Abstract
Macrophages are phagocytic cells that play essential roles in innate immunity and lipid homeostasis. The uptake of modified lipoproteins is an important early event in the development of atherosclerosis. We analyzed the ability of modified low-density lipoprotein (LDL) (oxidized and acetylated) to alter the expression and activity of arginases (ArgI and ArgII) in macrophages. We show that ArgI expression is potently induced by both oxidized and acetylated LDL in macrophages. We further show that this effect is mediated by peroxisome proliferator-activated receptors (PPAR). ArgI expression is highly responsive to agonists for PPARγ and PPARδ but not PPARα. Moreover, the induction of ArgI by both PPAR agonists and IL-4 is blocked in macrophages from PPARγ- and PPARδ-deficient mice. Functionally, PPAR activity induces macrophage activation toward a more Th2 immune phenotype in a model of Leishmania major infection. We show that PPARγ and -δ ligands promote intracellular amastigote growth in infected macrophages, and this effect is dependent on both PPAR expression and Arg activity. Collectively, our results strongly suggest that ArgI is a key marker of the alternative program triggered by PPAR in macrophages.
ATHEROSCLEROSIS IS A chronic inflammatory disease triggered by several factors including hypercholesterolemia (1). One of the initial events in the development of atherosclerosis is the uptake and oxidation of low-density lipoproteins (LDL) by resident macrophages in the arterial wall. In the face of hypercholesterolemia, an increased number of monocytes are recruited to the subendothelial space, where they take up LDL and become the so-called foam cells (2).
The uptake and internalization of oxidized LDL (oxLDL) and their lipid content induce important changes in macrophage gene expression, especially those that control cellular lipid homeostasis (3). At the same time, the conversion of these macrophages into foam cells triggers a dramatic change in their activation stage. Therefore, it is reasonable to hypothesize that the cellular response to modified lipids could alter the inflammatory properties of macrophages.
An important family of transcription factors that regulates the expression of genes linked to lipid metabolism is the peroxisome proliferator activated receptor (PPAR) subfamily of nuclear receptors. Peroxisome proliferator-activated receptors (PPAR) are ligand-dependent transcription factors that heterodimerize with the retinoid X receptor (RXR) (4). Early studies showed that PPARγ promotes macrophage gene expression and uptake of oxLDL (5). Several reports have also demonstrated that PPARγ inhibits the expression of proinflammatory genes, including cytokines and inducible nitric oxide synthase (iNOS) (6, 7).
Arginases catalyze the hydrolysis of l-arginine to l-ornithine and urea, and both isoforms are constitutive in resting mouse macrophages (8). Previous studies demonstrated that arginase I is induced by Th2-derived cytokines in macrophages (9) and this enzyme has therefore been considered one of the hallmarks of alternative macrophage activation (10). However, arginases are also coinduced with iNOS and other acute immune enzymes under proinflammatory challenge such as bacterial lipopolysaccharide (9).
The potential role of arginases in atherosclerosis is currently a subject of intense investigation. The majority of these studies have analyzed the contribution of arginase in vascular endothelial cells and smooth muscle cells. For example, Ignarro and colleagues (11) demonstrated that arginase I induction in aortic smooth muscle cells by Th2-derived cytokines promoted cell proliferation. Moreover, both arginases are expressed in atherosclerotic lesions as shown in models of hyperlipidemic rabbits (12). Arginase II is expressed predominantly in endothelial cells and reduces nitric oxide release in atherosclerotic mice (13). Remarkably, arginase II expression and activity are up-regulated by oxLDL in human aortic endothelial cells (14). In addition, arginase II was found to be a direct target of liver X receptor (LXR) in macrophages and contributes to some of the antiinflammatory effects observed with LXR agonists on macrophage activation (15).
The aim of the present work was to study the regulation of arginase expression by modified lipoproteins and its derivatives in macrophages. We show that modified LDL induce arginase I expression through PPARγ and -δ activation. The induction of arginase I by modified LDL is mimicked by PPARγ and -δ synthetic ligands and blocked by PPAR-specific antagonists. Moreover, response of arginase I expression to synthetic ligands is lost in PPARγ- and PPARδ-deficient cells. Finally, we show that arginase I induction by PPAR favors the growth of Leishmania major inside macrophages, suggesting that this enzyme contributes to an immune deactivation program triggered by PPAR.
RESULTS
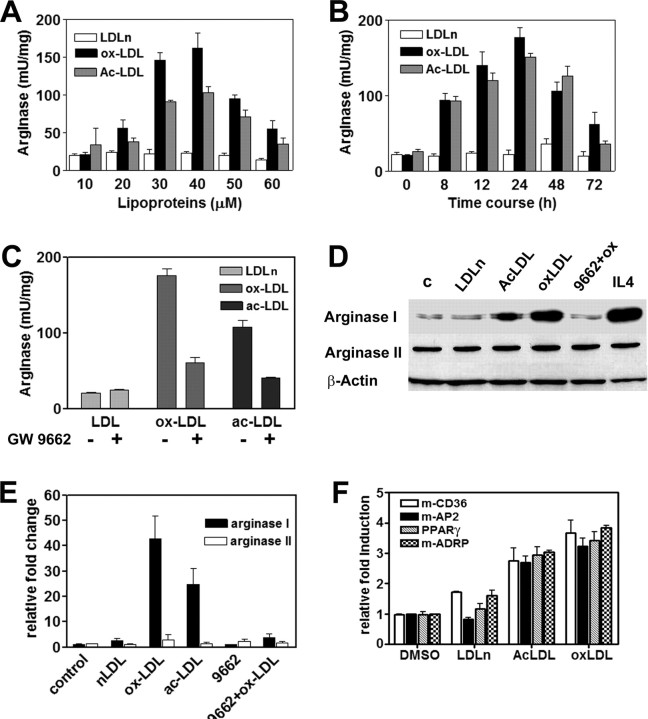
OxLDL Induces Arginase I in Bone-Marrow-Derived Macrophages
The metabolic fate of l-arginine in macrophages between NOS and arginase has been demonstrated to be regulated by the activation stage of the cell (16). In the context of atherosclerosis, the internalization of modified lipoproteins inhibits the inducible form of NOS (17), whereas in endothelial cells, arginase is up-regulated in response to oxLDL (14). To analyze the regulation of arginase isoforms in in vitro lipid-loaded cells, we measured arginase activity in macrophages treated with increasing concentrations of oxLDL and acetylated LDL (acLDL) (Fig. 1A). Both modified lipoproteins were able to increase arginase activity in a dose-dependent manner, with 30 μm and 24 h treatment being the optimal conditions (Fig. 1B). The response was specific for modified LDL because native LDL (nLDL) did not increase arginase levels significantly. Resting bone marrow-derived macrophages (BMDM) constitutively express arginase I and II (Fig. 1D). Modified lipoproteins selectively induced arginase I mRNA (Fig. 1E) and protein (Fig. 1D).
