Abstract
Fanconi anemia (FA) is a recessive disorder characterized by congenital abnormalities, progressive bone-marrow failure, and cancer susceptibility. Cells from FA patients are hypersensitive to agents that produce DNA crosslinks and, after treatment with these agents, have pronounced chromosome breakage and other cytogenetic abnormalities. Eight FANC genes have been cloned, and the encoded proteins interact in a common cellular pathway. DNA-damaging agents activate the monoubiquitination of FANCD2, resulting in its targeting to nuclear foci that also contain BRCA1 and BRCA2/FANCD1, proteins involved in homology-directed DNA repair. Given the interaction of the FANC proteins with BRCA1 and BRCA2, we tested whether cells from FA patients (groups A, G, and D2) and mouse Fanca–/– cells with a targeted mutation are impaired for this repair pathway. We find that both the upstream (FANCA and FANCG) and downstream (FANCD2) FA pathway components promote homology-directed repair of chromosomal double-strand breaks (DSBs). The FANCD2 monoubiquitination site is critical for normal levels of repair, whereas the ATM phosphorylation site is not. The defect in these cells, however, is mild, differentiating them from BRCA1 and BRCA2 mutant cells. Surprisingly, we provide evidence that these proteins, like BRCA1 but unlike BRCA2, promote a second DSB repair pathway involving homology, i.e., single-strand annealing. These results suggest an early role for the FANC proteins in homologous DSB repair pathway choice.
Keywords: double-strand break repair, FANC, homologous recombination, mammalian cells
Cellular DNA repair defects in a number of different pathways are associated with tumor susceptibility and developmental defects in humans and mice. Recent work has specifically implicated defects in homologous DNA repair in tumor predisposition in the hereditary breast cancer syndromes (1, 2). Mechanistically, pathways that use sequence homology for DNA repair are broadly characterized into two types based on whether homologous associations arise from DNA strand exchange or strand annealing (3). Homologous recombination, also termed homology-directed repair (HDR), utilizes strand exchange in a gene conversion reaction involving a single-strand and a DNA duplex, and is a major repair pathway in mammalian cells for DNA damage such as double-strand breaks (DSBs) (4).
The other DSB repair pathway using sequence homology is single-strand annealing (SSA), which involves the annealing of complementary single strands formed after resection at a DSB. The biological relevance of this pathway is uncertain, but it is a highly efficient mechanism of DSB repair in mammalian cells involving direct repeats (4). Because a large portion of the mammalian genome consists of repeat sequences, SSA could potentially be an important alternative pathway of homologous repair.
Although HDR and SSA involve a common intermediate (single strands formed after end resection), the subsequent strand exchange and strand-annealing steps, respectively, involve some distinct components. Proteins critical for HDR in mammalian cells include the strand exchange protein RAD51 (5, 6) and the products of the hereditary breast cancer susceptibility genes BRCA1 (7, 8) and BRCA2 (9, 10). BRCA2 directly interacts with RAD51, possibly to promote the strand invasion step of HDR (2, 11). Proteins involved in SSA in mammalian cells include RAD52 (6), which promotes strand annealing in vitro (12). Recent observations suggest a competitive interaction between HDR and SSA in mammalian cells: when either RAD51 or BRCA2 is disrupted, HDR is decreased and SSA is enhanced (6). Such a competitive interaction has also been reported in yeast for several HDR genes (13). However, some proteins may act in both pathways. For example, when BRCA1 function is disrupted, both HDR and SSA are decreased (6). This finding has led to the proposal that BRCA1 may have a role early in homologous repair before the branch point of the HDR and SSA pathways (6).
Cells from patients with Fanconi anemia (FA) have defects in DNA repair, because they are sensitive to DNA-damaging agents and exhibit chromosome aberrations (14, 15). The FANC proteins, which are disrupted in FA patients, have been implicated in HDR, although several reports are contradictory. Impaired DNA damage-induced RAD51 focus formation, which is often associated with HDR defects, has been reported in one study to be a characteristic of cells from the FA-D1 group (i.e., BRCA2) but not of cells from other FA complementation groups (16), whereas another study has reported attenuated RAD51 focus formation in cells from several FA groups (17). In apparent contradiction to these studies, patient fibroblasts representing several upstream FA groups have been reported to have highly elevated levels of homologous recombination between plasmids, suggesting that the FANC proteins suppress HDR (18). By contrast, however, studies in a highly recombinogenic chicken DT40 cell line have implicated the FANC proteins in promoting HDR, such that fancg or fancc mutant cells have either a severe or mild impairment of HDR, respectively (19, 20). Surprisingly, the fancc mutant chicken cells have an increased level of another indicator of recombination, that of sister chromatid exchange (20), which is not observed in cells from human FA complementation groups. To clarify the role of the FANC proteins in homologous repair mammalian cells, we have examined DSB repair in patient-derived FA cells from three complementation groups, as well as mouse cells with a targeted mutation, and report on these results.
