Significance
Nuclear domain 10 (ND10) bodies are both sensors and responders to many viruses that infect human cells. The key components of ND10 bodies are PML and SP100. Their importance in innate immune responses to infection is underscored by the observation that numerous viruses have evolved means to degrade, disperse, or at least inactivate PML and SP100. This report focuses on SP100, which, along with PML, is degraded by a viral E3 ligase in HSV-1–infected cells. The question posed here is the function of SP100 and PML in the course of HSV infection. The results indicate that the function of SP100 is different than, but complementary to, that of PML and ranks high in the cellular defense mechanisms against infection.
Keywords: ICP0, ICP8, replication compartments
Abstract
Nuclear domain 10 (ND10) bodies are small (0.1–1 μM) nuclear structures containing both constant [e.g., promyelocytic leukemia protein (PML), SP100, death domain-associated protein (Daxx)] and variable proteins, depending on the function of the cells or the stress to which they are exposed. In herpes simplex virus (HSV)-infected cells, ND10 bodies assemble at the sites of DNA entering the nucleus after infection. In sequence, the ND10 bodies become viral replication compartments, and ICP0, a viral E3 ligase, degrades both PML and SP100. The amounts of PML and SP100 and the number of ND10 structures increase in cells exposed to IFN-β. Earlier studies have shown that PML has three key functions. Thus, (i) the interaction of PML with viral components facilitates the initiation of replication compartments, (ii) viral replication is significantly less affected by IFN-β in PML−/− cells than in parental PML+/+ cells, and (iii) viral yields are significantly lower in PML−/− cells exposed to low ratios of virus per cell compared with parental PML+/+ cells. This report focuses on the function of SP100. In contrast to PML−/− cells, SP100−/− cells retain the sensitivity of parental SP100+/+ cells to IFN-β and support replication of the ΔICP0 virus. At low multiplicities of infection, wild-type virus yields are higher in SP100−/− cells than in parental HEp-2 cells. In addition, the number of viral replication compartments is significantly higher in SP100−/− cells than in parental SP100+/+ cells or in PML−/− cells.
Nuclear domain 10 (ND10), also known as promyelocytic leukemia (PML) bodies, are small nuclear structures 0.1–1 μm in diameter. ND10 structures form and reform in response to stress. They consist of an outer shell composed of PML, a set of constant proteins (e.g., CBP, Daxx, SP100 nuclear antigen), and various other proteins depending on the particular stress or function of the ND10 bodies (1–7). ND10 bodies have drawn attention recently, in large part because of increasing evidence suggesting that they sense and respond to infection (5, 8). For example, ND10 bodies have been shown to assemble de novo at herpes simplex virus 1 (HSV-1) DNA newly released from capsids at nuclear pores (9, 10).
Accruing evidence also indicates that viruses target ND10 for degradation, dispersal, or inactivation of their function. The most dramatic viral effect on ND10 bodies reported to date is that occurring in HSV-1–infected cells. In brief, infected cell protein 0 (ICP0), an α or immediate early protein, acts as a ubiquitin ligase, and in conjunction with ubiquitin-conjugating enzyme 5a (UbcH5a) degrades both PML and SP100 (11–15). The response of other viruses varies. In encephalomyocarditis virus-infected cells, PML is degraded by the viral protease 3C (16). Human cytomegalovirus redistributes PML and disrupts the ND10 bodies (17, 18). Disruption of ND10 bodies also occurs in the course of lytic infection with Epstein-Barr virus (19–21). Adenovirus-encoded E4 Orf3 relocalizes PML to nuclear fiber-like structures (22, 23). Several viruses relocate viral proteins into ND10 bodies to enhance their replication. Thus, SV-40 large T antigen colocalizes with PML and ND10 bodies, and papillomavirus minor capsid protein L2 colocalizes with PML and ND10 (24–26). It has been reported that one component of ND10, SP100, provides intrinsic immunity against papilloma viruses (27). A central question is why diverse viruses have evolved strategies designed to block the function of ND10 bodies.
Current data support the hypothesis that PML is an IFN effector and thus an appropriate target of viral gene products targeting cellular innate immune responses (1, 7, 8, 28). The evidence that PML is an effector of IFN rests on two observations. Exposure of cells to IFN results in significant increases in the amount of PML and the number of ND10 bodies (29, 30). Moreover, PML mediates the IFN-based antiviral activity, with HSV-1 replication significantly less affected in PML−/− cells pretreated with IFN compared with similarly treated sibling or parental cells (31, 32).
In an earlier study, we found that the replication of HSV-1 was unaffected in PML−/− cells infected at high ratios of virus per cell, but was significantly lower in cells exposed to low ratios of virus per cell (31). In addition, we noted delays in the degradation of SP100 mediated by ICP0, consistent with reduced colocalization of ICP0 with structures containing SP100 and Daxx (31). Because ND10 bodies appear to be predecessors of viral replication compartments in which viral DNA is made and packaged, the results suggest that ICP0 homes in on ND10 bodies to degrade PML and at the same time initiate the formation of replication compartments (33–37). In this report, we focus on SP100. The accumulation of SP100 is also enhanced by exposure of cells to IFN-β (31, 38, 39). SP100 appears to play a significant role in host defenses early in infection. Moreover, unlike PML−/− cells, SP100−/− cells retain their sensitivity to IFN-β.
Results
Generation and Properties of SP100−/−CL5-3 and PML/SP100−/−CL18-3 Derived from HEp-2 Cells and the ID2 PML−/− Cell Line.
SP100 locating at human chromosome 2q37 encodes at least four isoforms through mRNA splicing. These isoforms share their N-terminal 476 amino acids, encoded by the first 15 exons (GenBank: NG_029842.1). SP100−/− and PML/SP100−/− clones were derived from wild-type (WT) HEp-2 cells or PML−/− cell clone 1D2 (31) by transfection of a clustered regularly interspaced short palindromic repeats (CRISPR)/cas9 cassette targeting exon 3, 4, or 5 of human SP100. Experimental procedures are described in Materials and Methods. Single-cell–derived clones SP100−/−CL5-3 and PML/SP100−/−CL18-3 (simplified as SP100−/− and PML/SP100−/− hereinafter) were selected.
