimate of its cellular concentration. Considering its abundance and sequence-independent binding properties, the question is raised as to how mRNAs selectively escape MSY2 inactivation.
Fig. 1.
MSY2 is abundant in male germ cells. MSY2 was quantified in an extract from dissociated spermatogenic cells by using an affinity-purified antibody to MSY2. Enriched populations of dissociated germ cells were homogenized in RIPA buffer that solubilizes >85% of total MSY2. Recombinant MSY2 was used as a control calibration protein.
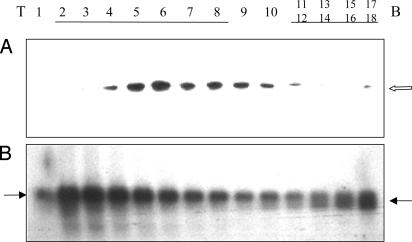
Immunoblotting of a polysomal testis gradient revealed MSY2 predominantly sediments in the nonpolysomal fraction (tubes 3–9) (Fig. 2A) as seen in Nycodenz gradients (21). The gradient was calibrated by hybridization with a P2 cDNA because the longer form of P2 mRNA is nonpolysomal (left arrow) whereas the shorter form is polysomal (right arrow) (18) (Fig. 2B).
Fig. 2.
Sucrose gradient fractionation of testis RNP particles. The gradients were fractionated into 18 tubes and divided for protein and RNA analysis (T, top; B, bottom). (A) Immunoblot of MSY2. The open arrow indicates the position of MSY2. (B) Northern blot of P2 mRNA. Gradients were centrifuged for 3 h at 100,000 × g rpm to maximally sediment the nonpolysomal RNPs. As a result, most of the polysomes are near the bottom (tubes 12–18). A 60% sucrose cushion prevented their pelleting.
Identifying Populations of mRNAs Bound or Not Bound to MSY2. Although MSY2 is an abundant RNA-binding protein in male germ cells, little is known of the populations of mRNAs it binds. To determine whether immunoprecipitation could fractionate mRNAs into MSY2-bound and -nonbound populations, real-time RT-PCR was used to compare twice immunoprecipitated nonpolysomal mRNAs to polysomal mRNAs. For a successful fractionation, we would expect that known targets of MSY2 such as the protamines (5) would be enriched in the bound fraction, whereas mRNAs expressed in somatic cells of the testis should not be bound. The P1 and P2 mRNAs were 10-fold-enriched in the MSY2-bound pellet, whereas the Sertoli cell-expressed clusterin mRNA and the somatic and early germ cell-expressed Gapdh mRNA were 10-fold-enriched in the polysomal fractions. Because MSY2 is expressed only in germ cells, the lack of binding to clusterin mRNA indicated that MSY2 was not binding to mRNAs adventitiously in extracts. These experiments demonstrated the feasibility to use immunoprecipitation to define mRNA populations in the MSY2-bound nonpolysomal and polysomal fractions.
To identify the in vivo targets of MSY2, we combined immunoprecipitation and SSH. cDNA inserts from randomly chosen clones of the MSY2-bound subtracted cDNA library (MSY2-bound) and the polysomal mRNA cDNA library (MSY2-nonbound) were amplified for differential screening confirmation (see Table 5, which is published as supporting information on the PNAS web site) and sequenced. Clones of genes expressed in cell types lacking MSY2, such as clusterin from Sertoli cells, were excluded from analysis.