Materials and Methods
Cell Lines. SV40-transformed FA fibroblasts (GM6914 FA-A, EUFA326 FA-G, and PD20 FA-D2) were grown in DMEM supplemented with 15% FBS. The previously described DR-GFP reporter (21, 22) was modified to contain a hygromycin resistance marker in place of a puromycin resistance marker, and then the vector DRGFPhygro was integrated into the genome of PD20 cells and hprtDRGFPhygro was integrated into the genome of GM6914 and EUFA326 cells. Clones with a single copy of DR-GFP were identified by Southern blotting. These clones were then used for infection with either an “empty” pMMP-puro retrovirus (vector) or a pMMP-puro retrovirus into which a FANC cDNA was inserted. Infected cells were selected in puromycin (generally 2–3 weeks) before subsequent manipulation. FANC expression was verified by Western blotting, immunofluorescence, and resistance to mitomycin C, as described (23). Two independent clones (C8 and H1) of mouse ES cells containing a targeted mutation in Fanca were constructed by sequentially deleting exons 37–39 (Y.-G.Y., M.D., and Z.-Q.W., unpublished results). The Fanca–/– cells, which contain no detectable Fanca protein, were subsequently targeted at the hprt locus with DR-GFP (22).
GFP Assays. To measure the repair of an I-SceI-generated DSB, 30 μg of the I-SceI expression vector pCBASce (21) or an empty vector was mixed with 5 × 106 cells suspended in 650 μl of opti-MEM medium (Invitrogen) in a 0.4-cm cuvette, followed by pulsing the cells at 270 V, 975 μF. To specifically determine the amount of HDR, the percentage of GFP-positive cells was quantitated by flow cytometric analysis 3 days after electroporation on a Becton Dickinson FACScan, as described (21).
PCR Assays. To determine the percentage of I-SceI site loss for each electroporation, genomic DNA was isolated 7 days after transfection. Genomic DNA (0.4 μg) was used as the template for PCR with primers in a reaction volume of 50 μl. The sequences of primers were as follows: DRGFP-F, CTGCTAACCATGTTCATGCC; DRGFP-R, AAGTCGTGCTGCTTCATGTG. PCRs were performed by using the PCR SuperMix (Invitrogen) with Mastercycler (Eppendorf). Amplification was done for 27 cycles with a 1-min elongation time. After amplification, PCR products were digested overnight with 10 units of I-SceI (Roche Molecular Biochemicals). After I-SceI digestion, products were purified by using GFX PCR DNA and Gel Band Purification Kit (Amersham Pharmacia Biosciences), and then half volume of products were digested with BcgI (NEB, Beverly, MA). The products digested with I-SceI and both I-SceI and BcgI were separated on a 1.2% agarose gel. The gel was stained with ethidium bromide, and the ethidium signals for the enzyme-resistant and enzyme-cleaved band were quantified by using nih image software.
To investigate SSA in the FA cell lines, we used a combined PCR-Southern blotting method. After electroporation, genomic DNA was used as PCR template for two pairs of PCR primers, either SA-F and SA-R1, or SA-F and SA-R2 (SA-F, TTTGGCAAAGAATTCAGATCC; SA-R1, CAAATGTGGTATGGCTGATTATG; SA-R2, ATGACCATGATTACGCCAAG. Amplification was for 20 cycles, which was determined to be in the linear range. After running the products on an 0.8% gel, DNA was transferred to nylon membrane and probed with 32P-labeled fragment from DR-GFP, which was cut with Hin-dIII/BamHI.
Results
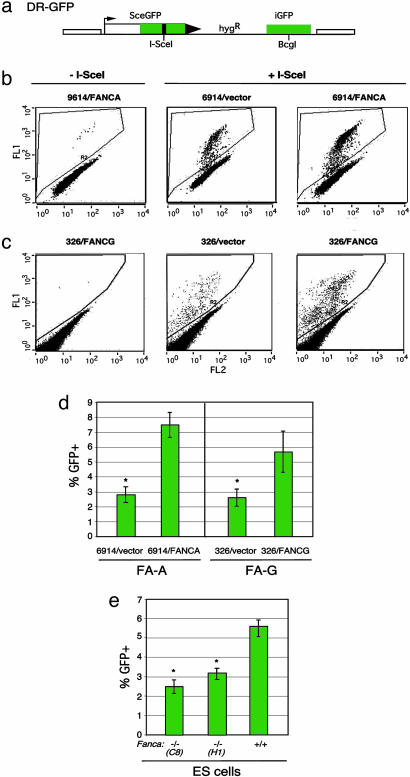
Upstream FANC Pathway Components Promote HDR. To determine whether the FA pathway has a role in HDR in human cells, we established patient-derived human cell lines from the FA-A (GM6914) and FA-G (EUFA326) complementation groups with an integrated copy of the DR-GFP reporter (21) (Fig. 1a). With this reporter, a DSB is introduced into the chromosome by expressing the I-SceI endonuclease, and, if HDR occurs, GFP is expressed, which is quantifiable by flow cytometry. After establishment of cell lines carrying the DR-GFP reporter, a retrovirus expression system was used to complement the specific FANC defect in the cells. Complementation was verified by Western blot analysis (data not shown) and increased survival after mitomycin C treatment (Fig. 6 a and b, which is published as supporting information on the PNAS web site), when cells infected with the FANC-expressing virus were compared with cells infected with a vector control virus.
Fig. 1.