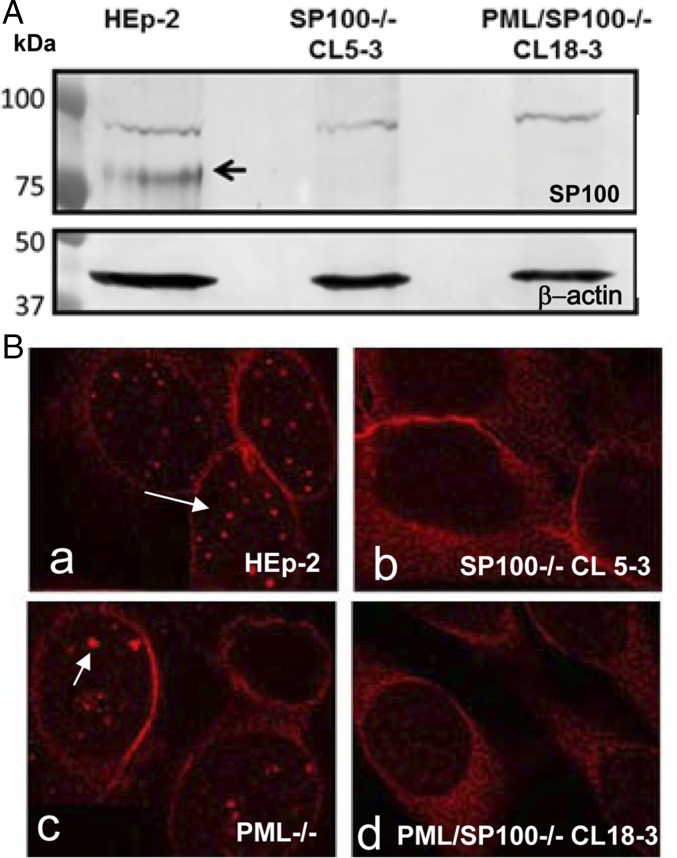
The knockout of SP100 from the two cell lines was validated in two sets of experiments. Because SP100 is an IFN-stimulated gene, all cell lines were exposed to IFN-β at 1,000 U/mL for 24 h to elevate SP100 expression. Cell lysates from HEp-2, SP100−/−, and PML/SP100−/− cells were solubilized, subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose sheet, and reacted with rabbit polyclonal antibody anti-SP100 and antibody recognizing β-actin (Fig. 1A). Although a nonspecific band running at 100 kDa was present in all three samples, an SP100 protein band at 80 kDa was detected only in WT HEp-2 cells.
Fig. 1.
Generation of SP100−/− and PML/SP100−/− single-cell–derived cell clones. Generation of single-cell–derived KO clones is described in Materials and Methods. The cells shown were pretreated with IFN-β at 1,000 U/mL for 24 h to facilitate detection and visualization of SP100. (A) Lysates of parental HEp-2, SP100−/−, and PML/SP100−/− cells were subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose membrane, and reacted with rabbit polyclonal antibody against SP100. The black arrow indicates the SP100 protein band. (B) HEp-2 (a), SP100−/− (b), PML−/− (c), and PML/SP100−/− (d) cells were fixed and reacted with rabbit polyclonal antibody against SP100 conjugated to Texas Red. The arrows point to aggregates of SP100 in HEp-2 and PML−/− cells. (Magnification: 100×.)
In the second set of experiments, cells were fixed and reacted with Texas Red-labeled polyclonal rabbit anti-SP100 antibodies (Fig. 1B). Uniform nuclear punctate structures (arrow in Fig. 1B, a) representing ND10 bodies were noted in HEp-2 cells, whereas PML−/− cells contained an array of structures of different sizes. The arrow in Fig. 1B, c points to dense bodies larger than the ND10 bodies in HEp-2 cells. Nuclear structures reacting with anti-SP100 antibody were not detected in the SP100−/− (Fig. 1B, b) or PML/SP100−/− (Fig. 1B, d) cell line.
PML Accumulation Is Reduced in SP100−/− Cells.
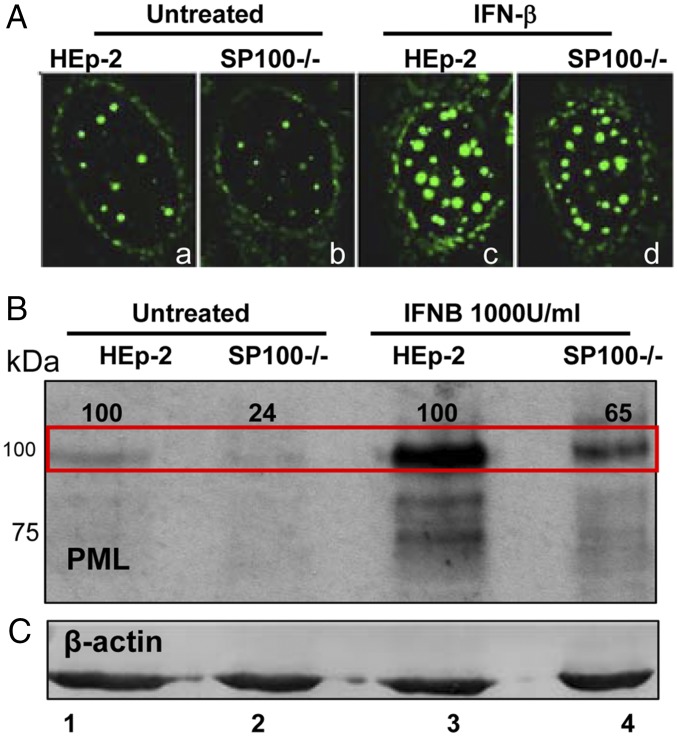
The results of two series of experiments suggest that the accumulation of PML is reduced in SP100−/− cells, but that PML is induced and accumulates to high levels on exposure to IFN in both HEp-2 and SP100−/− cells (39).
In the first series of experiments, replicate cultures of HEp-2 or SP100−/− cells were mock-treated or exposed to IFN-β (1,000 U/mL). After 24 h, the cells were harvested, processed as described above, and reacted with anti-PML antibody. The results, shown in Fig. 2B, suggest that the accumulation of PML was reduced by as much as fourfold in SP100−/− cells. Pretreatment of cells with IFN-β stimulated the accumulation of PML. Our previous study showed that IFN enhances the accumulation of SP100 mRNA in the absence of PML. The results presented here suggest that the synthesis of PML is stimulated by IFN in the absence of SP100, and thus both PML and SP100 are induced by IFN independently of each other, although the basal level of PML is affected by SP100.
Fig. 2.
PML protein levels are reduced in SP100−/− cells. As indicated, the cells were mock-treated or exposed to IFN-β (1,000 U/mL) for 24 h. (A) HEp2 (a and b) and SP100−/− (c and d) cells were reacted with mouse monoclonal antibody against PML and labeled with FITC. (Magnification: 100×.) Lysates of untreated or IFN-β–treated cells were subjected to electrophoresis in denaturing gels and reacted with rabbit polyclonal antibody against PML (B) or mouse monoclonal antibody against β-actin (C). The major PML band (marked by the red rectangle) of SP100−/− cells was quantified and normalized to β-actin and then to that of HEp2−/− cells under the same conditions (set as 100).