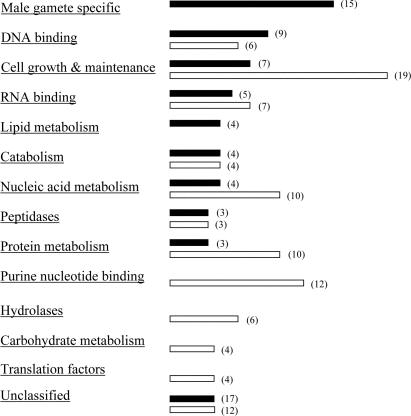
Forty-eight clones encoding MSY2-bound mRNAs and 50 clones of MSY2-nonbound mRNAs were identified (Table 1). Although we do not know which cell types express and whether they are posttranscriptionally regulated for all of the 98 clones, many germ cell-specific mRNAs known to undergo storage and translational delays, such as P1 and 2 (22), transition proteins (TPs) 1 and 2 (23), A kinase anchor protein 4 (24), and outer dense fiber 2 (25), were detected in the MSY2-bound group (Table 1). In contrast, many mRNAs in the MSY2-nonbound group, e.g., osmotic stress protein, cyclin A2 (26), calmodulin 2 (27), lactate dehydrogenase C4 (28), and tubulins are concomitantly transcribed and translated. Annotation analysis was performed by using the david database (which can be accessed at http://apps1.niaid.nih.gov/david) followed by gocharts, allowing classification into subsets according to biological processes and molecular function. Groups with three or more mRNAs were sorted and summarized (Fig. 3). Notably, 15 male gamete-specific mRNAs were detected in the MSY2-bound mRNAs, supporting the hypothesis that stored germ cell-specific mRNAs are MSY2-bound (Table 1 in bold). All of the male gamete mRNAs examined (eight) by using real-time RT-PCR were confirmed to be enriched in MSY2 RNPs (see Supporting Text, which is published as supporting information on the PNAS web site). The nonbound group was enriched for mRNAs that are not stored and often encoded constitutively expressed proteins involved in cell growth and maintenance (Table 1 in bold), purine nucleotide binding, nucleic acid, protein, and carbohydrate metabolism, and protein translation (Table 1).
Table 1. MSY2-bound/nonbound mRNAs and promoter analysis of genes identified from immunoprecipitation and suppressive subtractive hybridization.
| GenBank accession no. | MSY2-bound mRNAs | Y box | GenBank accession no. | MSY2-nonbound mRNAs | Y box |
|---|---|---|---|---|---|
| X14003 | Protamine 1 | + | NM_009828 | Cyclin A2 | + |
| NM_008933 | Protamine 2 | + | BC005778 | Proliferating cell nuclear antigen protein | + |
| NM_013694 | TP2 | + | NM_009446 | α-Tubulin 3 | + |
| AF087517 | A kinase anchor protein 4 (AKAP4) | + | AK011136 | ATP-dependent RNA helicase DDX56 | + |
| U31992 | Acrosomal protein SP-10 | + | Y00094 | Ras-related YPT1 protein | + |
| D00754 | Acrosin | + | AK033235 | ATP dependent RNA helicase DDX 19 | + |
| AF073954 | Y-box protein MSY2 | + | NM_008228 | Histone deacetylase 1 | + |
| NM_013615 | Outer dense fiber 2 (Odf2) | + | AB023062 | Actin-like 7b | + |
| BC008206 | GST, M5 | + | BC051444 | Calmodulin 2 | – |
| NM_009350 | Testis nuclear RNA binding protein (Tnbr) | + | BC002227 | ARP10 actin-related protein 10 | – |
| AB120716 | Spergen-1 | + | AF356520 | Axonemal dynein heavy chain 8 long form | – |
| NM_009407 | TP1 | – | NM_007628 | Cyclin A1 | – |
| AF246224 | Y-box protein MSY4 | – | BC050769 | α-Tubulin 7 | – |
| BC048680 | AKAP-like sperm protein | – | BC012401 | Translocating membrane protein 1 | – |
| AB116526 | Spetex-1 | – | NM_009211 | Actin regulator of chromatin Smarcc1 | – |
| NM_007808 | Cytochrome c, somatic | + | NM_011568 | RNA and export factor-binding protein 1 | – |
| M62867 | Y-box protein MSY1 | + | NM_011401 | Solute carrier family 2 member 3 | – |
| NM_198623 | Ubiquilin 3 | + | NM_138669 | Dead(Asp-Glu-Ala-Asp) box peptide 48 | – |
| NM_026449 | N-acetyl galactosaminyl transferase | + | AF294327 | Ran-binding protein/karyopherin beta3 | – |
| AK029613 | Peroxiredoxin 5 | + | BC027629 | NAD(P)H dehydrogenase, quinone 2 | + |
| NM_009611 | Actin-like 7a | + | BC049803 | Rho-interacting protein 3 | + |
| BC054413 | Ubiquitin A-52 ribosomal protein fusion form1 | + | NM_009840 | Chaperonin subunit 8 (θ) | + |
| BC043124 | Dipeptidylpeptidase 8 | + | BC050927 | Heat-shock 70-kDa protein 5 | + |
| BC052770 | Carnitine deficiency associated gene | + | BC016619 | Pyruvate kinase, muscle | + |