FA patient-derived cell lines have reduced HDR of a chromosomal DSB. (a) DSB reporter substrate. The recombination reporter DR-GFP is stably integrated into the genome of FA patient-derived cell lines. SceGFP is a GFP gene that contains an I-SceI endonuclease site within the coding region. Cleavage of the I-SceI site in vivo and repair by HDR directed by the downstream iGFP repeat results in GFP+ cells. (b and c) Flow cytometric analysis of FA-A cells (GM6914) and FA-G cells (EUFA326), demonstrating increased HDR after FANCA and FANCG complementation, respectively. FA cell lines were infected with the cognate FANC gene-expressing retrovirus or a nonexpressing virus (vector). Subsequently, they were transfected with an I-SceI expression vector (Center and Right) or an empty vector (Left). (d) HDR in FA-A and FA-G cells after I-SceI expression. The difference between the mutant and complemented FA-A or FA-G cells is statistically significant, as indicated by the asterisk (P = 0.001 and P = 0.006, respectively; unpaired t test). (E) HDR in Fanca–/– ES cell lines after I-SceI expression. The difference between the mutant and wild-type ES cells is statistically significant, as indicated by the asterisk (P = 0.0007 for clone C8 and P = 0.001 for clone H1).
In the absence of I-SceI expression, very few GFP+ cells were detected in any of the cell lines (Fig. 1 b and c). Transfection with the I-SceI expression vector into the uncomplemented GM6914 and EUFA326 cells resulted in ≈2.5–3% GFP+ cells (Fig. 1 b and c). By contrast, I-SceI expression in cell lines complemented with each wild-type cDNA resulted in an increased number of GFP+ cells (Fig. 1 b and c), i.e., a 2.7- and 2.2-fold increase, respectively (Fig. 1d). Transfection with an intact GFP expression vector gave a similar number of GFP+ cells for each of the cell lines, indicating that the increased number of GFP+ cells after I-SceI expression was not due to an increased transfection efficiency. These results imply that both the GM6914 and EUFA326 cell lines are deficient in the precise HDR pathway, indicating a role for both FANCA and FANCG in this pathway.
To rule out that the increased HDR was due to nonspecific effects rather that complementation of the specific FANC mutation, we also infected wild-type and noncognate FA cells with FANC-expressing retroviruses. For wild-type cells, either HEK-293 or U2OS cells were infected with FANCA- or FANCG-expressing retroviruses, respectively. No increase in HDR was observed in either cell line (Fig. 7a, which is published as supporting information on the PNAS web site, and data not shown), indicating that infection with the FANC-expressing retroviruses does not itself increase HDR. In addition, GM6914 cells were infected with the complementing FANCA-expressing retrovirus and the noncognate FANCG-expressing retrovirus. Infection with the noncognate FANCG-expressing retrovirus had no effect on HDR, whereas, as before, the FANCA-expressing retrovirus promoted HDR (Fig. 7b). Thus, the increase in HDR in the patient-derived FA cell lines upon infection with the cognate retrovirus is due to complementation of the FANC defect in the cells.
We also tested HDR in mouse ES cells containing a targeted mutation in Fanca (Y.-G.Y., M.D., and Z.-Q.W., unpublished results). Similar to the patient-derived FA-A cells, HDR is reduced in the Fanca–/– cells (Fig. 1e); comparing the two independently targeted Fanca–/– cell lines (C8 and H1) with the parental wild-type cell line, the reduction is 2-fold (2.8% vs. 5.5%, respectively). Thus, a mild reduction in HDR in cells with disrupted upstream FANC components is observed for both transformed human cell lines and mouse cell lines containing a newly derived mutation.
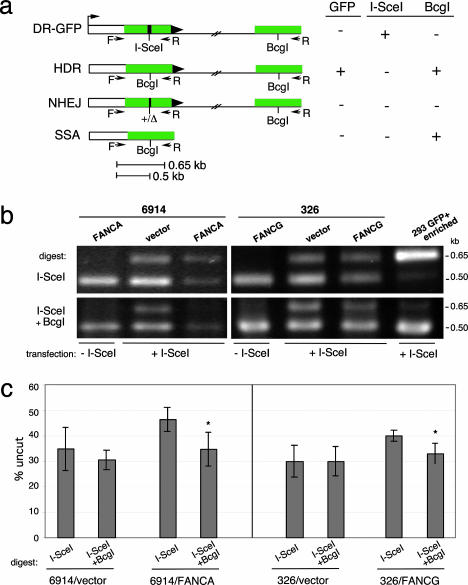
Nonhomologous End-Joining (NHEJ) Levels Are Not Altered in the FA Cell Lines. To determine whether overall DSB repair was substantially reduced in the patient-derived cell lines, we physically analyzed DSB repair products using a PCR assay. For this assay, we amplified the genomic region surrounding the I-SceI site after expression of I-SceI endonuclease with PCR primers that would amplify products from several types of repair: HDR, SSA, and NHEJ (Fig. 2a). To determine how much of the amplified fragment consisted of detectable DSB repair products, we cleaved the fragment with I-SceI, and, to specifically detect NHEJ products, we cleaved with I-SceI and with BcgI, which cleaves only the homologous repair product.
Fig. 2.
