Abstract
To maintain genome stability, the entire genome of a eukaryotic cell must be replicated once and only once per cell cycle. In many organisms, multiple overlapping mechanisms block rereplication, but the consequences of deregulating these mechanisms are poorly understood. Here, we show that disrupting these controls in the budding yeast Saccharomyces cerevisiae rapidly blocks cell proliferation. Rereplicating cells activate the classical DNA damage-induced checkpoint response, which depends on the BRCA1 C-terminus checkpoint protein Rad9. In contrast, Mrc1, a checkpoint protein required for recognition of replication stress, does not play a role in the response to rereplication. Strikingly, rereplicating cells accumulate subchromosomal DNA breakage products. These rapid and severe consequences suggest that even limited and sporadic rereplication could threaten the genome with significant damage. Hence, even subtle disruptions in the cell cycle regulation of DNA replication may predispose cells to the genomic instability associated with tumorigenesis.
INTRODUCTION
Eukaryotic DNA replication is tightly controlled such that every segment of the genome is replicated once and only once each cell cycle. This control is primarily exerted at the hundreds to thousands of replication origins where DNA replication initiates. Once an origin initiates in S phase, multiple mechanisms prevent it from reinitiating replication for the remainder of that cell cycle (Gopalakrishnan et al., 2001; Nguyen et al., 2001; Vas et al., 2001; Yanow et al., 2001; Vaziri et al., 2003). Such tight control suggests that even an occasional reinitiation event would be deleterious to cells, and it is readily apparent that, in principle, excessive synthesis of just small segments of the genome could eventually threaten its stable propagation. Nonetheless, a direct analysis of the consequences of rereplication is needed to understand whether and how rereplication contributes to genomic instability. S. cerevisiae provides a powerful genetic system for such an analysis, especially as there is considerable understanding of both the mechanisms regulating replication and those protecting genome stability in this organism.
Eukaryotic replication initiation can be divided into two fundamental stages (reviewed in Bell and Dutta, 2002). In the first stage, which occurs in early G1 phase, a prereplicative complex (pre-RC) is assembled at replication origins through the sequential loading of the initiation proteins origin recognition complex (ORC), Cdc6, Cdt1, and Mcm2-7. In the second stage, activation of two kinases, Dbf4-Cdc7 kinase and a cyclin-dependent kinase (CDK), triggers events that culminate in replication initiation and disassembly of the prereplicative complex: additional replication proteins are recruited to the origin, the DNA is unwound, and replisomes are assembled at two nascent replication forks.
In addition to triggering initiation, CDKs also prevent reinitiation of eukaryotic DNA replication (Broek et al., 1991; Dahmann et al., 1995; Sauer et al., 1995; Hua et al., 1997). CDKs do this in part by down-regulating multiple components of the pre-RC, thereby preventing reassembly of these complexes at origins that have initiated. In budding yeast, CDKs promote the nuclear exclusion of Mcm2-7 (Labib et al., 1999; Nguyen et al., 2000), inhibit CDC6 transcription (Moll et al., 1991) and promote its degradation (Drury et al., 1997; Elsasser et al., 1999; Drury et al., 2000), and they seem to inactivate ORC through phosphoryation (Nguyen et al., 2001). Making these three initiation factors refractory to CDK inhibition in metaphase-arrested cells allows a subset of origins to reinitiate and portions of the genome to rereplicate (Nguyen et al., 2001). The limited extent of reinitiation suggests that not all inhibitory mechanisms to block rereplication have been identified. Consistent with this, a recent study indicates that CDK binding to ORC provides an additional mechanism to inhibit pre-RC formation (Wilmes et al., 2004).
Analogous CDK-dependent mechanisms antagonizing Cdc6, ORC, and Cdt1 have been shown to inhibit rereplication in other eukaryotes (Jallepalli et al., 1997; Lopez-Girona et al., 1998; Nishitani et al., 2000; Vas et al., 2001; Wuarin et al., 2002; Zhong et al., 2003). Moreover, a CDK-independent mechanism to prevent pre-RC assembly has been identified in metazoans. Central to this mechanism is the protein Geminin (McGarry and Kirschner, 1998; Tada et al., 2001; Wohlschlegel et al., 2002), which binds to Cdt1 and is thought to sterically inhibit its ability to recruit Mcm proteins to replication origins (Lee et al., 2004; Saxena et al., 2004). Inactivation of geminin can lead to partial rereplication, confirming its role in preventing reinitiation of DNA replication (Quinn et al., 2001; Mihaylov et al., 2002; Melixetian et al., 2004; Zhu et al., 2004).
The partial extent of rereplication that we and others have observed suggests that these rereplicating forks are stalled or damaged before they can completely rereplicate the entire genome. Such insults to the rereplicating genome could trigger one or both of the checkpoint pathways that monitor genome integrity (reviewed in Melo and Toczyski, 2002; Nyberg et al., 2002). The replication stress pathway responds to slowed or stalled replication forks, such as those arising from inhibition of nucleotide incorporation. The DNA damage pathway responds to chromosomal insults such as double-stranded breaks generated by ionizing radiation or enzymatic cleavage. These pathways activate proteins that stabilize stalled replication forks and repair DNA damage, respectively. In addition, they provide critical time to complete the replication or repair of DNA by imposing arrests at key cell cycle transitions.