In the second series of experiments, we examined the distribution of PML in intranuclear bodies. Fig. 2A presents images of representative cells from cultures fixed and reacted with anti-PML antibody. The striking feature of these images is the reduced amount of PML and number of nuclear bodies containing PML (compare the cells in a and b). Cultures treated with IFN-β (1,000 U/mL for 24 h) exhibited a large increase in dense structures of varying size (cells c and d).
Replication of WT Virus in SP100−/− and SP100/PML−/− Cells.
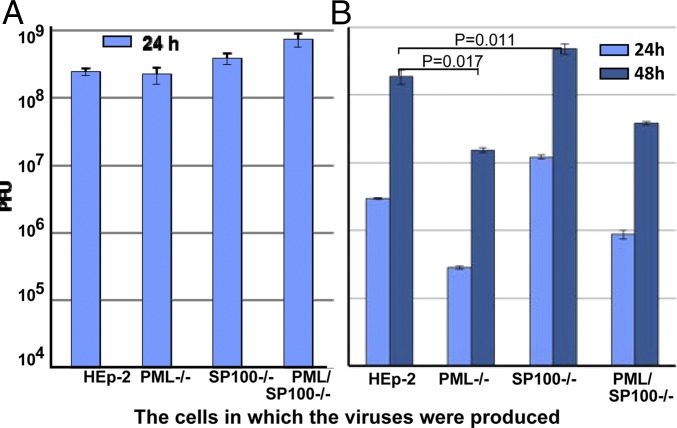
In this series of experiments, replicate cultures of HEp-2, PML−/−, SP100−/−, and PML/SP100−/− were exposed to either 0.01 or 5 PFU of HSV-1(F) per cell. The yields were titered on Vero cells. The results, shown in Fig. 3, can be summarized as follows. The virus harvested from cultures exposed to 5 PFU/cell were within a twofold range and were not significantly different (Fig. 3A). The yields from cultures exposed to 0.01 PFU/cell varied depending on cell genotype (Fig. 3B). As would be expected based on previously reported results, the yields from PML−/− cells were significantly lower than those obtained from HEp-2 cells [P = 0.017, comparison of yields obtained at 48 h postinfection (hpi)]. In contrast, the yields obtained from infected SP100−/− cells were twofold to ninefold higher than those obtained from infected HEp-2 cells in repeat experiments (P = 0.011 for the representative experiment shown in Fig. 3B). The positive and negative effects of depletion of PML and SP100 appear to balance out, given that the yields from infected PML/SP100−/− cells did not differ significantly from those obtained from HEp-2 cells.
Fig. 3.
At low multiplicity of infection, HSV-1(F) replicates to higher levels in cells devoid of SP100. HEp-2, PML−/−, SP100−/−, or PML/SP100−/− cells were exposed to 5 PFU (A) or 0.01 PFU (B) of HSV-1(F) per cell. The yields at indicated time points were determined in Vero cells. P values were calculated on virus yields at 48 hpi between HEp-2 and PML−/− cells, and between HEp-2 and SP100−/− cells.
These results suggest that in cells exposed to low ratios of virus per cell, a likely reflection of natural infections, the key constituents of ND10 bodies have contrasting roles. Thus, PML appears to be beneficial, whereas SP100 appears to have a negative effect on virus replication.
At a Low Ratio of Virus per Cell, ∆ICP0 Virus Replicates to Higher Titers in SP100−/− Cells Than in Parental HEp-2 Cells.
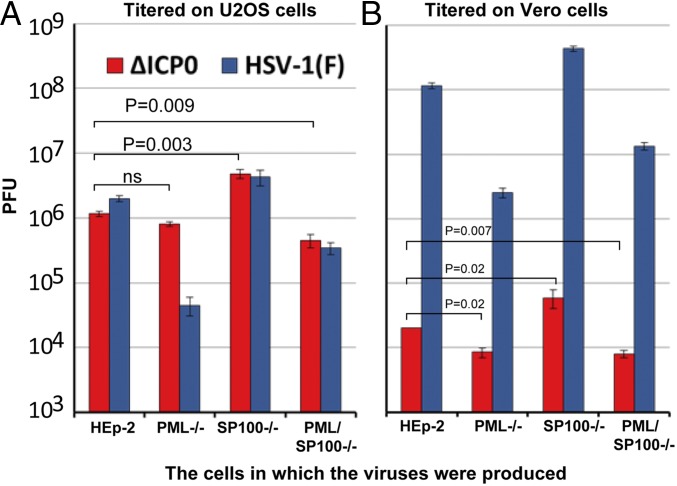
ICP0 is a major multifunctional protein created immediately after infection. Among its major functions are the recruitment of CLOCK histone deacetylase to the viral transcriptome (40), the dissociation of the CoREST/REST/LSD1 repressor complex from its cognate sites on viral DNA (41, 42) and the degradation of PML and SP100 (11, 15, 43). ∆ICP0 mutants replicate in U2OS cells but poorly in numerous cell lines, including human (e.g., HEp-2) and African green monkey kidney (Vero) cells (44–46). The question posed here is whether the replication of ∆ICP0 virus is affected by PML or SP100. In this series of experiments, cultures of HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were exposed to 0.01 PFU of HSV-1(F) and ∆ICP0 mutant R7910. The cells were harvested at 48 hpi and titered on both U2OS and Vero cells.
As illustrated in Fig. 4, human and primate cell lines differ with respect to their ability to support HSV replication and, by extension, the formation of plaques. Thus, the titers of the viruses grown in the HEp-2 cells and in mutant cell lines varied depending on the cell lines in which the titrations were done. The titers of WT [HSV-1(F)] virus were higher in Vero cells than in the U2OS cell line by as much as 30- to 100-fold. In contrast, the yields of ΔICP0 mutant were 10–100 to higher in U2OS cells than in Vero cells. These results, which were reproduced in several experiments, suggest that the U2OS cells are more permissive to ∆ICP0 virus and at the same time restrict the replication of HSV-1(F).
Fig. 4.
At low multiplicity of infection, ∆ICP0 virus replicates to higher levels in cell bereft of SP100 expression. HEp-2, PML−/−, SP100−/−, or PML/SP100−/− cells were exposed to 0.01 PFU of ∆ICP0 (R7910) or HSV-1(F) per cell. The yields at 48 hpi were determined in U2OS cells (A) and Vero cells (B). P values of comparison of the growth of ∆ICP0 (titered in U2OS cells) between HEp-2 and PML−/− cells, between HEp-2 and SP100−/− cells, and between HEp-2 and PML/SP100−/− cells were calculated. ns, not significant.