FA patient-derived cell lines have normal levels of chromosomal NHEJ. (a) PCR strategy to detect DSB repair products at the DR-GFP locus. In addition to HDR, other DSB repair pathways are NHEJ and SSA, although neither of these results in GFP+ cells. All three DSB repair pathways result in I-SceI site loss; the homologous repair pathways HDR and SSA additionally result in replacement of the I-SceI site with a BcgI restriction site. (b) PCR products (primers F,R) from mutant and complemented FA-A and FA-G cell lines transfected with or without an I-SceI expression vector. Amplified products from the same reaction were digested with I-SceI or with I-SceI and BcgI. Also shown is a control PCR product obtained from a HEK-293 DR-GFP cell line that had been transfected with an I-SceI expression vector and enriched by flow cytometry for GFP+ cells. As expected, the amplified product is digestible with BcgI but not I-SceI. (c) Quantification of the PCR results from cells transfected with the I-SceI expression vector demonstrating comparable NHEJ repair in FA mutant and complemented cells. The mutant FA-A and FA-G cells have a lower amount of total repair product (uncut by I-SceI) than that found for the complemented cells (P = 0.0281 and P = 0.0234), but the NHEJ repair product (uncut by I-SceI and BcgI) is similar for the mutant and complemented cells. The percentage uncut is the ratio of the 0.65-kb band to the 0.5-kb band. For both the FA-A and FA-G complemented cell lines, but not the mutant cells, there is statistical significance between the level of total repair product and the NHEJ repair product (asterisk), indicating a significant contribution of the homologous repair product (i.e., cleavable by BcgI) to total repair (FA-A, P = 0.0174; FA-G, P = 0.0172).
In cells that were not transfected with the I-SceI expression vector, the amplified product is completely cleaved by I-SceI (Fig. 2b) whereas, in cells that expressed I-SceI, a substantial portion of the amplified product was uncleaved (Fig. 2b), indicating in vivo cleavage by I-SceI followed by DSB repair, which results in I-SceI site loss (Fig. 2a). For the uncomplemented GM6914 and EUFA326 cells, I-SceI site loss was ≈30–35% of the amplified fragment, indicating that the chromosomal I-SceI site was cleaved and repaired in a substantial fraction of the cells (Fig. 2 b and c). Interestingly, in the FANCA- and FANCG-complemented cell lines, I-SceI site loss was ≈10% higher (Fig. 2 b and c), which is suggestive of enhanced DSB repair by one or more pathways.
To differentiate NHEJ from homologous repair, the amplified products were additionally treated with BcgI, because HDR and SSA products are both cleavable by this enzyme but NHEJ products are not (Fig. 2a). Thus, the amount of uncleaved DNA after digestion with both BcgI and I-SceI indicates the level of NHEJ. In all of the cell lines, whether uncomplemented or FANC-complemented, the BcgI/I-SceI noncleavable product was 30–35%. This result clearly demonstrates that the level of repair of a chromosomal DSB by NHEJ is not reduced by either FANCA or FANCG deficiency.
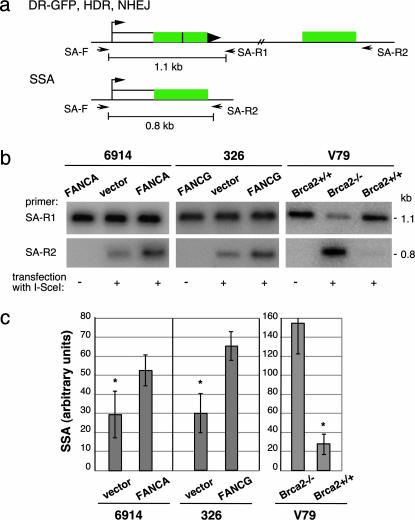
SSA Is Reduced in the FA Cell Lines. We noted in these experiments that the amount of the BcgI/I-SceI cleavable product differed in level in the uncomplemented versus complemented cells to a greater degree than expected from a decrease in the HDR product alone. This finding suggested that the other BcgI-cleavable product, i.e., deletions arising from SSA, may also be reduced by FANC deficiency. To examine SSA levels, we used a primer set that would specifically amplify the deletion product derived from SSA (Fig. 3a). This same deletion product would be obtained from an HDR pathway involving crossing over; however, crossovers have been measured in several systems to be rare outcomes of HDR, i.e., <2% of HDR events, so a significant contribution of crossovers to deletion events is unlikely (24–26). We also ruled out that long-tract gene conversions coupled to NHEJ contribute significantly to the PCR product (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 3.
FA patient-derived cell lines have reduced levels of DSB repair by SSA. (a) PCR strategy to quantify SSA. The 0.8-kb PCR fragment derived from primers SA-F and SA-R2 specifically detects the SSA repair product, whereas the 1.1-kb PCR fragment from primers SA-F and SA-R1 detects a structurally intact reporter, i.e., from HDR and NHEJ, as well as the parental DR-GFP reporter that retains the I-SceI site. (b) PCR products from mutant and complemented FA-A and FA-G cell lines transfected with or without an I-SceI expression vector. As a comparison, a similar analysis was performed on Brca2-deficient and control cells. Amplified products from parallel PCR reactions using primer sets SA-F/SA-R1 and SA-F/SA-R2 are shown. (c) Quantification of PCR results from mutant and complemented cells transfected with the I-SceI expression vector. Whereas the Brca2 mutant cells have an increased percentage of SSA relative to Brca2 wild-type cells, the FA-A and FA-G cell lines have a decreased percentage of SSA relative to their complemented counterparts. The level of SSA is derived from the ratio of the 0.8-kb band to the 1.1-kb band; because these bands are derived from independent PCR reactions, this is an arbitrary percentage and does necessarily not reflect the absolute level of SSA. For both the FA-A and FA-G cells, the difference between the amount of SSA for the mutant and complemented cells is statistically significant, as indicated by the asterisk (P = 0.0194 and P = 0.0086, respectively).