| J03750 | P9 DNA-binding protein | + | BC013509 | Succinate DH complex subunit B | + |
| NM_011462 | Spindlin | + | BC054386 | Aldehyde dehydrogenase family 1 | + |
| AF294328 | Ankyrin-like protein | + | BC025481 | Ribosomal protein F | + |
| AK005361 | Membrane-interacting protein RSG16 | + | BC009655 | Ribosomal protein L3 | + |
| AK015685 | DNA J-like protein | + | NM_027444 | HMG-box transcription factor BBX | + |
| BC043023 | Bernardinelli–Seip congenital lipodystrophy 2 | + | AK077878 | HepA-related protein | + |
| NM_019464 | SH3 domain GRB2-like B1 | + | AK015901 | Tubulin β-2 chain homolog | + |
| NM_029782 | Similar to calreticulin 3 | + | NM_008187 | Gene trap locus 3 | + |
| AK075758 | Similar to CLIP-170-related protein | + | NM_054004 | TBP-interacting protein 120A | + |
| AK077071 | Serine protease | + | BC016080 | Ribophorin 1 | + |
| AK038289 | GRAM domain-containing protein | + | BC050797 | Similar to T-complex protein 1 | + |
| AK007091 | Elongation factor-like protein | + | NM_011020 | Osmotic stress protein | – |
| BC034193 | Oncoprotein-induced transcript 1 | + | X04752 | Lactate dehydrogenase-C | – |
| NM_025356 | Ubiquitin-conjugating enzyme E2D3 | + | BC007152 | Translation elongation factor 2 | – |
| AK005938 | Similar to PNG protein | + | AK076067 | Translation initiation factor 4E-like protein | – |
| BC049963 | UBX domain-containing protein 2 | + | BC024899 | Translation initiation factor 5A | – |
| NM_199019 | Similar to SPPL2b; presenilin-like protein 1 | + | BC002072 | Cystatin C | – |
| NM_010191 | Farnesyl diphosphate farnesyl transferase 1 | + | AK013955 | Mitochondrial processing peptidase β | – |
| AK019488 | α/β hydrolase structure protein | + | BC052093 | Disintegrin and metalloproteinase domain 1a | – |
| AF234179 | Testis–brain RNA-binding protein (TB-RBP) | – | BC046233 | poly(A)-binding protein, cytoplasmic 1 | – |
| BC060971 | Synaptophysin-like protein, variant 1 | – | NM_021713 | Melanocyte proliferating gene | – |
| AK014926 | Hypothetical actin and actin-like protein | – | BC004651 | GM2 ganglioside activator protein | – |
| BC033444 | 1-Acylglycerol-3-phosphate O-acyltransferase 3 | – | BC019578 | Peroxiredoxin 4 | – |
| XM_356583 | Similar to solute carrier family 26 | * | AK077968 | Supernatant malic enzyme | – |
| XM_135742 | Similar to integral membrane transporter | – | |||
| D84391 | L1-repetitive element | – |
The genes in bold in the MSY2-bound and -nonbound groups represent male gamete-specific genes and cell growth- and maintenance-related genes, respectively. Sequences and sites of the Y boxes are presented in Supporting Text.
No promoter was found for the similar to solute carrier family 26 gene
Fig. 3.
Clustering of mRNAs bound and nonbound to MSY2 identified with SSH. The numbers in parentheses represent the number of mRNAs identified in the category. Proteins with multiple functions or involved in several biological processes were counted in multiple annotation categories. Black bars, MSY2-bound mRNAs; white bars, MSY2-nonbound mRNAs.
To confirm the differential expression we detected by SSH, quantitative real-time RT-PCR was performed. Although we did not assay all 98 of the mRNAs identified, of 16 mRNAs assayed, all (11 MSY2-bound mRNAs and 5 -nonbound mRNAs) were enriched in the nonpolysomal and polysomal fractions, respectively (see Fig. 5, which is published as supporting information on the PNAS web site). Analyzing the MSY2-bound mRNAs, we detected variations in enrichment with mRNAs encoding proteins such as P1, P2, A kinase anchor protein 4, SP-10, and testis–brain RNA-binding protein showing greater than a four-cycle enrichment, whereas MSY2, MSY4, and TP2 showed a two-cycle enrichment (see Fig. 5). Nonbound mRNAs, such as the osmotic stress protein, cyclin A1, calmodulin 2, and lactate dehydrogenase C, showed a four-cycle enrichment in the MSY2-nonbound fraction. Based on these distributions, we believe the SSH cDNA libraries are distinguishing between populations of MSY2-bound and -nonbound mRNAs.