Fig. 1.
Modified Lipoproteins Induce Arginase I in BDMD
A, Analysis of arginase activity in cells treated with increasing concentrations of oxLDL, acLDL, and nLDL for 24 h; B, time course of arginase induction in BMDM treated with 30 μm of each type of LDL; C, macrophages were pretreated overnight with 1 μm GW9662 and then stimulated with 30 μm modified LDL for 24 h; D, arginase I and II protein expression. Proteins were analyzed as described in Materials and Methods. β-Actin was used as loading control. IL-4, (2.5 ng/ml, 24 h) was used as positive control. c, Control; 9662+ox; GW9662 plus oxLDL. E, Arginase I and arginase II expression levels determined by TaqMan quantitative real-time PCR. The experimental conditions for D and E were identical to those in C. Data are expressed as the mean ± sd of triplicate measurements. F, Macrophages were loaded with LDL for 24 h and then used for RNA extraction. CD36, AP2, PPARγ, and ADRP gene expression were analyzed by real-time PCR SYBR Green assays and normalized to 36B4 gene expression.
It is well documented that uptake and internalization of oxidized lipoproteins activate PPARγ expression and activity in macrophages (18). Thus, to better understand the regulation of arginase I by modified lipoproteins, we treated lipid-loaded cells with the PPAR antagonist GW9662 (19). Our results show that pretreatment of macrophages with 1 μm GW9662 potently reduced the induction of arginase I activity by oxLDL and acLDL (Fig. 1C) and also blocked the induction of ArgI protein (Fig. 1D) and mRNA (Fig. 1E). Moreover, the induction of the enzyme by oxLDL was also inhibited by 1 μm of each, GW5393 (20) and T0070907 (21), two structurally different PPAR antagonists (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org).
Although the PPAR antagonists used here are known to be specific for PPARγ, at the concentrations used in this work (1 or 2 μm), they are inhibiting all PPAR isoforms, according to the references cited above. The mode of action of GW9662 and T0070907 is to covalently modify a cysteine residue in the ligand-binding domain of PPAR (19, 21), whereas GW5393 was designed as a PPAR ligand unable to recruit the coactivator cAMP response element binding protein-binding protein (CBP) to the receptor (20). Because murine macrophages constitutively express more than one PPAR isoform, we used these molecules as pan-antagonists to be sure that the induction of arginase I by modified LDL were dependent on PPAR activation.
Collectively, these results strongly suggest that the induction of arginase I in foam cells is mediated through a PPAR-dependent mechanism. Indeed, under the same experimental conditions, modified lipoproteins triggered the expression of PPARγ and others known PPAR target genes such as CD36, AP2 (22), or ADRP (adipose differentiation-related protein) (Fig. 1F). These genes have been found to be induced by oxLDL and up-regulated in atherosclerotic lesions (23, 24).
It is important to note that in Fig. 1, A and B, the levels of arginase-specific activity were calculated as a function of protein concentration. We paid special attention to this aspect because several reports have demonstrated that oxLDL induces macrophage proliferation (25). In our experimental conditions, we did not observe changes in protein concentration that could significantly affect enzyme-specific activity.
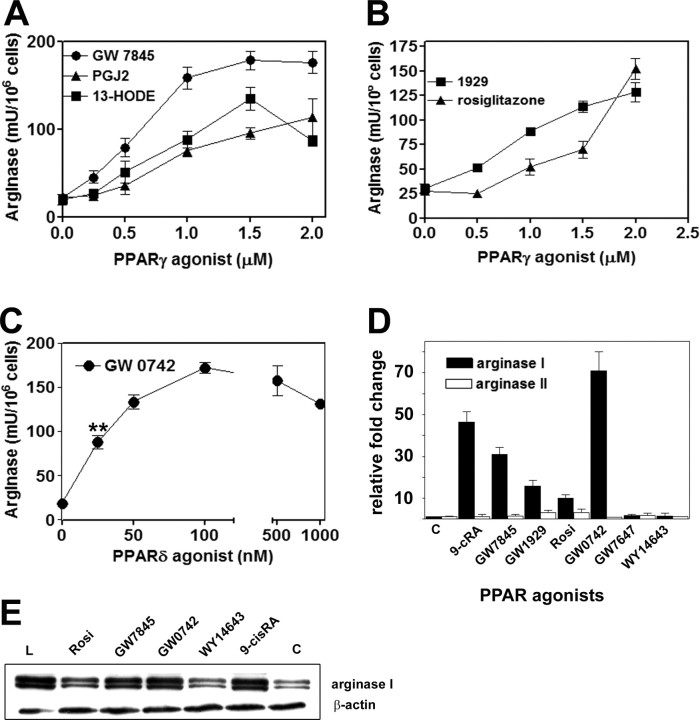
Arginase I Is Selectively Induced by PPARγ and -δ Agonists
To identify the PPAR isoforms responsible for arginase I expression, we next treated primary macrophages with different PPAR agonists. The results are presented in Fig. 2. Interestingly, the natural activators of PPAR, such as the eicosanoid 15dPGJ2 and the oxidized derivative of linoleic acid, (13-HODE), dose-dependently induced arginase I (Fig. 2A). We also used the thiazolidinedione rosiglitazone (Fig. 2B), a potent PPARγ synthetic activator and a widely used insulin sensitizer (26). This drug also induced arginase I and showed maximal effects at concentrations above 1 μm. Finally, GW7845 and GW1929, two non-thiazolidinedione, potent synthetic PPARγ agonists, also significantly increased arginase activity and expression.