We compared the level of the PCR product arising from SSA with the level of the PCR product obtained from a structurally intact DR-GFP reporter (Fig. 3a). Although the amount of this control product was similar in all of the cell lines (Fig. 3b), the FANCA-complemented GFM6914 and FANCG-complemented EUFA326 cells both demonstrated an ≈2-fold higher level of the SSA product relative to their uncomplemented counterparts (Fig. 3 b and c). This result suggests that these FANC proteins participate in both the HDR and SSA homologous repair pathways. Interestingly, this finding contrasts with a BRCA2 mutant cell line, in which SSA is increased rather than reduced (Fig. 3 b and c), even though HDR is also decreased in BRCA2-deficient cells (9).
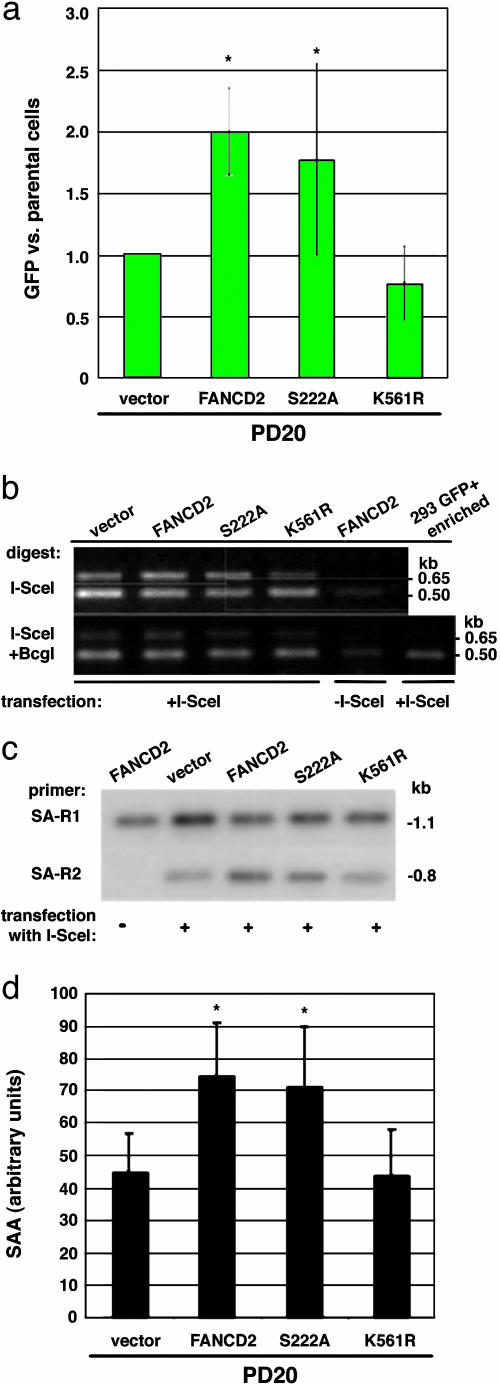
The Downstream FA Component, FANCD2, Participates in Homologous Repair: Role for FANCD2 Monoubiquitination. The FANC complex consists of the A, B, C, E, F, G, and L proteins, which apparently act upstream of FANCD2 (15, 27) and are necessary for FANCD2 monoubiquitination (28). To determine whether the results we obtained with FANC complex members are relevant to FANCD2 function, we performed similar experiments in the PD20 cell line, the reference FA-D2 patient-derived cell line (29). The baseline level of HDR in the uncomplemented PD20 cell line was higher than that obtained in either of the other FANC-deficient lines; however, complementation by expression of FANCD2 led to a similar 2-fold increased level of HDR (Fig. 4a). These results indicate a role for both upstream (FANCA, G) and downstream (FANCD2) FA pathway components in HDR.
Fig. 4.
FA-D2 mutant cells have impaired homologous repair. (a) HDR in FA-D2 cells (PD20) expressing wild-type or mutant FANCD2 proteins. HDR is expressed relative to the mutant FA-D2 cells. Cells expressing wild-type FANCD2 or the ATM phosphorylation site mutant FANCD2-S222A have a higher level of HDR than cells expressing the monoubiquitination site mutant FANCD2-K561R or mutant (vector) cells. The difference between the mutant and FANCD2-complemented FA-D2 cells is statistically significant (asterisk, P = 0.0069), as is the difference between mutant and FANCD2-S222A-expressing cells (P = 0.046), using a paired t test. (b) FA-D2 cells have normal levels of chromosomal NHEJ. See Fig. 2a for a description of the primers. Note that the ratio of the 0.65-kb band to the 0.5-kb band after I-SceI/BcgI digestion is similar in intensity for each of the mutant or wild-type-complemented PD20 cells, indicating normal levels of NHEJ in the FA-D2 cells. (c and d) FA-D2 cells have reduced SSA. As with HDR, FA-D2 cells complemented with wild-type or FANCD2-S222A have higher levels of SSA than FANCD2-K561R-expressing or mutant FA-D2 cells. PCR products are derived from the strategy shown in Fig. 3a. As with the FA-A and FA-G cells, the difference in SSA levels is ≈2-fold between mutant and complemented FA-D2 cells. The difference between the mutant and FANCD2 or FANCD2-S222A complemented FA-D2 cells is statistically significant (P = 0.0076 and P = 0.0051, respectively).
