ABSTRACT
Cells have evolved intricate mechanisms to maintain genome stability despite allowing mutational changes to drive evolutionary adaptation. Repetitive DNA sequences, which represent the bulk of most genomes, are a major threat to genome stability often driving chromosome rearrangements and disease. The major source of repetitive DNA sequences and thus the most vulnerable constituents of the genome are the rDNA (rDNA) repeats, telomeres, and transposable elements. Maintaining the stability of these loci is critical to overall cellular fitness and lifespan. Therefore, cells have evolved mechanisms to regulate rDNA copy number, telomere length and transposon activity, as well as DNA repair at these loci. In addition, non-canonical structure-forming DNA motifs can also modulate the function of these repetitive DNA loci by impacting their transcription, replication, and stability. Here, we discuss key mechanisms that maintain rDNA repeats, telomeres, and transposons in yeast and human before highlighting emerging roles for non-canonical DNA structures at these repetitive loci.
KEYWORDS: genome stability, rDNA (rDNA), repetitive DNA, retrotransposons, telomeres
Introduction
The maintenance of genome integrity is essential for the accurate inheritance of genetic information. Stress arising from endogenous or exogenous sources can threaten genome stability. During DNA synthesis, replication fork progression can be impeded by various interconnected obstacles including multidimensional structure-forming nucleic acid sequences, collisions between the replication and transcription machinery, and DNA-binding non-histone proteins.1,2 Impediments to replication fork progression can result in nucleic acid base mispairings, insertions, deletions and double strand breaks (DSBs), posing serious threats to genome stability. Therefore, cells have evolved various mechanisms to prevent or minimize the occurrence of such events.
However, these protective mechanisms are not capable of preventing all damage. Thus, cells also have an arsenal of intricate and evolutionarily conserved mechanisms to repair damaged DNA.3 The DNA damage response promotes cell survival by activating cell cycle checkpoints, allowing DNA repair to occur before resumption of DNA replication and further progression through the cell cycle. However, potential aberrant repair of certain types of loci such as those harbouring highly repetitive DNA sequences does constitute a major threat to genome integrity. In fact, chromosomal rearrangements driven by repetitive DNA sequences underlie many examples of genomic instability in both prokaryotes and eukaryotes.4, 5
Repetitive DNA sequences constitute a significant portion of genomes across species, making up over 70% of the human genome.6 Controversial references to “junk” DNA were put forward early on for many of these repeated sequences but it is now clear that numerous, if not most, such sequences are essential for genome function and stability. Repetitive DNA comprises both coding and non-coding sequences that can be tandemly arranged or interspersed throughout the genome. Tandemly arranged sequences can form a unique cluster positioned at a single locus or they can be organized into several clusters positioned across multiple loci on the same or different chromosomes.7 Template donor DNA is readily available for homologous recombination (HR) to repair a damaged unit within repetitive DNA.
Importantly, recombination events involving repetitive DNA sequences have to be very tightly controlled to avoid deleterious chromosome rearrangements. For instance, aberrant recombination between tandemly arranged sequences could result in the formation of toxic extrachromosomal circular DNA (eccDNA) or in a change in the number of repeats, therefore increasing genomic instability.8, 9 Similarly, DNA damage at interspersed repetitive sequences can be repaired by HR between non-allelic loci, potentially leading to gross chromosomal rearrangements (GCRs).10 As these loci are prone to deletion and insertion mutational errors during replication and/or repair, it is vital for cells to minimize DNA damage and/or regulate DNA repair at these sites. Interestingly, even though these loci can be major sources of genomic instability, many of them play key roles in cellular functions and are themselves major players in the maintenance of genome integrity. Examples of repetitive loci that can both promote and counter genome stability and inheritance include the rRNA genes (rDNA), telomeres and transposable elements.11, 12 Collectively, these loci constitute the majority of repetitive DNA sequences in numerous genomes including yeast and human.13
On another front, genomes have an abundance of secondary structure-forming DNA motifs that are thought to impact genome function and stability.5 At repetitive loci, non-canonical DNA structures such as R-loops and G-quadruplexes (GQs) can help modulate DNA replication, transcription, and protein sequestration.14 Although functionally important, these alternative DNA structures are often implicated in cancer and hereditary diseases as failure to resolve them can result in GCRs and mutagenesis.5, 14 For instance, DNA sequences capable of forming non-canonical structures at the human c-MYC and BCL2 loci, which co-localize with translocation breakpoints, undergo frequent DSBs in mammalian cells.15, 16 Moreover, the structural organization of chromatin can be altered by these non-canonical DNA structures, affecting DNA repair efficiency and therefore further increasing genome instability.15
Here, we review some of the mechanisms that modulate the stability of repetitive DNA loci. We first discuss the structure and function of rDNA, telomeres, and transposable elements and cover how cells ensure the stability at these loci. We then explore the effect of non-canonical DNA structures on the stability and function of repetitive loci.
rDNA repeats
rDNA organization
The rDNA repeats are essential housekeeping genes and are the most abundant genes in the eukaryotic genome. In the model organism Saccharomyces cerevisiae, an rDNA unit comprises 2 open reading frames, 35S and 5S, transcribed by RNA polymerase I (RNA Pol I) and III (RNA Pol III) respectively, and 2 intergenic spacers, IGS1 and IGS2, (Fig. 1a).11, 17 A replication fork barrier (RFB) and a bi-directional origin of replication called ribosomal autonomous replication sequence (rARS) are respectively located within the IGS1 and IGS2 regions.18 Transcription of the rDNA loci is an active and ongoing process due to the cell's exigence for ribosomes, the critical machinery required for protein synthesis. To cope with these demands, cells across species contain multiple copies of rDNA repeats ranging from 7 in Escherichia coli to 12,000 in Zea mays.11 In eukaryotes, these multiple copies are arranged as clusters of tandem repeats and are spatially isolated from the rest of the DNA, making up the bulk of the major nuclear compartment called the nucleolus. Roughly 150–200 rDNA units are all tandemly arranged on chromosome XII of S. cerevisiae, while ∼350 human rDNA units are arranged on chromosomes 13, 14, 15, 21 and 22, with roughly 70 tandem repeats per chromosome (Fig. 1b).11, 17 Although such an organization of rDNA repeats is necessary to meet the cells demands, it can also compromise genome stability especially in the absence of the tight controls that typically limit the ability of HR to access or modulate these repeats.
Figure 1.
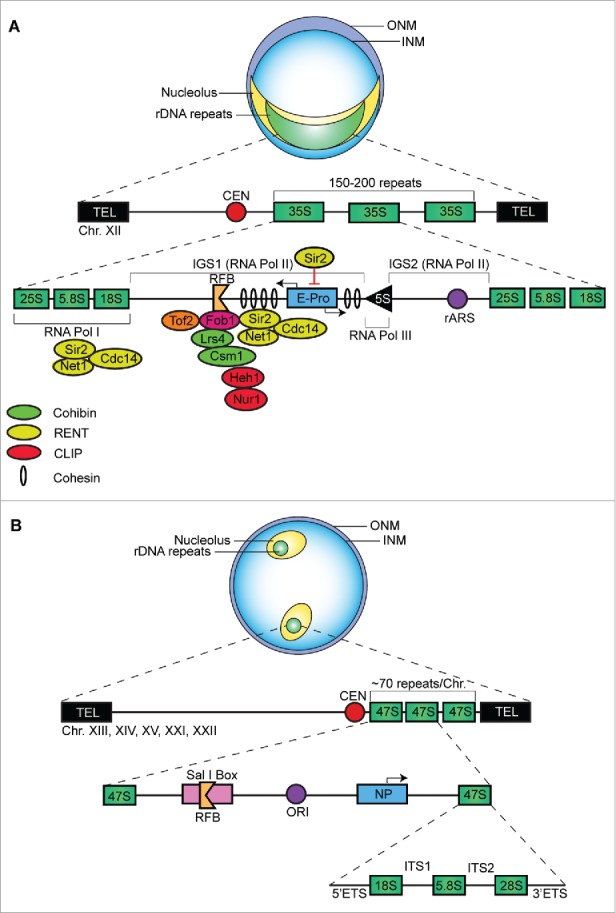
Organization of rDNA repeats in yeast and human cells. (A) Organization S. cerevisiae rDNA repeats. 150–200 repeats are tandemly arranged on Chr XII. A single unit comprises 35S (25S, 5.8S and 18S) and 5S genes as well as the intergenic spacers (IGS1 & IGS2) respectively harbouring the replication fork barrier (RFB) and a bi-directional autonomous origin of replication (rARS). The non-coding RNA promoter, E-Pro, is located within IGS1. The fork-blocking protein 1 (Fob1) binds the RFB stalling the advancing DNA replication forks and creating a recombination hotspot. Additionally, Fob1 interacts with the regulator of nucleolar silencing and telophase exit (RENT) complex (comprised of the Sir2, Net1, and Cdc14 proteins), topoisomerase-related factor (Tof2) and the cohibin complex (comprised of Lrs4 and Csm1) to promote rDNA silencing. The Sir2 protein also regulates the activity of the E-Pro; Sir2 inhibition of the E-Pro allows the cohesion complex to associate with intergenic rDNA regions thereby promoting equal sister-chromatid exchange. (B) Organization of homo sapiens rDNA repeats. About 350 repeats are tandemly arranged on Chr. XIII, XIV, XV, XXI and XXII, with ∼70 repeats per Chr. A single unit comprises the 47S (18S, 5.8S and 28S) gene and an intergenic spacer containing an RFB and origin of replication (ORI).
Controlled HR at rDNA repeats
Although organisms contain many rDNA units, only about half are actively transcribed by RNA Pol I.19 In other words, rDNA repeats typically harbour both active and inactive units. Additionally, within a given rDNA unit, IGS regions can be subject to mechanisms that suppress RNA polymerase II (RNA Pol II)-dependent transcription and promote repeat stability.20 Cells use multiple rDNA-regulatory mechanisms to maintain this repeat stability including: sister chromatid alignment, replication fork progression, IGS transcriptional silencing, and localization of the rDNA repeats. In S. cerevisiae, rDNA units associate with condensin, a 5-subunit protein complex that condenses rDNA and helps align sister-chromatids to warrant accurate HR-dependent repair of damaged rDNA sites and maintain an optimal number of rDNA units.19, 21 When rDNA copy numbers are at sub-optimal levels as in cells lacking condensin, cells are hyper-sensitive to DNA damaging agents.19 Additionally, the shared cellular longevity-sustaining factors are distributed between rDNA and the rest of the genome to ensure genome stability; this distribution is intimately influenced by the size of the rDNA repeat tract.22 Thus, maintenance of an optimal or wild-type number of rDNA units is critical to efficient ribosome biogenesis, cellular longevity, overall genome function, and survival under stress.