Fig. 2.
PPARγ and -δ Agonists Induce Arginase I in BMDM
A and B, Dose response of arginase induction with different PPARγ agonists. Cells were treated with agonists for 24 h and enzyme activity measured as described in Materials and Methods; C, arginase activity in cells treated with increasing concentrations of PPARδ agonist GW0742 for 24 h; D, arginase I and arginase II mRNA expression levels from cells treated with the indicated ligands for 24 h. The relative fold induction was calculated by using 18S mRNA as internal control and TaqMan real-time quantitative PCR; E, protein expression by Western blot. The concentrations of agonists used in D or E were 1 μm PPARγ agonists rosiglitazone (Rosi), GW1929, or GW7845; 100 nm PPARδ agonist GW0742; 2 μm of the PPARα agonists GW7647 and WY14643, and 0.5 μm 9-cRA. Lm Liver extract, used as positive control for arginase I. β-Actin was used as loading control. Data represent the mean values and sd of replicate cultures of four independent experiments. **, P < 0.01 by the Student’s t test at 25 nm, compared with the control value.
We also treated cells with PPARδ and PPARα agonists under the same experimental conditions. Remarkably, the specific PPARδ agonist GW0742 (27) induced arginase activity very efficiently, showing significant induction at 25–100 nm concentrations (Fig. 2C). In contrast, treatment with low micromolar concentrations of two different PPARα ligands (GW7647 and WY14643) did not result in significant changes in arginase activity (data not shown). These results are consistent with the minor expression of PPARα in mouse macrophages, in agreement with previous reports (28). Both RNA expression levels measured by real-time quantitative PCR (Fig. 2D) and protein expression (Fig. 2E) confirmed the induction of arginase I by PPARγ and -δ ligands, whereas the two PPARα activators did not increase protein or mRNA levels. Together, these results demonstrate that arginase I expression and activity are specifically modulated by PPARγ and -δ activators in primary macrophages.
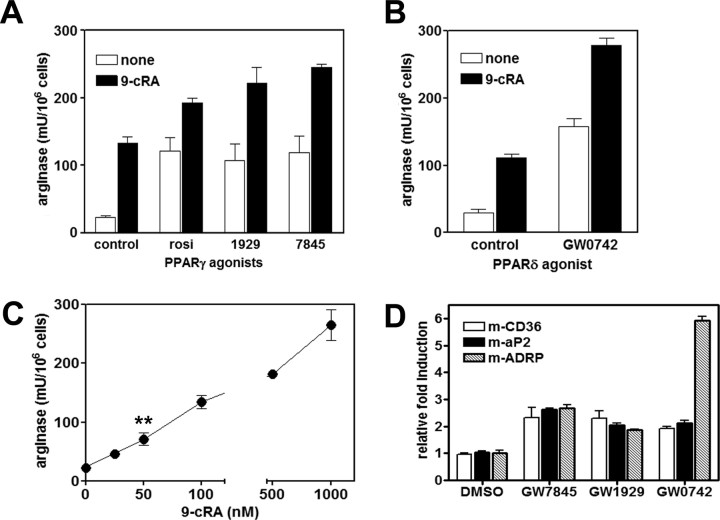
The Induction of Arginase by PPARγ and -δ Ligands Is Increased by RXR Agonists
Because PPAR/RXR heterodimers can be activated through both the PPAR and the RXR arms of the complex, we analyzed arginase activity using the RXR ligand 9-cis-retinoic acid (9-cRA), in the presence or absence of different PPAR agonists. The data of Fig. 3 demonstrate an additive effect on arginase activity by cotreatment with PPARγ and -δ activators (Fig. 3, A and B). On the other hand, cis-RA alone also induced arginase activity at concentrations from 50 nm (Fig. 3D). As a control, we verified that PPAR ligands increased the expression of the known target genes CD36, AP2, and ADRP (24, 29, 30) by real-time PCR (Fig. 3E).
Fig. 3.
The Induction of Arginase by PPARγ and -δ Ligands Is Increased in the Presence of the RXR Ligand 9-cRA
A, Arginase activity of cells treated with PPARγ agonists, added at the same dose as in Figs. 1 and 2 (white bars) or in combination with 100 nm 9-cRA for 24 h; B, arginase activity in cells treated with 100 nm GW0742 with or without 100 nm 9-cRA for 24 h; C, dose response of arginase induction by 24 h treatment with increasing concentrations of 9-cRA; E, mRNA relative induction of PPAR target genes CD36, AP2, and ADRP. BMDM were treated for 24 h with 1 μm concentration of the indicated PPAR agonists, and relative gene expression was analyzed by real-time PCR SYBR Green assays, normalized to 36B4 expression, as previously described. **, P < 0.01 by the Student’s t test at 50 nm, compared with the control value.