FANCD2 is monoubiquitinated on lysine 561 in response to DNA damage and during S phase, and monoubiquitination is necessary for normal levels of resistance to DNA damaging agents (28) (Fig. 6c). Moreover, FANCD2 is phosphorylated by the ATM kinase on serine 222, which is responsible for the S-phase checkpoint after DNA damage but is not essential for DNA repair (23). We tested the role of these modifications on FANCD2 function in the HDR assay. We found that an intact FANCD2 monoubiquitination site was necessary to promote HDR, but that an intact ATM phosphorylation site was not (Fig. 4a).
We also performed the I-SceI site-loss assay in the various PD20 derivatives and found that levels of the NHEJ product were similar in all cell lines (Fig. 4b), indicating that, as with the upstream components of the FA pathway, NHEJ levels are not affected by the FANCD2 status of the cells. Finally, we also examined SSA in the PD20 cells and found that it was increased nearly 2-fold when cells were complemented by FANCD2 (Fig. 4 c and d). As with HDR, the complementation depended upon an intact monoubiquitination site, because the FANCD2-K561R mutant showed similar levels of SSA as the vector control. Like wild-type FANCD2, SSA was increased by expression of FANCD2-S222A, indicating that, as for HDR, ATM phosphorylation at S222 is not essential for normal levels of homologous repair, such that its role may be limited to S-phase checkpoint control (Fig. 4d) (23).
Discussion
In this article, we determined that upstream and downstream components of the FANC pathway promote HDR in human cells (Fig. 5). An intact monoubiquitination site in FAND2 is required to promote HDR, although the FANCD2-S222 ATM phosphorylation site is not, which parallels the requirement for normal levels of resistance to crosslinking agents (23). Importantly, the level of HDR impairment in FA patient-derived cell lines is mild, especially when compared with the severe recombination defects found in BRCA1 (30), BRCA2 (9), and RAD51 mutants (6), or even RAD51 paralog mutants (21, 31). We obtained a similarly mild defect in HDR in mouse ES cells containing a targeted mutation in Fanca, suggesting this is a general feature of mammalian FANC mutants. Thus, our results imply that, although FANC proteins promote HDR, they are not essential components of the HDR machinery. It still remains possible, however, that they have a crucial role in the repair of a particular subset of DNA lesions.
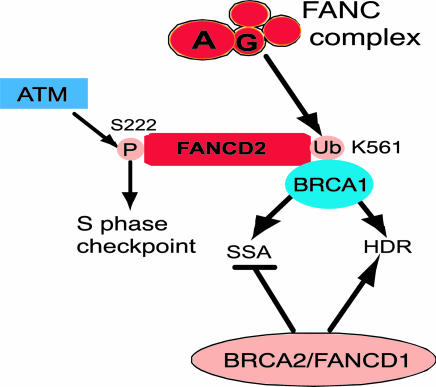
Fig. 5.
Both upstream and downstream components of the FA pathway promote homologous repair by HDR and SSA, similar to BRCA1 but in contrast to BRCA2, which promotes HDR but suppresses SSA. See Discussion for further details.
This mild HDR impairment found with FANC deficiency is consistent with the viability of FA patients and mice, but distinct from the more severe defect observed in fancg mutant chicken DT40 cells (19). Although the recombinogenic DT40 B cells may magnify repair defects that are substantially milder in mammalian cells, it should be noted that fancc mutant DT40 cells also show only a mild HDR defect (20). Nevertheless, the mild impairment could certainly be causative for the increased frequencies of chromosome aberrations, spontaneous HPRT deletions (32), and loss of heterozygosity (33) observed in FA cells, as well as for the tumor predisposition of patients (14, 15). Mice with weak hypomorphic alleles of Brca1 (8) and Brca2 (9) have only small reductions in HDR (i.e., 5-fold), yet they manifest chromosomal instability, developmental defects, and cancer predispositon (34–36).
Unlike HDR, we observed that FA cells have levels of NHEJ that are comparable with those of complemented cells. Preliminary analysis of NHEJ junctions in FA-A cells also indicates that the junction sequences are comparable (K.N. and M.J., unpublished results). Previous reports have been contradictory about a role for FANC proteins in NHEJ. One group reported that NHEJ of a plasmid DSB containing cohesive overhangs, which are also present after I-SceI cleavage, is not impaired in FA cells (37), although another group reported an NHEJ defect in FA cells using a similar plasmid assay (38). It is not clear what accounts for the different results obtained in these plasmid assays. However, because plasmids are susceptible to degradation and other processing events (39), the chromosomal assays using I-SceI may better reflect DSB repair.