Y-Box Sequences Are Present in the Promoter Regions of Genes Whose mRNAs are MSY2-Bound. The distinct populations of MSY2-bound and -nonbound mRNAs in male germ cells raises the question as to why specific mRNAs are recognized by MSY2. The answer we believe lies, in part, in the multifunctionality of MSY2. MSY2 is both a DNA-binding protein recognizing the consensus sequence (CTGATTGGC/TC/TAA) and a mostly sequence-independent RNA-binding protein. By using genomatix software, we analyzed the Y-box sequences in 800 bp of the promoters of the genes we identified. Analysis of 97 genes revealed a greater frequency of Y-box sequences in the promoter regions of the MSY2-bound mRNAs (82.9%) than in promoter regions of -nonbound mRNAs (48.9%) (see Table 1 and Supporting Text). When we compare the male gamete-specific mRNAs to the cell and growth and maintenance mRNAs (Fig. 3), Y-box sequences are present in the promoters of 12 of 15 (80%) MSY2-bound mRNAs versus 8 of 19 (42%) of the MSY2-nonbound mRNAs. Considering that we do not know which Y-box sequences are functional in cells and many of the Y-box containing genes may not be expressed in germ cells, these numbers can only be interpreted as representing a trend to be experimentally tested with additional assays (see below).
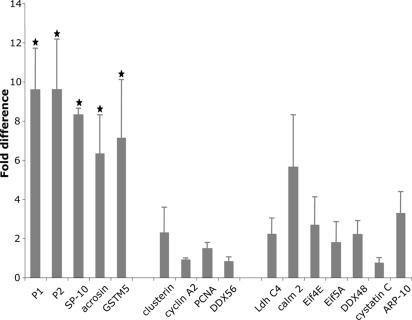
ChIP Reveals MSY2 Binds to Promoters Containing a Y-Box Sequence. To establish which Y-boxes are bound by MSY2 in vivo, we used ChIP to assay MSY2 binding to a representative subset of promoters from Table 1 that contain or lack the consensus Y-box motif. As positive controls, we know that there are two Y-box sequences in the -200-bp region of the P2 promoter that are functional in in vitro assays (17) and P1 and P2 mRNAs are bound by MSY2 (5). The promoter fragments for P1 and P2 and three other genes whose mRNAs are bound by MSY2 (SP-10, acrosin, and GSTM5) were significantly enriched after precipitation with anti-MSY2 (Fig. 4). In contrast, clusterin, cyclin A2, PCNA, and DDX56, genes whose promoters contain Y-boxes but whose mRNAs are not bound by MSY2, were not enriched in the ChIP-MSY2 precipitate. Other genes whose promoters lack Y boxes including lactate dehydrogenase C, calmodulin 2, Eif4E, Eif5A, DDX48, cystatin C, and actin-related protein-10 were also not selectively precipitated. Of particular note, although Sertoli cells do not express MSY2, the promoter for clusterin, a Sertoli cell protein that contains a Y-box sequence (at -206 to -192), was not preferentially precipitated by MSY2. As a negative control, the precipitated promoters were not immunoprecipitated with anti-HSF-3 β antibodies (data not shown). Thus, by analyzing a subset of the genes from Table 1, we find a statistically significant in vivo association between MSY2 and promoter regions of genes whose mRNAs are bound by MSY2, which is consistent with MSY2 linkage of transcription and mRNA storage in vivo.
Fig. 4.
ChIP products were analyzed by using quantitative real-time RT-PCR. Twenty-one-day-old CD1 mice were used to maximize ease of chromatin sonication after DNA-protein crosslinking. Identical results were obtained with sexually mature CD-1 mice. The promoter sequences from 16 genes expressed in testis were analyzed. The data from three independent assays represent the average of six determinations ± SEM and are presented as the fold difference of target genes from the DNA immunoprecipitated with anti-MSY2 compared with controls where no antibody was added (the control level for each promoter was set as a value of 1) after normalization with input DNA amplification. Assigning each sample a rank based on fold enrichment, differences between ranks were analyzed by using the Kruskal–Wallis test. The five genes marked with stars were significantly different from the others (P < 0.001).
