Abstract
MOF was first identified in Drosophila melanogaster as an important component of the dosage compensation complex. As a member of MYST family of histone acetyltransferase, MOF specifically deposits the acetyl groups to histone H4 lysine 16. Throughout evolution, MOF and its mammalian ortholog have retained highly conserved substrate specificity and similar enzymatic activities. MOF plays important roles in dosage compensation, ESC self-renewal, DNA damage and repair, cell survival, and gene expression regulation. Dysregulation of MOF has been implicated in tumor formation and progression of many types of human cancers. This review will discuss the structure and activity of mammalian hMOF as well as its function in H4K16 acetylation, DNA damage response, stem cell pluripotency, and carcinogenesis.
Keywords: MOF, H4K16, histone acetylation, MSL, NSL
1. Introduction
In eukaryotes, the vast amount of genetic material must fit within a relatively small nucleus. To achieve this, genomic DNA along with histone proteins are tightly packed together to form a highly dynamic structure called chromatin [1]. Nucleosome, the basic structural unit of chromatin, is composed of ~147-bp of double stranded DNA wrapped around an octamer core with two sets of histone proteins H2A, H2B, H3 and H4. The N-terminal tail of each histone protein protrudes from the nucleosome and serves as a platform for various chemical modifications, including methylation, acetylation, phosphorylation, and many others [2]. Among these posttranslational modifications, acetylation of histone H4 at lysine 16 (H4K16) is particularly important to higher-order chromatin structure. Residing in the basic N-terminal tail, H4K16 acetylation not only directly influences the formation of compact higher-order chromatin, but also modulates the functional interaction between non-histone proteins and chromatin fiber [3,4]. Given its important role, two multiprotein complexes responsible for the deposition of an acetyl group on H4K16 are highly conserved from fruit flies to mammals [5,6]. MOF, Males absent on the first, is the only acetyltransferase shared by these two complexes [6,7].
MOF was identified in Drosophila melanogaster as an important component of the dosage compensation complex [8,9]. In metazoans, females have one extra X-chromosome compared to males. Dosage compensation is a mechanism in ensuring equal expression of X-chromosome-related genes in both sexes [10]. In Drosophila, this compensation is achieved by doubling the amount of male X-chromosome transcripts via a multiprotein dosage-compensation complex (DCC), also known as male specific lethal (MSL) complex. The binding of MSL complex to the male X chromosome is accompanied by increased acetylation of histone H4 lysine 16 (H4K16) as well as a 2-fold transcription activation of X-linked genes [8,11]. MOF is the only acetyltransferase in the MSL complex. A point mutation in the MOF enzyme activity domain not only abolishes the histone H4 acetylation in the male X-chromosome, but also leads to the death of male flies (male-specific lethal). Thus, MOF plays a fundamental role in male specific gene dosage compensation via acetylation of histone H4K16. In addition to the MSL complex, MOF is also a part of the non-specific lethal (NSL) complex, in which gene disruption by P element can lead to lethality in both sexes [6]. Indeed, NSL complex acetylates autosomal H4K16 in both male and female [6].
The human ortholog of Drosophila MOF (hMOF) exhibits highly conserved substrate specificity and enzymatic activity [5,12]. Both MSL and NSL complexes are found in mammalian cells, which are responsible for acetylation of H4K16 in the nucleosome [5]. However, mammals employ a very different approach to achieve the balance of gene expression between different sexes. Rather than doubling transcripts from the male X-chromosome as seen in Drosophila, one of the X-chromosome is inactivated in the mammalian female [13,14]. The function of hMOF and its complexes in mammals may differ from those in flies, which is supported by the observation that MOF knockout in either sex of mice is lethal during the early period of embryonic development [15,16]. Moreover, hMOF and histone H4K16 acetylation play important roles in DNA damage response, cell cycle regulation, and gene expression. Dysregulation of MOF has been reported in many types of human cancer. This review will discuss the structure of mammalian hMOF as well as its role in H4K16 acetylation, DNA damage response, stem cell pluripotency, and carcinogenesis.
2. MOF is a member of MYST family of acetyltransferase
2.1. Structure characteristics
MOF belongs to the MYST family of histone acetyltransferase. The name MYST stands for the first four founding members: MOZ, Ybf2 (Sas3), Sas2, and Tip60. Other important family members include MOF, Esa1, MORF, and HBO1 [17,18]. The common characteristic of this family is the catalytic MYST domain that transfers an acetyl group from acetyl-CoA to the lysine side chain of histone tail. Within the MYST family, MOF is closely related to Tip60 as they both contain a chromo-like domain adjacent to the N-terminal of the MYST domain.