Unexpectedly, we observed that the FA patient-derived cell lines from the three FA groups tested have a mild defect in another DSB repair pathway, SSA. By contrast to what we observed in the FA cells, mutations in BRCA2, which has been considered to be an FANC gene (40), increase levels of SSA in cells (6, 10). The results obtained with the FA cells, however, parallel those reported recently when BRCA1 is disrupted, in which both homologous repair pathways are disrupted (6) (Fig. 5). They also oppose the results obtained with the NHEJ mutant Ku70, in which both HDR and SSA are elevated (6). Although the biological relevance of SSA is uncertain, the effect of HDR in relation to SSA in mutants provides insight into the role of homologous repair pathway components. Thus, whereas RAD51 and BRCA2 are both key components of the strand invasion steps of HDR (11, 41), BRCA1 and the FANC proteins may regulates step(s) common to both HDR and SSA pathways; Ku70 may oppose this step.
One step common to HDR and SSA is DNA strand resection, which generates the single strands important for strand invasion and strand annealing, respectively. An effect on resection could be direct or indirect, for example, by antagonizing Ku (6, 22, 42). Alternatively, the FANC proteins may affect some other step common to the homologous repair pathways. It is interesting to note that the chicken FANCC has been implicated recently in an additional DNA repair pathway, that of translesion synthesis (20). This observation, together with our results, may again indicate a role for the FANC proteins at a common step in DNA damage repair pathways, perhaps in processing or stabilizing intermediates for multiple pathways, including homologous repair and translesion synthesis. Further speculation will require confirmation of a role for the mammalian proteins in translesion synthesis.
In summary, our results provide evidence that the FA pathway promotes homologous repair in mammalian cells. The consistently mild defect we observed in FANC mutants implies a role for these components that is distinct from central HDR components, yet which may be sufficient to account for the chromosomal abnormalities observed in FA cells.
Supplementary Material
Acknowledgments
We thank members of the M.J. laboratory, especially Nicole Christ, Mingguang Han, Jeremy Stark, Hiroshi Saeki, and Mary Ellen Moynahan, for technical help and intellectual contributions. This work was supported by the Fanconi Anemia Research Fund, the Julie Laub Fund, the Hecksher Foundation, the Fritz-Thyssen-Stiftung (to M.D.), National Institutes of Health Grant P01 CA94060, and National Institutes of Health Grant GM54668 (to M.J.).
Author contributions: K.N., A.J.P., M.D., Z.-Q.W., and M.J. designed research; K.N., Y.-G.Y., and A.J.P. performed research; K.N., Y.-G.Y., A.J.P., T.T., M.D., and A.D.D. contributed new reagents/analytic tools; K.N., Y.-G.Y., and Z.-Q.W. analyzed data; and K.N. and M.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FA, Fanconi anemia; DSB, double-strand break; HDR, homology-directed repair/homologous recombination; SSA, single-strand annealing; NHEJ, nonhomologous end-joining.
References
- 1.Scully, R. & Livingston, D. M. (2000) Nature 408, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jasin, M. (2002) Oncogene 21, 8981–8993. [DOI] [PubMed] [Google Scholar]
- 3.West, S. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 435–445. [DOI] [PubMed] [Google Scholar]
- 4.Liang, F., Han, M., Romanienko, P. J. & Jasin, M. (1998) Proc. Natl. Acad. Sci. USA 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stark, J. M., Hu, P., Pierce, A. J., Moynahan, M. E., Ellis, N. & Jasin, M. (2002) J. Biol. Chem. 277, 20185–20194. [DOI] [PubMed] [Google Scholar]
- 6.Stark, J. M., Pierce, A. J., Oh, J., Pastink, A. & Jasin, M. (2004) Mol. Cell. Biol. 24, 9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. (1999) Mol. Cell 4, 511–518. [DOI] [PubMed] [Google Scholar]
- 8.Moynahan, M. E., Cui, T. Y. & Jasin, M. (2001) Cancer Res. 61, 4842–4850. [PubMed] [Google Scholar]
- 9.Moynahan, M. E., Pierce, A. J. & Jasin, M. (2001) Mol. Cell 7, 263–272. [DOI] [PubMed] [Google Scholar]
- 10.Tutt, A., Bertwistle, D., Valentine, J., Gabriel, A., Swift, S., Ross, G., Griffin, C., Thacker, J. & Ashworth, A. (2001) EMBO J. 20, 4704–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, H., Jeffrey, P. D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N. H., Zheng, N., Chen, P. L., Lee, W. H. & Pavletich, N. P. (2002) Science 297, 1837–1848. [DOI] [PubMed] [Google Scholar]