GFP mRNA Partitions in the MSY2-Bound Fraction if It Is Transcribed from a Y-Box Promoter. To test directly the hypothesis that a Y box could direct an exogenous mRNA into the MSY2-bound population of mRNAs in a functional assay, we assayed three different lines of transgenic mice expressing GFP. Because the promoter of acrosin contains a Y box sequence at -69 to -58 bp and acrosin mRNA is bound by MSY2 (Table 1 and Supporting Text), we analyzed transgenic mice expressing GFP from a 2.4-kb acrosin promoter (29). Like P2 and acrosin mRNAs (data not shown), GFP mRNA was enriched in the MSY2-bound fraction (Table 2). Control mRNAs encoding clusterin and Gapdh were enriched in the nonbound fractions.
Table 2. Real-time RT-PCR analysis of GFP mRNA distribution in transgenic mice.
| mRNA | MSY2-bound | MSY2-nonbound |
|---|---|---|
| Acrosin-GFP-transgenic mice | ||
| Protamine 2 | 1 | 0.13 ± 0.01 |
| Clusterin | 1 | 525 ± 77.78 |
| GAPDH | 1 | 8.2 ± 4.2 |
| GFP | 1 | 0.5 ± 0.28 |
| SP-10-GFP-transgenic mice (–266/+28 promoter) | ||
| Protamine 2 | 1 | 0.14 ± 0.01 |
| Clusterin | 1 | 189 ± 74.95 |
| GAPDH | 1 | 32.45 ± 10.67 |
| GFP | 1 | 1.85 ± 0.54 |
| SP-10-GFP-transgenic mice (–408/+28 promoter) | ||
| Protamine 2 | 1 | 0.16 ± 0.01 |
| Clusterin | 1 | 222 ± 38.89 |
| GAPDH | 1 | 17.7 ± 11.75 |
| GFP | 1 | 0.59 ± 0.03 |
Quantification was performed with the ΔΔ cycle of threshold method normalizing the amount of each mRNA to the MSY2-bound fraction. The bold values indicate the GFP-enriched fractions.
To evaluate more critically the relationship between Y boxes and mRNA storage, two lines of transgenic mice expressing GFP from the same germ cell-specific SP-10 promoter (-408/+28 and -266/+28) were examined (30, 31). The SP-10 promoter has a Y-box consensus element at -338 to -327, allowing us to examine the distribution of GFP mRNA from the same promoter with or without a Y box. GFP mRNA was enriched in the nonbound fraction when driven by the -266/+28 promoter, whereas the -408/+28 shifted the GFP mRNA into the nonpolysomal fraction (Table 2). This finding suggests that the subcellular translational fate of the exogenous GFP mRNA is determined by the presence or absence of a Y box in its promoter.
Discussion
Messenger RNA storage plays an especially important role in regulating maternal mRNAs in oocytes. This process is also essential in meiotic and postmeiotic spermatogenic cells, because transcription terminates in the early stages of haploid germ cell differentiation. MSY2 is one of the most abundant DNA/RNA-binding proteins, constituting ≈2% and 0.7% of total protein in oocytes (15) and spermatogenic cells, respectively (Fig. 1). The Xenopus ortholog of MSY2, FRGY2a/b, functions in large RNP complexes to stabilize, store, and suppress mRNA translation (2, 3). In mouse testis, it is estimated that up to 75% of polyadenylated RNA is complexed with MSY4 and MSY2 (11).
Here, we have combined immunoprecipitation with SSH to obtain populations of mRNAs bound or not bound to MSY2. The association of MSY2 with specific mRNAs is complex, and for MSY2-bound mRNAs highly transient, because all are presumably translated in later-stage cells. To minimize the complexity of changing associations of MSY2 binding/nonbinding as cells differentiate, we compared the MSY2-bound mRNAs immunoprecipitated from a nonpolysomal fraction to the mRNAs in polysomes, a subcellular fraction with little, if any, MSY2. This comparison provided an initial approach to the identification of MSY2 bound mRNA and comparison with a steady-state population of unbound mRNAs. Future studies will be needed examine the populations of bound and nonbound mRNAs from individual populations of germ cells as well as compare the bound/unbound mRNAs in RNPs.