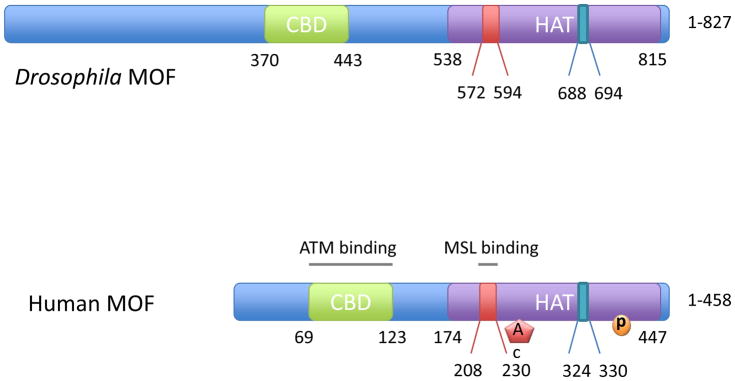
The structure of human and Drosophila MOF are quite similar, sharing 50% identity and 69% similarity (Figure 1). The histone acetyltransferase (HAT) domain of MOF consists of a central core that mediates acetyl-CoA binding and an atypical C2HC-type zinc finger within the flanking N-terminal region that mediates substrate recognition. Within the central core, a glutamate (E350) residue is crucial for the HAT activity, as the E350Q mutation essentially abolishes the MOF HAT activity [19]. A unique C2HC-type zinc finger (CxxCx12HxxxC) directly interacts with the globular part of the nucleosome and the histone H4 tail, and is crucial for HAT activity and substrate specificity [20]. Point mutations in the highly conserved cysteine or histidine residues completely abolish HAT activity as well as its interaction with the nucleosome [20].
Figure 1.
Structure diagram showing the functional domains of Drosophila and Human MOF.
Another common feature of the MOF/Tip60/Esa1 subfamily is the chromobarrel domain, named for its beta-barrel structure. Unlike canonical chromodomains that exhibit the structure of alpha + beta folds and bind to methylated lysine residues, the chromobarrel domains in MOF/Tip60/Esa1 lack the aromatic cage and consequently mediate the interaction between protein and nucleic acids [9,21]. Disruption of the chromobarrel domain compromises MOF HAT activity, resulting in a global reduction of H4K16 acetylation [22].
2.2. MOF and histone H4K16 acetylation
Multiple lines of evidence indicate that MOF is a H4K16 specific acetyltransferase. First, MOF is the only subunit in Drosophila MSL complex that has acetyltransferase activity [8,9]. Binding of MSL complex to the male X chromosome is correlated with a significant increase of H4K16 acetylation in that chromosome [8,23]. In addition, similar to its Drosophila ortholog, hMOF is a part of the human MSL complex, which is responsible for the majority of H4K16 acetylation in mammalian cells [24]. Moreover, knockdown of hMOF in HeLa and HepG2 cells leads to a strong decrease in H4K16 acetylation but other histone lysine acetylation remains unchanged [24,25]. Furthermore, the level of H4K16 acetylation is nearly undetectable in MOF knockout cells [15,16], while other histone acetylation marks including H4K5ac, H4K8ac, H4K12ac, H3K9ac and H3K14ac are not affected [16]. However hMOF is able to acetylate other histone lysines in in vitro assays, indicating the importance of protein complex, such as MSL or NSL, in regulating histone acetylation in cells. Finally, expressing MOF in cells where this gene was knocked out resulted in the recovery of H4K16 acetylation [26]. Taking together, these results indicate that MOF specifically and exclusively targets nucleosomal H4K16.
2.3. Non-histone targets of hMOF
Besides acting as a histone acetyltransferase, hMOF has been shown to acetylate the lysine residue in several non-histone proteins, including the subunits of multiprotein complexes (Msl3, TIP5) and crucial transcription factors (p53, Nrf2).
Drosophila MOF can acetylate MSL3 (a major component of the MSL complex) at lysine residue (K116) that is adjacent to one of its chromodomains [27]. MSL3 acetylation leads to a temporary loss of interaction between MSL3 and RNA, therefore contributing to the fine-tuning of dosage compensation levels in Drosophila [27]. However, it is not clear whether this function is conserved in the mammalian MSL complex. Interestingly, mammalian MOF is able to acetylate TIP5, the largest subunit of NoRC chromatin-remodeling complex, at lysine 633 that is adjacent to the TIP5 RNA-binding domain [28]. Acetylation of TIP5 also weakens its interaction with promoter-associated RNA and is required for NoRC-mediated rDNA silencing [28].
Transcription factors are also the target of MOF. As a master regulator of cellular response to genotoxic stress, p53 controls cells undergoing either cell cycle arrest or apoptosis by triggering different sets of downstream target genes [29,30]. It is reported that hMOF as well as its closely related MYST member TIP60, are capable of acetylating p53 at K120, a lysine residue located in the DNA-binding domain of p53. Acetylation of p53 K120 modulates its DNA-binding preference, leading to increased transcription of pro-apoptotic genes: PUMA and BAX [31]. Further studies show that p53K120 acetylation regulates cell apoptosis by both transcription-dependent and -independent mechanisms [31,32].
P53 is not the only transcription factor that can be targeted by MOF. A recent study on human non-small cell lung cancer reported the acetylation of NF-E2-related factor 1 (Nrf2) by hMOF [33]. Nrf2 is a crucial factor protecting cells against oxidative stress, for example, cancer cells require activated NrF2 in order to survive high levels of oxidative stress that exist in this disease [34]. hMOF is able to bind and acetylate Nrf2 at lysine 588, which facilitates its nuclear translocation and increases transcription of Nrf2 downstream genes. Nrf2 protein levels remain the same in the presence or absence of hMOF, suggesting that hMOF affects the Nrf2 transcription activity rather than its expression level [33].