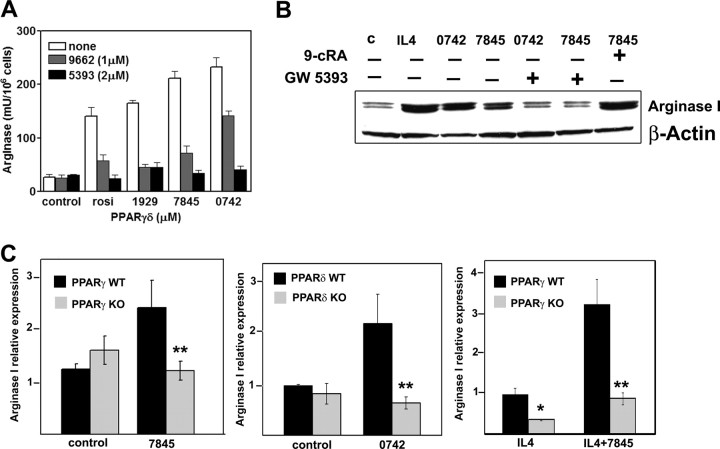
Arginase I Induction Is Inhibited by PPAR Antagonists and Reduced in Cells from PPARγ and PPARδ Knockout Mice
To confirm that the induction of arginase I expression by PPAR agonists was receptor dependent, we used two different experimental approaches. First, arginase activity and protein levels were significantly inhibited by two structurally unrelated PPAR antagonists, GW9662 and GW5393 (Fig. 4, A and B). Second, we analyzed macrophages with conditional disruption of PPARγ and macrophages from PPARδ-deficient mice. Macrophages from C57BL/6 wild-type (WT) mice showed increased mRNA expression of arginase I when treated for 12 h with 100 nm PPARγ agonist GW7845 or with 100 nm GW0742 (Fig. 4C). However, arginase I induction was completely abrogated in cells from either PPARγ0 or PPARδ-deficient mice. Moreover, because arginase I is induced by IL-4 in murine macrophages, we used this Th2-derived cytokine as a positive control in all our experiments.
Fig. 4.
Arginase I Induction by PPARγ and -δ Is Inhibited by PPAR Antagonists and Reduced in Macrophages from PPARγ- and -δ-Deficient Mice
A, BMDM were pretreated overnight with the PPAR antagonists GW9662 or GW5393 and then triggered with the corresponding agonists for 24 h. After this time, arginase activity was measured in cell lysates. B, Immunoblots show arginase I protein inhibition by PPAR antagonists and increased amount of protein in the presence of PPAR-RXR heterodimer. IL-4 is shown as positive control. C, Arginase I relative expression in PPARγ WT and KO peritoneal macrophages with or without 100 nm GW7845 for 12 h or PPARδ WT and KO cultures treated with 100 nm 0742 for 12 h. Finally, the relative induction of arginase I to IL-4 was analyzed in cells from PPARγ WT and KO pretreated overnight with 2.5ng/ml IL-4 and then with the PPARγ ligand for another 12 h. **, P < 0.01 by the Student’s t test for arginase I expression by PPARγ and -δ agonists in experiments with macrophages from PPARγ- and PPARδ-deficient mice vs. the WT mice; *, P < 0.05 for arginase I expression by IL-4 in cells from PPARγ WT cells compared with those from the knockout.
It has also been documented that IL-4 induces PPARγ expression in macrophages (31). Therefore we checked whether arginase induction by IL-4 could be inhibited by PPAR antagonists. Pretreatment with either GW9662 or GW5393 resulted in more than 50% inhibition of enzyme activity (supplemental Fig. 1). As expected, expression of arginase I mRNA by IL-4 was highly increased by cotreatment with GW7845. Furthermore, the synergistic effect of GW7845 on IL-4-induced arginase I expression was completely suppressed in PPARγ-deficient macrophages.
Together, these results demonstrate that arginase I is specifically induced by activation of PPARγ and -δ isoforms in macrophages.
PPAR Ligands Promote L. major Growth in Macrophages in an Arginase-Dependent Manner
The mouse model of L. major infection between the susceptible BALB/c and resistant C57BL/6 strains is one of the best characterized examples in which macrophage arginase I, induced in the context of a predominant Th2 response in susceptible mice, is triggering the growth of parasites inside macrophages (32). To determine the functional impact of PPAR-dependent arginase regulation for immune responses, we treated Leishmania-infected BMDM with PPAR ligands. The results presented in Fig. 5 clearly demonstrate that both GW7845 and GW0742, when added in vitro to infected macrophages, significantly increased the growth of intracellular Leishmania amastigotes (Fig. 5, A and B, and supplemental Fig. 2). Moreover, Leishmania growth was prevented by pretreatment with 2 μm PPAR antagonist GW5393 or by adding Nω-hydroxy-nor-l-arginine (nor-NOHA), the specific arginase inhibitor, together with either the PPARδ agonist GW0742 (Fig. 5, A and B) or PPARγ ligand GW7845 (supplemental Fig. 2). We also show the potent induction of parasitic growth achieved in the presence of IL-4 as a positive control for arginase I induction. Intracellular amastigote growth was strictly correlated with the levels of macrophage arginase I activity. In cells cotreated with PPAR/RXR ligands (supplemental Fig. 2), the amount of intracellular parasites was similar to those infected in the presence of IL-4, and both effects were reverted by Nor-NOHA.