The key result is the similar patterns of replication of HSV-1(F) in the four cell lines observed in Vero cells and U2OS cells. Specifically, the highest yields were obtained in SP100−/− cells, and the lowest yields were obtained in PML−/− cells. Although the yields of ΔICP0 measured in Vero cells were orders of magnitude lower than those of HSV-1(F), the basic patterns were similar. Again, the highest yields were obtained in SP100/−/− cells. In U2OS cells, the yields were highest in SP100−/− cells and nearly equal in HEp-2 and PML−/− cells.
PML and SP100 Do Not Have Significant Roles in the Transcription of Viral mRNAs.
A central and as-yet unresolved question is whether PML or SP100 plays a role in the early transcription of viral genes after infection (14, 42, 47–51). Both PML and SP100 are degraded within the first 6 hpi, and thus the major effect of PML or SP100 on the transcription of viral DNA would occur during that period (11). The experiments designed to answer this question are based on the observation that the first new progeny viruses become apparent within the first 6 hpi. Implicit in this observation is that measurements done during the first 6 h after exposure of the virus to the cell would detect only transcripts accumulating during the first round of replication and, moreover, would capture the accumulation of transcripts made both early and late in the replicative cycle.
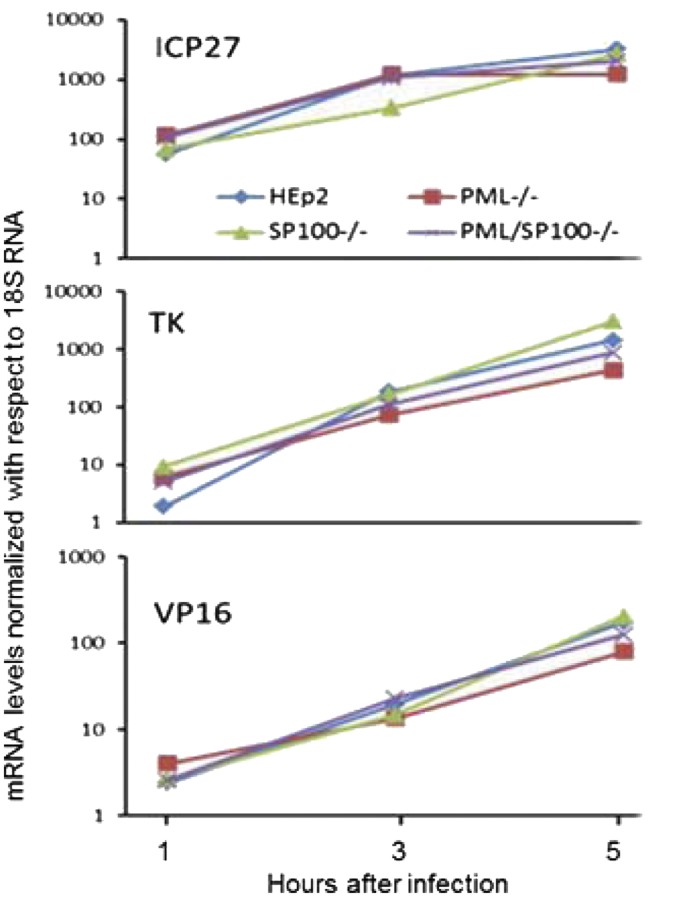
In this series of experiments, cultures of HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were exposed to 0.1 PFU of HSV-1(F) per cell. At 1, 3, and 5 hpi, the cells were harvested, and the mRNAs representative of α, (ICP27), β (TK), and ϒ (VP16) genes were assayed as described in Materials and Methods. The results, shown in Fig. 5, did not show any significant differences in the accumulation of various classes of mRNAs during the early stages of infection. These results indicate that the differences in virus replication in the cell lines studied in this report are not due to variation in the rates of accumulation of viral mRNAs.
Fig. 5.
No significant changes in viral transcription levels are seen in HSV-1(F)-infected KO clones. Early after infection, the accumulation of representative α (IC27), β (TK), or γ (VP16) mRNAs in PML−/−, SP100−/−, or PML/SP100−/− cells could not be differentiated from those accumulating in HEp-2 cells.
Formation of Viral Replication Compartments Is Hampered in the Absence of PML and Facilitated in the Absence of SP100.
Viral DNA synthesis and some early stages of virion assembly occur in nuclear structures and, at least in the initial stages of formation, at ND10 bodies (9, 52, 53). In this series of experiments, we used immunostaining with an antibody to ICP8 to visualize the presence of replication compartments. ICP8 is viral protein that binds DNA and is essential for viral DNA synthesis (54–57).
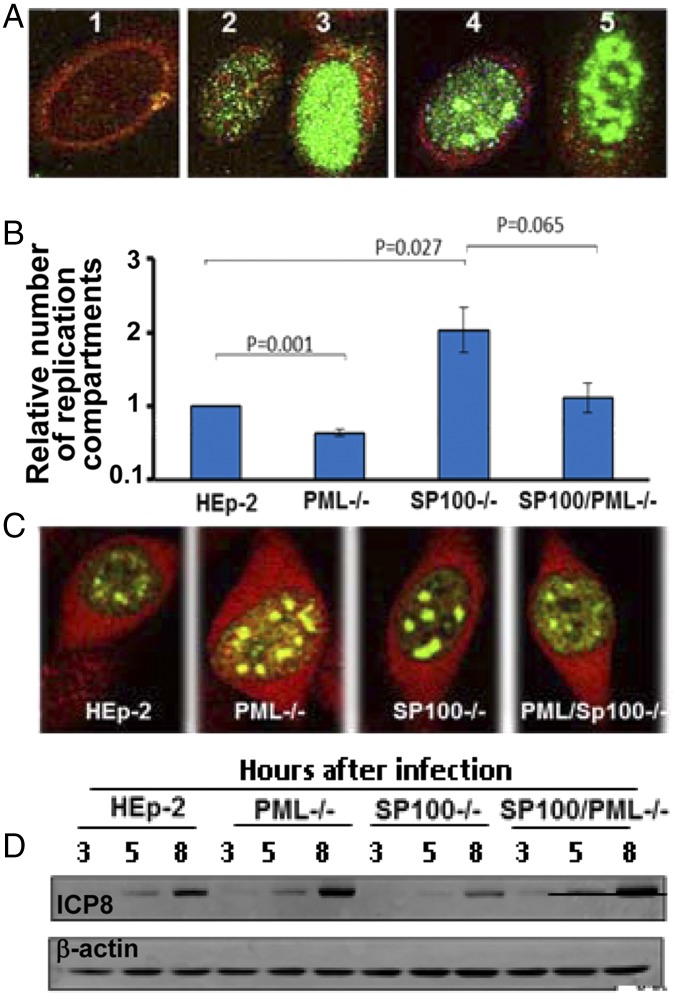
To facilitate analyses of infected cells, we classified the appearance of ICP8 in infected cells into four categories, as shown in Fig. 6A. These images, collected at 6 hpi, were classified into uninfected or very early stage of infection (cell 1) or diffuse distribution of ICP8 from very faint to dense (cells 2 and 3) or forming very dense aggregates (cells 4 and 5). The aggregates seen in cells 4 and 5 were classified as replication compartments (34, 52, 58, 59).