To maintain optimal rRNA levels when rDNA copy numbers decrease, cells activate typically inactive rDNA units increasing RNA Pol I localization to the rDNA loci.19,23 Increased RNA Pol I localization consequently limits the association of condensin with rDNA thereby interfering with condensin/HR-dependent repair of potential DNA damage.19,23 However, cells are equipped with a compensatory copy number reconfiguration mechanism that can gradually restore optimal rDNA copy numbers. At the heart of this copy number reconfiguration system is a protein called fork-blocking protein 1 (Fob1). Fob1 physically binds to the rDNA intergenic RFB regions, creating a unidirectional block that stalls advancing DNA replication forks (Fig. 1A).24,25 Stalling of the replication fork leads to the formation of DSBs creating a recombination hotspot. Such DSBs can then be repaired by intra- or inter-chromosomal recombination events that alter rDNA copy number.24 During intra-chromosomal recombination, DSBs are repaired by strand invasion between rDNA units located on the same chromosome, potentially resulting in the loss of repeats located between the break site and the donor site. As such, intra-chromosomal recombination at rDNA loci is responsible for the production of potentially toxic extrachromosomal rDNA circles (ERCs) that can result in subsequent contraction of the rDNA array via the loss of repeats that comprise ERCs.24 Alternatively, repair via inter-chromosomal recombination can occur at DSBs either by equal sister chromatid exchange maintaining rDNA copy number or by unequal sister chromatid exchange (USCE) giving rise to an expanded or contracted rDNA copy number.26 In the context of a short rDNA repeat tract and increased local transcriptional activity, USCE will favor rDNA copy number expansion.26 A key player in this process is a non-coding RNA Pol II-dependent promoter that is located within the IGS1 regions of rDNA and known as the EXP Promoter (E-Pro).26 During the S phase of the cell cycle, the cohesin protein complex associates with intergenic rDNA regions and helps align sister chromatids during replication to ensure equal sister-chromatid exchange (Fig. 1A). However, activation of E-Pro leads to the transcription of long intergenic non-coding RNA molecules effectively limiting local cohesion association and favoring USCE events.26 Cells with an optimal or wild-type rDNA copy number have a selective advantage and optimal copy number will be restored over several cell divisions. E-Pro activity is in turn regulated by the NAD+-dependent histone deacetylase Sir2.27 When rDNA copy number is at optimal levels, Sir2 represses E-Pro activity altering the chromatin structure and allowing cohesin to associate at the rDNA array. In contrast, when rDNA copy numbers decrease, Sir2 inhibition of the E-Pro is compromised resulting in a loss of cohesin association at the rDNA loci culminating in USCE with consequential amplification until wild-type optimal copy numbers are reached.27 Importantly, Sir2 is part of a larger molecular network that helps silence intergenic rDNA transcription and maintain rDNA repeat stability.
Although recombination at rDNA is a mechanism used to maintain optimal rDNA units, hyper-recombination events can lead to genomic instability and must therefore be inhibited. To prevent hyper-recombination within the rDNA repeats, yeast cells assemble IGS DNA into silent chromatin structures via a process known as rDNA silencing, which implicates both Sir2-dependent and Sir2-indepednent mechanisms.20,28 The RENT complex (regulator of nucleolar silencing and telophase exit) comprises the Sir2, Net1 and Cdc14 proteins.29 The RENT complex induces rDNA silencing via the association of both Sir2 and Net1 with coding (Pol I promoter) and non-coding (IGS1/RFB) regions of rDNA (Fig. 1a).28 DNA-bound Fob1 physically associates with the RENT complex trough Net1 and Sir2, favoring their association at the RFB. RENT also localizes around the RNA Pol I promoter via Fob1-independent physical interaction of RNA Pol I with Sir2 and Net1 (Fig. 1a).28 RENT promotes functional interactions between Sir2 and the N-terminal tails of histones H3 and H4. Sir2-dependent deacetylation of lysine residues within the N-terminal tails of H3 and H4 helps establish a more closed or silent chromatin structure that limits access to RNA Pol II within the intergnic rDNA regions.30 In addition to Sir2, the silencing of rDNA is further promoted by other chromosomal factors including the cohibin complex, comprised of Lrs4 and Csm1, and topoisomerase-related factor 2 (Tof2).20, 31 Similar to the RENT complex, cohibin and Tof2 are recruited to the IGS1 region in a Fob1-dependent fashion and are required for rDNA silencing (Fig. 1a).31 Silencing of the rDNA loci through these mechanisms safeguards against hyper-recombination, thus preserving rDNA integrity.
In addition to rDNA silencing, yeast cells prevent excessive aberrant recombination and maintain the stability of rDNA repeats by regulating their localization within the nucleus. It is well established that rDNA is physically sequestered within the major subnuclear compartment known as the nucleolus, which in yeast is crescent shaped and abuts the nuclear envelope. The subnuclear localization of rDNA plays an important role in rDNA stability, as Sir2-dependent silencing alone is not sufficient to inhibit USCE.20 Within the nucleolus, rDNA is tethered to the inner nuclear membrane (INM) via interaction of the rDNA-associated cohibin complex and the nuclear envelope-embedded chromosome linkage INM proteins (CLIP), namely Heh1 (helix extension helix 1) and Nur1 (nuclear rim 1) (Fig. 1A).20
CLIP and cohibin suppress overall recombination at rDNA while also specifically limiting aberrant recombination events. First, the solid support provided by the nuclear envelope-embedded CLIP allows cohibin to align sister chromatids during rDNA replication, thereby promoting equal and suppressing unequal recombination events.20 Additionally, perinuclear tethering of rDNA by CLIP sequesters the repeats within the recombination-suppressive environment of the nucleolus, shielding the repeats from functional Rad52 recombination proteins enriched in the nucleoplasm.20, 32
In addition, to further prevent uncontrolled recombination between the highly repetitive rDNA repeats, cells temporarily localize rDNA units experiencing DSBs outside of the nucleolus, possibly with at least one other intact donor rDNA unit.32 To warrant a compartment-specific delocalization of HR processes, the Rad52 recombination protein is excluded from the nucleolus by the Smc5-Smc6 and the MRX complexes, and by sumoylation of the Rad52 protein itself.32 Upon induction of a DSB within a given rDNA unit, the MRX complex, comprised of the Mre11, Rad50, and the Xrs2 proteins, recognizes and localizes to the DSB site within the nucleolus. The MRX complex recruits the Smc5-Smc6 complex, whose Mms2 subunit possesses sumoylation activity.33 Interestingly, although the Smc5-Smc6 complex is required for the exclusion of Rad52 from the nucleolus, it is not required for its sumoylation. Therefore, the role of Smc5-Smc6 in the exclusion of functional Rad52 from the nucleolus could be indirect, possibly involving the sumoylation of other recombination proteins.32 After resection of the MRX-bound DSB, it is relocated from the nucleolus to the nucleoplasm. MRX dissociation is then followed by the recruitment of unsumoylated, and thus functional, Rad52 to the DSB site via RPA proteins, allowing access to the HR machinery downstream of Rad52 and mediating recombinational repair.32
The various pathways that regulate recombination within the rDNA repeats highlight the importance of regulating repetitive DNA loci. Although HR is critical to the maintenance of genome stability, it can also be a potential source of GCRs and overall genome instability. The action of HR and other specialized mechanisms at telomeres, another type of conserved and repetitive DNA loci located at the ends of linear chromosomes, is also critical to overall genome function and stability.
Telomeres
Introduction to telomeres
Across species, the ends of linear chromosomes are constituted of telomeres, which are repetitive DNA sequences that help counter natural chromosome shortening, prevent chromosome end recognition by the DNA repair machinery, and avoid destabilizing chromosome end-to-end fusions (Fig. 2).34, 35 Telomeres are comprised of non-coding double-stranded and single-stranded DNA (ssDNA) sequences consisting of tandem G-rich repeats such as the TG1–3 of budding yeast and human T2AG3.34 The ssDNA regions, known as G-tails, serve as substrates for several DNA-binding proteins whose roles are to preserve telomere length and structure.36 Telomere length varies across species with 300–400 repeats in yeast to tens of thousands in higher eukaryotes.36 Maintaining telomeres at optimal lengths is pivotal to genome stability and health. Disruption of this equilibrium can lead to catastrophic genome instability and telomere-related diseases.
Figure 2.