3. Regulating MOF activity
MOF alone is able to acetylate free histone H4K16. However, acetylation of nucleosomal H4K16 occurs only in the presence of multiprotein complexes [35], suggesting that the associated protein complex tightly regulates MOF activity and substrate specificity. There are two MOF-containing protein complexes, and MOF functions differently depending on the substrate specificity of the complex. In addition, MOF can be post-translationally modified, and these modifications also modulate its HAT enzyme activity.
3.1. MOF-containing protein complex
MOF exists in two distinct but highly conserved multiprotein complexes: MSL and NSL. MOF is best known as a major HAT in Drosophila MSL complex. Drosophila MSL complex contains five protein subunits and two non-coding roX RNAs [13,36]. Four other components include male-specific lethal 1-3 (MSL1, MSL2, MSL3) and the ATP-dependent RNA/DNA helicase maleless (MLE). While MSL1 and MSL3 are crucial for MOF activity and substrate specificity, MSL2, MLE and RNA components recruit MOF to specific chromatin locations [13,36]. Mammalian orthologs for these protein subunits have been identified in human, but not the RNA components (roX1/2). Interestingly, human MSL complex consisting only of MSL1-3 and MOF seem sufficient for acetylation of nucleosomal H4K16 [24]. Recently, MOF was found as a key component of another chromatin-modifying complex, nonspecific lethal (NSL) complex [6,24,37,38]. All the components of the NSL complex are highly conserved, consisting of at least 7 subunits: MOF/hMOF, NSL1/KANSL1, NSL/KANSL2, NSL3/KANSL3, WDS/WDR5, MCRS2/MCRS1, dHCF/HCFC1, OTG/OTG1 and MBD-R2/PHF20, in Drosphila and mammals, respectively [37,38].
MOF is the only common subunit shared by the MSL and NSL complexes. Both complexes are able to acetylate H4K16 in varied chromatin contexts, suggesting that MOF activity and substrate specificity are tightly controlled by the associated proteins in the complex. This is supported by the fact that Drosophila MSL complex is only associated with male X-chromosome as MSL2 is exclusively expressed in male cells [13,39]. On the other hand, the NSL complex exists in both male and female cells, and is responsible for global H4K16 acetylation [38]. In addition, while mammalian MSL complex only acetylates nucleosomal histone H4K16, the NSL complex can acetylate nucleosomal H4K5 and H4K8 as well as non-histone targets (p53 K120) [37]. Moreover, recent genome-wide studies have revealed different distribution patterns of the two complexes. In Drosophila, the MSL complex is localized in the coding region or 3′ end of X-linked genes, and the level correlates to gene transcription. The NSL complex is primarily located in the promoter region of more than 4000 genes and controls H4K16 acetylation and gene expression [38,40,41]. In mammals, the NSL complex is globally associated with the promoter region of housekeeping genes [40,42]. Interestingly, mammalian MSL complex binds to both promoter and gene bodies and regulates many embryonic stem cells-specific and bivalent genes [42,43].
3.2. Autoacetylation and phosphorylation
In 2011, three groups reported that MOF is capable of autoacetylation at lysine 274, a highly conserved lysine residue in the C2HC zinc-finger domain [19,44,45]. Two groups studied the crystal structure of MOF and found that a strong electron density at the tip of the K274 side chain could be modeled as an acetyl group [19,44]. The acetylation of K274 is critical for the proper position of the hairpin structure and affects MOF substrate specificity and HAT activity, which is supported by a lack of enzyme activity in various MOF K274 mutants (K274A, K274R, and K274Q) [19,44]. The third group [45] studied MOF autoacetylation based on its similarity to Tip60, where autoacetylation enhances its interaction with substrates [46]. Lu et al reported that both in vitro and in vivo MOF autoacetylation reduced its ability to bind to nucleosomes. Sirt1, a class III histone deacetylase, is able to remove the acetyl group from hMOF and thereby restore its interaction with nucleosomal substrates [45]. On the other hand, Yuan and his colleagues reported that K274 autoacetylation is required for MOF mediated acetylation of p53 and histone H4K16 [47].
In addition to acetylation, phosphorylation may also play a role in regulating MOF activity. A recent study found that the DNA double-strand break (DSB) induced ATM-dependent MOF phosphorylation at threonine 392 [48]. Phosphorylated MOF co-localized with γ-H2Ax, ATM, and 53BP1 foci, and is required for the recruitment of homologous repair proteins to DNA damaged sites during S/G2 phase.
4. Cellular function of hMOF
4.1. hMOF and ATM
ATM (Ataxia Telangiectasia Mutated) protein is a serine protein kinase that has sequence homology to PI3K (Phosphoinositide 3-kinase) [49]. ATM gene was originally identified by position cloning of an altered protein in patients with Ataxia Telangiectasia, a neuron degeneration disorder characterized by cerebellar degeneration and neuromoter dysfunction. Later, numerous studies have established its critical role in DNA damage response and cell cycle checkpoint [49]. In the absence of DNA damage stress, ATM is presented as an inactive dimer or oligomer [50]. Following DNA damage, ATM autophosphorylates at serine 1981 and converts from an inactive dimer to an active monomer, which in turn phosphorylates and activates downstream effectors, such as p53 and checkpoint kinase 2 (Chk2).
