Abstract
Histone modifications influence gene expression in complex ways. The RNA interference (RNAi) machinery can repress transcription by recruiting histone-modifying enzymes to chromatin, although it is not clear whether this is a general mechanism for gene silencing or whether it requires repeated sequences such as long terminal repeats (LTRs). We analyzed the global effects of the Clr3 and Clr6 histone deacetylases, the Clr4 methyltransferase, the zinc finger protein Clr1, and the RNAi proteins Dicer, RdRP, and Argonaute on the transcriptome of Schizosaccharomyces pombe (fission yeast). The clr mutants derepressed similar subsets of genes, many of which also became transcriptionally activated in cells that were exposed to environmental stresses such as nitrogen starvation. Many genes that were repressed by the Clr proteins clustered in extended regions close to the telomeres. Surprisingly few genes were repressed by both the silencing and RNAi machineries, with transcripts from centromeric repeats and Tf2 retrotransposons being notable exceptions. We found no correlation between repression by RNAi and proximity to LTRs, and the wtf family of repeated sequences seems to be repressed by histone deacetylation independent of RNAi. Our data indicate that the RNAi and Clr proteins show only a limited functional overlap and that the Clr proteins play more global roles in gene silencing.
Histone-modifying enzymes alter the chemical and structural properties of nucleosomes, thus influencing gene expression, DNA repair, recombination, and the general organization of chromosomes. Two particularly widespread modifications are the acetylation and methylation of lysine residues, which can occur in a variety of patterns leading to the formation of distinct types of chromatin (for reviews, see references 45 and 47).
The Schizosaccharomyces pombe (fission yeast) Clr3, Clr6, and Clr4 enzymes possess histone deacetylase (Clr3 and Clr6 [8]) and histone methyltransferase (Clr4 [30]) activities. Their genes were identified in searches for mutants that were defective in transcriptional silencing (11, 13, 39). The three proteins facilitate heterochromatin formation at centromeres, telomeres, and the mating-type region, and Clr3 and Clr4 mediate the silencing of PolII-transcribed marker genes inserted in the rRNA gene repeats (1, 8, 11, 13, 39, 42). Their mode of action and the subsequent events leading to heterochromatin formation are understood in some detail. Clr3 belongs to the class II family of histone deacetylases, like the Saccharomyces cerevisiae Hda1 enzyme, and appears to specifically deacetylate lysine 14 of histone H3 (8). Clr6 belongs to the class I family of histone deacetylases, like the S. cerevisiae Rpd3 enzyme, and displays a broader substrate specificity than Clr3 (8). Clr4 is a member of the Su(var)3-9 family of methyltransferases, whose catalytic activity resides in a so-called SET domain (30). Like other members of this family, Clr4 methylates lysine 9 of histone H3, creating a binding site for the chromodomain protein Swi6 (26, 30).
Marker genes are repressed when they are introduced into any of the characterized heterochromatic regions of S. pombe. However, the influence of heterochromatic histone modifications on the transcription of endogenous genes is undocumented, with the exception of genes located in the mating-type region. In that region, heterochromatin represses transcription of the mat2-P and mat3-M loci, preventing an aberrant form of meiosis that leads to cell death (41).
Little is known about the DNA sequences or events that initiate heterochromatin formation. One pathway of transcriptional silencing involving components of the RNA interference (RNAi) machinery was recently discovered (31, 48). An RNase III helicase (Dcr1), an RNA-dependent RNA polymerase (Rdr1), and a protein of the Argonaute family (Ago1) act in that pathway. These proteins mediate the formation of small interfering RNAs (siRNAs) originating from centromeric repeats and are required for heterochromatin formation, suggesting that siRNAs recruit Clr3, Clr6, and/or Clr4 to centromeric regions. In RNAi-deficient cells, centromeres do not function optimally due to defects in their heterochromatic structure, and such cells therefore lose chromosomes at a relatively high rate (29, 49).
Although it is critical to centromeric heterochromatin, the RNAi pathway is not essential for silencing in the mating-type region (14, 48). The establishment of silencing in that region has been proposed to be slower in RNAi mutants than in wild-type strains based on the observation that silencing is not established for several generations when a clr4 mutation is crossed out of a strain in an RNAi-defective background (14). However, a long phenotypic gap was also observed when a clr4 mutation was crossed out in a wild-type background (G. Thon, unpublished observations), raising the possibility that the contribution of RNAi to the establishment of silencing in the mating-type region is not as strong as originally perceived. In the absence of RNAi, silencing is eventually established to a level that is indistinguishable from that of the wild type and is maintained stably thereafter (14), indicating that pathways other than RNAi can recruit silencing factors and histone-modifying enzymes to the mating-type region in a manner that is possibly redundant with RNAi. cis-acting elements whose silencing properties were identified by chromosomal deletion analysis are likely to participate in this recruitment (3, 39, 40). In addition, trans-acting factors other than those mentioned above may play a role in the process. One such silencing factor is Clr1, a zinc finger protein (41; G. Thon and A. Klar, unpublished observation). Silencing near centromeres and telomeres, in the mating-type region, and in rRNA gene clusters is reduced to similar levels in clr1 and clr3 mutants (1, 11, 13, 8, 40; Thon, unpublished observations). Furthermore, mutations in either clr1 or clr3, with the exclusion of mutations in any other known silencing factor, have a strong synergistic effect with a mutated clr6 allele or with a deletion of the IR-R boundary element in the mating-type region (13, 35). These similarities suggest that Clr1 and Clr3 act in concert for gene silencing.
We determined the patterns of global gene expression in clr1, clr3, clr4, and clr6 mutants to test whether genes outside of the known regions are regulated by these factors. We also examined the gene expression profiles of dcr1, rdp1, and ago1 mutants to compare and contrast gene regulation by the RNAi machinery and that by silencing factors. The comparison revealed some overlap but also showed intriguing differences between the functions of the silencing and RNAi machineries in fission yeast.
MATERIALS AND METHODS
Strains.
The following published S. pombe alleles were used: clr1-5 (41), clr3-735 and clr4-681 (40), clr6-1 (13), clr4Δ (18), and dcr1Δ, ago1Δ, and rdp1Δ (48). A kanR replacement of the clr3 open reading frame (ORF), clr3Δ::kanR, was created with the pFA6a-kanMX6 plasmid (5) by cloning PCR products containing regions upstream and downstream of the clr3 ORF next to the kanR gene; the oligonucleotides GGCAGCTGTCGACAAGGGTGAATGTTTCCGGG and GGAGATCTCCCGGGTCGTTTTCTATCGCCC were used to amplify the clr3 upstream region, and the oligonucleotides GGGAGCTCGAATTCGTCACACATTTAACCTCC and GGCCGCGGACTAGTTTCTACCAGTTGCTAACACC were used to amplify the clr3 downstream region. The first 2 kb of the clr1 ORF were replaced with the S. pombe ura4 gene to create clr1Δ::ura4; the oligonucleotides GGGGTACCGCTAGCCAAAGTTATATGATGGAGGAGG and CCATCGATACCGGTATCTAACAACGAATTGCATGC were used to amplify a DNA fragment immediately upstream of the clr1 ORF which was cloned to one side of the ura4 gene in Bluescript SKII(−) (Stratagene), and a 2-kb BamHI-PstI fragment containing the clr1 downstream region was cloned at the other side of ura4. Correct chromosomal integration of the clr3Δ::kanR and clr1Δ::ura4 constructs was verified by Southern blotting. To avoid secondary effects due to the derepression of mat2-P and mat3-M in silencing mutants, we used strains that contained solely M mating-type information, as follows: SPK10 (h−S), SPK11 (h−S clr1-5), SPK13 (h−S clr3-735), SPK14 (h−S clr4-681), SPK16 (h−S clr1Δ::ura4 ura4-D18), SPK19 (h−S clr3Δ::kanR), SPK20 (h−S clr4Δ::ura4 ura4-D18), SPK28 (h−S clr6-1), and SPK27 (h−S clr6-1 clr3Δ::kanR).