Fig. 6.
Replication compartments assemble with different efficiencies in PML−/− or SP100−/− cells. Replicate cultures of HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were exposed to 20 PFU of HSV-1(F)/cell (A–C) or to 5 PFU of HSV-1(F)/cell (D). The cells shown in A and C were fixed at 6 hpi, reacted with FITC-labeled monoclonal antibody against ICP8 and Texas Red- labeled rabbit antibody against ICP0 (A and C). (A) Nuclear pattern of ICP8 in the infected cells were classified into image 1 (no detectable accumulation), images 2 and 3 (no apparent replication compartment), and images 4 and 5 (multiple replication compartments). (Magnification: 100×.) (B) Relative efficiency of replication factories assembly in different cell types was quantified as described in Materials and Methods. P values for comparisons between HEp-2 and PML−/− cells, between HEp-2 and SP100−/− cells, and between SP100−/−and PML/SP100−/− cells were calculated. (C) Images of replication compartments assemble in various cells lines. (Magnification: 100×.) (D) Lysates of infected cells were collected at indicated time points and immunoblotted with antibodies against ICP8 and β-actin.
We next quantified the frequency of occurrence of replication compartments in the various cell lines. In these experiments, cultures of HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were exposed to 20 PFU of HSV-1(F) per cell. At 6 hpi, the cells were fixed and reacted with antibodies against ICP0 (red) and ICP8 (green). At least 20 different fields comprising on average 200 positive cells for ICP8 in each of the four cell lines were photographed and classified as described above. Error bars were calculated based on data obtained in two independent experiments in which the cells were counted and classified three times. As shown in Fig. 6C, infected cells exhibiting replication compartments were readily detected in all four cell lines. The number of replication compartments shown in this figure is not representative of the cultures.
To combine three independent calculations from two sets of independently conducted experiments, the exact percentage of replication compartment-positive cells out of the total number of ICP8-positive cells of each cell line was normalized to that of HEp-2 cells (set as 1), combined, and plotted as shown in Fig. 6B. The data show that ablation of PML hampered the efficient formation of replication compartments (P = 0.001), whereas knockout of SP100 facilitated this process (P = 0.027). The difference between SP100−/− and SP100/PML−/− did not reach statistical significance (P = 0.065).
One hypothesis that could account for the results is that ICP8 accumulates at different rates in the four cell lines. To test this hypothesis, we exposed cultures of HEp-2, PML−/−, SP100−/−, and PML/ SP100−/− cells to 5 PFU of HSV-1(F) per cell. The cells were harvested at 3, 5, and 8 hpi, processed as described above, and immunoblotted with antibody to ICP8 and β-actin. The results (Fig. 6D) indicate no correlation between the accumulation of ICP8 and the assembly of replication compartments in these cells.
The Degradation of ICP0 Early in Infection Is Unrelated to Its Role in the Degradation of SP100 or PML.
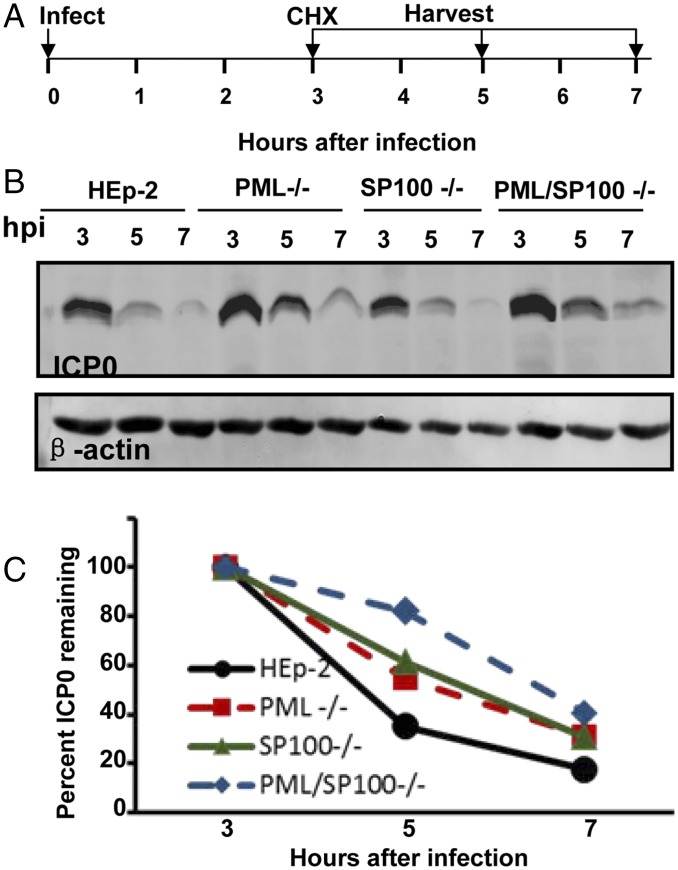
ICP0 functions as a ubiquitin ligase that in conjunction with ubiquitin-conjugating enzyme UbcH5a degrades PML and SP100 (11, 15, 43). A curious property of ICP0 is that the protein made early in infection coincident with the time course of degradation of SP100 and PML has a relatively short half-life, whereas that made at later times after infection is stable (60). The objective of the experiments reported herein was to test the hypothesis that the decay of ICP0 early in infection is related to its activity as an ubiquitin ligase in the degradation of PML or SP100. The design of these studies is illustrated schematically in Fig. 7A. In brief, replicate cultures of the four cell lines were exposed to 10 PFU of HSV-1(F) per cell for 2 h. At 3 hpi, the cells were rinsed and overlaid with medium containing cycloheximide (100 μg/mL). The cells were harvested at 3, 5, or 7 hpi, solubilized and processed as described above, and then immunoblotted with antibody against ICP0 and β-actin. Fig. 7B shows the accumulation of ICP0 in the four cell lines. Fig. 7C shows the amounts of ICP0 normalized with respect to loading control (β-actin) and the amount of ICP0 at 3 hpi (set as 100%). The results show that the rates of decay of ICP0 vary but that ultimately, by 7 hpi, the residual amounts of ICP0 are similar. The results do not support the hypothesis that the degradation of ICP0 is causally related to the degradation of PML, SP100, or both.
Fig. 7.