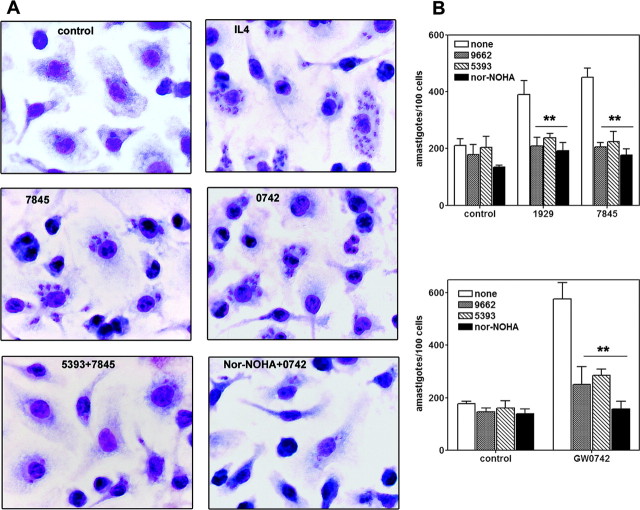
Fig. 5.
PPARγ/δ-Dependent Arginase I Induction Promotes the Growth of L. major Inside Macrophages
A, Micrographs of L. major-infected BMDM. Cells were treated with PPAR agonists (1 μm 7845 or 100 nm 0742) for 2 h or alternatively pretreated with 1 μm concentration of each antagonist for 24 h and PPAR agonist during 2 h and finally infected with stationary phase L. major promastigotes for 24 h. After this time, cultures were stained as described in Materials and Methods. Original magnification, ×100. B, Enumeration of intracellular amastigotes in infected BMDM. Data are the results of counting the parasites from micrographs of A, where the errors bars represent results obtained from three independent macrophage infections performed in triplicate. White bars are control infected cultures or cells pretreated 2 h with PPAR agonists, infected, and counted 24 h after infection. PPAR antagonists were added 24 h before the agonists, and 100 μm Nor-NOHA (black bars) was added at the same time as the agonists. **, P < 0.01 by the Student’s t test of intracellular parasite counts of infected cells treated with PPAR ligands alone compared with those pretreated with PPAR antagonists of nor-NOHA.
Together, these results establish a role for PPARγ and PPARδ in arginase I expression and alternative macrophage activation.
DISCUSSION
The role of arginase in the context of vascular pathology has recently become an area of intense investigation (33). Both arginase isoforms are present in human and mouse atherosclerotic lesions (34), and recent studies have established a direct correlation between arginase I polymorphisms and the risk of myocardial infarction (35). However, the mechanisms that regulate arginase expression in macrophages are not completely understood.
Here we have analyzed the expression of arginase isoforms in lipid-loaded macrophages. We showed that low doses of modified LDL (oxLDL and acLDL) induce arginase I expression and that this effect is mediated by activation of PPAR signaling. Finally, we showed that induction of arginase expression by the PPAR pathway renders macrophages more susceptible to L. major infection. These studies demonstrate that regulation of cellular metabolism by PPAR have a direct consequence for immune responses.
Previous studies have shown that PPARγ as well as arginase I expression is induced by the Th2-derived cytokine IL-4 (31, 9), providing another link between the two genes. Furthermore, PPARγ and arginase I may also be connected in the context of inflammation.
Activation of PPARγ in macrophages leads to transcriptional repression of a cohort of inflammatory genes induced by external insults, including iNOS, the inducible form of NOS (36). Arginase and iNOS share l-arginine as a substrate, and both enzymes are believed to play reciprocal roles when both are present in immune cells (37). Finally, arginase II has been shown to be a target of LXR (15), another nuclear receptor previously shown to be up-regulated by PPAR ligands in human macrophages (38).
Previous studies have also demonstrated that arginase II is regulated by oxLDL in human aortic endothelial cells (14). Arginase II induction in the Ryoo et al. (14) model contributed to endothelial dysfunction by down-regulation of NO production. In contrast, in our experimental conditions, the addition of oxLDL to resting macrophages did not change either iNOS arginase II expression or nitric oxide release (17), and thus, arginase I is the only arginine-metabolizing enzyme induced in our system.
The demonstration that arginase I expression is regulated by PPARγ/δ in lipid-loaded cells reinforces previous studies showing that PPAR contributed to the modification of macrophage functions and the reprogramming of their activation status. Interestingly, arginase I induction has been proven to be a major component of this alternative status, triggered by a Th2-predominant immune response (16). Our results indicate that macrophage PPAR activity impacts the balance of Th1/Th2 responses through specific induction of arginase I expression and activity. During the preparation of the present manuscript, Chawla and colleagues (39) reported the identification of a peroxisome proliferator response element in the promoter region of arginase I. Their results also suggest that PPAR activity participates in transcriptional programs that switch the macrophage toward an alternative activation state.
We showed here using an L. major infection model that arginase induction by PPAR changes macrophage functions to a more permissive stage that is suitable for parasite growth. This activation status closely resembles the one triggered by a Th2 response, because the magnitude of parasite proliferation in the presence of PPAR ligands is comparable to that achieved in the presence of IL-4. Inhibition of parasite growth by both PPAR antagonists and arginase inhibition clearly demonstrates that these nuclear receptors are key factors in macrophage alternative activation. Our results are consistent with previous studies by Kopf and colleagues (40) in which dyslipidemia inhibited protective Th1-type immunity and increased host susceptibility to L. major infection.
Recently, macrophage arginase I has been identified as candidate gene for atherosclerosis resistance (41). Arginase I is highly expressed in rabbits genetically resistant to atherosclerosis. PPAR agonists exert anti-atherogenic roles in murine models of atherosclerosis. We hypothesize that arginase I induction in foam cells may inhibit atherogenic inflammation. Arginases compete for arginine with NOSs. Therefore, the induction of arginase in foam cells may counter the accumulation of reactive nitrogen intermediates that contribute to the inflammatory process during plaque development. Future studies will be required to test the potential roles of macrophage arginase I activity in atherosclerosis.