RNA extraction, labeling, and reverse transcription-PCR (RT-PCR).
RNAs were extracted and labeled as described previously (23; http://www.sanger.ac.uk/PostGenomics/S_pombe/). Briefly, overnight cultures of wild-type and mutant cells were diluted approximately 60-fold, to an optical density at 600 nm of 0.06 to 0.09, in 50 ml of EMM2 and then were propagated to an optical density of 0.3 to 0.5 (approximately 8 × 106 cells/ml). The cells were harvested by a 5-min centrifugation at 3,000 rpm in a Heraeus rotor 8155 and were immediately frozen in liquid N2. Total RNAs were extracted according to a hot phenol protocol. Ten to 20 μg of RNA was reverse transcribed into labeled cDNA with Superscript II (Invitrogen), Cy3- or Cy5-dCTP, and oligo(dT) as a primer.
RT-PCRs were conducted as described by Volpe et al. (48) by use of a one-step RT-PCR kit (Qiagen) and the following conditions in an Applied Biosystems Gene Amp PCR system 2700: 30 min at 50°C; 15 min at 95°C; 27 cycles of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C; and a final step of 10 min at 72°C. Serial dilutions of RNA preparations spanning a range of 25 to 100 ng per reaction were used as templates, and 1 μCi of [α-32P]dCTP (3,000 Ci/mmol) was added to each reaction to allow for quantification of the PCR products with a PhosphorImager and the ImageQuant software package (Molecular Dynamics).
Hybridization to microarrays, data acquisition, and quality control.
We used DNA microarrays displaying probes for >99% of all known and predicted genes of S. pombe spotted in duplicate onto glass slides, performing hybridization and initial data processing as previously described (23). Four independent biological experiments with RNA preparations and subsequent hybridizations were performed for the clr1-5 and clr3-735 mutants, three were performed for the clr4-681 mutant, and two were performed for the Δclr1, Δclr3, Δclr4, clr6-1, clr6-1 Δclr3, Δago1, Δdcr1, and Δrdp1 mutants. Dye swaps were performed for each strain to exclude dye-specific artifacts. The normalized data were analyzed with GeneSpring software (Silicon Genetics) and Excel (Microsoft). An average mutant/wild type ratio was calculated for each gene. Relatively low cutoff values (1.5-fold) were used for comparisons between mutants and for subsequent analyses. Although low cutoff values can increase the occurrence of false-positive results in the lists of selected genes, they seemed preferable here because of the high reproducibility between independently performed experiments and an emerging coherent picture of the subtle changes in gene expression caused by the silencing mutants. Gene annotations were downloaded from S. pombe GeneDB (http://www.genedb.org/). The entire processed data sets and supplemental tables (Tables S1 to S10) are available in the supplemental material.
Comparison with expression profiles of S. cerevisiae.
A list of S. pombe and S. cerevisiae homologs provided by Valerie Wood (Sanger Center, Cambridge, United Kingdom) was used to compare the list of genes affected by Clr3 (or Clr6) in S. pombe with those affected by Hda1 (or Rpd3) in S. cerevisiae (7). These lists are presented in the supplemental material (Table S6). A comparison of the genes repressed by the histone class II deacetylase Clr3 in S. pombe with those repressed by the histone class II deacetylase Hda1 in S. cerevisiae is shown in Table S7 in the supplemental material. An annotated list of S. pombe genes repressed by the histone class II deacetylase Clr3 that have an S. cerevisiae homolog repressed by the histone class II deacetylase Hda1 is shown in Table S9 in the supplemental material. A comparison of the genes repressed by the histone class I deacetylase Clr6 in S. pombe with those repressed by the histone class I deacetylase Rpd3 in S. cerevisiae is shown in Table S10 in the supplemental material. An annotated list of S. pombe genes repressed by the histone class I deacetylase Clr6 that have an S. cerevisiae homolog repressed by the histone class I deacetylase Rpd3 is also available in the supplemental material. P values for Venn diagram overlaps were calculated with kzr.cmd software (IBM).
RESULTS
Gene expression patterns in cells with mutations in silencing and RNAi factors.
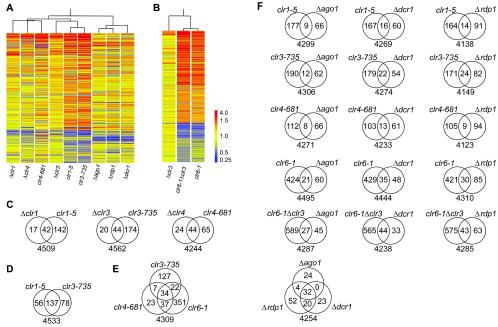
We compared the RNA expression profiles of S. pombe cells with mutations in silencing or RNAi factors with the expression profiles of wild-type cells. The comparison was performed by hybridization to microarrays displaying >99% of all known and predicted ORFs and a few noncoding sequences. Strains containing either a point mutation (clr1-5, clr3-735, clr4-681, or clr6-1) or a null allele (Δclr1, Δclr3, Δclr4, Δago1, Δdcr1, or Δrdp1) and a double mutant (clr6-1 Δclr3) were examined. A global picture of the relationship between the mutants is presented in Fig. 1. Figure 1A presents all strains except those containing the clr6-1 allele. The clr6-1 mutation, which affected a much larger number of genes than the other mutations, is displayed in a separate panel (Fig. 1B). Two main branches were observed in a cluster tree of the expression profiles, one comprising the silencing factor mutants and one comprising the three RNAi factor mutants. The numbers and overlaps of affected genes are shown in Fig. 1C to F. P values reflecting the similarities between gene lists are presented in Table 1.
FIG. 1.
Relationship between transcriptomes of silencing and RNAi factor mutants. (A and B) Spearman correlation coefficients between data sets were used to hierarchically cluster the genes. Genes whose expression was regulated ≥1.5-fold in at least two of the nine mutants (A) or in at least one of the three mutants (B) displayed in each panel are represented by horizontal bars. Two hundred sixty-two genes are displayed in panel A, and 962 genes are displayed in panel B. (C to F) The numbers of genes that were upregulated ≥1.5-fold in the indicated mutants are presented in Venn diagrams. The total number of genes considered for each comparison is indicated below the diagrams. This number corresponds to genes for which an expression value was obtained for all mutants presented in a specific diagram.
TABLE 1.
Comparison of silencing and RNAi mutant expression profiles
| Mutant | Probability of similar expression profilea
|
|||||
|---|---|---|---|---|---|---|
| clr3-735 | clr4-681 | clr6-1 | Δago1 | Δdcr1 | Δrdp1 | |
| clr1-5 | <1 × 10−40 | 3.6 × 10−16 | 2.4 × 10−8 | >0.05 | 3.2 × 10−7 | >0.05 |
| clr3-735 | 1.6 × 10−29 | 2.8 × 10−11 | >0.05 | 2.5 × 10−5 | 4.4 × 10−4 | |
| clr4-681 | <1 × 10−40 | >0.05 | 3.7 × 10−5 | >0.05 | ||
| clr6-1 | 5.8 × 10−3 | 1.8 × 10−9 | 6.8 × 10−5 | |||
| Δago1 | 2.9 × 10−40 | 1.2 × 10−32 | ||||
| Δdcr1 | <1 × 10−40 | |||||
The data reported are the probabilities that similarities between the lists of genes that were upregulated ≥1.5-fold in the indicated mutants occurred by chance.