- 12.Singleton, M. R., Wentzell, L. M., Liu, Y., West, S. C. & Wigley, D. B. (2002) Proc. Natl. Acad. Sci. USA 99, 13492–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov, E. L., Sugawara, N., Fishman-Lobell, J. & Haber, J. E. (1996) Genetics 142, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grompe, M. & D'Andrea, A. (2001) Hum. Mol. Genet. 10, 2253–2259. [DOI] [PubMed] [Google Scholar]
- 15.D'Andrea, A. D. & Grompe, M. (2003) Nat. Rev. Cancer 3, 23–34. [DOI] [PubMed] [Google Scholar]
- 16.Godthelp, B. C., Artwert, F., Joenje, H. & Zdzienicka, M. Z. (2002) Oncogene 21, 5002–5005. [DOI] [PubMed] [Google Scholar]
- 17.Digweed, M., Rothe, S., Demuth, I., Scholz, R., Schindler, D., Stumm, M., Grompe, M., Jordan, A. & Sperling, K. (2002) Carcinogenesis 23, 1121–1126. [DOI] [PubMed] [Google Scholar]
- 18.Donahue, S. L., Lundberg, R., Saplis, R. & Campbell, C. (2003) J. Biol. Chem. 278, 29487–29495. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto, K., Ishiai, M., Matsushita, N., Arakawa, H., Lamerdin, J. E., Buerstedde, J. M., Tanimoto, M., Harada, M., Thompson, L. H. & Takata, M. (2003) Mol. Cell. Biol. 23, 5421–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niedzwiedz, W., Mosedale, G., Johnson, M., Ong, C. Y., Pace, P. & Patel, K. J. (2004) Mol. Cell 15, 607–620. [DOI] [PubMed] [Google Scholar]
- 21.Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. (1999) Genes Dev. 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce, A. J., Hu, P., Han, M. G., Ellis, N. & Jasin, M. (2001) Genes Dev. 15, 3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taniguchi, T., Garcia-Higuera, I., Xu, B., Andreassen, P. R., Gregory, R. C., Kim, S. T., Lane, W. S., Kastan, M. B. & D'Andrea, A. D. (2002) Cell 109, 459–472. [DOI] [PubMed] [Google Scholar]
- 24.Richardson, C., Moynahan, M. E. & Jasin, M. (1998) Genes Dev. 12, 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dronkert, M. L., Beverloo, H. B., Johnson, R. D., Hoeijmakers, J. H., Jasin, M. & Kanaar, R. (2000) Mol. Cell. Biol. 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark, J. M. & Jasin, M. (2003) Mol. Cell. Biol. 23, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joenje, H. & Patel, K. J. (2001) Nat. Rev. Gen. 2, 446–457. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Higuera, I., Taniguchi, T., Ganesan, S., Meyn, M. S., Timmers, C., Hejna, J., Grompe, M. & D'Andrea, A. D. (2001) Mol. Cell 7, 249–262. [DOI] [PubMed] [Google Scholar]
- 29.Jakobs, P. M., Sahaayaruban, P., Saito, H., Reifsteck, C., Olson, S., Joenje, H., Moses, R. E. & Grompe, M. (1996) Somatic Cell Mol. Genet. 22, 151–157. [DOI] [PubMed] [Google Scholar]
- 30.Westermark, U. K., Reyngold, M., Olshen, A. B., Baer, R., Jasin, M. & Moynahan, M. E. (2003) Mol. Cell. Biol. 23, 7926–7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, R. D., Liu, N. & Jasin, M. (1999) Nature 401, 397–399. [DOI] [PubMed] [Google Scholar]
- 32.Papadopoulo, D., Guillouf, C., Mohrenweiser, H. & Moustacchi, E. (1990) Proc. Natl. Acad. Sci. USA 87, 8383–8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sala-Trepat, M., Boyse, J., Richard, P., Papadopoulo, D. & Moustacchi, E. (1993) Mutat. Res. 289, 115–126. [DOI] [PubMed] [Google Scholar]
- 34.Deng, C. X. & Wang, R. H. (2003) Hum. Mol. Genet. 12, R113–R123. [DOI] [PubMed] [Google Scholar]
- 35.McAllister, K. A., Bennett, L. M., Houle, C. D., Ward, T., Malphurs, J., Collins, N. K., Cachafeiro, C., Haseman, J., Goulding, E. H., Bunch, D., et al. (2002) Cancer Res. 62, 990–994. [PubMed] [Google Scholar]
- 36.Donoho, G., Brenneman, M. A., Cui, T. X., Donoviel, D., Vogel, H., Goodwin, E. H., Chen, D. J. & Hasty, P. (2003) Genes Chromosomes Cancer 36, 317–331. [DOI] [PubMed] [Google Scholar]
- 37.Escarceller, M., Buchwald, M., Singleton, B. K., Jeggo, P. A., Jackson, S. P., Moustacchi, E. & Papadopoulo, D. (1998) J. Mol. Biol. 279, 375–385. [DOI] [PubMed] [Google Scholar]
- 38.Donahue, S. L. & Campbell, C. (2002) J. Biol. Chem. 277, 46243–46247. [DOI] [PubMed] [Google Scholar]
- 39.Liang, F. & Jasin, M. (1996) J. Biol. Chem. 271, 14405–14411. [DOI] [PubMed] [Google Scholar]
- 40.Howlett, N. G., Taniguchi, T., Olson, S., Cox, B., Waisfisz, Q., de Die-Smulders, C., Persky, N., Grompe, M., Joenje, H., Pals, G., et al. (2002) Science 297, 606–609. [DOI] [PubMed] [Google Scholar]
- 41.Sung, P., Krejci, L., Van Komen, S. & Sehorn, M. G. (2003) J. Biol. Chem. 278, 42729–42732. [DOI] [PubMed] [Google Scholar]
- 42.Lee, S. E., Moore, J. K., Holmes, A., Umezu, K., Kolodner, R. D. & Haber, J. E. (1998) Cell 94, 399–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.