Despite the limitations of using total testes extracts and designating the polysomes as the unbound fraction, we identified a sizable number of MSY2-bound marker mRNAs by using two complementary but different techniques. We obtained similar distributions of MSY2-bound and -nonbound mRNAs by using SSH with testis extracts or by immunoprecipitation of extracts from transgenic mice expressing GFP (Fig. 4). In both cases, many of the mRNAs bound to MSY2 were gamete-specific meiotic and postmeiotic transcripts that are known to be stored and critical for germ cell development. In contrast, many genes involved in cell growth and general metabolism whose mRNAs are immediately translated were not bound by MSY2. Interestingly, MSY2 protein binds to its own mRNA, suggesting its autoregulation as seen for other RNA-binding proteins such as α-CP2 GFP mRNA (32) and PABP (33). Immunoprecipitation and cDNA microarrays have been used to define specific populations of mRNAs complexed with the RNA-binding proteins, HuB (34) and α-CP2 (32). Relating clusters of structurally or functionally related mRNAs that are bound by MSY2 to clusters recognized by other RNA-binding proteins may provide insight into a new level, coordinating gene expression regulation in mammalian cells.
In Xenopus egg extracts, FRGY2 facilitates in vitro transcription from oocyte-specific promoters containing Y-box sequences (17). Transfection of FRGY2 into somatic cells activates transcription from the Xenopus heat shock protein 70-kDa promoter and herpes simplex virus tk promoter, creating mRNAs masked from translation (35). Specific protein–RNA interactions and the splicing process within the nucleus influence the fate of mRNAs in the cytoplasm (10, 36). Overexpression of FRGY2 protein in Xenopus oocytes selectively represses translation of mRNAs transcribed in vivo, but does not affect translation of mRNAs microinjected into the oocyte cytoplasm (37). Many endogenous mRNAs that show translational repression in Xenopus oocytes are synthesized from intronless genes (36). The relief of this translational repression by injection of anti-FRGY2 into nuclei suggests mRNAs undergoing splicing evade this translational repression.
We find that in vivo the mammalian germ cell Y-box protein, MSY2, also links transcription and mRNA storage/stabilization in male germ cells. Many of the genes whose mRNAs are bound to MSY2 contain Y-box DNA sequences in their promoters. The binding of MSY2 to DNA appears selective because ChIP assays demonstrate that transcripts from genes that contain Y boxes in their promoters (P1 and P2, SP-10, acrosin, and GSTM5) are in MSY2 complexes, whereas promoters from genes such as lactate dehydrogenase, calmodulin 2, and cystatin C that lack these sequences are not precipitated and their mRNAs are not bound. Moreover, although the promoters of genes such as clusterin, PCNA, and cyclin A2 contain Y-box sequences, they are not precipitated and their mRNAs are not bound by MSY2. Real-time RT-PCR assays of total testis RNA from wild-type and germ cell-less (c-kit) mice indicates that some of the genes we identified as containing a Y-box such as aldehyde dehydrogenase (family 1) are enriched in somatic cells of the testis (data not shown), which is consistent with their fractionation into the MSY2-nonbound fraction (Table 1).
Our transgenic mice studies that specifically test whether the fate of a marker mRNA is determined by the presence/absence of a Y-box sequence were consistent with a direct MSY2-mediated linkage of transcription and mRNA inactivation in mouse male germ cells. Clearly, this is only one level of many regulatory mechanisms controlling gene expression in the testis because many nonbound mRNAs contain Y boxes in their promoters and a few translationally delayed mRNAs such as TP1 lack the sequences (at least in the first 800 bp of their promoters). Although MSY2 is predominately cytoplasmic in germ cells, low levels are detected in male germ cell nuclei and FRGY2 stimulates transcription from the mouse protamine promoter in vitro (17). Moreover, the association of the somatic Y-box protein, YB-1, with Balbiani ring pre-mRNAs in the nucleus is maintained in the cytoplasm (38). The selective marking by MSY2 of mRNAs transcribed from promoters containing Y-box sequences helps explain how a population of mRNAs is preferentially bound or escapes complexing with MSY2 and provides a mechanism whereby an RNA-binding protein with apparently low binding specificity gains specificity.