In 2005, Gupta and colleague reported a direct physical interaction between hMOF and ATM [51]. This interaction was mediated by hMOF chromobarrel domain and the leucine zipper domain in ATM. How does this interaction affect hMOF or ATM function? The experimental data suggests that MOF is able to influence ATM function. First, ionizing radiation (IR) induced ATM-independent but MOF-dependent increase of H4K16 acetylation. In addition, inactivation of MOF using either MOF mutant or MOF knockdown resulted in a significant decrease of IR-induced ATM autophosphorylation. ATM kinase activity and phosphorylation of downstream effector Chk2 were also reduced. Interestingly, an increase in H4K16 acetylation using histone deacetylase inhibitors (trichostatin A or sodium butyrate) had no effect on ATM activity, suggesting that H4K16 acetylation alone is not sufficient to influence ATM function. These data suggest that hMOF is able to affect the early stages of DNA damage signaling through modulating ATM activation [51]. However, a recent study from the same group reported that ATM is able to phosphorylate hMOF at threonine 392, which is critical for cell survival and damage repair in the S and G2 phase [48]. Therefore, hMOF may act both upstream and downstream of ATM. It is worth noting that mice deficient in Purkinje-specific MOF displayed an ataxia-telangiectasia-like neurological phenotype [52], suggesting a functional interaction between MOF and ATM in vivo.
4.2. DNA damage response
Mammalian cells are constantly exposed to genotoxic stress from both internal and external sources. To protect genomic integrity, cells have developed a highly sophisticated defense response (DNA damage response, DDR) that consists of multiple key components: damage sensors, signal transducers, mediators, and effectors [53]. Successful DNA repair requires functional DDR to recruit specific DNA repair complexes as well as chromatin remodeling machineries to control DNA accessibility [53].
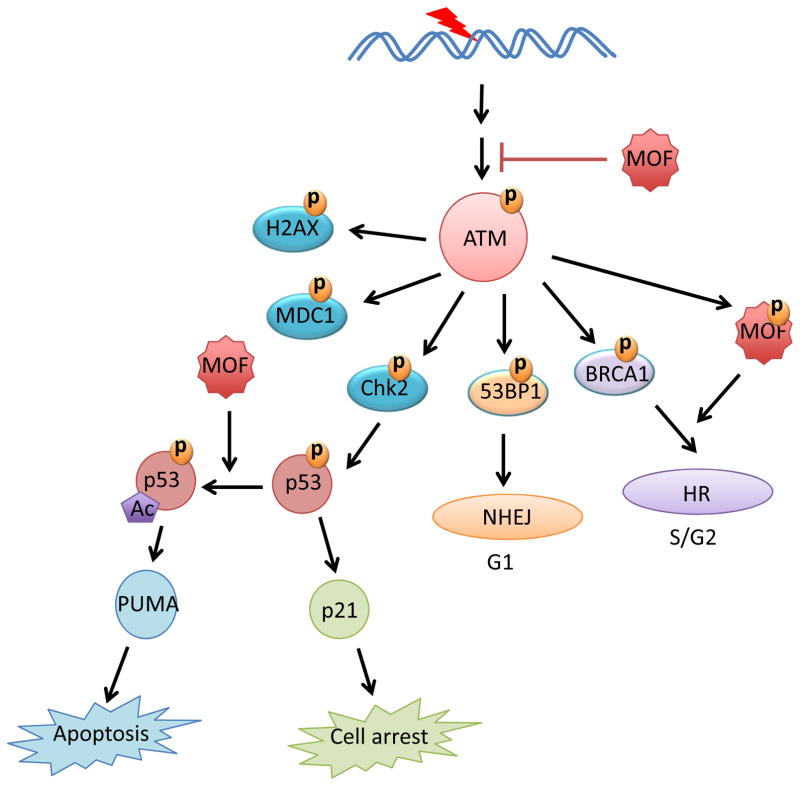
Given its fundamental role in modulating higher-order chromatin structure, several studies have investigated the involvement of hMOF in each step of DDR. MOF induced acetylation of H4K16 after DNA damage is subsequently modulated by the internucleosomal interaction between the acidic pocket of H2A-H2B surface and the basic patch on histone H4 tail [26]. In addition, it was reported that MOF is associated with ATM and the DNA-dependent protein kinase subunits (DNA-PKcs). Depletion of MOF reduced ATM autophosphorylation and abrogated ATM-dependent phosphorylation of Chk2 and DNA-PKcs [51,54]. Moreover, IR-induced recruitment of repair mediator Mdc1, 53BP1 and BRCA1 to DNA damage foci was completely abolished in MOF conditional knockout mouse embryonic fibroblasts (MEFs) [26]. These results indicate that hMOF is involved in nearly every step of DDR, and actively modulates DDR signaling as well as recruitment of DDR metiators and effectors (Figure 2). It is worth noting that early DNA damage sensing and ATM-mediated signaling were not affected in MOF conditional knockout MEFs [26]. This discrepency may result from the different ways that MOF was depleted or the type of cells used.
Figure 2.