The gene expression patterns in the clr1, clr3, and clr4 mutants were highly correlated, suggesting that Clr1, Clr3, and Clr4 act together in many instances. These factors tend to repress transcription, since most of the affected genes were upregulated in the mutant strains. Twenty-six genes were commonly upregulated ≥1.5-fold in the clr1-5, clr3-735, and clr4-681 mutants, and 18 were upregulated in the Δclr1, Δclr3, and Δclr4 mutants. The clr1-5, clr3-735, and clr4-681 point mutations affected larger numbers of genes than the corresponding deletions (Fig. 1C). Point mutations can have a stronger penetrance than deletions when the mutated protein physically blocks a redundant activity. Consistent with this, the clr1-5, clr3-735, and clr4-681 mutations are codominant with the wild-type alleles (40), and the clr4-681 mutant encodes an enzyme with a reduced activity (26). A particularly high similarity was observed for the expression profiles of the clr1-5 and clr3-735 mutants (Fig. 1D; Table 1). Δclr1 and Δclr3 cells also had highly similar expression profiles, and the lists of genes that were upregulated ≥1.5-fold in either mutant relative to the wild type were correlated, with a P value of <10−40.
The three RNAi mutants formed a subcluster that was separate from the clr mutants (Fig. 1A). Accordingly, Δago1, Δrpd1, and Δdcr1 cells displayed related expression patterns (Table 1), with 26 genes being upregulated and 16 being downregulated ≥1.5-fold in all three mutants. Although these expression patterns differed from those of the clr mutants, some correlations between gene lists were observed, most notably between the genes regulated by the Clr proteins and those regulated by Dcr1 (Table 1). The expression of 39 genes was increased ≥1.5-fold in both an RNAi mutant and a clr1, clr3, or clr4 mutant. Among the transcripts with increased abundance in both types of mutants were those originating from centromeric regions; moreover, the expression of Tf2 retrotransposons increased in RNAi and clr6 mutants (see below).
The expression profiles of the two strains containing the clr6-1 allele (clr6-1 and clr6-1 Δclr3) appeared strikingly different from those of the other mutants because a much larger part of the genome was affected (Fig. 1B). However, positive correlations between the lists of genes regulated by Clr6 and those of genes regulated by Clr1, Clr3, or Clr4 were observed (Table 1). The clr6-1 allele, like the other clr mutations, led mostly to an increase in the RNA levels of the affected genes, indicating that Clr6 generally represses transcription.
Finally, a large overlap was observed between the lists of genes that were regulated by histone methylation and those that were regulated by histone deacetylation (Fig. 1E). Five of the 23 genes that were upregulated ≥1.5-fold in clr4-681 but not in clr3-735 or clr6-1 cells were upregulated ≥1.5-fold in the clr6-1 Δclr3 double mutant. Hence, the silencing of nearly all genes that are regulated by the Clr4 methylase also depends on deacetylation, while many genes that are regulated by deacetylation appear to be independent of Clr4.
Chromosomal location of genes that are repressed by Clr proteins.
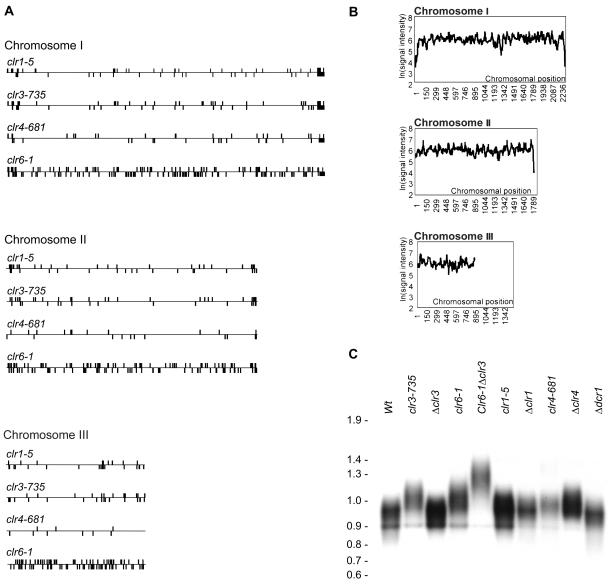
Many of the genes whose mRNA levels increased in the clr1, clr3, clr4, or clr6 mutant were located in subtelomeric regions, with their clustering in these regions being higher than expected from a random distribution (Fig. 2A). In addition, the absolute signal intensities for these genes in wild-type strains were low, as expected for silenced genes (Fig. 2B). Large rRNA gene clusters are adjacent to the telomeric repeats of chromosome III that are not present on the microarrays. Accordingly, we could not observe silencing or derepression of chromosome III telomeric genes.
FIG. 2.
Chromosomal location of affected genes and telomere length. (A) The three S. pombe chromosomes are represented by horizontal bars. Genes whose expression was upregulated ≥1.5-fold in the indicated mutants are represented by vertical bars above or below the chromosomes depending on their direction of transcription. (B) The median microarray signal intensities within a running window of 15 neighboring genes were plotted as a function of their chromosomal locations. Absolute microarray signal intensities were measured after the hybridization of samples of RNA isolated from vegetatively growing wild-type cells, providing an approximation of gene expression levels. (C) DNAs extracted from the indicated strains were digested with EcoRI and hybridized to a probe made with the 300-bp ApaI-EcoRI fragment from pNSU70, which consists almost exclusively of telomeric repeats (37).
Increased transcription of subtelomeric regions is observed during nitrogen starvation (24), a natural prelude to meiosis in S. pombe. This raised the possibility that the Clr proteins repress genes that are expressed during nitrogen starvation, which was indeed the case (see below). The effects on subtelomeric gene expression also suggested that silencing factors perform a structural role at the end of chromosomes, e.g., in the stabilization of specialized protein complexes associated with these regions. We therefore measured the length of telomeric repeats in vegetatively growing silencing and RNAi factor mutant cells and found that several mutants had elongated telomeres (Fig. 2C). The most extreme effect was observed for the clr6-1 Δclr3 double mutant, whose telomeric repeats were approximately 400 bp longer than those of wild-type cells. In contrast, Δdcr1 cells had telomeres of the same length as the wild type, indicating that silencing factors, but not the RNAi machinery, contribute to the telomere structure.
Centromeric transcripts.
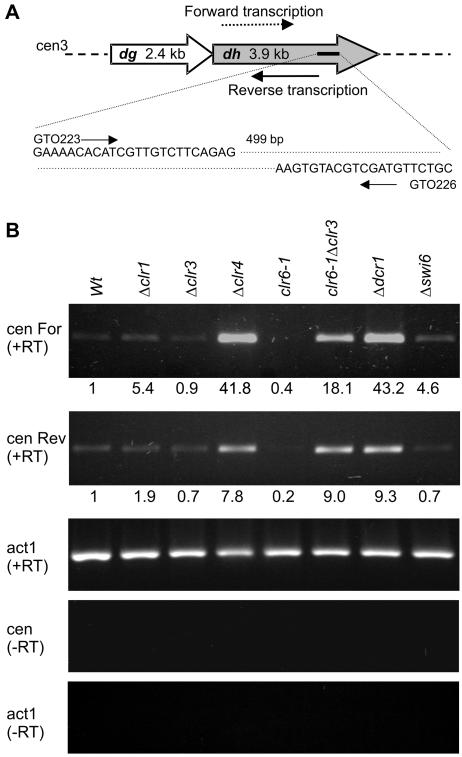
Ago1, Rdp1, and Dcr1 influence transcription of the S. pombe centromeric dg and dh repeats, possibly leading to the formation of small double-stranded RNAs (31, 48). Complementary transcripts originating from the dh repeats are referred to as forward and reverse transcripts (Fig. 3A). A current model proposes that the transcription of centromeric reverse transcripts occurs at a very low level in wild-type S. pombe cells, while the transcription of forward transcripts is repressed by silencing factors (48). An occasional failure in silencing would lead to an accumulation of forward transcripts and the formation of double-stranded RNAs which, after processing by the RNAi machinery, could reestablish silencing.