Although we demonstrate that a Y-box sequence in the promoter of genes expressed in male germ cells can direct their transcripts into MSY2 complexes, this is likely to be only one of many ways that MSY2 regulates mRNA translation. In the cytoplasm, Y-box proteins form large RNP particles that allow long-term mRNA storage and could restrict the recruitment of mRNA to the translational machinery. Although MSY2 generally binds RNA in a sequence-independent manner in both male and female germ cells (21, 39), certain RNA sequences such as UCCAUCA can be preferentially recognized by MSY2 (5) and Selex assays indicate preferential binding to the sequence AACAUC (4). We, however, have not detected a differential enrichment of these sequences or any specific secondary structure in our populations of MSY2-bound or -nonbound mRNAs. Because MSY2 appears capable of binding most, if not all, mRNAs, perturbations in the cellular levels of MSY2 will likely prove useful in investigating how it fine tunes its selective binding to mRNA. Reduction of MSY2 levels in mouse oocytes reduces fertility (16), and overexpression in mouse testis of another Y-box protein, MSY4, severely disrupts spermatogenesis (40). Preliminary studies suggest that a targeted deletion of the MSY2 gene in mice causes infertility in both males and females, providing an additional model to define the mechanism linking this highly abundant DNA/RNA-binding protein and selective mRNA expression in germ cells.
Supplementary Material
Acknowledgments
We thank G. Gerton (University of Pennsylvania, Philadelphia) for generously providing the acrosin-GFP-transgenic mice and C. Williams for assistance with statistical analysis. This work was supported by National Institutes of Health Grant HD44449 (to R.M.S. and N.B.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SSH, suppressive subtractive hybridization; ChIP, chromatin immunoprecipitation; Pn, protamine n; TPn, transition protein n; RNP, ribonucleoprotein.
References
- 1.Eddy, E. M. (2002) Recent Prog. Horm. Res. 57, 103-128. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto, K. & Wolffe, A. P. (1998) Trends Cell Biol. 8, 318-323. [DOI] [PubMed] [Google Scholar]
- 3.Sommerville, J. (1999) BioEssays 21, 319-325. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet, P., Matsumoto, K. & Wolffe, A. P. (1995) J. Biol. Chem. 270, 28297-28303. [DOI] [PubMed] [Google Scholar]
- 5.Giorgini, F., Davies, H. G. & Braun, R. E. (2001) Mol. Cell. Biol. 21, 7010-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno, K., Izumi, H., Uchiumi, T., Ashizuka, M. & Kuwano, M. (2003) BioEssays 25, 691-698. [DOI] [PubMed] [Google Scholar]
- 7.Davydova, E. K., Evdokimova, V. M., Ovchinnikov, L. P. & Hershey, J. W. (1997) Nucleic Acids Res. 25, 2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minich, W. B., Maidebura, I. P. & Ovchinnikov, L. P. (1993) Eur. J. Biochem. 212, 633-638. [DOI] [PubMed] [Google Scholar]
- 9.Minich, W. B. & Ovchinnikov, L. P. (1992) Biochimie 74, 477-483. [DOI] [PubMed] [Google Scholar]
- 10.Braddock, M., Muckenthaler, M., White, M. R., Thorburn, A. M., Sommerville, J., Kingsman, A. J. & Kingsman, S. M. (1994) Nucleic Acids Res. 22, 5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, H. G., Giorgini, F., Fajardo, M. A. & Braun, R. E. (2000) Dev. Biol. 221, 87-100. [DOI] [PubMed] [Google Scholar]
- 12.Gu, W., Tekur, S., Reinbold, R., Eppig, J. J., Choi, Y. C., Zheng, J. Z., Murray, M. T. & Hecht, N. B. (1998) Biol. Reprod. 59, 1266-1274. [DOI] [PubMed] [Google Scholar]