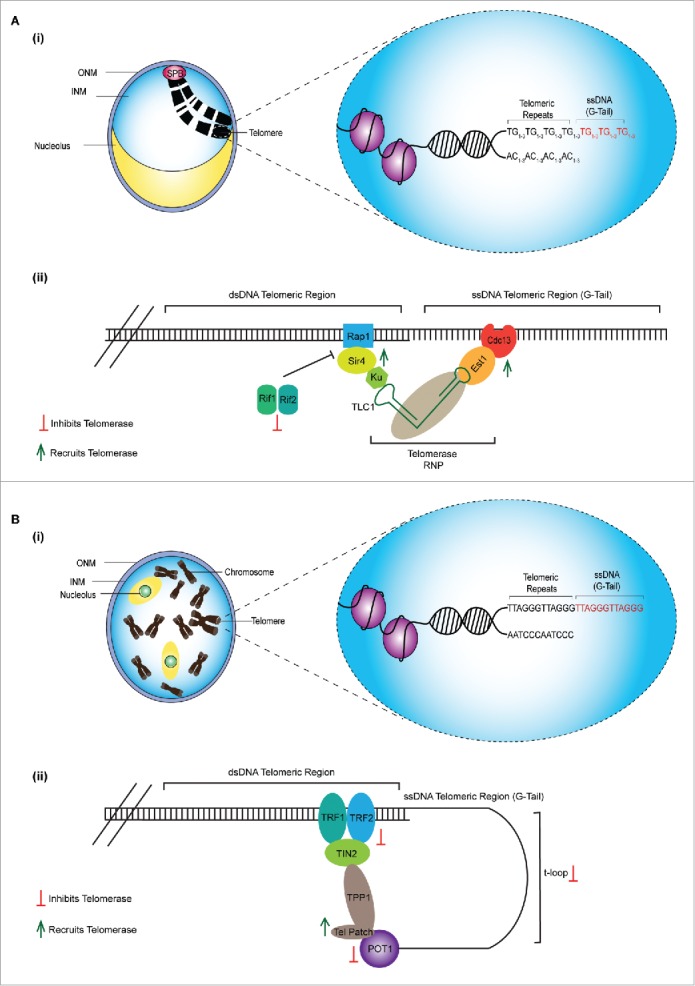
Organization of telomeres. (A) (i) Chromosome termini of S. cerevisiae are capped with TG1–3 DNA sequence repeats known as telomeres. The extreme ends are comprised of a single stranded G-rich 3′ overhang, known as G-tails. (ii) In the budding yeast, S. cerevisiae, telomerase is recruited via the interaction between Cdc13, a single-stranded DNA (ssDNA) telomeric-binding protein and the telomerase factor Est1. Additionally, the telomerase RNA (TLC1), binds Ku, which in turns binds the Sir4 protein associated with the double-stranded DNA (dsDNA) binding protein Rap1. This interactions enhances the Est1-Cdc13 interaction at the ssDNA G tail. Telomerase access is restricted by the Rif1 and Rif2 proteins which compete against Sir4 for Rap1 binding. (B) (i) H. sapiens telomeres are comprised of TTAGGG DNA sequence repeats whose extreme ends are also made up of G-tails (ii) In mammalian cells, telomerase is recruited via interaction between hTERT telomerase component and the TEL patch of TPP1, a component of the shelterin complex. TRF1 and TRF2, bind the dsDNA telomeric region inducing the formation of a telomeric loop (t-loop) which physically blocks telomerase. The t-loop structure is further stabilized by the ssDNA telomeric-binding protein POT1, which itself also physically blocks telomerase access.
Telomere-linked diseases include cancer and premature aging syndromes, collectively called telomeropathies, which have overlapping symptoms and incompletely penetrant phenotypes, with a highly variable age of onset.37-39 The DNA replication machinery cannot copy the very ends of linear chromosomes thus the number of protective telomeric repeats should naturally decrease with each round of DNA replication and cell division.40, 41 However, an optimal length is maintained through an interplay of shortening and elongation mechanisms. It is imperative that such an interplay be highly regulated to maintain homeostatic telomeric length and subsequently avert the occurrence of telomeropathies.
Telomerase-dependent telomere length maintenance
Telomere length is maintained through the activity of a ribonucleoprotein complex called telomerase, which is recruited to short telomeres to add telomeric DNA repeats to chromosome ends.42 The components of telomerase vary across species with the exception of telomerase reverse transcriptase (TERT) and telomerase RNA (TERC). TERC, an RNA subunit, not only serves as a template for reverse transcription but also provides scaffolding to allow telomerase protein components to form the telomerase RNP holoenzyme.43 Although TERC length and sequence varies across species, it contains a conserved catalytic core that is required for substrate binding and telomerase activity.43 If telomerase were left to its own devices, the outcome would be an undesirable increase in telomere length rendering cells immortal which could potentially give rise to cancerous cells.44 As such, in addition to regulating telomerase levels, cells also use various factors that highly regulate telomerase recruitment and access to telomeres.
Telomerase recruitment and the frequency/extent of telomere elongation is tightly regulated in cis by the activities of both single- and double-stranded DNA binding proteins.45 In yeast, in vivo/in vitro studies reveal that the telomerase accessory factor Est1 directly interacts with the recruitment domain of the ssDNA telomeric-binding protein Cdc13, thereby mediating telomerase access to telomeres.46, 47 Additionally, telomerase is recruited to telomeres through a Ku-mediated mechanism; Ku binds both the telomerase RNA (TLC1) and the telomeric silencing protein Sir4, which in turn interacts with the telomeric dsDNA-binding protein Rap1.48 Moreover, this interaction subsequently enhances the Cdc13-Est1 interaction at the ssDNA G tail. Recruitment of telomerase by Sir4-Ku interaction is inhibited by Rif1 and Rif2, which compete against Sir4 for Rap1 binding (Fig. 2A(ii)).48-50 In this setting, Cdc13-Est1-mediated telomerase recruitment is still possible but at a decreased efficiency.48 In mammalian cells, telomerase is recruited via a member of the shelterin complex, TPP1.51 Specifically, TPP1s oligosaccharide binding (OB) fold known as the TEL patch interacts with the TEN-domain of the hTERT component of telomerase (Fig. 2B(ii)).52, 53 Thus, various telomere-binding factors help recruit telomerase to chromosome ends.
Multiple telomere-binding proteins can also inhibit telomeric elongation by obstructing telomerase accessibility. Depletion of the Rap1/Rif1/Rif2 complex in budding yeast, or components of the shelterin complex, TRF1, TRF2 and POT1, in mammalian cells, increases telomere length, indicating that these proteins act as negative regulators of telomere length.50,54-56 In yeast, Rap1 binds telomeres and recruits Rif1/Rif2, which are responsible for inhibiting telomeric elongation (Fig. 2A(ii)).50 The N-terminus of Rif2 is able to suppress telomere lengthening in rif1Δ, rif2Δ and rif1Δ rif2Δ mutants in a telomerase-dependent fashion. This suggests that the N-terminal domain of Rif2, termed the BAT domain (Blocks Addition of Telomeres), negatively regulates telomere length by inhibiting their elongation by telomerase.50 In mammalian cells, early studies revealed that TRF1 and TRF2, members of the shelterin complex, bind dsDNA regions and inhibit telomere elongation by inducing the formation of a telomeric loop, t-loop, which can physically block telomerase access.54, 55 Additionally POT1 binding at the ssDNA region, can physically block telomerase access as well as stabilize the t-loop (Fig. 2B(ii)).56
Telomerase-independent elongation of telomeres
In addition to telomerase-mediated elongation, telomeres can also be lengthened by other mechanisms. In mammalian cells, this can occur by a recombination-driven amplification mechanism known as alternative lengthening of telomeres (ALT). This recombination-dependent but telomerase-independent mechanism is a hallmark of many mesenchymal-cancerous cells and occurs in approximately 10–15% of human cancers.57, 58 ALT gives rise to abnormally longer telomeres with various lengths.59 ALT is able to bypass mechanisms that regulate telomerase potentially rendering cells immortal. Two different mechanisms that rely on the recombination machinery have been proposed for ALT: unequal telomere sister chromatid exchange (T-SCE) and lengthening via break induced replication.60, 61
In telomerase-null yeast cells, a small population of cells is able to maintain telomere elongation via 2 distinct Rad52-dependent pathways, type I and type II, which are collectively known as RTE (RAD52-dependent recombinational telomere elongation). Type II RTE closely resembles mammalian ALT.62,63 Thus, HR-based mechanisms allowing cells to bypass telomerase-dependent telomere length control are evolutionarily conserved.
To prevent the fusion of telomeres on different chromosomes or chromosome arms to each other, various telomere capping factors ensure the local inhibition of non-homologous end-joining (NHEJ). Telomere-binding proteins cap telomeres thereby shielding them from DNA damage response pathways such as NHEJ. This is crucial as a single NHEJ event at telomeres can give rise to GCRs resulting in genome instability. In budding yeast, Rap1 is required for NHEJ inhibition and several studies have shown that the loss of Rap1 results in a significant increase in NHEJ events, leading to telomere fusions.64 As a single NHEJ event at telomeres can be catastrophic to genome stability, cells have used different Rap1-mediated pathways to inhibit such an occurrence. The Rap1 C-terminal domain inhibits NHEJ through distinct Sir4-dependent and Rif2-dependent pathways.64 Additionally, the Rap1 central domain inhibits NHEJ in a Sir4 and Rif2 independent manner.64 In mammalian cells, Rap1 and TRF2 protect telomeres from being processed by ALT via suppression of telomeric localization of the HR factors: PARP1 and SLX4.65 In the absence of Rap1 and TRF2, the accumulation of HR factors at telomeres leads to telomere resection, loss, and ultimately chromosomal fusions.65
Taken together, evolutionarily conserved interplays between telomerase, telomeres, their various binding/capping factors, and the basic DNA repair/recombination machineries are critical to genome function and stability.
Transposable elements
Introduction to transposable elements
Transposable elements (TEs), arguably the greatest source of repetitive DNA, are widely distributed across the eukaryotic kingdom. They are commonly known as “jumping genes,” due to their ability to move around within the genome.66 TEs are comprised of 2 main classes reflecting the mechanism by which they move around in the genome: class I TEs, retrotransposons, move in a copy and paste manner by way of an RNA intermediate, and class II TEs, DNA transposons, move in a cut and paste manner, independent of an RNA intermediate.67 Retrotransposons are comprised of 2 sub-classes: long-terminal repeat (LTR) and non-LTR retrotransposons, based on the presence or absence of flanking long-terminal repeat sequences. Retrotransposons replicate with each round of transposition and comprise about 3% and 45% of the yeast and human genomes respectively. Transposon dysregulation has been linked to natural aging, neurologic disorders and various cancers.13,68-71
In budding yeast, retrotransposons are termed transposons of yeast (Ty). They are flanked with LTRs and are comprised of 5 families, Ty1–5, interspersed across the genome, with Ty1 being the most abundant and active full length Ty element.68 In humans, the non-LTR retrotransposons, Long Interspersed Element (LINE-1 or L1), Alu (the most abundant Short Interspersed Element – SINE) and SVA (SINE-VNTR-Alu) are the only active TEs.72 Ty and LINE-1 are autonomous elements containing 2 open reading frames, TYA/ORF1 and TYB/ORF2 which encode proteins that are required for retrotransposition (Fig. 3A).72, 73 The retrotransposition cycles of both yeast Ty and human LINE-1 elements include reverse transcription of the RNA and integration/insertion of the resulting cDNA product into new genome loci (Fig. 3B-C). As retrotransposons are capable of replicating and inserting themselves into new genomic loci, they can restructure the genome, alter gene expression and trigger deleterious chromosomal rearrangements. Therefore, retrotransposons are considered a major source of potential genome instability. Importantly, to maintain genome integrity, cells have adopted multiple mechanisms to regulate retrotransposition at each stage of the retrotransposon life cycle.
Figure 3.