MOF and DNA damage response
In addition to DDR signaling, hMOF is also crucial for DNA damage repair. DNA double-strand breaks can be repaired by either homologous recombination (HR) or nonhomologous end-joining (NHEJ) pathways, depending on the cell cycle phase as well as the opposing activity of 53BP1 and BRCA1 [55,56]. NHEJ is more active in G1 as 53BP1 inhibits DNA end resection of DSBs. On the other hand, HR is mostly active in the S/G2 phase due to inhibition of 53BP1 by BRCA1 [56]. Results from both MOF knockout and knockdown cells indicate that MOF is required for both HR and NHEJ [26,51,54]. Interestingly, phosphorylation of MOF by ATM facilitates the recruitment of BRCA1 as well as HR-related repair proteins to DNA damage sites in the S/G2 phase [48].
4.3. Cell survival and death
Since its identification, MOF is believed to play an important role in the life or death decision of cells and organisms. In Drosophila, mutation of MOF results in male-specific lethality [8]. In mammals, depletion of MOF leads to peri-implantation death in both males and females [15,16]. Additionally, it is impossible to derive MOF-deficient cell lines from MOF null embryo, as all MOF−/− cells died by apoptosis [15,16]. Moreover, Purkinje cell-specific MOF deficient mice exhibits A-T like phenotype as well as a significant loss of Purkinje cells [52]. Similarly, T-cell-specific deletion of MOF leads to significantly reduced T-cell number as well as defective T-cell maturation and differentiation [57]. These in vivo data suggest that, under normal condition, MOF is indispensible for cell viability.
However, MOF can be pro-apoptotic when cells are subject to ionizing radiation induced stress. MOF acetylates p53 at K120, promotes the expression of BAX and PUMA genes, and thereby induces apoptosis following radiation [31].
A recent study reported that MOF and H4K16 acetylation regulates the outcome of autophagy, a cellular process devoted to stress adaptation [58]. As a catabolic process in which the lysosome degrades damaged organelles, proteins, and other cell contents [59,60], autophagy normally protects cells from apoptosis. However in some cases, autophagy can also contribute to cell death [61,62]. MOF and H4K16 acetylation are down-regulated in cell autophagy induced by various stimuli, which subsequently lead to reduced expression of autophagy-related genes [58]. In addition, overexpression of MOF or inhibition of H4K16 deacetylation increased autophagy flux and can lead to cell death. These results support the existence of a feedback regulatory loop in cells to control autophagy flux. MOF, as a major player in this feedback loop, prevents excessive autophagy flux and fine-tunes survival versus death decision upon induction of autophagy.
4.4. Gene Expression
Acetylation of histones in nucleosomes can lead to the opening of the chromatin structure and increase its accessibility for other proteins to alter transcription. MOF has been shown to be involved in regulating gene expression by increasing H4K16 acetylation. On the Drosophila male X chromosome, MOF is found on both the 5′ and 3′ end of genes, suggesting that the MSL complex and H4K16 acetylation may increase gene expression through the transcriptional elongation process [23]. The NSL complex in Drosophila has also been reported to regulate gene expression through H4K16 acetylation on the non- X chromosome. The complex binds to the promoter region of over 4000 genes, of which 70% of these genes are actively transcribed [38]. Additionally, the loss of NSL as well as MOF binding led to a striking reduction of gene expression. Regulation of gene expression by MOF is also found in humans. Similar to its Drosophila othorlog, hMOF accumulates at both gene promoters and coding regions [63,64]. The NSL complex has recently been found to be a co-activator of c-Jun, a proto-oncogene. Knockdown of NSL complex component negatively affected c-Jun downstream gene expression while knockdown of MSL complex had little to no effect [65]. hMOF and H4K16 were also found at active enhancer sites [64,66]. Studies on ESCs further demonstrate that hMOF and H4K16 define a new set of enhancer elements that are independent of H3K27ac and EP300 [66]. Two recent studies investigated two different MOF complexes and gene expression in ESCs [42,43]. The MSL complex regulates expression of genes required for ESCs self-renewal and pluripotency, while the NSL complex regulates housekeeping genes as well as genes associate with growth and proliferation. All these studies suggest that MOF, whether in the MSL or NSL complex, is able to influence gene expression.
5. MOF and Embryonic stem cells
Embryonic stem cells (ESCs) are pluripotent stem cells derived from the inner cell mass of early mammalian embryo. Pluripotency (the ability to differentiate into any cell type) and self-renewal (the ability to indefinitely reproduce themselves) are two key features of ESCs, and both of these features depend on MOF activity. Depletion of MOF in mice leads to peri-implantation lethality [15,16], which indicates the important role of MOF in early embryonic development. However, no ESC line can be established from MOF−/− embryos, suggesting an integral role of MOF in the ES cell self-renewal process. Using conditional MOF knockout ES cells lines, Li et al. reported that MOF depleted ESCs exhibited reduced H4K16 acetylation, loss of self-renewal, and impaired embryoid body formation [64]. Further studies show that MOF is bound to the promoter region of ESC core transcription factors such as Nanog and Oct4, and is crucial for their expression. Interestingly, Genome-wide analysis of MOF binding sites demonstrated that MOF is enriched in TSS but is also distributed evenly in gene-coding regions. Nearly 50% of MOF peaks are enriched in the coding region of ES cells, while only less than 20% is observed in human CD4+ cells, suggesting it as a unique feature for ESCs. Surprisingly, some genes exhibiting MOF binding in their coding regions were up-regulated in MOF null ESCs despite a loss of MOF binding and H4K16 acetylation. Thus MOF binding to the promoter and coding region can modulate the expression of target genes [64].