Degradation of ICP0 is not dependent on SP100 or PML. (A) Experimental design of the experiments. HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were exposed to 10 PFU of HSV-1(F) per cell. At 3 h after exposure to the virus, the cells were rinsed and incubated in medium containing cycloheximide (CHX) (100 μg/mL). Replicate infected cell cultures were harvested at 3, 5, or 7 hpi. (B) Electrohoretically separated lysates of the infected cells were immunoblotted with antibodies to ICP0 and β-actin. (C) Band intensity of ICP0 was normalized to its loading control (β-actin), and the degradation rate of ICP0 was plotted. The value of ICP0/β-actin of each cell line at 3 hpi was set as 100%.
Viral Replication Is Suppressed in SP100−/− Cells Pretreated with IFNβ.
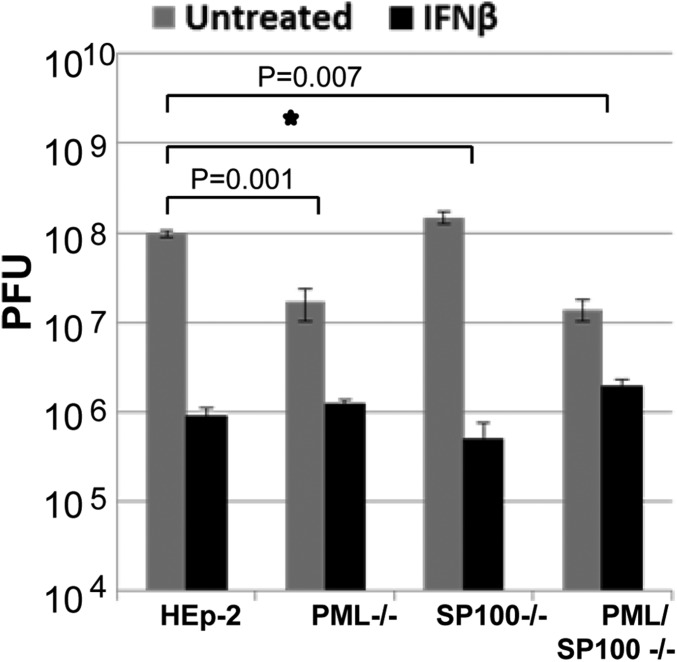
Previous studies have shown that PML is a potent effector of IFN in that it is induced by IFN and that the suppression of viral replication in PML−/− cells is significantly reduced compared with that observed in parental PML+/+ cells (5, 31, 32, 61). Because SP100 is independently induced by IFN, the question arose concerning its role as an effector of IFN. In these experiments, replicate cultures of HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were incubated for 24 h in medium only or medium containing IFN-β (1,000 U/mL), then rinsed and exposed to 0.1 PFU of HSV-1(F). Fig. 8 shows the virus yields from cells harvested at 24 hpi. As expected, the reduction in virus yields was significantly lower in IFN-β–pretreated PML−/− cells compared with parental HEp-2 cells. The reduction in virus yields from IFN-pretreated SP100−/− cells was not significantly different from that seen in parent HEp-2 cells. These results suggest a different role of SP100 as an IFN effector compared with PML.
Fig. 8.
Viral replication is suppressed in SP100−/− cells pretreated with IFN-β. HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were pretreated with IFN-β (1,000 U/mL) for 24 h and then exposed to 0.1 PFU of HSV-1(F). The cultures were harvested at 24 hpi, and virus yields were titered in Vero cells. The inhibitory effect of IFN-β treatment is represented by the fold reduction of virus titers in IFN-β–treated groups, and P values between HEp-2 and PML−/− cells or between HEp-2 and PML/SP100−/− cells are shown. The star indicates no statistic difference.
Discussion
PML and SP100 are key structural components of ND10 bodies (38). Both are induced by IFN, and both are subject to degradation by ICP0 in conjunction with UbcH5a (11, 15, 43). The studies described here and in our previous report (31) focused on the respective roles of PML and SP100 on the course of HSV replication. The salient features of the results may be summarized as follows.
Our data reported here show significantly lower levels of PML in cells depleted of SP100. The results obtained to date suggest that SP100 impacts positively on the synthesis of PML. Previous studies have shown that exposure of cells to IFN results in increased levels of both PML and SP100 (38). The results reported here show that exposure of cells to IFN enhances the accumulation of PML or SP100 independent of each other.
The fundamental questions posed in this report are the mechanisms by which PML and SP100 affect viral replication. In principle, to initiate the replicative cycle, the entering DNA must bind VP16, HCF1, and associated transcriptional factors to initiate the transcription of viral genes (62). To define the roles of PML and SP100, we examined the accumulation of viral transcripts starting at 1 hpi. The meaningful data are those collected while PML and SP100 are still abundant in infected cells. Additional arguments favoring the validity of this approach is that viral DNA synthesis begins as early as 3 hpi, and the earliest progeny can be detected by 6 hpi. Thus, infected cells could be expected to begin the accumulation of mRNAs of genes expressed both early and late after infection. The results shown in Fig. 6 suggest that PML and SP100 do not play a significant role in regulating the transcription of viral genes within the time frame of persistence of PML and SP100 in infected cells.
Previously published data suggest that concurrent with their dissolution the ND10 bodies evolve into replication compartments in which viral DNA is synthesized with the participation of a set of viral proteins that include ICP8 and packaged into capsids (9, 52, 53). In early stages, they appear as small dense bodies (Fig. 6A, cell 4) that ultimately grow in size and coalesce to partly fill the nucleus (Fig. 6A, cell 5). As reported in Results, even at high multiplicities of infection, only a fraction of the infected cells exhibited structures characteristic of dense bodies. In a large fraction of the infected cells, ICP8 was dispersed uniformly throughout the nucleus, suggesting that the synthesis of viral DNA and virus assembly was dispersed, was delayed, or had failed. The significant observation is that the number of replication compartments as defined above was highest in SP100−/−-infected cells. The relative number of the dense bodies containing ICP8 correlates with virus yields and suggests that SP100 plays a role in down-regulating the formation of replication compartments.
In our previous study, we found that the replication of HSV-1 is unaffected in PML−/− cells exposed to high ratios of virus per cell, whereas at low ratios of virus per cell, PML−/− cells produce significantly lower yields (31). In the present work, we reproduced our earlier results but also found higher viral yields in SP100−/− cells exposed to low ratios of virus per cell compared with parental HEp-2 cells.
A central issue explored in this study is the replication of ΔICP0 mutant in cells lacking PML, SP100, or both PML and SP100. Our results lead to two observations. First, ΔICP0 mutants replicate poorly in cells other than U2OS (44–46). Because one function of ICP0 is to mediate the degradation of PML and SP100, an obvious question is whether ΔICP0 virus replicates better in cells that do not express these proteins. Based on titrations in both the Vero and SP100 cell lines, the ΔICP0 mutant, like the WT parent, replicated to higher titers of SP100−/− cells. The results suggest that SP100 is a repressor of HSV replication and explain, at least in part, the evolution of viral functions that led to its degradation.