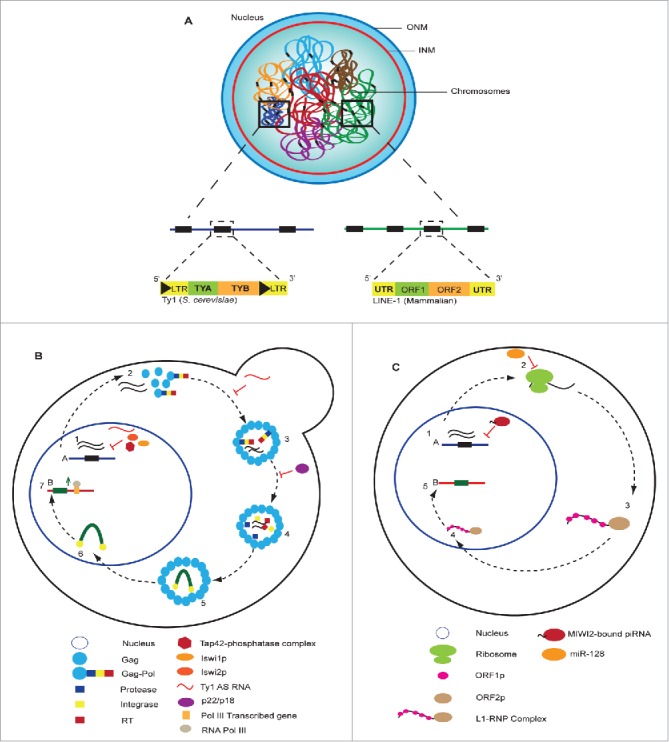
Structure of interspersed retrotransposons. (A) Retrotransposons are interspersed throughout the genome across multiple chromosomes. The Ty1 LTR-retrotransposons of yeast and the mammalian LINE1 non-LTR-retrotransposon are active autonomous elements that contain 2 open reading frames (ORFs), TYA1/ORF1 and TYB1/ORF2, which encode for proteins required for retrotransposition. (B) In S. cerevisiae, Ty1 retrotransposition cycle begins with (1) the transcription of chromosomal Ty1 elements. Ty1 transcripts exit the nucleus into the cytoplasm where they are (2) translated into Gag and Gag-Pol proteins. Gag and Gag-Pol proteins form a (3) Virus-like particle (VLP) encapsulating Ty1 transcipts. Upon VLP maturation (4), Ty1 transcripts are reverse transcribed into cDNA (5) that then associates with integrase to form the preintegration complex (PIC). The PIC is imported back into the nucleus (6) where integrase interacts with RNA Pol III to facilitate integration of the Ty1 cDNA upstream of RNA Pol III transcribed genes (7), giving rise to a new and full-length Ty1 chromosomal element. The Tap42-phosphatase complex, Iswi1p, Iswi2p and antisense Ty1 (Ty1 AS) RNA are able to suppress Ty1 transcription thereby inhibiting retrotransposition. Post-transcriptional inhibition is also achieved through Ty1 AS transcripts, that also decrease Ty1 protein levels, and via a truncated for of Gag known as p22 (unprocessed) and p18 (processed), which inhibit Gag maturation thereby inhibiting retrotransposition. (C) In mammalian cells, LINE-1 retrotransposition also begins with (1) transcription of chromosomal LINE-1. LINE-1 transcripts exit the nucleus into the cytoplasm where they are (2) translated into ORF1p and ORF2p that associate with the LINE-1 transcript (3) giving rise to the formation of LINE-1 ribonucleoprotein (L1-RNP) complex. The L1-RNP is imported back into the nucleus (4) where it is reverse transcribed and inserted into a new genomic locus (5). In germ cells, LINE-1 is silenced by the MIWI2-bound piRNA that orchestrates DNA methylation at LINE-1 by DNMT3. Post-transcriptionally, the micro RNA miR-128, inhibits LINE-1 by binding to the ORF2 regions of the LINE-1 transcripts, thereby targeting LINE-1 for degradation.
Stage-specific regulation of retrotransposition
The first stage at which cells regulate retrotransposition is at the level of transcription. In yeast, studies using a TYA1:URA3 reporter showed that Ty1 transcription is subject to a negative feedback control known as Ty1 transcriptional co-suppression.74 Specifically, in Ty1-less mutants harbouring either a single-copy or multi-copy plasmid of Ty1 elements, only the latter was able repress a Ty1 containing reporter plasmid. This suggests that in the presence of a high or low copy number of Ty1 elements, these jumping genes are either silenced or expressed respectively.74 A subsequent report proposed that Ty1 transcriptional co-suppression is independent of Ty1 homology, a hallmark of co-suppression.75 This opened the idea that the ability to translate Ty1 mRNA could be driving Ty1 transcriptional silencing in this co-suppression mechanism. Wu and Jiang, 2008, found that Ty1 transcriptional silencing is initiated by non-translatable Ty1 mRNA, mRNA molecules that are poor templates for translation. Over-production of non-translatable Ty1 mRNA results in the inactivation of the nuclear cap-binding complex (CBC) leading to the hyperstimulation of the TOR pathway, which in turn further silences Ty1 transcription through the Tap42-phosphatase complex.75,76 We note that Ty1 co-suppression was not observed when a TYA1:GFP reporter was used, raising the possibility that it may be affected by some reporters or that it may be an artificial effect of other reporters.77 Additionally, Ty1 transcription is also generally inhibited by the chromatin remodelling ATPases Isw1p and Isw2p (Fig. 3B).78 In mammalian cells, LINE-1 is silenced through methylation and hypomethylation increases LINE-1 levels and is a key hallmark of many cancers.79 In addition, TE silencing is especially critical in germ cells as any TE-related genome instability can be transmitted to the next generation or even potentially decrease fertility. Key to germ line TE silencing are single-stranded noncoding RNAs known as PIWI-interacting RNAs (piRNAs).80 piRNA-dependent silencing of LINE-1 requires the PIWI Argonoute protein, MIWI2, which orchestrates DNMT3-dependent DNA methylation at TEs. In a model by Pezic et al 2014, piRNA-dependent methylation of LINE1 is targeted to the promoter-containing 5′ region of LINE-1. In this model, MIWI2/piRNA complexes recognize and bind nascent LINE-1 transcripts allowing MIWI2 to then recruit DNMT3 ensuring the trimethylation of H3K9 and silencing of active LINE-1 elements (Fig. 3C). Thus, cells can repress TE activity via transcriptional processes.
Cells also have an arsenal of post-transcriptional mechanisms allowing for the regulation and repression of retrotransposition. In yeast, Ty1 retrotransposition events are rare despite the relatively high levels of Ty1 RNA, which can be as much as 0.8% of total RNA, indicating that inhibition of Ty1 retromobility exists at one or more post-transcriptional levels.73,81 Indeed, posttranscriptional control of Ty1 mobility does occur via modulation of each of the stages of the retrotransposition life-cycle including translation (protein synthesis), reverse transcription (cDNA synthesis), and integration into new genomic loci.
In human somatic cells, which lack the piRNA activity found in germ cells, LINE-1 activity is inhibited post-transcriptionally by the micro-RNA miR-128.82 miR-128 directly binds a site within the ORF2 region of LINE-1 transcripts resulting in the formation of a LINE-1-miR-128-RISC complex, thereby targeting LINE-1 transcripts for degradation and consequently inhibiting retrotransposition (Fig. 3C).82 Although S. cerevisiae lacks RNAi activity, it is able to make use of antisense and non-coding RNAs to inhibit retrotransposition. Ty1 retromobility is post-transcriptionally inhibited by an RNA-dependent gene silencing mechanism involving a non-coding antisense Ty1 cryptic unstable transcript (CUT), termed Ty1-RTL.83 RNA Pol II-dependent and Set1 histone methyltransferase-regulated Ty1-RTL transcription is initiated within Ty1 and inhibits Ty1 expression in trans by acting on newly synthesized Ty1 transcripts via an RNA-dependent silencing mechanism. This in turn leads to a decrease in RNA Pol II occupancy at Ty1 further promoting silencing (Fig. 3B).83 These studies highlight mechanisms by which both human and yeast cells inhibit retrotransposition post-transcriptionally.
Cells have also adopted RNA-mediated mechanisms to modulate retrotransposition at the post-translational level. In S. cerevisiae, this may partially be achieved via the action of antisense Ty1 transcripts. Ty1 mRNA levels generally increase as chromosomal Ty1 copy numbers increase, although this is not always accompanied by increases in retrotransposition.84 This is because cells with more chromosomal Ty1 copies have also been shown to have greater levels of antisense Ty1 transcripts, Ty1-AS-RNA, that can inhibit retrotransposition.84 There are various size-based classes of Ty1-AS-RNAs. Transcription of Ty1-AS-RNAs is independent of full-length Ty1 transcription, as these antisense transcripts are still present in mutants where Ty1 transcription is inhibited. Ty1-AS-RNA sequences are complementary to the copy number control (CNC) region of Ty1 and are required for post-transcriptional copy number control.84 Ty1-AS-RNAs are trans-acting and associate within cytoplasmic virus-like particles (VLPs), thereby suppressing the levels of Ty1 mRNA-encoded proteins (Fig. 3B). Several mechanisms are envisaged through which Ty1-AS-RNAs may be decreasing Ty1 protein levels: 1) base pairing of Ty1-AS-RNA with Ty1 mRNA may prevent Ty1 mRNA dimerization within VLPs; 2) Ty1-AS-RNAs may inhibit processing of the Pol proteins; and 3) Ty1-AS-RNA may block tRNAmet (which acts as a primer for RT/cDNA synthesis) from binding to Ty1 mRNA.84 Ty1-AS-RNAs represent one class of Ty1 RNA species that are able to regulate retrotransposition.