FIG. 3.
Centromeric transcription. (A) Locations and sequences of primers used for RT-PCRs to detect forward or reverse dh transcripts. (B) RT-PCRs were performed with RNAs from wild-type cells or from the indicated mutants. First-strand cDNA synthesis was primed with GTO226, allowing the detection of forward transcripts [cenFor (+RT)], or with GTO223, allowing the detection of reverse transcripts [cenRev (+RT)]. Both primers were subsequently used for PCR amplification. act1 (+RT), control in which the actin transcript was reverse transcribed and amplified. RT-PCR controls lacking reverse transcription are also displayed [cen (−RT) and act1 (−RT)]. Each reported value represents the average of two PhosphorImager measurements of a mutant/wild-type ratio normalized to act1 (+RT).
As expected, the microarrays revealed an accumulation of centromeric transcripts in the ago1, rdp1, and dcr1 mutants. In addition, high levels of centromeric transcripts were detected in the clr4 and clr6-1 Δclr3 mutants. Consistent with the observations of Volpe et al. (48), the signal intensities for all centromeric transcripts were low in wild-type cells. We further checked for the presence of forward and reverse dh transcripts by using RT-PCR. The results obtained with the silencing mutants are presented in Fig. 3B, and Δdcr1 and Δswi6 cells were processed in parallel for comparison. The dh transcript levels in Δclr4 or clr6-1 Δclr3 cells were in the same range as the levels in Δdcr1 cells. We found approximately twofold more dh transcript in the Δclr4 mutant than in the clr6-1 Δclr3 double mutant when measuring transcript levels by RT-PCR, whereas the opposite ratio was observed for microarray measurements. This difference might reflect the fact that cDNA synthesis was primed differently for the microarray [oligo(dT) priming] and for RT-PCR (sequence-specific primer). However, this difference does not impinge on the conclusion that Clr4 and the histone deacetylases Clr3 and Clr6 significantly limit the abundance of centromeric dh transcripts in wild-type cells, with Clr3 and Clr6 acting in an overlapping fashion.
The Clr3 and Clr6 histone deacetylases are partially redundant.
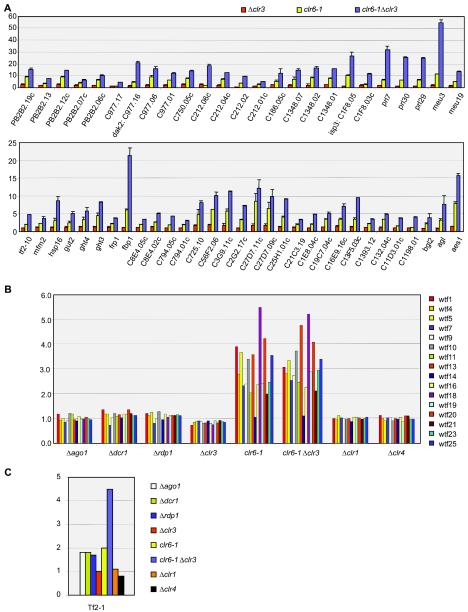
The Clr6 histone deacetylase acts redundantly with Clr3 to repress transcription in the mating-type and centromeric regions. We tested whether this might be more generally the case by comparing transcript levels in Δclr3, clr6-1, and clr6-1 Δclr3 cells. In many instances, the clr6-1 Δclr3 double mutant displayed a more pronounced phenotype than either single mutant (Table 2). Examples of genes for which a cumulative or synergistic effect was observed are displayed in Fig. 4A. These included (i) telomeric and subtelomeric genes whose expression was three- to fourfold higher in the clr6-1 mutant than in the Δclr3 mutant and increased an additional twofold in the double mutant; (ii) Tf2 retrotransposable elements which were upregulated in clr6-1 and clr6-1 Δclr3 mutants (∼2.0- and ∼4.5-fold, respectively) but not in the clr1, clr3, or clr4 mutant (see below); and (iii) several RNAs that were not associated with an ORF (prl7, prl29, prl30, meu3, and meu19 [50, 51]) (Fig. 4A). The most extreme cumulative effect was observed for the meu3 RNA (meu3), which was upregulated ∼5- and ∼28-fold in the clr6-1 Δclr3 mutant compared to the clr6-1 or Δclr3 mutant, respectively. Synergistic effects between clr6-1 and Δclr3 were also observed for telomere length (Fig. 2C) and in response to salt stress (see below).
TABLE 2.
Upregulated genes in histone deacetylase mutants
| Mutant | No. of genes whose expression increased by the indicated factor
|
||
|---|---|---|---|
| 1.5 | 5 | 10 | |
| Δclr3 | 66 | 3 | 1 |
| clr6-1 | 448 | 44 | 7 |
| clr6-1 Δclr3 | 592 | 84 | 41 |
FIG. 4.
Effects of histone deacetylation and RNAi on selected genomic elements. (A) The Clr3 and Clr6 histone deacetylases redundantly repress telomeric genes and noncoding RNAs (top) and nontelomeric genes (bottom). The genes displayed were selected for having a clr6-1 Δclr3/Δclr3 ratio of ≥3 and a clr6-1 Δclr3/clr6-1 ratio of ≥1.4. (B) Mutant/wild-type expression ratios of 16 wtf elements spotted on our arrays were plotted for the indicated mutants. Nomenclature is used as described previously (9). wtf1, SPAC2E12.05; wtf4, SPCC548.03c; wtf5,SPCC794.02; wtf7, SPCC736.05; wtf9, SPCC970.11c; wtf10, SPCC1183.10; wtf11, SPCC1281.08 (meu24); wtf13, SPCC162.04c; wtf14, SPCC663.02; wtf16, SPCC1450.08c; wtf18, SPCC285.07C; wtf19, SPCC1906.03; wtf20, SPCC1906.04; wtf21, SPCC1739.15; wtf23, SPCC1620.02; wtf25, SPCC1919.06c. (C) RNAi and silencing factor mutations affect Tf2 transcript levels differently. Values obtained with a single representative element, Tf2-1, are displayed. Additional elements represented on the microarrays gave highly similar hybridization patterns, but they are not shown here because of the high probability of cross-hybridization.
Histone modifications and stress response.
Histone-modifying enzymes might be expected to regulate a wide variety of genes. Consistent with this, we found that genes involved in various biological processes were aberrantly expressed in the mutants examined here. However, a few classes of genes were particularly affected in some mutants, pointing to specific biological functions.