The different patterns of MOF enrichment in ESCs were further addressed in two recent studies [42,43]. Ravens et al. reported that both MSL and NSL complexes exist in ESCs. While NSL complex is mainly enriched at the promoter region of housekeeping genes and mediates cell proliferation, MSL complex is bound to both promoters and gene bodies and is the main acetyltransferase complex for H4K16 in ESCs [42]. Chelmicki et al. showed that both NSL and MSL complexes could function in concert to ensure proper regulation of gene expression [43].
6. MOF and cancer
Given its pivotal role in DNA damage response and cell cycle regulation, it is not surprising that dysregulation of hMOF has been extensively implicated in human carcinogenesis. The first link of MOF and cancer came from the finding that the levels of H4K16 acetylation were reduced in many different types of primary tumor tissues and tumor cell lines [67]. Furthermore, the levels of hMOF were frequently down regulated in primary breast carcinoma and medulloblastoma [68], which is associated with lower survival rates and has been used as a biomarker for clinical outcome in these tumors. Recently, reduced MOF levels have been observed in many primary tumors and tumor cell lines, including breast cancer [68,69], renal cell carcinoma [70,71], gastric cancer [70,72], ovarian cancer [73,74], hepatocellular carcinoma [75], and colorectal carcinoma [70]. Moreover, modulating hMOF levels and activity in tumor or cell line can significantly impact tumor cell growth both in vivo and in vitro [75] (Table 1).
Table 1.
Dysregulation of hMOF expression in human cancers
| Type of cancer | Sample number | Method | MOF changes | References |
|---|---|---|---|---|
| Breast cancer | N=298 | IHC | Reduced mRNA and protein in tumor samples | 68 |
| Breast cancer | N=127 | RT-PCR | Low mRNA levels in tumor samples | 69 |
| Renal cell carcinoma | N=47 | RT-PCR | Reduced mRNA (> 2-fold) in 35/47 samples (74%) | 70 |
| Renal cell carcinoma | N=21 | RT-PCR/IHC | Reduced mRNA and protein in 19/21 samples (90.5%) | 71 |
| Gastric cancer | N=16 | RT-PCR | Reduced mRNA (> 2-fold) in 15/16 samples (94%) | 70 |
| Gastric cancer | N=52 | RT-PCR | Reduced mRNA (> 2-fold) in 42/52 samples (81%) | 72 |
| Ovarian cancer | N=30 | RT-PCR, immunoblot | Reduced mRNA and protein in tumor samples | 73 |
| Ovarian cancer | N=47 | RT-PCR | Down regulation in 81% of the patients Up regulated in 13% of the patients |
74 |
| Hepatocellular carcinoma | N=70 | RT-PCR, immunoblot, IHC | Reduced mRNA and protein in tumor samples | 75 |
| Colorectal carcinoma | N=44 | RT-PCR | Reduced mRNA (> 2-fold) in 25/44 samples (57%) | 70 |
| Non small cell lung cancer | N=20 | RT-PCR | Increased mRNA in 10/20 samples (50%) | 76 |
| Non small cell lung cancer | N=43 | IHC | High expression in 14/43 cases (32.6%) | 77 |
| Non small cell lung cancer | N=54 | RT-PCR | Increased mRNA and protein in 26/28 patients | 33 |
| Medulloblastoma | N=180 | IHC | Reduced mRNA and protein | 68 |
On the contrary, hMOF and H4K16 acetylation are frequently increased in non-small cell lung cancers (NSCLC). In one study, overexpression of hMOF was observed in 3 out of 5 primary NSCLC cell lines, 10 out of 20 frozen lung cancer tissues, as well as 37.6% (184/489) of paraffin-embedded NSCLC tissues [76]. Within NSCLCs, the incidence of hMOF overexpression is higher in squamous cell carcinomas than that of adenocarcinomas. The levels of H4K16 acetylation ware also increased in NSCLCs compared to adjacent normal lung tissues [76] [77]. Skp2, an important gene for the G1 to S phase progression, exhibited increased enrichment of hMOF and H4K16 acetylation in the promoter region of NSCLCs [77]. A recent study reported that hMOF is able to acetylate Nrf2 to promote its nuclear translocation and transcriptional activity, which subsequently facilitates anti-drug response in NSCLCs [33] (Table 1).
7. Conclusion
This review summarizes recent studies on the function and activity of MOF. As a H4K16 specific histone acetyltransferase, MOF plays important roles in dosage compensation, ESC self-renewal, DNA damage and repair, cell survival, and gene expression. Dysregulation of MOF has been implicated in tumor formation and progression of many types of human cancers. MOF is a component of two different protein complexes and requires protein partners to control its HAT activity and substrate specificity. Future challenges will include understanding how the MSL and NSL complexes, as well as different components in the complex, modulate MOF activity and specificity. In addition, it will be important to identify new downstream targets of MOF particularly the non-histone targets. Moreover, increasing evidence suggests the involvement of hMOF in carcinogenesis. Knowing that inhibited MOF activity can lead to the hypoacetylation of H4K16, it becomes important for future studies to investigate what types of external factors, such as environmental toxins, are capable of impeding MOF activity. Although dysregulation of hMOF expression has been found in many human cancers, little is known regarding the cis-regulatory element or trans-regulator of MOF, which indicates that far more research is needed in the upstream regulators of hMOF as well as their changes in human cancer. It is worth noting that the role of hMOF in tumor formation and progression may vary in different types of cancer. Further studies on hMOF and cancer will not only help us understand the mechanism underlying hMOF related cancer, but also provide valuable insights for future cancer therapy.