In addition to Ty1-AS-RNAs, S. cerevisiae cells can inhibit Ty1 retrotransposition through an internal sense transcript that codes for a trans-dominant protein; the internal Ty1 sense-strand located at the 5′ region, termed Ty1i-RNA, is transcribed from an internal promoter within TYA, about 800 nucleotides downstream of the full-length start site.85 It encodes an altered (N-terminally truncated) form of Gag capsid protein (Gag), known as p22 (unprocessed) or p18 (p22 processed by Ty1 Protease), that mediates Ty1 CNC.85 p22/p18 complexes interact with Ty1 VLPs changing their spherical morphology and rendering them open or incomplete thereby inhibiting VLP maturation, cDNA synthesis and retrotransposition (Fig. 3B). Inhibition by p22/p18 is partly due to the direct interaction between Gag and p22, as mutations in GAG open reading frames (ORFs) confers resistance to p22, allowing for mature VLP assembly.86 Specifically, the N-terminal of p18 interacts with the C-terminal region (CTR) of Gag and inhibits its nucleic acid chaperone (NAC) activity, which is required for Ty1 retrotransposition. Additionally, p18 simultaneously binds Ty1 transcripts with high affinity thus competing with Gag for Ty1 RNA binding. p22/p18 also prevent Ty1 mRNA dimerization and packaging and renders Ty1 mRNA more prone to nuclease degradation, thereby inhibiting retrotransposition.87
Even under normal conditions, retrotransposition occurs in “healthy” wild-type cells. To ensure that base-line retrotransposition does not give rise to genome instability, cells target retrotransposon cDNAs to gene-poor regions, resulting in non-deleterious insertions. Ty1 cDNA preferentially integrates upstream of Pol III transcribed genes, mainly the tRNA genes.88 Prior to integration, Ty1 cDNA exists as a preintegration complex (PIC) that comprises a cDNA element and an integrase; Ty1 PIC is targeted to regions upstream of RNA Pol III transcribed genes via interaction between the C-terminus of integrase and the AC40 subunit of RNA Pol III (Fig. 3B).89 Although RNA Pol I also contains the AC40 subunit, Ty1 does not integrate upstream of RNA Pol I-transcribed genes, suggesting that some other RNA Pol III cofactor is required for the interaction between integrase and the AC40 subunit of RNA Pol III. When this interaction is lost, Ty1 cDNA targets telomeric and subtelomeric heterochromatic regions.89 Although the mechanism behind this secondary targeting has not been elucidated, it could plausibly be mediated by interaction between integrase and telomeric proteins. The insertion of retrotransposition into these hotspots ensures non-deleterious integration of Ty1 cDNA.
Although retrotransposon insertions are highly regulated, an increase in the number of repeated interspersed sequences in the genome, a result of retrotransposition, may lead to an increase in non-allelic HR (NAHR) frequency. In other words, an augmentation in the number of retrotransposons, interspersed throughout the genome, might lead to undesirable NAHR events, or ectopic recombination, as a consequence of DSB repair or of the restarting of stalled replication forks. As such, aberrant recombination events at retrotransposons could be detrimental to cells, potentially giving rise to GCRs and genome instability. In budding yeast, DSBs at Ty elements have been shown to be repaired by HR between Ty elements dispersed in the genome.90 Moreover, recent studies in human cells, demonstrated that NAHR events between LINE-1 elements occur frequently and that such events can cause structural genomic rearrangements, which can contribute to genome instability and give rise to human genetic disorders.91, 92
Our review thus far illustrates the multitude of mechanisms and factors used by cells to regulate and maintain the integrity of repetitive DNA loci such as rDNA repeats, telomeric repeats, and retrotransposons. In the remainder of this review, we discuss how the regulation of these repetitive DNA loci can intersect in either beneficial or deleterious fashions with different and abundant non-canonical nucleic acid structures.
Non-canonical DNA structures at repetitive loci
In addition to the Watson and Crick double-helical structure, DNA can also adopt various non-standard, or non-canonical, DNA conformations. Such non-canonical structures are dependent on various DNA sequence motifs, ionic environments, and DNA-protein interactions. Of particular interest, 2 types of non-canonical DNA structures known as R-loops (harbouring RNA-DNA hybrids) and G-quadruplexes (GQ) are especially abundant within repetitive DNA loci.5 The formation of these non-canonical DNA structures within repetitive loci can be beneficial or deleterious depending on the physiologic or pathological context.
R-loops
R-loops are 3-stranded nucleic acid structures comprised of an RNA-DNA hybrid and a displaced ssDNA.93 They are often by-products of transcription, arising when nascent transcribed transcripts anneal to the DNA template strand (Fig. 4B). R-loops are potentially DNA damaging structures that have been shown to pause progression of the replication fork, leading to DSBs that may be repaired by HR. Moreover, R-loops are closely connected with GQs, which can be often formed on the displaced ssDNA of R-loops.93 Therefore, the prevention and resolution of R-loops at various loci has to be carefully orchestrated and an arsenal of proteins, including RNase H and Topoisomerase enzymes, play a central role in this process.94-96
Figure 4.
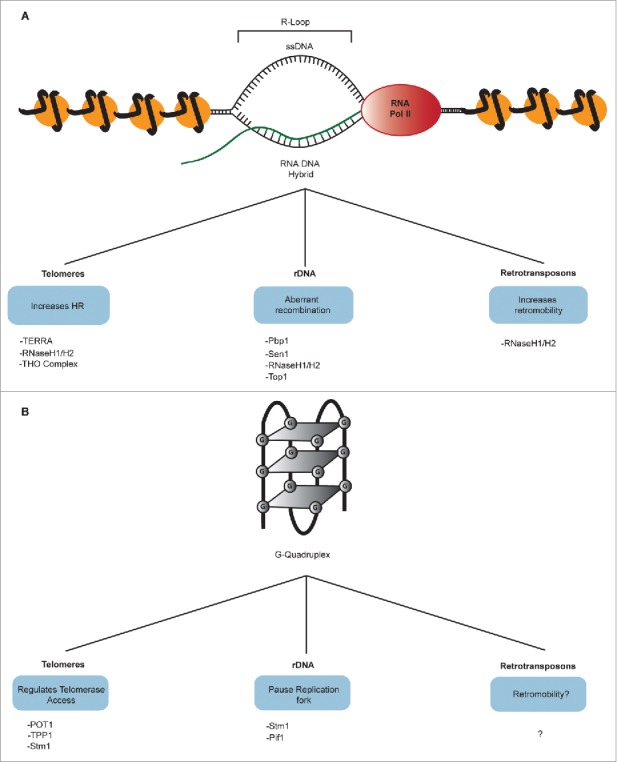
Non-canonical R-loop and G-quadruplex DNA structures. (A) R-loops are 3-stranded nucleic acid structures comprised of an RNA-DNA hybrid and a displaced single-strand of DNA. TERRA-associated R-loops at telomeres gives rise to an increase in HR-mediated elongation of telomeres, thus their accumulation is inhibited by the RNase H enzymes in yeast and mammalian cells and the THO complex in yeast cells. R-loop accumulation at yeast rDNA causes aberrant recombination and is therefore inhibited by various proteins including the yeast Ataxin-2 protein Pbp1 and Topoisomerase 1 (Top1). RNA:DNA hybrid accumulation at Ty1 retrotransposons increase Ty1 retromobility and are also inhibited by the RNase H enzymes. (B) G-Quadruplex (GQ) structures are formed via G4 motifs (GNxGNxGNx)4. Guanine residues form Hoogsteen bonds with 2 bordering guanines resulting in G-tetrads. The enclosed nucleotides, N, form loops that vary in size. GQs promote telomere stability and function and are regulated by POT1/TTP1, components of human shelterin complex, and are stabilized by the yeast Stm1 protein. G4 motifs are abundant at rDNA and GQ accumulation impedes progression of the replication fork progression. The yeast protein, Stm1 stabilizes GQs leading to R-loop accumulation but this action is counteracted by the conserved 5′-3′ DNA helicase Pif1, which binds and unwinds GQs. Potential GQ forming sequences have been identified in mammalian retrotransposons but little is known about GQ formation and function at these loci.
R-loops naturally occur in vivo, and have many important biologic functions such as immunoglobulin class switching and the regulation of gene expression.97, 98 Despite their biologic importance, R-loop accumulation poses a threat to genome integrity and is implicated in several cancers and syndromes. For example, mutations in amyotrophic lateral sclerosis (ALS)-linked factors can give rise to R-loop accumulation resulting in aberrant transcript accumulations.99
Studies have shown that the long non-coding telomeric repeat-containing RNA (TERRA), a transcript conserved across species, is implicated in R-loop formation/accumulation at telomeres in both yeast and human cells.94,100-102 Loss of the RNA-DNA hybrid-suppressing RNase H enzymes (RNaseH1 and RNaseH2), which degrade the RNA component of RNA-DNA hybrids, leads to R-loops accumulations at telomeres delaying cellular senescence.94,100 When the RNase H enzymes are deleted in telomerase deficient yeast cells, recombination at telomeres is increased giving rise to longer telomeres and consequently a delay in cellular senescence.100 This suggests that R-loop accumulation at telomeres leads to an increase in telomere length in an HR-mediated elongation pathway. In contrast, in HR-deficient yeast cells, R-loop accumulation at telomeres leads to premature cellular senescence.100 In human ALT cells, RNase H1 is highly enriched at telomeres and knock-down of this enzyme gives rise to an increase in TERRA-associated hybrids, which in turn increase the recombinogenic nature of ALT cells.94 These yeast and human studies suggest that telomeric R-loops can both inhibit and accelerate senescence, potentially giving rise to cancerous cells or premature aging, respectively (Fig. 4A).
Therefore, cells tightly regulate R-loop levels at these loci and use various factors, in addition to RNase H, to inhibit their undesirable accumulation at telomeres; such factors include members of the THO complex: Hpr1, Thp2, and Tho2 in yeast cells (Fig. 4A).101, 102 The THO complex is required for the assembly and export of mRNA.103 In telomerase deficient cells, THO mutants give rise to both premature senescence and telomere elongation through Rad53-dependent telomeric recombination.102 In these mutants, TERRA-associated R-loop accumulation triggers the Rad53-dependent DNA damage response (DDR).102 The induction of DDR in this setting is most likely due to the accumulation of DSBs caused by stalled replication forks at TERRA-harbouring R-loops.
As R-loops are transcriptional by-products capable of inducing DSBs, due to their ability to stall replication forks, it is plausible to assume that their accumulation at the highly transcribed rDNA loci may have detrimental effects. Indeed, R-loops have been shown to accumulate at rDNA IGS regions, particularly at E-pro, in the absence of the yeast Ataxin-2 protein Pbp1 or Topoisomerase 1 (Top1).95,96 This accumulation leads to aberrant recombination events at rDNA, resulting in genomic instability and culminating in a decrease in replicative life span (Fig. 4A). This mechanism seems to be evolutionarily conserved as human ATXN2, the ortholog of yeast Pbp1, was recently shown to prevent R-loop accumulations within nucleoplasmic and nucleolar regions of the nucleus in human cell culture.104 It remains an intriguing but untested possibility that physiologic R-loops at coding or non-coding rDNA regions may regulate various aspects of rDNA repeats in wild-type cells.