It is important to note that arginase I expression is triggered efficiently by PPARγ and PPARδ activity. PPAR have been recently associated with protective pathways that control cellular repair (reviewed in Ref. 42). PPARγ activation potentiates antiinflammatory and antifibrotic actions, whereas PPARδ activation induces cellular proliferation and enhances the expression of fibrotic markers. Therefore, we speculate that induction of arginase I by PPAR could have different consequences depending on the stage of atherosclerosis development. At early stages, arginase I and II isoforms, triggered by PPARγ and LXR receptors, respectively, could counteract inflammation by competing with iNOS. However, as macrophages change their activation programs and as atherosclerosis progresses, the ornithine generated by arginase could be used for cell proliferation and collagen synthesis, both processes essential for tissue remodeling but also profibrotic in pathogenic conditions.
MATERIALS AND METHODS
Reagents and Chemicals
Lipoproteins, oxLDL, acLDL, and nLDL, were from Biomedical Technologies, Inc. (Stoughton, MA) and used according to the manufacturer’s instructions. The RXR agonist 9-cRA; PPARα agonists WY14643 and GW7647, PPARγ agonist GW1929, the PPAR antagonist GW9662 (2-chloro-5-nitrobenzanilide), and the natural PPAR ligands 15dPGJ2 and 13-HODE were purchased from Sigma-Aldrich (Madrid, Spain), whereas the PPARδ agonist GW0742, PPARγ agonist GW7845, and the PPAR antagonist GW5393 (1,3,4-oxadiazole) were provided by Tim Willson and Jon Collins (GlaxoSmithKline, Research Triangle Park, NC). The PPAR antagonist T0070907 (2-chloro-5-nitro-N-4-pyridinyl-benzamide) and rosiglitazone were purchased from Cayman Chemical (Ann Arbor, MI). PPAR and RXR ligands were dissolved in dimethylsulfoxide before use. Recombinant murine IL-4 was obtained from PeproTech, EC Ltd. (London, UK) and used in complete culture media at a concentration of 2.5 ng/ml.
Cell Culture
BMDM were obtained by flushing the femurs from 6-wk-old BALB/c mice (Harlam Iberica, Barcelona, Spain) and cultured in hydrophobic Teflon bags (Cell Genix, Freiburg, Germany) in DMEM containing 10% inactivated fetal calf serum, 2 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (all from Sigma, Spain), and 15% of L929 supernatant as a source of macrophage colony-stimulating factor.
Measurement of Arginase Activity
Arginase activity was measured in macrophages lysates as previously described (43). One unit of enzyme activity is defined as the amount of enzyme that catalyzes the formation of 1 μmol urea/min.
RNA and Protein Analysis
RNA was isolated using TRIzol reagent (Sigma-Aldrich, Spain). Reverse transcription was made by using the high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. TaqMan or SYBR Green real-time quantitative assays were performed using an Applied Biosystems 7500 sequence detector system. The relative expression of arginase I (Arg1) and arginase II (Arg2) genes were analyzing by using TaqMan Gene Expression Assays, and the relative expression in each experiment was compared by the 2−ΔΔCt calculation method as described (44). The culture conditions used in this study did not alter the expression of 18S rRNA, thus validating its use as endogenous control. Primers and probes for arginase I and II and 18S rRNA were obtained from Applied Biosystems.
The rest of the genes were analyzed using SYBR Green gene expression assays. In these experiments, each sample was run in duplicate and was normalized to 36B4, as previously described (45). Primers and probes are published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org (supplemental Table 1).
Protein expression was assayed by Western blot using macrophage lysates with the same protein content (assayed by the Bradford method; Bio-Rad, Spain). Briefly, Proteins were separated by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membranes, and then incubated with anti-arginase I (1:1000; BD Transduction Laboratories, Lexington, KY), anti-arginase II (1:5000; Santa Cruz Biotechnology, Santa Cruz, CA), or anti-β-Actin (1:3000; Sigma) which served as loading control. Goat antirabbit IgG-horseradish peroxidase secondary antibody (1:10000; Santa Cruz Biotechnology) was used against arginase II and β-actin, and goat antimouse IgG (1:3000; Pierce, Rockford, IL) were used against arginase I. Detection was made with and ECL-Plus western blotting detection system (Amersham Biosciences, Uppsala, Sweden).
Leishmania Infection
L. major promastigotes (MHOM/IL/80/Friedlin) were cultured as previously described (31). Cells were cultured onto round slides inside 24-well plates. PPAR antagonists were added to macrophage cultures 24 h before infection, and PPAR agonists were added 2 h before the infection. Then, stationary-phase promastigotes were added to BMDM at a parasite to cell ratio of 3:1. After 24 h infection, slides were mounted and stained with Diff-Quik (Panreac, Barcelona, Spain). Results are presented as the number of intracellular amastigotes per 100 cells.
Arginase I Expression in Macrophages from PPARγ- and PPARδ-Deficient Mice
Primary peritoneal macrophages were elicited from thioglycollate-injected mice. Cells were derived from floxed PPARγ-MxCre−/+ mice and PPARδ−/− mice (C57BL/6 background). The PPARγ fl/fl mice were provided by Ron Evans, and the PPARδ−/− mice were provided by Frank Gonzalez. Mice were maintained on standard chow diet in pathogen-free conditions. The cells were flushed and cultured in DMEM with 10% fetal bovine serum (Omega Scientific, Tarzana, CA) and 1% penicillin/streptomycin (Cellgro, Lawrence, KS). BMDM were derived by flushing cells from the femur and tibia of floxed PPARγ-MxCre−/+ and PPARδ−/− mice. Cells were then differentiated using DMEM, 20% fetal bovine serum, 1% penicillin/streptomycin, and 30% L929-conditioned media. Cells were activated by ligand for 12 h before harvesting total RNA using TRIzol reagent (Invitrogen, Carlsbad, CA). Alternatively, cells were pretreated with 2.5 ng/ml IL-4 for 12 h before activating with ligand for 24 h. Total RNA was harvested after 24 h using TRIzol reagent (Invitrogen). Expression of arginase I was analyzed by quantitative real-time PCR (SYBR Green) using an Applied Biosystems 7900 sequence detector.