A puzzling observation reported earlier is that ICP0 made early in infection has a relatively short half-life, whereas that made after 6–8 hpi appears to be stable (60). A central question hitherto unresolved is whether the instability observed early in infection reflects the function of ICP0 as an E3 ligase involved in the degradation of PML and SP00. The results reported here indicate that the instability of ICP0 seen early in infection is not related to its function as an E3 ligase for the degradation of PML or SP100.
Finally, both PML and SP100 are independently induced by IFN-β and by definition are IFN-stimulated genes. The function of PML related to its function as an IFN effector is apparent from studies showing that IFN is less effective in suppressing viral replication in the absence of PML. This report shows that SP100−/− cells retain the sensitivity of the parental cells to IFN-β but exhibit enhanced formation of replication compartments and yield higher levels of both WT and ΔICP0 viruses.
Materials and Methods
Cells and Viruses.
The sources and maintenance of HEp-2, Vero, and U20S cells have been reported elsewhere (31). PML−/− cells, generated from HEp-2 cells using CRISPR/cas9 (31), were maintained in 5% FBS DMEM (Gibco) plus 2 μg/mL puromycin. HSV-1(F) is the prototype strain used in our laboratory, and ∆ICP0 mutant (R7910) was generated by deletion of both copies of ICP0 from HSV-1(F) (45). HSV-1(F) was amplified in HEp-2 cells, and ∆ICP0 mutant was propagated in U20S cells. For determination of virus yields, all infected cells were collected, resuspended in 2% milk, briefly sonicated to release all viral particles, and then titrated on Vero or U2OS monolayer cell cultures.
RNA Isolation and Real-Time PCR.
RNA extraction and assays were performed according to the manufacturer’s protocol (Thermo Fisher Scientific). mRNA levels of HSV-1(F) viral genes ICP27, VP16, and TK were measured using gene-specific primers as described previously (31). All targets were normalized to host internal RNA 18s (Ambion).
Generation of the SP100−/− Cell Line Using CRISPR/cas9.
Three sets of CRISPR/cas9 systems were designed to target the N-terminal exons (exons 3, 4, and 5) of the human SP100 gene: gRNA 5′-TTGTGATGAGATCACGATCA-3′ and gRNA 5′-GCAGCCTGTCATCTACACCC-3′ on exon 3, gRNA 5′-TTGTACACCACTCTCTGTAC-3′ and gRNA 5′-GTACAGAGAGTGGTGTACAA-3′ on exon 4, and gRNA 5′-CTCCAACTAAGTCTTGAACA-3′ and gRNA 5′-TACCTTGTTCAAGACTTAGT-3′ on exon 5. Generation of SP100−/− single-cell–derived lines were generated as described previously (31). In brief, parental HEp-2 or PML−/− cells were cotransfected with one set of gRNAs (two gRNAs) and their donor vectors containing a selection cassette expressing GFP-blasticidine flanked by sequences homologous to the target site of the gRNAs. Surviving cells under blasticidine selection (2–10 μg/mL) were serially diluted to form single-cell–derived colonies. The colonies were further screened for expression of SP100 by immunoblotting and immunofluorescence staining.
Antibodies and Drugs.
The antibodies used in this study included monoclonal anti-PML (Santa Cruz Biotechnology; sc-966), rabbit polyclonal anti-PML (Abcam; 179466), rabbit polyclonal anti-SP100 (H-60) (Santa Cruz Biotechnology; sc-25568), rabbit polyclonal anti-SP100 (Abcam; ab4315), mouse monoclonal anti-Daxx (Abcam; ab9091), mouse monoclonal anti–β-actin (Thermo Fisher Scientific; MA5-15739), rabbit polyclonal anti-ICP0 (laboratory stock), and mouse monoclonal anti-ICP8, -US11, and -ICP4 (laboratory stock). The drugs used in this study included blasticidine (Sigma-Aldrich; 15205-25MG), puromycin (Sigma-Aldrich; p8833), and cycloheximide (Sigma-Aldrich; C7698-5G).
Immunoblot Analyses.
At the indicated times, the cells were lysed, heat-denatured, electrophoretically separated in denaturing gels, transferred to PVDF membranes, and probed with the appropriate primary antibody as described previously (31). The membranes were then incubated with alkaline phosphatase-conjugated goat anti-mouse or goat anti-rabbit secondary antibody (Sigma-Aldrich) and visualized with BCIP/NBT Western Blotting Detection Reagent (GE Healthcare); to detect PML protein, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse (Sigma-Aldrich) and developed using ECL western blotting detection reagent (GE Healthcare).
ImageJ Quantification and Statistics.
ImageJ software was used to quantify band intensity. To quantify the relative PML protein level in HEp-2 and SP100−/− cells shown in Fig. 2B, the intensity of the major band of PML (marked by a rectangle) was normalized to the loading control (β-actin), and the relative protein amount of PML in SP100−/− cells was normalized to that in HEp-2 (set as 100) under each condition (untreated and treated with IFN-β). For measuring ICP0 protein stability in early infection, the ICP0 protein band intensity in Fig. 7B was quantified, normalized to β-actin, and normalized to the relative ICP0 level at 3 h postinfection (hpi) (set as 100%).
Statistics.
Error bars represent SE/SD of the mean. Two-tailed P values were calculated using a standard t test.
Immunofluorescence Staining and Confocal Microscopy.
In brief, cells in four-well slides were fixed and permealized, and then blocked and incubated with primary antibodies at 4 °C overnight. The next day, they were rinsed three times with blocking solution and then subjected to reaction with FITC-conjugated goat anti-mouse (Invitrogen) or/and Texas Red-conjugated goat anti-rabbit (Sigma-Aldrich) secondary antibodies. Images were obtained with a Leica SP5 II STED-CW super-resolution laser scanning confocal microscope.
Assessment of Replication Compartment Formation.
HEp-2, PML−/−, SP100−/−, and PML/SP100−/− cells were seeded onto microslides at 24 h before infection, then exposed to 20 PFU of HSV-1(F) per cell. At 6 hpi, the cells were fixed and reacted with anti-ICP8 and anti-ICP0 antibodies. On average, 20 different fields were photographed at random at either 40× or 60× magnification for each of the four cell lines. Approximately 200 ICP8-positive cells were counted for each cell line, and three independent countings were conducted for two independent experiments. The percentage of replication compartments forming cells out of the total number of ICP8-positive cells was calculated according to the classification scheme shown in Fig. 6A and normalized to that of HEp-2 cells. Error bars represent SE/SD of the mean.