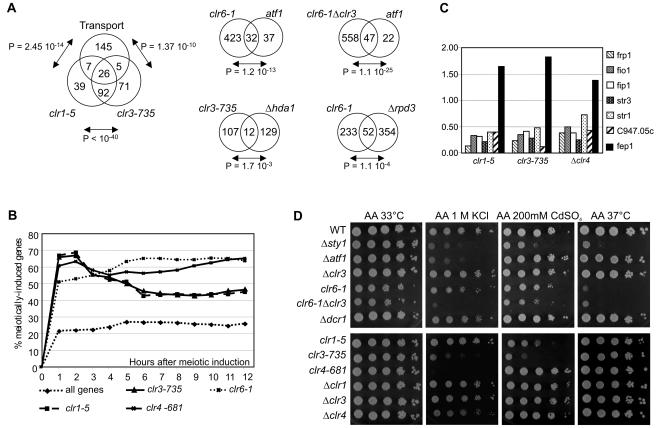
Genes encoding membrane proteins with known or predicted functions in transport were preferentially affected in clr1 or clr3 mutants, with their representation in lists of genes that were upregulated ≥1.5-fold in these mutants being considerably higher than expected from a random distribution (Fig. 5A; also see Table S5 in the supplemental material). An abundance of metabolic enzymes was also found among the upregulated genes (see Table S3 in the supplemental material). A similar pattern of effects has been described for S. cerevisiae cells lacking HDA1, the ortholog of clr3 (7), and Hda1 preferentially deacetylates the promoter regions of genes involved in carbohydrate or drug transport (32). This prompted us to compare the changes in expression profiles for S. pombe clr3 mutant cells with those for S. cerevisiae hda1 mutant cells (7) (Fig. 5A; also see Tables S6 and S7 in the supplemental material). S. cerevisiae homologs were identified for 120 genes that were upregulated ≥1.5-fold in the clr3-735 mutant. Among those, only 12 were upregulated ≥1.5-fold in Δhda1 cells, indicating a large degree of divergence between Hda1- and Clr3-regulated genes. However, 6 of these 12 genes encoded predicted or known transporters, and 1 encoded a membrane protein.
FIG. 5.
Biological functions of Clr-regulated genes and stress sensitivity of clr mutants. (A) Lists of genes that were induced ≥1.5-fold in the indicated clr mutants were compared with S. pombe genes encoding predicted or known transporters, S. pombe genes regulated by the Atf1 transcription factor, and S. cerevisiae genes whose expression was increased ≥1.5-fold in Δhda1 or Δrpd3 cells. A table linking 3,187 S. pombe genes to their 3,584 S. cerevisiae sequence homologs was used for these comparisons. (B) clr-regulated genes are defined here as genes whose expression was upregulated ≥1.5-fold in a clr mutant compared with the wild type. The data displayed are the percentages of genes whose expression increased ≥1.5-fold at the indicated meiotic time points among all genes probed on the arrays (diamonds), clr1-regulated genes (clr1-5; squares), clr3-regulated genes (clr3-735; triangles), clr4-regulated genes (clr4-681; crosses), or clr6-regulated genes (clr6-1; stars). Meiotic data are for nitrogen starvation-induced meiosis (24). (C) Mutant versus wild-type expression ratios of six genes encoding iron-siderophore transport components (frp1, fio1, fip1, str1, str3, and C947.05c) and the gene encoding their transcriptional repressor (fep1). (D) Tenfold serial dilutions of cell suspensions were spotted on the indicated plates.
The expression of transporters and metabolic enzymes is generally regulated as a function of the environmental conditions in which cells are placed. We examined whether Clr1, Clr3, or any of the other factors examined here influenced the expression of genes that are known to respond to external stimuli. Genome-wide analyses of gene expression have been performed by the use of microarrays with fission yeast that have been subjected to nitrogen starvation, which induces meiosis (24), or to various environmental stresses (10).
We observed that 60 to 67% of the genes that were repressed by Clr1, Clr3, or Clr4 and 50% of the genes that were repressed by Clr6 were upregulated ≥1.5-fold in wild-type cells after 1 h of nitrogen starvation (Fig. 5B). Clr6-regulated genes, and to a lesser extent, Clr4-regulated genes also tended to be expressed later during meiotic differentiation, with approximately 65% of the Clr6-regulated genes and 55 to 65% of the Clr4-regulated genes being upregulated ≥1.5-fold 6 to 12 h after the induction of meiosis. Mata et al. (24) observed that a high density of nitrogen starvation-responsive genes is found near telomeres. This raised the possibility that the overrepresentation of nitrogen starvation-induced genes among Clr-regulated genes simply reflected the chromosomal localization bias of Clr-regulated genes. However, this was not the case, as the overrepresentation was still observed after the elimination of all genes located within 80 kb of the ends of chromosomes 1 and 2 from the lists of Clr-regulated genes (data not shown). We concluded that the Clr factors are involved in repressing genes that are activated during nitrogen starvation and meiotic differentiation.
The Atf1 transcription factor is a central mediator of the S. pombe stress response (reviewed in reference 43), and the expression of 69 genes increases twofold or more under various stresses in atf1+ cells, but not in cells with an atf1 deletion (10). A remarkable overlap was observed between these 69 genes and those regulated by the Clr6 histone deacetylase (Fig. 5A; also see Table S8 in the supplemental material). An even larger proportion of Atf1-activated genes might conceivably be controlled by histone deacetylation if this modification affects the expression of certain genes only under stress, which we did not assay.
We compared genes that were affected by Clr6 with those affected by its S. cerevisiae ortholog, Rpd3. There was a significant overlap between the list of homologs whose expression was increased ≥1.5-fold in S. pombe clr6-1 mutant cells and that for homologs whose expression was increased in S. cerevisiae Δrpd3 mutant cells (Fig. 5A; also see Tables S9 and S10 in the supplemental material).
The deletion of a clr gene predominantly resulted in the upregulation of certain messages, but a few genes were also downregulated (see Table S4 in the supplemental material). Among the 10 genes whose expression was most downregulated in the clr1-5 mutant were six genes encoding components of the iron-siderophore system (frp1, fio1, fip1, str1, str3, and c947.05c) (Fig. 5C). These were also among the genes whose expression was most severely reduced in the clr3-735 and Δclr4 mutants (Fig. 5C; also see Table S4 in the supplemental material). In the three mutants, the downregulation correlated with an increased abundance of fep1 mRNA, a repressor of iron uptake genes (Fig. 5C) (reference 28 and references therein), indicating that the action of Clr1, Clr3, and Clr4 may be indirect in this case, with the three proteins most likely repressing fep1 in wild-type cells.
The fact that transporters and/or stress-responsive genes are aberrantly expressed in silencing factor mutants suggests that the stress response may not proceed normally in these mutants. We examined the ability of mutant cells to form colonies under salt or heavy metal stress or at a high temperature (Fig. 5D). Neither the Δclr3 nor the clr6-1 strain proved particularly sensitive to high concentrations of KCl. However, the clr6-1 Δclr3 double mutant grew poorly on 1 M KCl, with a sensitivity similar to that of a strain lacking the atf1 gene (Fig. 5D). Cells with the clr3-735 mutant allele were much more sensitive to salt than were those with the Δclr3 mutation, consistent with the higher penetrance of the clr3-735 allele seen in the array analyses and with Clr6 possibly being able to substitute more efficiently for Clr3 in Δclr3 cells than in clr3-735 cells. Similarly, cells with the clr4-681 point mutation were more sensitive to a high salt concentration than were cells with a disruption of clr4. Mutations in clr1 did not affect the sensitivity of cells to 1 M KCl, indicating that Clr1 does not act together with Clr3 in this instance. On the other hand, point mutations in either clr1 or clr3 increased the sensitivity to CdSO4, a stress that is handled by cells via an Atf1-independent pathway (44) (Fig. 5D). The lack of dcr1 failed to sensitize the cells to any stress to which they were subjected.
Regulation by RNAi or silencing factors and proximity to LTRs.