Acknowledgments
The work was supported by NIH center grant number R03 ES023028, P30 ES000260 R01 ES 022935 and R01 ES 034174.
Footnotes
Conflict of interest
All authors declare no conflicts of interest in this paper.
References
- 1.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Shogren-Knaak M, Ishii H, Sun JM, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 4.Avvakumov N, Cote J. Functions of myst family histone acetyltransferases and their link to disease. Subcell Biochem. 2007;41:295–317. [PubMed] [Google Scholar]
- 5.Rea S, Xouri G, Akhtar A. Males absent on the first (MOF): from flies to humans. Oncogene. 2007;26:5385–5394. doi: 10.1038/sj.onc.1210607. [DOI] [PubMed] [Google Scholar]
- 6.Mendjan S, Taipale M, Kind J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Mellert HS, McMahon SB. hMOF, a KAT(8) with many lives. Mol Cell. 2009;36:174–175. doi: 10.1016/j.molcel.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, et al. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 10.McElroy KA, Kang H, Kuroda MI. Are we there yet? Initial targeting of the Male-Specific Lethal and Polycomb group chromatin complexes in Drosophila. Open Biol. 2014;4:140006. doi: 10.1098/rsob.140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bone JR, Lavender J, Richman R, et al. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 12.Neal KC, Pannuti A, Smith ER, et al. A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim Biophys Acta. 2000;1490:170–174. doi: 10.1016/s0167-4781(99)00211-0. [DOI] [PubMed] [Google Scholar]
- 13.Keller CI, Akhtar A. The MSL complex: juggling RNA-protein interactions for dosage compensation and beyond. Curr Opin Genet Dev. 2015;31:1–11. doi: 10.1016/j.gde.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Brockdorff N, Turner BM. Dosage compensation in mammals. Cold Spring Harb Perspect Biol. 2015;7:a019406. doi: 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta A, Guerin-Peyrou TG, Sharma GG, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas T, Dixon MP, Kueh AJ, et al. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Utley RT, Cote J. The MYST family of histone acetyltransferases. Curr Top Microbiol Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- 18.Thomas T, Voss AK. The Diverse Biological Roles of MYST Histone Acetyltransferase Family Proteins. Cell Cycle. 2014;6:696–704. doi: 10.4161/cc.6.6.4013. [DOI] [PubMed] [Google Scholar]
- 19.Kadlec J, Hallacli E, Lipp M, et al. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol. 2011;18:142–149. doi: 10.1038/nsmb.1960. [DOI] [PubMed] [Google Scholar]
- 20.Akhtar A, Becker PB. The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep. 2001;2:113–118. doi: 10.1093/embo-reports/kve022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen PR, Nietlispach D, Buscaino A, et al. Structure of the chromo barrel domain from the MOF acetyltransferase. J Biol Chem. 2005;280:32326–32331. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- 22.Conrad T, Cavalli FM, Holz H, et al. The MOF chromobarrel domain controls genome-wide H4K16 acetylation and spreading of the MSL complex. Dev Cell. 2012;22:610–624. doi: 10.1016/j.devcel.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 24.Smith ER, Cayrou C, Huang R, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taipale M, Rea S, Richter K, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Corsa CA, Pan PW, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buscaino A, Kocher T, Kind JH, et al. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell. 2003;11:1265–1277. doi: 10.1016/s1097-2765(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Schmitz KM, Mayer C, et al. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 29.Bueno-Perez R, Martin-Calvo A, Gomez-Alvarez P, et al. Enantioselective adsorption of ibuprofen and lysine in metal-organic frameworks. Chem Commun (Camb) 2014;50:10849–10852. doi: 10.1039/c4cc03745f. [DOI] [PubMed] [Google Scholar]
- 30.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 31.Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes SM, Stanek TJ, Frank A, et al. Acetylation of the DNA binding domain regulates transcription-independent apoptosis by p53. J Biol Chem. 2009;284:20197–20205. doi: 10.1074/jbc.M109.026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Ye X, Tang N, et al. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. Br J Pharmacol. 2014;171:3196–3211. doi: 10.1111/bph.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales V, Straub T, Neumann MF, et al. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 2004;23:2258–2268. doi: 10.1038/sj.emboj.7600235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rea S, Akhtar A. MSL proteins and the regulation of gene expression. Curr Top Microbiol Immunol. 2006;310:117–140. doi: 10.1007/3-540-31181-5_7. [DOI] [PubMed] [Google Scholar]