Acknowledgments
We thank Lindsay Smith for invaluable assistance. This study was supported by a grant from the Joseph Regenstein Foundation.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 2.Dellaire G, Bazett-Jones DP. PML nuclear bodies: Dynamic sensors of DNA damage and cellular stress. BioEssays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 3.Ishov AM, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong S, et al. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 5.Scherer M, Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. 2016;90:5850–5854. doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang M, et al. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123:392–400. doi: 10.1242/jcs.053496. [DOI] [PubMed] [Google Scholar]
- 7.Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res. 2011;31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 9.Everett RD, Sourvinos G, Orr A. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J Virol. 2003;77:3680–3689. doi: 10.1128/JVI.77.6.3680-3689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79:5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100:8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu H, Zheng Y, Roizman B. Interaction of herpes simplex virus ICP0 with ND10 bodies: A sequential process of adhesion, fusion, and retention. J Virol. 2013;87:10244–10254. doi: 10.1128/JVI.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuchet-Lourenço D, Vanni E, Glass M, Orr A, Everett RD. Herpes simplex virus 1 ubiquitin ligase ICP0 interacts with PML isoform I and induces its SUMO-independent degradation. J Virol. 2012;86:11209–11222. doi: 10.1128/JVI.01145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett RD, et al. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelbi-Alix MK, de Thé H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 16.El McHichi B, et al. SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J Virol. 2010;84:11634–11645. doi: 10.1128/JVI.01321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly C, Van Driel R, Wilkinson GW. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 18.Ahn JH, Hayward GS. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szekely L, et al. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamson AL, Kenney S. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell P, Lieberman PM, Maul GG. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J Virol. 2000;74:11800–11810. doi: 10.1128/jvi.74.24.11800-11810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puvion-Dutilleul F, et al. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp Cell Res. 1995;218:9–16. doi: 10.1006/excr.1995.1125. [DOI] [PubMed] [Google Scholar]
- 23.Leppard KN, Everett RD. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J Gen Virol. 1999;80:997–1008. doi: 10.1099/0022-1317-80-4-997. [DOI] [PubMed] [Google Scholar]
- 24.Tang Q, Bell P, Tegtmeyer P, Maul GG. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol. 2000;74:9694–9700. doi: 10.1128/jvi.74.20.9694-9700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day PM, Baker CC, Lowy DR, Schiller JT. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci USA. 2004;101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bund T, et al. An L2 SUMO interacting motif is important for PML localization and infection of human papillomavirus type 16. Cell Microbiol. 2014;16:1179–1200. doi: 10.1111/cmi.12271. [DOI] [PubMed] [Google Scholar]
- 27.Stepp WH, Meyers JM, McBride AA. Sp100 provides intrinsic immunity against human papillomavirus infection. MBio. 2013;4:e00845–e13. doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Stadler M, et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- 30.Chelbi-Alix MK, et al. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033. [PubMed] [Google Scholar]
- 31.Xu P, Mallon S, Roizman B. PML plays both inimical and beneficial roles in HSV-1 replication. Proc Natl Acad Sci USA. 2016;113:E3022–E3028. doi: 10.1073/pnas.1605513113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkham J, Coen DM, Weller SK. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uprichard SL, Knipe DM. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 35.Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 37.Maroui MA, et al. Latency entry of herpes simplex virus 1 is determined by the interaction of its genome with the nuclear environment. PLoS Pathog. 2016;12:e1005834. doi: 10.1371/journal.ppat.1005834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grötzinger T, Sternsdorf T, Jensen K, Will H. Interferon-modulated expression of genes encoding the nuclear dot-associated proteins Sp100 and promyelocytic leukemia protein (PML) Eur J Biochem. 1996;238:554–560. doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- 39.Negorev DG, Vladimirova OV, Ivanov A, Rauscher F, 3rd, Maul GG. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J Virol. 2006;80:8019–8029. doi: 10.1128/JVI.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci USA. 2010;107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu H, Roizman B. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J Virol. 2009;83:181–187. doi: 10.1128/JVI.01940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boutell C, et al. A viral ubiquitin ligase has substrate preferential SUMO-targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011;7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao F, Schaffer PA. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sacks WR, Schaffer PA. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berscheminski J, Groitl P, Dobner T, Wimmer P, Schreiner S. The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription. J Virol. 2013;87:965–977. doi: 10.1128/JVI.02023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Catez F, et al. HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons. PLoS Pathog. 2012;8:e1002852. doi: 10.1371/journal.ppat.1002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutrieux J, et al. PML/TRIM19-dependent inhibition of retroviral reverse-transcription by Daxx. PLoS Pathog. 2015;11:e1005280. doi: 10.1371/journal.ppat.1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Negorev DG, Vladimirova OV, Maul GG. Differential functions of interferon-upregulated Sp100 isoforms: Herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J Virol. 2009;83:5168–5180. doi: 10.1128/JVI.02083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor JL, Unverrich D, O’Brien WJ, Wilcox KW. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J Interferon Cytokine Res. 2000;20:805–815. doi: 10.1089/10799900050151076. [DOI] [PubMed] [Google Scholar]
- 52.Taylor TJ, McNamee EE, Day C, Knipe DM. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology. 2003;309:232–247. doi: 10.1016/s0042-6822(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 53.Burkham J, Coen DM, Hwang CB, Weller SK. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J Virol. 2001;75:2353–2367. doi: 10.1128/JVI.75.5.2353-2367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darwish AS, Grady LM, Bai P, Weller SK. ICP8 filament formation is essential for replication compartment formation during herpes simplex virus infection. J Virol. 2015;90:2561–2570. doi: 10.1128/JVI.02854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bryant KF, Yan Z, Dreyfus DH, Knipe DM. Identification of a divalent metal cation binding site in herpes simplex virus 1 (HSV-1) ICP8 required for HSV replication. J Virol. 2012;86:6825–6834. doi: 10.1128/JVI.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boehmer PE, Craigie MC, Stow ND, Lehman IR. Association of origin binding protein and single-strand DNA-binding protein, ICP8, during herpes simplex virus type 1 DNA replication in vivo. J Biol Chem. 1994;269:29329–29334. [PubMed] [Google Scholar]
- 57.de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA-binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 58.Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 59.Lukonis CJ, Burkham J, Weller SK. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: Only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Z, Du T, Zhou G, Roizman B. The stability of herpes simplex virus 1 ICP0 early after infection is defined by the RING finger and the UL13 protein kinase. J Virol. 2014;88:5437–5443. doi: 10.1128/JVI.00542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 62.Fields BN, Knipe DM, Howley PM. 2013. Fields Virology (Lippincott Williams & Wilkins, Philadelphia, PA)