RNAi and chromatin modifications have both been proposed to influence retrotransposons in other organisms. Furthermore, RNAi was proposed to induce heterochromatin formation in the vicinity of long terminal repeats (LTRs) in fission yeast, thereby regulating the expression of nearby genes (33). We tested whether our microarray data supported such a model by specifically examining the effects of RNAi and silencing factor mutations on the expression of wtf and Tf2 elements, which are both associated with LTRs, and by measuring the distances separating the genes that were most highly affected by RNAi from their nearest LTRs (Fig. 4B and C and Table 3).
TABLE 3.
Localization of RNAi-controlled transcripts relative to LTRs and ORFs
| Transcript | Ratio of mutant transcripts to wild-type transcripts
|
Distance to nearest LTR (kb)a
|
Distance to nearest ORF (kb)a
|
||||
|---|---|---|---|---|---|---|---|
| Δago1 | Δdcr1 | Δrdp1 | Upstream | Downstream | Upstream | Downstream | |
| ptr2 | 2.7 | 1.7 | 2 | 66.6 | 122.6 | 3.7 | 0.7 |
| C417.10 | 2.2 | 2.2 | 2.6 | 10.8 | 19 | 1.3 | 0.9 |
| C1683.06c | 2.4 | 1.5 | 1.9 | 305.7 | 15.3 | 0.8 | 1.4 |
| isp4 | 2.5 | 2 | 2.1 | >45.2 | 17.5 | 5.4 | 1.1 |
| cta3 | 26.5 | 17.6 | 11.4 | 14 | 65.1 | 3.8 | 1.4 |
| C4F6.17c | 2 | 1.6 | 2.2 | 11.9 | 86.1 | 1.7 | 0.8 |
| C1773.03c | 2.1 | 1.7 | 1.7 | 171.1 | 149.5 | 2.2 | 1.5 |
| C63.05 | 1.7 | 1.5 | 1.7 | 61.8 | 109.1 | 0.4 | 0.5 |
| C965.11c | 1.9 | 1.9 | 2 | 14.2 | 73.4 | 0.6 | 1.4 |
| pB21E7.08 | 4.2 | 2.3 | 2.3 | 9.9 | 10.9 | 1.3 | 2.9 |
| C26H5.15 | 1.6 | 1.8 | 2.5 | 14.6 | 131.6 | 3 | 0.7 |
| C1773.12 | 4.8 | 2.9 | 2.9 | 171.6 | 148.5 | 1.7 | 0.8 |
| C1685.05 | 1.6 | 1.8 | 1.6 | 47.1 | 84.9 | 2.1 | 0.8 |
| hsp16 | 3.3 | 7.4 | 9.4 | 75.6 | 23.3 | 6.5 | 1.1 |
| PB24D3.07c | 3.4 | 3.3 | 2.9 | 84.5 | 15.7 | 3.2 | 1.0 |
| C338.12 | 1.5 | 1.8 | 1.6 | 45.0 | 58.6 | 0.8 | 1.1 |
| Tf2-1,2,5,6,7,9,10 | 1.6-1.8 | 1.7-1.9 | 1.6-1.8 | NA | NA | NA | NA |
| C1E8.04c | 1.7 | 1.9 | 1.8 | NA | NA | NA | NA |
| C794.05c | 1.6 | 1.8 | 1.7 | NA | NA | NA | NA |
| dg.cen.repeat | 3.2 | 4.5 | 3.9 | NA | NA | NA | NA |
| dh.cen.repeat | 11.8 | 17.1 | 11.5 | NA | NA | NA | NA |
NA, not available.
The expression of wtf elements was strikingly up-regulated by the clr6-1 mutation but by none of the other mutations tested (Fig. 4B). Fourteen of the 16 wtf elements spotted on the array are within 700 bp of an LTR, indicating that proximity to an LTR is not sufficient to place a gene under the control of RNAi or heterochromatic silencing. The abundance of Tf2 RNA was slightly increased in the three RNAi factor mutants and in the clr6-1 mutant and was more significantly increased in the clr6-1 Δclr3 double mutant (Fig. 4C). The nine Tf2 probes spotted on our microarrays produced nearly identical hybridization patterns. Cross-hybridization was expected between the various probes because of the high degree of similarity shared by S. pombe Tf2 elements, and the results obtained with a single probe are displayed in Fig. 4C.
A model based on RNAi or silencing mediated via LTRs predicts that some of the genes whose expression is increased in RNAi mutants are close to an LTR. Table 3 presents an analysis of the chromosomal regions flanking the genes whose expression was most strongly affected in the three RNAi mutants we examined. These genes are not closely linked to LTRs. Furthermore, ORFs located between the affected genes and their neighboring LTRs were not affected by mutations in the RNAi or silencing pathway, indicating that they are not controlled by an RNA or heterochromatic structure spreading from the LTR. Schramke and Allshire (32) presented evidence that seven meiotically induced genes located close to an LTR were affected by RNAi. Six of those genes were present on our arrays. Surprisingly, the expression of these six genes did not seem to be affected in RNAi mutants beyond the 1.5-fold cutoff that we used. We investigated whether the cause for this discrepancy was a detection problem in the microarray experiment. RT-PCRs conducted to detect the meu6, meu8, or meu28 transcript revealed that the expression of these genes was not significantly altered in our RNAi mutant strains compared to the wild type, indicating that the problem was not merely detection on the microarrays (see the supplemental material). We are currently investigating other possible causes for the discrepancy.
DISCUSSION
We have determined the global effects on mRNA abundance of mutations in silencing and RNAi factors in fission yeast. The observed effects are likely to reflect both structural and regulatory functions of heterochromatin. Clr1, Clr3, and Clr4 repress overlapping sets of genes, consistent with the phenotypes of mutants of these factors. Many of the genes whose expression is affected by Clr1, Clr3, or Clr4 are also affected by Clr6, which in addition influences the expression of a large number of other genes and genomic elements. Some transcripts are under the dual control of RNAi and silencing factors. However, regulation by silencing factors does not show an extensive correlation with the regulation of transcription by RNAi, indicating that transcriptional silencing does not systematically require RNAi for its establishment or maintenance.
Concerted action of histone-modifying enzymes.
Histone modifications are believed to be interdependent, either indirectly when the modification of one residue affects the modification of another or directly when the modification of one residue prevents a different modification on that same residue. We found that the repression of nearly all genes that were regulated by the histone methyltransferase Clr4 also depended on the histone deacetylase Clr3 or Clr6. On the other hand, many genes that were regulated by Clr3 or Clr6 were not affected by Clr4. This indicates that methylation is not required for deacetylation and is consistent with deacetylation being a prerequisite for methylation. Furthermore, this indicates that histone deacetylation is a more common way of repressing gene expression in fission yeast than histone methylation.
Overlapping pathways and partially redundant functions are often observed in eukaryotic cells and are a determinant of robustness. Histone-modifying enzymes can be used to illustrate this point. Three classical histone deacetylases are present in S. pombe cells, specifically Clr3 and Clr6, which are nuclear enzymes, and a cytoplasmic enzyme, HdaI (8). We found that Clr3 and Clr6 tend to repress transcription, with Clr6 controlling a larger number of loci than Clr3. The two enzymes acted together or could substitute for each other in several cases, leading to stronger levels of derepression in the clr3 clr6 double mutant than in either the clr3 or clr6 single mutant (Table 2 and Fig. 3 and 4). Other classes of genes were controlled solely by Clr3 or Clr6. More complex patterns of gene expression were also observed, reminiscent of the patterns seen for the mating-type region, for which transcriptional silencing is increased in clr6 mutants, alleviated in clr3 mutants, and totally lost in clr3 clr6 double mutants (13). Such complex patterns of expression suggest that Clr3 and Clr6 occasionally compete to deacetylate histones. Clr3 specifically deacetylates lysine 14 of histone H3, in contrast to Clr6, which shows broad in vivo substrate specificity (8). Thus, Clr3 may establish a tighter repression than that observed in wild-type cells when it is allowed to operate alone in some chromosomal regions (e.g., SPBC337.02c, SPCC569.01c, SPCC794.06, and SPAC1B3.17).