- 37.Cai Y, Jin J, Swanson SK, et al. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raja SJ, Charapitsa I, Conrad T, et al. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell. 2010;38:827–841. doi: 10.1016/j.molcel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 39.Rattner BP, Meller VH. Drosophila male-specific lethal 2 protein controls sex-specific expression of the roX genes. Genetics. 2004;166:1825–1832. doi: 10.1534/genetics.166.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feller C, Prestel M, Hartmann H, et al. The MOF-containing NSL complex associates globally with housekeeping genes, but activates only a defined subset. Nucleic Acids Res. 2012;40:1509–1522. doi: 10.1093/nar/gkr869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam KC, Muhlpfordt F, Vaquerizas JM, et al. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8:e1002736. doi: 10.1371/journal.pgen.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravens S, Fournier M, Ye T, et al. Mof-associated complexes have overlapping and unique roles in regulating pluripotency in embryonic stem cells and during differentiation. Elife. 2014:3. doi: 10.7554/eLife.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chelmicki T, Dundar F, Turley MJ, et al. MOF-associated complexes ensure stem cell identity and Xist repression. Elife. 2014;3:e02024. doi: 10.7554/eLife.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun B, Guo S, Tang Q, et al. Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274. Cell Res. 2011;21:1262–1266. doi: 10.1038/cr.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L, Li L, Lv X, et al. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res. 2011;21:1182–1195. doi: 10.1038/cr.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;285:11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan H, Rossetto D, Mellert H, et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012;31:58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta A, Hunt CR, Hegde ML, et al. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep. 2014;8:177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 50.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 51.Gupta A, Sharma GG, Young CS, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar R, Hunt CR, Gupta A, et al. Purkinje cell-specific males absent on the first (mMof) gene deletion results in an ataxia-telangiectasia-like neurological phenotype and backward walking in mice. Proc Natl Acad Sci U S A. 2011;108:3636–3641. doi: 10.1073/pnas.1016524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sulli G, Di Micco R, d’Adda di Fagagna F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer. 2012;12:709–720. doi: 10.1038/nrc3344. [DOI] [PubMed] [Google Scholar]
- 54.Sharma GG, So S, Gupta A, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 56.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 57.Gupta A, Hunt CR, Pandita RK, et al. T-cell-specific deletion of Mof blocks their differentiation and results in genomic instability in mice. Mutagenesis. 2013;28:263–270. doi: 10.1093/mutage/ges080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fullgrabe J, Lynch-Day MA, Heldring N, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fullgrabe J, Klionsky DJ, Joseph B. Histone post-translational modifications regulate autophagy flux and outcome. Autophagy. 2013;9:1621–1623. doi: 10.4161/auto.25803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marino G, Niso-Santano M, Baehrecke EH, et al. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X, Li L, Pandey R, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Long Y, Xing Z, et al. C-Jun recruits the NSL complex to regulate its target gene expression by modulating H4K16 acetylation and promoting the release of the repressive NuRD complex. Oncotarget. 2015;6:14497–14506. doi: 10.18632/oncotarget.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor GC, Eskeland R, Hekimoglu-Balkan B, et al. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 2013;23:2053–2065. doi: 10.1101/gr.155028.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 68.Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122:1207–1213. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- 69.Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol. 2011;31:1594–1609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao L, Zhu L, Yang J, et al. Correlation of low expression of hMOF with clinicopathological features of colorectal carcinoma, gastric cancer and renal cell carcinoma. Int J Oncol. 2014;44:1207–1214. doi: 10.3892/ijo.2014.2266. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Zhang R, Wu D, et al. Epigenetic change in kidney tumor: downregulation of histone acetyltransferase MYST1 in human renal cell carcinoma. J Exp Clin Cancer Res. 2013;32:8. doi: 10.1186/1756-9966-32-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu L, Yang J, Zhao L, et al. Expression of hMOF, but not HDAC4, is responsible for the global histone H4K16 acetylation in gastric carcinoma. Int J Oncol. 2015;46:2535–2545. doi: 10.3892/ijo.2015.2956. [DOI] [PubMed] [Google Scholar]
- 73.Cai M, Hu Z, Liu J, et al. Expression of hMOF in different ovarian tissues and its effects on ovarian cancer prognosis. Oncol Rep. 2015;33:685–692. doi: 10.3892/or.2014.3649. [DOI] [PubMed] [Google Scholar]
- 74.Liu N, Zhang R, Zhao X, et al. A potential diagnostic marker for ovarian cancer: Involvement of the histone acetyltransferase, human males absent on the first. Oncol Lett. 2013;6:393–400. doi: 10.3892/ol.2013.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Liu H, Pan H, et al. The histone acetyltransferase hMOF suppresses hepatocellular carcinoma growth. Biochem Biophys Res Commun. 2014;452:575–580. doi: 10.1016/j.bbrc.2014.08.122. [DOI] [PubMed] [Google Scholar]
- 76.Song JSCS, Lee JY, Kim DK, Kim YH, Jang SJ. The Histone Acetyltransferase hMOF is Overexpressed in Non-small Cell Lung Carcinoma. J Pathol Transl Med. 2011;45:386–396. [Google Scholar]
- 77.Zhao L, Wang DL, Liu Y, et al. Histone acetyltransferase hMOF promotes S phase entry and tumorigenesis in lung cancer. Cell Signal. 2013;25:1689–1698. doi: 10.1016/j.cellsig.2013.04.006. [DOI] [PubMed] [Google Scholar]