Recent studies have shown that RNA-DNA hybrids accumulate at Ty1 elements in the absence of the yeast RNase H enzymes (Rnh1 and Rnh201).105, 106 Whether hybrids accumulate at genomic Ty1 elements or at Ty1 cDNA is unclear: while one study reports that hybrids accumulate at chromosomal Ty1, another reports that hybrids primarily accumulate at Ty1 cDNA resulting in an increase in the frequency of retrotransposition (Fig. 4A).105,106 Yet another study proposed a model in which RNA-DNA hybrids exist in newly integrated Ty1 cDNA.107 Based on our current knowledge of R-loops, and the effect that their accumulation has at telomeres and rDNA repeats, one can postulate that the buildup of these structures at chromosomal retrotransposons might lead to DSBs repairable by non-allelic HR using other retrotransposon elements as template. Considering that retrotransposons are interspersed throughout the genome, HR repair at these loci can potentially result in GCRs. Indeed, Stamenova et al., proposed a model in which the Rrm3 helicase, a member of the Pif1 family, prevents the formation of DSBs within RNA-DNA hybrids present in newly integrated cDNA. In the absence of Rrm3, hybrid accumulations lead to an increase in GCRs, retrotransposition and the formation of tandem arrays of Ty1 cDNA.107 In these mutants, there was an increased recombination of Ty1 cDNA known as Ty1 multimers; high levels of Ty1 multimers increases the numbers of Ty1 cDNA incorporated into the genome per round of retrotransposition; this in turn gives rise to an increase in GCRs.107 As hybrids have been shown to affect telomere length, rDNA stability and Ty1 retromobility, it is imperative that cells keep these potentially damaging structures in check.
G-quadruplexes
DNA sequences harbouring (GNxGNxGNx)4 or G4 motifs have the capacity to form the tetrameric structure known as a G-quadruplex or GQ (Fig. 4A). GQs comprise 4 guanine nucleotides forming non-canonical Hoogsteen bonds with 2 bordering guanines resulting in a conformation of stacked square planar structures, or G-tetrads.108 The enclosed nucleotides form loops that vary in size, with smaller loops giving rise to relatively more stable structures. GQs can be classified based on 2 main features: 1) Intramolecular or intermolecular formation respectively via one or multiple strands; 2) parallel or antiparallel orientation of constituent DNA strands. The presence of a monovalent cation at the center of the G-tetrads stabilizes the GQ structure (for a full review on GQ structures please see108). G4 rich motifs that have the potential to form GQs are scattered throughout the human genome and are in abundance particularly within repetitive DNA loci.
G4 motifs with GQ-forming potential are enriched at telomeres due to the inherent local concentration of G-rich double-stranded DNA and single-stranded 3′ overhangs. There is mounting evidence for functional roles of GQs at telomeres. For example, telomeric GQs can promote telomere stability by allowing POT1/TPP1, components of the shelterin complex, to outcompete RPA binding to telomeres.109 ssDNA of telomeric 3′ overhangs interacts with POT1/TPP1 and RPA.109 RPA initiates the ataxia telangiectasia and Rad3 (ATR) DDR, a key player in the cell cycle checkpoint that induces cell cycle arrest in response to DNA damage.110 If the ATR response is prematurely/incorrectly initiated at telomeres, it can prematurely trigger cell cycle checkpoint and arrest. To avoid such an incorrect activation of this cell cycle checkpoint response, telomeric GQs enhance the binding of POT1 to ssDNA overhangs.109 Subsequent recruitment of TPP1 blocks RPA binding, protecting telomeres from premature ATR activation. In yeast cells, GQs have also been shown to act as telomere caps when standard capping is compromised.111 For instance, in telomere capping mutants, overexpression of the GQ stabilizing protein Stm1 stabilizes telomeric GQs allowing them to act as rudimentary telomere caps.111 Studies have also shown that GQs at telomeric 3′ overhangs can block and prevent telomere extension by Oxitricha telomerase in vitro (Fig. 4A).112 Intriguingly, human telomerase is capable of binding and partially unwinding parallel, but not anti-parallel, intermolecular GQs allowing for telomeric extension.113 These findings suggest that in addition to the mere presence of GQs, the specific conformations adopted by these structures are also important to the ultimate effect that they have on telomere length and stability. Thus, these studies show that GQs can indeed impact telomere stability and function.
G4 motifs are also abundant within rDNA repeats, which are the most actively transcribed loci in the genome and are paramount to overall cellular function. Thus, it is reasonable to postulate that formation of GQs must be highly regulated at these sites. The accumulation of GQ within rDNA repeats can impede progression of replication forks directly via GQ structures and indirectly via stabilization of GQ-containing R-loops (Fig. 4B).96,114 Indeed overexpression of the yeast GQ-stabilizing protein Stm1 leads to R-loop accumulation within rDNA repeats.104 In both yeast and humans, a conserved 5′-3′ DNA helicase termed Pif1, maintains the replication fork barrier at rDNA, promoting rDNA integrity.115 A common function of Pif1 is to bind and unwind GQs; in the absence of Pif1, GQ accumulations stall replication forks triggering DSBs that are repaired via aberrant recombination.114 GQs can therefore directly and indirectly impact rDNA repeat stability.
Potential GQ forming sequences have also been identified within the 3′ ends of LINE-1 and plant LTRs116,117 Although little is known about the role and/or effect that GQs have on retrotransposition, it is an intriguing hypothesis that the formation of GQs at these loci can regulate retrotransposition itself. GQs at the 3′ ends of cDNA may act as a barrier to reverse transcriptase (RT) thus blocking synthesis of second strand cDNA. Additionally, chromosomal formation of GQs on the sense strand can impede access to transcription machinery, transcriptionally silencing retrotransposition (Fig. 4B). Future work in the field should experimentally test these possibilities.
Concluding remarks
It is clear that eukaryotic cells must ensure the integrity of repetitive DNA loci to preserve genome function and stability. Here, we have highlighted key molecular mechanisms allowing cells to maintain rDNA repeats, telomeres, and transposons (Table 1). We have also reviewed emerging evidence supporting the intersection of these repetitive DNA loci and non-canonical DNA structures such as R-loops and GQs. These intersections can be beneficial or detrimental. We anticipate that future work characterizing these intersections will reveal secrets of eukaryotic genomes as well as provide unique insights into human health and disease.
Table 1.
Standard and non-canonical mechanisms regulating repetitive DNA loci.
|
rDNA REPEATS | |||
|---|---|---|---|
| Standard | |||
| Expansion & Contraction of array | Fob-1; Cohesin; Sir2 | Allows HR repair of damaged DNA and to maintain rRNA levels. | 24-27 |
| rDNA silencing | Fob-1; RENT; cohibin; Tof2 | Prevents hyper-recombination of rDNA repeats. | 20,28,31 |
| Nuclear localization | Cohibin, INM, | Inhibits USCE. | 20 |
| Delocalization of DSBs | Smc5-Smc6; MRX. | Prevents aberrant recombination events during DNA repair thereby inhibiting USCE. | 32,33 |
| Non-canonical | |||
| R-loop accumulation | Pbp1, ATXN2, Top1 | R-loop accumulation is implicated in aberrant rDNA recombination | 95,96,104 |
| G4 accumulation |
Stm1, Pif1 |
Stabilizes and destabilizes G4 respectively |
104,114 |
|
TELOMERES | |||
| Standard | |||
| Promoting telomerase-dependent elongation | Telomerase; Cdc13; Sir4; Ku; TPP1 | Prevents premature cellular senescence. | 46-48,51-53 |
| Inhibiting telomerase-dependent elongation | Rif1/Rif2; TRF1/TRF2; POT1 | Prevents survival and proliferation of cancerous cells. | 48-50,54-56 |
| NHEJ Inhibition | Rap1; Sir4; Rif2; Trf2 | Prevents end-to-end telomeric fusions. | 64,65 |
| Non-canonical | |||
| R-loops | TERRA, RNase H, THO Complex | R-loop accumulation increases HR-mediated elongation of telomeres | 94,100-102 |
| G-quadruplex |
hTelomerase, POT1, TPP1, Stm1 |
Regulation of telomere length, Protection against premature ATR activation, provides supplemental telomere capping |
109,111-113 |
|
RETROTRANSPOSONS | |||
| Standard | |||
| Transcriptional silencing | Tap42-phosphatase complex; Isw1p/Isw2p; MIWI2 | Represses retrotransposition thereby consequently inhibiting increase in genomic retroelement copy numbers. | 75,76,78, 80 |
| Post-transcriptional repression | miR-128; Ty1 RTL; Ty1 AS; p22/18 | Represses retrotransposition thereby consequently inhibiting increase in genomic retroelement copy numbers. | 82-87 |
| Targeted integration | RNA Pol III & Integrase | Ensures non-deleterious integrations. | 89 |
| Non-canonical | |||
| R-loops | RNase H, Rrm3 | R-loop accumulation increases Ty1 cDNA multimers, retrotransposition and GCRs | 105-107 |
| G-Quadruplex | Unknown | Unknown | |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
We thank members of the Mekhail laboratory for fruitful discussions.
Funding
A.C.H. is supported by an Ontario Graduate Scholarship (OGS). This work is funded by a grant from the Canadian Institutes of Health Research to K.M. who holds the Canada Research Chair in Spatial Genome Organization.