Effects of silencing factors in telomeric and subtelomeric regions.
We found that silencing is not restricted to the close proximity of telomeric repeats, where it had previously been observed after the insertion of reporter genes (27), but that it affects many subtelomeric genes located >50 kb away from the chromosome ends. Remarkably, expression of the same subtelomeric genes is highly increased during the early steps of meiosis triggered by nitrogen starvation (24). This increased expression might be facilitated by structural changes connected with the association of telomeres with the spindle pole body during meiotic prophase (reviewed in reference 53), a process in which the association of silencing factors with subtelomeric regions may be perturbed. In support of silencing factors performing a structural role at telomeres, we observed that telomeric repeats were elongated in silencing mutants, particularly in the clr6-1 Δclr3 double mutant. In addition to this structural role, our data indicate that Clr1, Clr3, Clr4, and Clr6 regulate transcription per se in subtelomeric regions. Several permeases and metabolic enzymes are encoded by subtelomeric genes (17). Their derepression following nitrogen starvation might be part of an adaptive response. Similarly, S. cerevisiae subtelomeric regions contain many genes whose expression in response to nutrient depletion or stressful conditions facilitates survival. These regions are hypoacetylated and repressed in an Hda1-dependent manner (32). Hence, a remarkable organization of subtelomeric gene clusters that are responsive to nutritional stress and repressed by histone deacetylation appears to have been conserved during the evolution of these two types of yeast. Orthologous enzymes, Clr3 and Hda1, mediate the repression of these regions.
Biological functions of regulated genes.
Many of the genes that are regulated by Clr1, Clr3, Clr4, or Clr6 were induced by a particular stress, nitrogen starvation. These include the previously mentioned clusters of subtelomeric genes as well as additional nontelomeric genes. Notably, the derepression of nitrogen starvation- and meiosis-related genes observed in the clr1, clr3, clr4, and clr6 mutants was substantially lower than the increase observed after nitrogen starvation, by which mRNA levels can be induced as much as 100-fold (24). This suggests that nitrogen-regulated activators of transcription provide an additional level of induction to these genes. This mode of regulation would be similar to that operating in the mating-type region, where transcription of the mat2-P and mat3-M loci in silencing factor mutants requires nitrogen-responsive transcription factors such as the Ste11 protein. Dual controls of gene expression of this type might more generally account for the small amplitude of the effects observed in the clr mutants.
The clr1, clr3, clr4, and clr6 mutants were originally identified because of their aberrant mating and sporulation phenotypes (11, 13, 40, 41). The transcriptional repression they exert in the mating-type region is central to mating-type switching and homothallism. It allows clonal populations of cells to form spores when nutrients are depleted, ensuring survival under conditions in which vegetative cells would die. Our present study suggests that the same silencing factors mediate cell adaptation to adverse environmental conditions in a more general fashion. This is indicated by the abundance of transporters and metabolic enzymes that are regulated by Clr1 and Clr3 and by the overlap observed between Clr6- and Atf1-affected genes. We cannot rule out that some of the effects that occurred in the clr6-1 cells were indirect and caused by stress in sick mutant cells. However, a direct role of Clr6 and Clr3 in the regulation of stress-inducible genes is supported by the observation that several of the clr mutations examined were sensitive to various stresses.
Expression profiles in silencing and RNAi factor mutants.
Some similarities were observed when we compared the expression profiles of the silencing factor mutants with those of the RNAi factor deletion strains (Table 1), supporting a model in which changes in chromatin structure occasionally accompany RNAi in S. pombe. Among the common effects observed for RNAi and silencing factor mutants were the increased abundance of transcripts originating from centromeric repeats and the increased abundance of Tf2 transcripts in RNAi and clr6-1 mutants (discussed below). However, striking differences were also apparent between the two classes of mutants. For example, mutations in the RNAi pathway had no general effect in telomeric or subtelomeric regions. This lack of effect was consistent with a previously isolated ago1 mutant that derepresses transcription in centromeric but not telomeric regions (12) and with Chp1, a chromodomain protein that mediates transcriptional repression at centromeres but not telomeres (42), being part of an RNAi protein complex (46). Furthermore, many nontelomeric genes that were regulated by the clr genes were not affected in the RNAi mutants, raising the possibility that alternative pathways trigger them for silencing. The fact that the clr1 and clr3 mutants displayed similar expression profiles suggests that Clr1 may be involved in the recruitment of Clr3. Similarly, our data strongly suggest that Atf1 can recruit the histone deacetylase Clr6. The histone deacetylation of Atf1 target genes by Clr6 would mediate a reversible repression of transcription in unstressed cells, with the phosphorylation of Atf1 in response to stress leading to different protein associations and transcriptional activation. Noticeably, Atf1 binding sites are also found in a silencing element near the mat3-M locus (39), and Atf1 contributes to silencing of the mating-type region (19), consistent with its attracting Clr6 there as well.
Centromeric repeat transcripts.
Centromeric transcripts originate from both strands of the dg and dh repeats (48). The lack of a silencing factor, the chromodomain protein Swi6, increases the amount of forward transcript without significantly affecting the reverse transcript, suggesting that the transcription of one DNA strand only is repressed by silencing (48) (Fig. 3). Our observation that both forward and reverse dh transcripts accumulated in clr4 and clr6-1 Δclr3 mutant cells supports the notion that heterochromatin affects the transcription of both strands. Formation of the reverse transcript might be repressed by proteins other than Swi6, e.g., additional chromodomain proteins which remain associated with centromeres in the swi6 mutant but not in the clr4 or clr6 clr3 mutant.
LTRs, Tf2 retrotransposons, and wtf elements.
The genome of the sequenced S. pombe strain contains 13 integrated full-length copies of Tf2, a retrotransposon related to the Drosophila gypsy elements, ∼250 solo LTRs, and 25 wtf elements, a family of genes of unknown function that are tightly associated with LTRs (4, 9). Schramke and Allshire (33) proposed that RNAi operates via LTRs to control the transcription of nearby genes, with repressive chromatin being nucleated at the LTR and spreading to adjacent DNA. Fission yeast LTRs can be grouped into nine families according to their sequences. RNAi can be expected to distinguish among the different families, and its acting independently on each of them may provide a mechanism for the coregulation of gene sets. We tested whether genes whose expression depended on RNAi in our microarray analysis were in close proximity to LTRs. However, we could not detect any correlation between genes that were modulated by RNAi and neighboring LTRs (Table 3), which suggests that LTRs are not the major determinant of RNAi regulation in S. pombe.