Statistical Analysis
All data were processed by using GraphPad Prism 4.0 software and are the result of at least three independent experiments performed in triplicate. Errors bars were calculated as sd of the data, and when necessary, we have analyzed significance by the Student’s t test (paired t test).
Acknowledgments
We thank Ronald Evans and Frank Gonzalez for the PPARγ- and PPARδ-deficient mice. respectively. We also thank Jon Collins and Tim Willson for the GW7845, GW0742, and GW5393.
NURSA Molecule Pages:
Ligands: 15-Deoxy-Δ12 | 9-cis-Retinoic acid | GW 1929 | GW 7647 | GW 9662 | GW0742X | Pirinixic acid | Rosiglitazone;
Nuclear Receptors: PPARα | PPARγ | PPARδ.
Footnotes
This work was supported by Grants from Spanish Ministry of Health (FIS: PI051383) to I.C. and by the Ministry of Education and Science (SAF2005–03270), Fundación “La Caixa” BM05-228, Funcis 67/05, and Fundación Ramón Areces to A.C. A.G.-S. is a recipient of a Ministry of Health training grant. P.T. is an Investigator of the Howard Hughes Medical Institute.
Disclosure Statement: The authors have nothing to disclose.
First Published Online March 6, 2008
Abbreviations: acLDL, Acetylated LDL; BMDM, bone marrow-derived macrophages; 9-cRA, 9-cis-retinoic acid; iNOS; inducible nitric oxide synthase; LDL, low-density lipoprotein; LXR, liver X receptor; oxLDL, oxidized LDL; nLDL, native LDL; nor-NOHA, Nω-hydroxy-nor-l-arginine; PPAR, peroxisome proliferator-activated receptors; RXR, retinoid X receptor; WT, wild type.
References
- 1.Steinberg D 2002. Atherogenesis in perspective: hypercholesterolemia and inflammation as partners in crime. Nat Med 8:1211–1217 [DOI] [PubMed] [Google Scholar]
- 2.Ross R 1993. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature 362:801–809 [DOI] [PubMed] [Google Scholar]
- 3.Castrillo A, Tontonoz P 2004. Nuclear receptors in macrophage biology: at the crossroads of lipid metabolism and inflammation. Annu Rev Cell Dev Biol 20:455–480 [DOI] [PubMed] [Google Scholar]
- 4.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM 1998. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93:241–252 [DOI] [PubMed] [Google Scholar]
- 6.Ricote M, Huang JT, Welch JS, Glass CK 1999. The peroxisome proliferator-activated receptor (PPARγ) as a regulator of monocyte/macrophage function. J Leukoc Biol 66:733–739 [DOI] [PubMed] [Google Scholar]
- 7.Jiang C, Ting AT, Seed B 1998. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature 391:82–86 [DOI] [PubMed] [Google Scholar]
- 8.Luis CA, Reichner JS, Henry WL, Mastrofrancesco B, Gotoh T, Mori M, Albina JE 1998. Distinct arginase isoforms expressed in primary and transformed macrophages: regulation by oxygen tension. Am J Phisiol Regul Integr Comp Physiol 274:R775–R782 [DOI] [PubMed]
- 9.Corraliza IM, Soler G, Eichmann K, Modolell M 1995. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem Biophys Res Commun 206:667–673 [DOI] [PubMed] [Google Scholar]
- 10.Gordon S 2003. Alternative activation of macrophages. Nat Rev Immunol 3:23–35 [DOI] [PubMed] [Google Scholar]
- 11.Wei LH, Wu G, Morris Jr SM, Ignarro LJ 2001. Elevated arginase I expression in rat aortic smooth muscle cells increases cell proliferation. Proc Natl Acad Sci USA 98:9260–9264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi T, Esaki T, Sumi D, Mukherjee T, Iguchi A, Chaudhuri G 2006. Modulating role of estradiol on arginase II expression in hyperlipidemic rabbits as an atheroprotective mechanism. Proc Natl Acad Sci USA 103:10485–10490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Hein TW, Wang W, Chang CI, Kuo L 2001. Constitutive expression of arginase in microvascular endothelial cells counteracts nitric oxide-mediated vasodilatory function. FASEB J 15:1264–1266 [DOI] [PubMed] [Google Scholar]
- 14.Ryoo S, Lemmon CA, Soucy KG, Gupta G, White AR, Nyhan D, Shoukas A, Romer LH, Berkowitz DE 2006. Oxidized low-density lipoprotein-dependent endothelial arginase II activation contributes to impaired nitric oxide signaling. Circ Res 99:951–960 [DOI] [PubMed] [Google Scholar]
- 15.Marathe C, Bradley MN, Hong C, Lopez F, Ruiz de Galarreta CM, Tontonoz P, Castrillo A 2006. The arginase II gene is an anti-inflammatory target of liver X receptor in macrophages. J Biol Chem 281:32197–32206 [DOI] [PubMed] [Google Scholar]
- 16.Modolell M, Corraliza I, Link F, Soler G, Eichmann K 1995. Reciprocal regulation of the nitric oxide synthase/arginase balance in mouse bone marrow-derived macrophage by Th1 and Th2 cytokines. Eur J Immunol 25:1101–1104 [DOI] [PubMed] [Google Scholar]
- 17.Hamilton TA, Major JA, Chisolm GM 1995. The effects of oxidized low density lipoproteins on inducible mouse macrophage gene expression are gene and stimulus dependent. J Clin Invest 95:2020–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM 1998. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell 93:229–240 [DOI] [PubMed] [Google Scholar]
- 19.Leesnitzer LM, Parks DJ, Bledsoe RK, Cobb JE, Collins JL, Consler TG, Davis RG, Hull-Ryde EA, Lenhard JM, Patel L, Plunket KD, Shenk JL, Stimmel JB, Therapontos C, Willson TM, Blanchard SG 2002. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry 41:6640–6650 [DOI] [PubMed] [Google Scholar]