References
- [1].Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie 2005; 87(7):591-602; PMID:15989976; http://dx.doi.org/ 10.1016/j.biochi.2004.10.020 [DOI] [PubMed] [Google Scholar]
- [2].Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 2007; 71(1):13-35; PMID:17347517; http://dx.doi.org/ 10.1128/MMBR.00030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature 2001; 411(6835):366-74; PMID:11357144; http://dx.doi.org/ 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- [4].Bzymek M, Lovett ST. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A 2001; 98(15):8319-25; PMID:11459970; http://dx.doi.org/ 10.1073/pnas.111008398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci 2010; 67(1):43-62; PMID:19727556; http://dx.doi.org/ 10.1007/s00018-009-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7(12):e1002384; PMID:22144907; http://dx.doi.org/ 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ames D, Murphy N, Helentjaris T, Sun N, Chandler V. Comparative analyses of human single- and multilocus tandem repeats. Genetics 2008; 179(3):1693-704; PMID:18562644; http://dx.doi.org/ 10.1534/genetics.108.087882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cohen S, Mechali M. A novel cell-free system reveals a mechanism of circular DNA formation from tandem repeats. Nucleic Acids Res 2001; 29(12):2542-8; PMID:11410662; http://dx.doi.org/ 10.1093/nar/29.12.2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene 1997; 14(8):977-85; PMID:9050997; http://dx.doi.org/ 10.1038/sj.onc.1200917 [DOI] [PubMed] [Google Scholar]
- [10].George CM, Alani E. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit Rev Biochem Mol Biol 2012; 47(3):297-313; PMID:22494239; http://dx.doi.org/ 10.3109/10409238.2012.675644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kobayashi T. Ribosomal RNA gene repeats, their stability and cellular senescence. Proc Jpn Acad Ser B Phys Biol Sci 2014; 90(4):119-29; PMID:24727936; http://dx.doi.org/ 10.2183/pjab.90.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lopez-Flores I, Garrido-Ramos MA. The repetitive DNA content of eukaryotic genomes. Genome Dyn 2012; 7:1-28. PMID:22759811 [DOI] [PubMed] [Google Scholar]
- [13].Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al.. Initial sequencing and analysis of the human genome. Nature 2001; 409(6822):860-921; PMID:11237011; http://dx.doi.org/ 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- [14].Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 2013; 502(7471):389-92; PMID:24025772; http://dx.doi.org/ 10.1038/nature12584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang G, Vasquez KM. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst) 2014; 19:143-51; PMID:24767258; http://dx.doi.org/ 10.1016/j.dnarep.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci U S A 2006; 103(1):135-40; PMID:16371466; http://dx.doi.org/ 10.1073/pnas.0509691102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 2010; 11(5):317-28; PMID:20414256; http://dx.doi.org/ 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kobayashi T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci 2011; 68(8):1395-403; PMID:21207101; http://dx.doi.org/ 10.1007/s00018-010-0613-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 2010; 327(5966):693-6; PMID:20133573; http://dx.doi.org/ 10.1126/science.1179044 [DOI] [PubMed] [Google Scholar]
- [20].Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 2008; 456(7222):667-70; PMID:18997772; http://dx.doi.org/ 10.1038/nature07460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu N, Yu H. The Smc complexes in DNA damage response. Cell Biosci 2012; 2:5; PMID:22369641; http://dx.doi.org/ 10.1186/2045-3701-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Salvi JS, Chan JN, Pettigrew C, Liu TT, Wu JD, Mekhail K. Enforcement of a lifespan-sustaining distribution of Sir2 between telomeres, mating-type loci, and rDNA repeats by Rif1. Aging Cell 2013; 12(1):67-75; PMID:23082874; http://dx.doi.org/ 10.1111/acel.12020 [DOI] [PubMed] [Google Scholar]
- [23].French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 2003; 23(5):1558-68; PMID:12588976; http://dx.doi.org/ 10.1128/MCB.23.5.1558-1568.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 1998; 12(24):3821-30; PMID:9869636; http://dx.doi.org/ 10.1101/gad.12.24.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 2003; 23(24):9178-88; PMID:14645529; http://dx.doi.org/ 10.1128/MCB.23.24.9178-9188.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 2005; 309(5740):1581-4; PMID:16141077; http://dx.doi.org/ 10.1126/science.1116102 [DOI] [PubMed] [Google Scholar]
- [27].Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 2004; 117(4):441-53; PMID:15137938; http://dx.doi.org/ 10.1016/S0092-8674(04)00414-3 [DOI] [PubMed] [Google Scholar]
- [28].Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 2003; 17(17):2162-76; PMID:12923057; http://dx.doi.org/ 10.1101/gad.1108403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 1999; 97(2):233-44; PMID:10219244; http://dx.doi.org/ 10.1016/S0092-8674(00)80733-3 [DOI] [PubMed] [Google Scholar]
- [30].Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 2002; 22(12):4167-80; PMID:12024030; http://dx.doi.org/ 10.1128/MCB.22.12.4167-4180.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huang J, Brito IL, Villén J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 2006; 20(20):2887-901; PMID:17043313; http://dx.doi.org/ 10.1101/gad.1472706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragón L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 2007; 9(8):923-31; PMID:17643116; http://dx.doi.org/ 10.1038/ncb1619 [DOI] [PubMed] [Google Scholar]
- [33].Eckert-Boulet N, Lisby M. Regulation of rDNA stability by sumoylation. DNA Repair (Amst) 2009; 8(4):507-16; PMID:19261548; http://dx.doi.org/ 10.1016/j.dnarep.2009.01.015 [DOI] [PubMed] [Google Scholar]
- [34].Blackburn EH. Structure and function of telomeres. Nature 1991; 350(6319):569-73; PMID:1708110; http://dx.doi.org/ 10.1038/350569a0 [DOI] [PubMed] [Google Scholar]
- [35].Stewart JA, Chaiken MF, Wang F, Price CM. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res 2012; 730(1-2):12-9; PMID:21945241; http://dx.doi.org/ 10.1016/j.mrfmmm.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Vega LR, Mateyak MK, Zakian VA. Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol 2003; 4(12):948-59; PMID:14685173; http://dx.doi.org/ 10.1038/nrm1256 [DOI] [PubMed] [Google Scholar]
- [37].Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet 2012; 13(10):693-704; PMID:22965356; http://dx.doi.org/ 10.1038/nrg3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet 2008; 73(2):103-12; PMID:18005359; http://dx.doi.org/ 10.1111/j.1399-0004.2007.00923.x [DOI] [PubMed] [Google Scholar]
- [39].Holohan B, Wright WE, Shay JW. Cell biology of disease: Telomeropathies: an emerging spectrum disorder. J Cell Biol 2014; 205(3):289-99; PMID:24821837; http://dx.doi.org/ 10.1083/jcb.201401012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Watson JD. Origin of concatemeric T7 DNA. Nat New Biol 1972; 239(94):197-201; PMID:4507727; http://dx.doi.org/ 10.1038/newbio239197a0 [DOI] [PubMed] [Google Scholar]
- [41].Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 1973; 41(1):181-90; PMID:4754905; http://dx.doi.org/ 10.1016/0022-5193(73)90198-7 [DOI] [PubMed] [Google Scholar]
- [42].Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 2004; 117(3):323-35; PMID:15109493; http://dx.doi.org/ 10.1016/S0092-8674(04)00334-4 [DOI] [PubMed] [Google Scholar]
- [43].Niederer RO, Zappulla DC. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA 2015; 21(5):1053. PMID:25883220 [PMC free article] [PubMed] [Google Scholar]
- [44].Lee KM, Choi KH, Ouellette MM. Use of exogenous hTERT to immortalize primary human cells. Cytotechnology 2004; 45(1-2):33-8; PMID:19003241; http://dx.doi.org/ 10.1007/10.1007/s10616-004-5123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cifuentes-Rojas C, Shippen DE. Telomerase regulation. Mutat Res 2012; 730(1-2):20-7; PMID:22032831; http://dx.doi.org/ 10.1016/j.mrfmmm.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science 1999; 286(5437):117-20; PMID:10506558; http://dx.doi.org/ 10.1126/science.286.5437.117 [DOI] [PubMed] [Google Scholar]
- [47].Wu Y, Zakian VA. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci U S A 2011; 108(51):20362-9; PMID:21969561; http://dx.doi.org/ 10.1073/pnas.1100281108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hass EP, Zappulla DC. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. Elife 2015; 4; PMID:26218225; http://dx.doi.org/ 10.7554/eLife.07750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 2002; 16(12):1528-39; PMID:12080091; http://dx.doi.org/ 10.1101/gad.988802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kaizer H, Connelly CJ, Bettridge K, Viggiani C, Greider CW. Regulation of Telomere Length Requires a Conserved N-Terminal Domain of Rif2 in Saccharomyces cerevisiae. Genetics 2015; 201(2):573-86; PMID:26294668; http://dx.doi.org/ 10.1534/genetics.115.177899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 2010; 30(12):2971-82; PMID:20404094; http://dx.doi.org/ 10.1128/MCB.00240-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 2012; 492(7428):285-9; PMID:23103865; http://dx.doi.org/ 10.1038/nature11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schmidt JC, Dalby AB, Cech TR. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife 2014; 3; http://dx.doi.org/ 10.7554/eLife.03563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature 1997; 385(6618):740-3; PMID:9034193; http://dx.doi.org/ 10.1038/385740a0 [DOI] [PubMed] [Google Scholar]
- [55].Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 2000; 20(5):1659-68; PMID:10669743; http://dx.doi.org/ 10.1128/MCB.20.5.1659-1668.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature 2003; 423(6943):1013-8; PMID:12768206; http://dx.doi.org/ 10.1038/nature01688 [DOI] [PubMed] [Google Scholar]
- [57].Dilley RL, Greenberg RA. ALTernative Telomere Maintenance and Cancer. Trends Cancer 2015; 1(2):145-156; PMID:26645051; http://dx.doi.org/ 10.1016/j.trecan.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 1997; 3(11):1271-4; PMID:9359704; http://dx.doi.org/ 10.1038/nm1197-1271 [DOI] [PubMed] [Google Scholar]
- [59].Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 1995; 14(17):4240-8. PMID:7556065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bailey SM, Brenneman MA, Goodwin EH. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res 2004; 32(12):3743-51; PMID:15258249; http://dx.doi.org/ 10.1093/nar/gkh691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet 2000; 26(4):447-50; PMID:11101843; http://dx.doi.org/ 10.1038/82586 [DOI] [PubMed] [Google Scholar]