Transposable elements in other organisms can be packaged into structures with the features of heterochromatin, and in some cases RNAi contributes to their repression (2, 15, 16, 20, 21, 22, 25, 34, 38, 54). This has been viewed as an organismal defense against the deleterious spread of parasitic DNA in the genome. We presented evidence that in S. pombe, mutations in the RNAi machinery result in small increases in the abundance of transcripts originating from Tf2 retrotransposons, while these same mutations do not affect the transcription of wtf elements (Fig. 4). Some S. pombe LTRs are associated with Swi6 and with histone H3 methylated at lysine 9 (33). Whether Tf2 elements are packaged similarly is not known, but neither Tf2 nor wtf transcript levels were elevated in clr4 mutant cells. In contrast, the transcription of Tf2 and wtf elements clearly increased in histone deacetylase mutants, indicating that the transcription of these elements is normally inhibited by hypoacetylation. Since S. pombe retrotransposons tend to integrate in intergenic regions near promoters (6, 36), one possibility is that individual transposons are subject to the regulation of nearby genes by histone deacetylation. However, it is equally likely that the transposon itself attracts histone-modifying enzymes. Retrotransposon LTRs contain strong outward-reading promoters, and in the budding yeast strain Ty1 LTRs regulate neighboring genes via chromatin remodeling and histone deacetylation (52). In S. pombe, Tf2 elements may occasionally repress the transcription of adjacent genes via a pathway utilizing Clr6. The pronounced effect of histone deacetylation on Tf2 RNA levels is strikingly reminiscent of the situation in Arabidopsis, in which histone deacetylase mutants activate transposons that are not activated in RNAi or histone methyltransferase mutants (22). The contribution of RNAi to the repression of Tf2 elements as revealed by our microarray data is relatively minor, but this does not exclude the possibility that RNAi plays a more predominant role in establishing the silencing of transposons once they are activated by other means.
Supplementary Material
Acknowledgments
We thank Stanley Brown and members of the Department of Genetics, University of Copenhagen, for comments on the manuscript and the experiments, Janne Verhein-Hansen for technical assistance, Nefeli Nikdaidou-Katsaridou and Al Ivens for array printing, Valerie Wood for help with data analysis, and Derek Goto for RT-PCR analysis of meu transcripts and for general resourcefulness.
We acknowledge the support of the National Science Foundation (grant DBI0077774 to R.M.), the National Institutes of Health (grant GM067014 to R.M.), Cancer Research UK (J.B.), the Novo Nordisk Foundation, and the Danish Research Council (G.T.). T.V. was supported by a postdoctoral fellowship from the National Cancer Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Aravin, A. A., N. M. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozovsky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. [DOI] [PubMed] [Google Scholar]
- 3.Ayoub, N., I. Goldshmidt, R. Lyakhovetsky, and A. Cohen. 2000. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics 156:983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bähler, J., and V. Wood. 2003. How to get the best from fission yeast genome data, p. 282-288. In R. Egel (ed.), The molecular biology of Schizosaccharomyces pombe: genetics, genomics, and beyond. Springer-Verlag, Berlin, Germany.
- 5.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 6.Behrens, R., J. Hayles, and P. Nurse. 2000. Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res. 28:4709-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjerling, P., R. A. Silverstein, G. Thon, A. Caudy, S. Grewal, and K. Ekwall. 2002. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22:2170-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowen, N. J., I. K. Jordan, J. A. Epstein, V. Wood, and H. L. Levin. 2003. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 13:1984-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bähler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekwall, K., and T. Ruusala. 1994. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekwall, K., G. Cranston, and R. C. Allshire. 1999. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153:1153-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grewal, S. I., M. J. Bonaduce, and A. J. S. Klar. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirochika, H., H. Okamoto, and T. Kakutani. 2000. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt, C., K. Moore, Z. Xiang, S. M. Hurst, R. C. McDougall, M. A. Rajandream, B. G. Barrell, R. Gwilliam, V. Wood, M. H. Lyne, and S. J. Aves. 2001. Subtelomeric sequence from the right arm of Schizosaccharomyces pombe chromosome I contains seven permease genes. Yeast 18:355-361. [DOI] [PubMed] [Google Scholar]
- 18.Ivanova, A. V., M. J. Bonaduce, S. V. Ivanov, and A. J. Klar. 1998. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 19:192-195. [DOI] [PubMed] [Google Scholar]
- 19.Jia, S., K. Noma, and S. I. Grewal. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304:1971-1976. [DOI] [PubMed] [Google Scholar]
- 20.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNase D. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 21.Lindroth, A. M., X. Cao, J. P. Jackson, D. Zilberman, C. M. McCallum, S. Henikoff, and S. E. Jacobsen. 2001. Requirement of chromomethylase 3 for maintenance of CpXpG methylation. Science 292:2077-2080. [DOI] [PubMed] [Google Scholar]
- 22.Lippman, Z., B. May, C. Yordan, T. Singer, and R. Martienssen. 2003. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici, D. Chen, C. Langford, D. Vetrie, and J. Bähler. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mata, J., R. Lyne, G. Burns, and J. Bähler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 25.Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada, and T. Kakutani. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212-214. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Nimmo, E. R., G. Cranston, and R. C. Allshire. 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 13:3801-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelletier, B., J. Beaudoin, C. C. Philpott, and S. Labbe. 2003. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 31:4332-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provost, P., R. A. Silverstein, D. Dishart, J. Walfridsson, I. Djupedal, B. Kniola, A. Wright, B. Samuelsson, O. Radmark, and K. Ekwall. 2002. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. USA 99:16648-16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 31.Reinhart, B. J., and D. B. Bartel. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831. [DOI] [PubMed] [Google Scholar]
- 32.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 33.Schramke, V., and R. Allshire. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301:1069-1074. [DOI] [PubMed] [Google Scholar]
- 34.Singer, T., C. Yordan, and R. A. Martienssen. 2001. Robertson's mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene decrease in DNA methylation (DDM1). Genes Dev. 15:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, G., and A. J. Klar. 2002. The 2.1-kb inverted repeat DNA sequences flank the mat2,3 silent region in two species of Schizosaccharomyces and are involved in epigenetic silencing in Schizosaccharomyces pombe. Genetics 162:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton, T. L., and H. L. Levin. 2002. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryot. Cell 1:44-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugarawa, N. 1989. DNA sequences at the telomeres of the fission yeast S. pombe. Ph.D. thesis. Harvard University, Cambridge, Mass.
- 38.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 39.Thon, G., K. P. Bjerling, and I. S. Nielsen. 1999. Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics 151:945-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thon, G., A. Cohen, and A. J. Klar. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thon, G., and A. J. Klar. 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131:287-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thon, G., and J. Verhein-Hansen. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155:551-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toone, W. M., and N. Jones. 2004. Stress responses in S. pombe, p. 57-72. In R. Egel (ed.), The molecular biology of Schizosaccharomyces pombe: genetics, genomics, and beyond. Springer-Verlag, Berlin, Germany.
- 44.Toone, W. M., S. Kuge, M. Samuels, B. A. Morgan, T. Toda, and N. Jones. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 46.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermaak, D., K. Ahmad, and S. Henikoff. 2003. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 15:266-274. [DOI] [PubMed] [Google Scholar]
- 48.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 49.Volpe, T., V. Schramke, G. L. Hamilton, S. A. White, G. Teng, R. A. Martienssen, and R. C. Allshire. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11:137-146. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe, T., K. Miyashita, T. T. Saito, K. Nabeshima, and H. Nojima. 2002. Abundant poly(A)-bearing RNAs that lack open reading frames in Schizosaccharomyces pombe. DNA Res. 9:209-215. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, T., K. Miyashita, T. T. Saito, T. Yoneki, Y. Kakihara, K. Nabeshima, Y. A. Kishi, C. Shimoda, and H. Nojima. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29:2327-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winston, F., and P. Sudarsanam. 1998. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp. Quant. Biol. 63:553-561. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto, A., and Y. Hiraoka. 2001. How do meiotic chromosomes meet their homologous partners?: lessons from fission yeast. Bioessays 23:526-533. [DOI] [PubMed] [Google Scholar]
- 54.Zilberman, D., X. Cao, and S. E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716-719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.