- 13.Tafuri, S. R., Familari, M. & Wolffe, A. P. (1993) J. Biol. Chem. 268, 12213-12220. [PubMed] [Google Scholar]
- 14.Tekur, S., Pawlak, A., Guellaen, G. & Hecht, N. B. (1999) J. Androl. 20, 135-144. [PubMed] [Google Scholar]
- 15.Yu, J., Hecht, N. B. & Schultz, R. M. (2001) Biol. Reprod. 65, 1260-1270. [DOI] [PubMed] [Google Scholar]
- 16.Yu, J., Deng, M., Medvedev, S., Yang, J., Hecht, N. B. & Schultz, R. M. (2004) Dev. Biol. 268, 195-206. [DOI] [PubMed] [Google Scholar]
- 17.Yiu, G. K. & Hecht, N. B. (1997) J. Biol. Chem. 272, 26926-26933. [DOI] [PubMed] [Google Scholar]
- 18.Yang, J., Chennathukuzhi, V., Miki, K., O'Brien, D. A. & Hecht, N. B. (2003) Biol. Reprod. 68, 853-859. [DOI] [PubMed] [Google Scholar]
- 19.Wu, X. Q., Lefrancois, S., Morales, C. R. & Hecht, N. B. (1999) Biochemistry 38, 11261-11270. [DOI] [PubMed] [Google Scholar]
- 20.Oko, R., Korley, R., Murray, M. T., Hecht, N. B. & Hermo, L. (1996) Mol. Reprod. Dev. 44, 1-13. [DOI] [PubMed] [Google Scholar]
- 21.Herbert, T. P. & Hecht, N. B. (1999) Nucleic Acids Res. 27, 1747-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleene, K. C., Distel, R. J. & Hecht, N. B. (1984) Dev. Biol. 105, 71-79. [DOI] [PubMed] [Google Scholar]
- 23.Heidaran, M. A. & Kistler, W. S. (1987) J. Biol. Chem. 262, 13309-13315. [PubMed] [Google Scholar]
- 24.Morales, C. R., Lefrancois, S., Chennathukuzhi, V., El-Alfy, M., Wu, X., Yang, J., Gerton, G. L. & Hecht, N. B. (2002) Dev. Biol. 246, 480-494. [DOI] [PubMed] [Google Scholar]
- 25.Morales, C. R., Oko, R. & Clermont, Y. (1994) Mol. Reprod. Dev. 37, 229-240. [DOI] [PubMed] [Google Scholar]
- 26.Ravnik, S. E. & Wolgemuth, D. J. (1996) Dev. Biol. 173, 69-78. [DOI] [PubMed] [Google Scholar]
- 27.Cruzalegui, F. H., Kapiloff, M. S., Morfin, J. P., Kemp, B. E., Rosenfeld, M. G. & Means, A. R. (1992) Proc. Natl. Acad. Sci. USA 89, 12127-12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroft, T. L., Li, S., Doglio, L. & Goldberg, E. (2003) J. Androl. 24, 843-852. [DOI] [PubMed] [Google Scholar]
- 29.Nakanishi, T., Ikawa, M., Yamada, S., Parvinen, M., Baba, T., Nishimune, Y. & Okabe, M. (1999) FEBS Lett. 449, 277-283. [DOI] [PubMed] [Google Scholar]
- 30.Reddi, P. P., Flickinger, C. J. & Herr, J. C. (1999) Biol. Reprod. 61, 1256-1266. [DOI] [PubMed] [Google Scholar]
- 31.Reddi, P. P., Shore, A. N., Acharya, K. K. & Herr, J. C. (2002) J. Reprod. Immunol. 53, 25-36. [DOI] [PubMed] [Google Scholar]
- 32.Waggoner, S. A. & Liebhaber, S. A. (2003) Mol. Cell. Biol. 23, 7055-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornstein, E., Harel, H., Levy, G. & Meyuhas, O. (1999) FEBS Lett. 457, 209-213. [DOI] [PubMed] [Google Scholar]
- 34.Tenenbaum, S. A., Carson, C. C., Lager, P. J. & Keene, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranjan, M., Tafuri, S. R. & Wolffe, A. P. (1993) Genes Dev. 7, 1725-1736. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto, K., Wassarman, K. M. & Wolffe, A. P. (1998) EMBO J. 17, 2107-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouvet, P. & Wolffe, A. P. (1994) Cell 77, 931-941. [DOI] [PubMed] [Google Scholar]
- 38.Soop, T., Nashchekin, D., Zhao, J., Sun, X., Alzhanova-Ericsson, A. T., Bjorkroth, B., Ovchinnikov, L. & Daneholt, B. (2003) J. Cell Sci. 116, 1493-1503. [DOI] [PubMed] [Google Scholar]
- 39.Yu, J., Hecht, N. B. & Schultz, R. M. (2002) Biol. Reprod. 67, 1093-1098. [DOI] [PubMed] [Google Scholar]
- 40.Giorgini, F., Davies, H. G. & Braun, R. E. (2002) Development (Cambridge, U.K.) 129, 3669-3679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.