- [62].Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19(12):8083-93; PMID:10567534; http://dx.doi.org/ 10.1128/MCB.19.12.8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 2001; 21(5):1819-27; PMID:11238918; http://dx.doi.org/ 10.1128/MCB.21.5.1819-1827.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. Multiple pathways inhibit NHEJ at telomeres. Genes Dev 2008; 22(9):1153-8; PMID:18451106; http://dx.doi.org/ 10.1101/gad.455108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rai R, Chen Y, Lei M, Chang S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun 2016; 7:10881; PMID:26941064; http://dx.doi.org/ 10.1038/ncomms10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mc CB. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A 1950; 36(6):344-55; PMID:15430309; http://dx.doi.org/ 10.1073/pnas.36.6.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Munoz-Lopez M, Garcia-Perez JL. DNA transposons: nature and applications in genomics. Curr Genomics 2010; 11(2):115-28; PMID:20885819; http://dx.doi.org/ 10.2174/138920210790886871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 1998; 8(5):464-78. PMID:9582191 [DOI] [PubMed] [Google Scholar]
- [69].Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 2014; 24(7):1053-63; PMID:24823667; http://dx.doi.org/ 10.1101/gr.163659.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One 2012; 7(9):e44099; PMID:22957047; http://dx.doi.org/ 10.1371/journal.pone.0044099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sedivy JM, Kreiling JA, Neretti N, De Cecco M, Criscione SW, Hofmann JW, Zhao X, Ito T, Peterson AL. Death by transposition - the enemy within? Bioessays 2013; 35(12):1035-43; PMID:24129940; http://dx.doi.org/ 10.1002/bies.201300097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hancks DC, Kazazian HH Jr.. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 2012; 22(3):191-203; PMID:22406018; http://dx.doi.org/ 10.1016/j.gde.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Curcio MJ, Lutz S, Lesage P. The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae. Microbiol Spectr 2015; 3(2):MDNA3-0053-2014; PMID:26104690; http://dx.doi.org/ 10.1128/microbiolspec.MDNA3-0053-2014 [DOI] [PubMed] [Google Scholar]
- [74].Jiang YW. Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes Dev 2002; 16(4):467-78; PMID:11850409; http://dx.doi.org/ 10.1101/gad.923502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wu X, Jiang YW. Overproduction of non-translatable mRNA silences. The transcription of Ty1 retrotransposons in S. cerevisiae via functional inactivation of the nuclear cap-binding complex and subsequent hyperstimulation of the TORC1 pathway. Yeast 2008; 25(5):327-47; PMID:18435413; http://dx.doi.org/ 10.1002/yea.1591 [DOI] [PubMed] [Google Scholar]
- [76].Jiang YW. An essential role of Tap42-associated PP2A and 2A-like phosphatases in Ty1 transcriptional silencing of S. cerevisiae. Yeast 2008; 25(10):755-64; PMID:18949822; http://dx.doi.org/ 10.1002/yea.1631 [DOI] [PubMed] [Google Scholar]
- [77].Scholes DT, Kenny AE, Gamache ER, Mou Z, Curcio MJ. Activation of a LTR-retrotransposon by telomere erosion. Proc Natl Acad Sci U S A 2003; 100(26):15736-41; PMID:14673098; http://dx.doi.org/ 10.1073/pnas.2136609100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev 2001; 15(5):619-26; PMID:11238381; http://dx.doi.org/ 10.1101/gad.190301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xiao-Jie L, Hui-Ying X, Qi X, Jiang X, Shi-Jie M. LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med 2016; 18(5):431-9; PMID:26334179; http://dx.doi.org/ 10.1038/gim.2015.119 [DOI] [PubMed] [Google Scholar]
- [80].Pezic D, Manakov SA, Sachidanandam R, Aravin AA. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 2014; 28(13):1410-28; PMID:24939875; http://dx.doi.org/ 10.1101/gad.240895.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Curcio MJ, Hedge AM, Boeke JD, Garfinkel DJ. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet 1990; 220(2):213-21; PMID:2157950; http://dx.doi.org/ 10.1007/BF00260484 [DOI] [PubMed] [Google Scholar]
- [82].Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, Pedersen IM. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol 2015; 22(10):824-31; PMID:26367248; http://dx.doi.org/ 10.1038/nsmb.3090 [DOI] [PubMed] [Google Scholar]
- [83].Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 2008; 22(5):615-26; PMID:18316478; http://dx.doi.org/ 10.1101/gad.458008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A 2009; 106(37):15657-62; PMID:19721006; http://dx.doi.org/ 10.1073/pnas.0908305106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Saha A, Mitchell JA, Nishida Y, Hildreth JE, Ariberre JA, Gilbert WV, Garfinkel DJ. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol 2015; 89(7):3922-38; PMID:25609815; http://dx.doi.org/ 10.1128/JVI.03060-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tucker JM, Larango ME, Wachsmuth LP, Kannan N, Garfinkel DJ. The Ty1 Retrotransposon Restriction Factor p22 Targets Gag. PLoS Genet 2015; 11(10):e1005571; PMID:26451601; http://dx.doi.org/ 10.1371/journal.pgen.1005571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nishida Y, Pachulska-Wieczorek K, Błaszczyk L, Saha A, Gumna J, Garfinkel DJ, Purzycka KJ. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res 2015; 43(15):7414-31; PMID:26160887; http://dx.doi.org/ 10.1093/nar/gkv695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev 1996; 10(5):620-33; PMID:8598291; http://dx.doi.org/ 10.1101/gad.10.5.620 [DOI] [PubMed] [Google Scholar]
- [89].Bridier-Nahmias A, Tchalikian-Cosson A, Baller JA, Menouni R, Fayol H, Flores A, Saïb A, Werner M, Voytas DF, Lesage P. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science 2015; 348(6234):585-8; PMID:25931562; http://dx.doi.org/ 10.1126/science.1259114 [DOI] [PubMed] [Google Scholar]
- [90].Parket A, Inbar O, Kupiec M. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics 1995; 140(1):67-77. PMID:7635309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Robberecht C, Voet T, Zamani Esteki M, Nowakowska BA, Vermeesch JR. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res 2013; 23(3):411-8; PMID:23212949; http://dx.doi.org/ 10.1101/gr.145631.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Startek M, Szafranski P, Gambin T, Campbell IM, Hixson P, Shaw CA, Stankiewicz P, Gambin A. Genome-wide analyses of LINE-LINE-mediated nonallelic homologous recombination. Nucleic Acids Res 2015; 43(4):2188-98; PMID:25613453; http://dx.doi.org/ 10.1093/nar/gku1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell 2012; 46(2):115-24; PMID:22541554; http://dx.doi.org/ 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- [94].Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 2014; 5:5220; PMID:25330849; http://dx.doi.org/ 10.1038/ncomms6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 2010; 24(14):1546-58; PMID:20634320; http://dx.doi.org/ 10.1101/gad.573310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Salvi JS, Chan JN, Szafranski K, Liu TT, Wu JD, Olsen JB, Khanam N, Poon BP, Emili A, Mekhail K. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev Cell 2014; 30(2):177-91; PMID:25073155; http://dx.doi.org/ 10.1016/j.devcel.2014.05.013 [DOI] [PubMed] [Google Scholar]
- [97].Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 2008; 28(1):50-60; PMID:17954560; http://dx.doi.org/ 10.1128/MCB.01251-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Castellano-Pozo M, Santos-Pereira JM, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, García-Muse T, Aguilera A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 2013; 52(4):583-90; PMID:24211264; http://dx.doi.org/ 10.1016/j.molcel.2013.10.006 [DOI] [PubMed] [Google Scholar]
- [99].Salvi JS, Mekhail K. R-loops highlight the nucleus in ALS. Nucleus 2015; 6(1):23-9; PMID:25587791; http://dx.doi.org/ 10.1080/19491034.2015.1004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 2013; 20(10):1199-205; PMID:24013207; http://dx.doi.org/ 10.1038/nsmb.2662 [DOI] [PubMed] [Google Scholar]
- [101].Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J 2013; 32(21):2861-71; PMID:24084588; http://dx.doi.org/ 10.1038/emboj.2013.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Yu TY, Kao YW, Lin JJ. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proceedings of the National Academy of Sciences of the United States of America 2014; 111(9):3377-3382; PMID:24550456; http://dx.doi.org/ 10.1073/pnas.1307415111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Luna R, Rondon AG, Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim Biophys Acta 2012; 1819(6):514-20; PMID:22207203; http://dx.doi.org/ 10.1016/j.bbagrm.2011.11.012 [DOI] [PubMed] [Google Scholar]
- [104].Abraham KJ, Chan JN, Salvi JS, Ho B, Hall A, Vidya E, Guo R, Killackey SA, Liu N, Lee JE, et al.. Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA-DNA hybrids. Nucleic Acids Res 2016; 44(18):8870-8884; PMID:27574117; http://dx.doi.org/ 10.1093/nar/gkw752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet 2014; 10(10):e1004716; PMID:25357144; http://dx.doi.org/ 10.1371/journal.pgen.1004716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 2014; 10(4):e1004288; PMID:24743342; http://dx.doi.org/ 10.1371/journal.pgen.1004288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Stamenova R, Maxwell PH, Kenny AE, Curcio MJ. Rrm3 protects the Saccharomyces cerevisiae genome from instability at nascent sites of retrotransposition. Genetics 2009; 182(3):711-23; PMID:19414561; http://dx.doi.org/ 10.1534/genetics.109.104208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 2012; 13(11):770-80; PMID:23032257; http://dx.doi.org/ 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc Natl Acad Sci U S A 2014; 111(8):2990-5; PMID:24516170; http://dx.doi.org/ 10.1073/pnas.1321436111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300(5625):1542-8; PMID:12791985; http://dx.doi.org/ 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
- [111].Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou MP, Abramowitz L, et al.. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 2011; 18(4):478-85; PMID:21399640; http://dx.doi.org/ 10.1038/nsmb.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature 1991; 350(6320):718-20; PMID:2023635; http://dx.doi.org/ 10.1038/350718a0 [DOI] [PubMed] [Google Scholar]
- [113].Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, Lovrecz GO, Beck JL, Bryan TM. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun 2015; 6:7643; PMID:26158869; http://dx.doi.org/ 10.1038/ncomms8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011; 145(5):678-91; PMID:21620135; http://dx.doi.org/ 10.1016/j.cell.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 2000; 100(4):479-89; PMID:10693764; http://dx.doi.org/ 10.1016/S0092-8674(00)80683-2 [DOI] [PubMed] [Google Scholar]
- [116].Lexa M, Kejnovský E, Steflová P, Konvalinová H, Vorlícková M, Vyskot B. Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res 2013; 42(2):968-78; PMID:24106085; http://dx.doi.org/ 10.1093/nar/gkt893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lexa M, Ullrich-Lüter E, Ortega-Martinez O, Dupont S, Arnone MI, Mallefet J, Flammang P. Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics 2014; 15:1035; PMID:25429842; http://dx.doi.org/ 10.1186/1471-2164-15-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]