- 20.Willson T 2003. Chemical genomics of orphan nuclear receptors. Ernst Schering Res Found Workshop 42:29–42 [DOI] [PubMed] [Google Scholar]
- 21.Lee G, Elwood F, McNally J, Weiszmann J, Lindstrom KA, Nakamura M, Miao S, Cao P, Learned M, Chen JL, Li Y 2002. T0070907, a selective ligand for peroxisome proliferator-activated receptor γ, functions as an antagonist of biochemical and cellular activities. J Biol Chem 277:19649–19657 [DOI] [PubMed] [Google Scholar]
- 22.Hummasti S, Tontonoz P 2006. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol Endocrinol 20:1261–1275 [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Reape TJ, Li X, Rayner K, Webb CL, Burnand KG, Lysko PG 1999. Induced expression of adipophilin mRNA in human macrophages stimulated with oxidized low-density lipoprotein and in atherosclerotic lesions. FEBS Lett 462:145–150 [DOI] [PubMed] [Google Scholar]
- 24.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, Linton MF 2001. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 7:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yui S, Sasaki T, Miyazaki A, Horiuchi S, Yamazaki M 1993. Induction of murine macrophage growth by modified LDLs. Arterioscler Thromb 13:331–337 [DOI] [PubMed] [Google Scholar]
- 26.Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ). J Biol Chem 270:12953–12956 [DOI] [PubMed] [Google Scholar]
- 27.Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, LeGrumelec C, Xu HE, Montana VG, Lambert MH, Willson TM, Oliver WR, Sternbach DD 2003. Novel selective small molecule agonists for peroxisome proliferator-activated receptor δ (PPARδ): synthesis and biological activity. Bioorg Med Chem Lett 13:1517–1521 [DOI] [PubMed] [Google Scholar]
- 28.Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM 2005. A nuclear receptor atlas: macrophage activation. Mol Endocrinol 19:2466–2477 [DOI] [PubMed] [Google Scholar]
- 29.Tontonoz P, Hu E, Spiegelman BM 1995. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor γ. Curr Opin Genet Dev 5:571–576 [DOI] [PubMed] [Google Scholar]
- 30.Pelton PD, Zhou L, Demarest KT, Burris TP 1999. PPARγ activation induces the expression of the adipocyte fatty acid binding protein gene in human monocytes. Biochem Biophys Res Commun 261:456–458 [DOI] [PubMed] [Google Scholar]
- 31.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D, Glass CK 1999. Interleukin-4-dependent production of PPAR-γ ligands in macrophages by 12/15-lipoxygenase. Nature 400:378–382 [DOI] [PubMed] [Google Scholar]
- 32.Iniesta V, Gomez-Nieto LC, Corraliza I 2001. The inhibition of arginase by Nω-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J Exp Med 193:777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris Jr SM 2005. Arginine metabolism in vascular biology and disease. Vasc Med 10:S83–S87 [DOI] [PubMed]
- 34.Getz GS, Reardon CA 2006. Arginine/arginase NO NO NO. Arterioscler Thromb Vasc Biol 26:237–239 [DOI] [PubMed] [Google Scholar]
- 35.Dumont J, Zureik M, Cottel D, Montaye M, Ducimetiere P, Amouyel P, Brousseau T 2007. Association of arginase 1 gene polymorphisms with the risk of myocardial infarction and common carotid intima media thickness. J Med Genet 44:526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK 1998. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 391:79–82 [DOI] [PubMed] [Google Scholar]
- 37.Morris SM Jr 2006. Arginine: beyond protein. Am J Clin Nutr 83:508S–512S [DOI] [PubMed] [Google Scholar]
- 38.Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, Teissier E, Minnich A, Jaye M, Duverger N, Brewer HB, Fruchart JC, Clavey V, Staels B 2001. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med 7:53–58 [DOI] [PubMed] [Google Scholar]
- 39.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Eagle AR, Vats D, Brombacher F, Ferrante AW, Chawla A 2007. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447:1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shamshiev AT, Ampenberger F, Ernst B, Rohrer L, Marsland BJ, Kopf M 2007. Dyslipidemia inhibits Toll-like receptor-induced activation of CD8α-negative dendritic cells and protective Th1 type immunity. J Exp Med 204:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teupser D, Burkhardt R, Wilfert W, Haffner I, Nebendahl K, Thiery J 2006. Identification of macrophage arginase I as new candidate gene of atherosclerosis resistance. Arterioscler Thromb Vasc Biol 26:365–371 [DOI] [PubMed] [Google Scholar]
- 42.Michalik L, Wahli W 2006. Involvement of PPAR nuclear receptors in tissue injury and wound repair. J Clin Invest 116:598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corraliza IM, Campo ML, Soler G, Modolell M 1994. Determination of arginase activity in macrophages: a micromethod. J Immunol Methods 174:231–235 [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 45.Bookout AL, Mangelsdorf DJ 2003. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 1:e012 [DOI] [PMC free article] [PubMed]