Distinguishing whether the replication stress and/or DNA damage pathway is activated is an important first step in understanding the immediate molecular response to re-replication. This distinction is difficult because many checkpoint proteins and events are shared between the two pathways. For example, in metazoans, both types of genomic insults lead to the induction of p21, p53, and PIG3 protein levels; the phosphorylation of histone H2AX, p53, Cdc2, and the checkpoint kinases Chk1 and Chk2; and the organization of H2AX and Rad51 into subnuclear foci (Haaf et al., 1995; Gottifredi et al., 2001; Saintigny et al., 2001; Ward and Chen, 2001; Brown and Baltimore, 2003). In a few of these responses, the kinetics or degree of change may vary between the two pathways, but overall the events considered to be hallmarks of DNA damage also are observed with replication stress. Complicating the distinction between these two responses is the potential for stalled forks to degenerate into damaged forks, particularly if the stalled forks are not properly stabilized (reviewed in Nyberg et al., 2002).
Two groups have recently reported that the induction of rereplication in human cells induces a checkpoint response. The first group initially reported that rereplication induced by overexpression of Cdc6 and Cdt1 activates a DNA damage response (Vaziri et al., 2003), but they have subsequently observed that overexpression of Cdc6 alone can induce this response in the absence of any detectable rereplication (Zhu et al., 2004). Instead, they now report that rereplication induced by geminin depletion leads to what they suspect is a stalled fork response (Zhu et al., 2004). Thus, they no longer assert that rereplication generates DNA damage. A second group observes similar events during geminin depletion, which they attribute to either a DNA damage or replication stress response (Melixetian et al., 2004). Thus, although a clear assignment of pathways was not possible, the data are consistent with rereplication generating a replication stress-like response.
In Saccharomyces cerevisiae, the DNA damage and replication stress responses can be genetically distinguished, because the DNA damage pathway is primarily dependent on the BRCA1 C-terminus checkpoint protein Rad9p (reviewed in Toh and Lowndes, 2003), whereas the replication stress pathway is primarily dependent on Mrc1p (Alcasabas et al., 2001; Osborn and Elledge, 2003). In this work, we take advantage of this genetic distinction to unambiguously determine which response is activated upon rereplication. We present evidence that rereplication leads to significant invi-ability and the activation of a RAD53 (budding yeast Chk2)-dependent checkpoint response. The RAD9 dependence of the signaling pathway suggests that rereplication is triggering a DNA damage response and is not inducing a replication stress pathway. Moreover, we present the first direct evidence for the accumulation of chromosomal damage as a consequence of rereplication. These data indicate that rereplication induces DNA damage and poses an immediate threat to both cell viability and genome integrity.
MATERIALS AND METHODS
Strain Construction
All strains (Table 1) with the exception of YJL310 were derived from YJL1737 (orc2-cdk6A orc6-cdk4A leu2 ura3-52 trp1-289 ade2 ade3 bar1Δ::LEU2). The orc2-cdk6A and orc6-cdk4A alleles encode mutant proteins in which alanine is substituted for the phosphoacceptor serines or threonines in CDK consensus phosphorylation sites (S/T-P-X-K/R). For orc2-cdk6A, residues 16, 24, 70, 174, 188, and 206 were mutated and for orc6-cdk4A, residues 106, 116, 123, and 146 were mutated. The following plasmids were digested and integrated as follows: pJL806 (pGAL1, URA3/StuI; Nguyen et al., 2001), pJL1489 (pGAL1- Δntcdc6, URA3/StuI; Nguyen et al., 2001), pRS304-Rad53-HA-HIS (RAD53-HA-HIS, TRP1/HpaI; Emili, 1998), YIp22 (pMET3-HA3-CDC20, TRP1/MscI; Uhlmann et al., 2000), and pBO1555 (pMET3-HA3-CDC20, NatMX4/MscI). pJL1206 (MCM7-2NLS, URA3/AspI; Nguyen et al., 2001) was used to replace MCM7 with MCM7-2NLS by two-step gene replacement. The plasmid pBO1555 was generated by subcloning a BglII to SalI pMET3-HA3-CDC20 fragment from YIp22 into pAG25 (Goldstein and McCusker, 1999) cut with BglII and SalI.
Table 1.
Strains used in this study
| Strain | Source | Genotype |
|---|---|---|
| YJL310 | Detweiler and Li (1998) | leu2-3112 ura3-52 trp1-289 bar1Δ::LEU2 |
| YJL3244 | Nguyen et al. (2001) | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3) trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} |
| YJL3248 | Nguyen et al. (2001) | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} |
| YJL3604 | This study | rad53Δ::kanMX6 sml1Δ::TRP1 orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 |
| YJL3607 | This study | rad53Δ::kanMX6 sml1Δ::TRP1 orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 |
| YJL5048 | This study | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} |
| YJL5055 | This study | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} |
| YJL5060 | This study | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} rad9Δ::kanMX |
| YJL5065 | This study | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} rad9Δ::kanMX |
| YJL5085 | This study | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} mrc1Δ::kanMX |
| YJL5087 | This study | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} mrc1Δ::kanMX |
| YJL5132 | This study | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 ddc2::{DDC2-GFP, kanMX} |
| YJL5135 | This study | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 ddc2::{DDC2-GFP, kanMX} |
| YJL5408 | This study | rad53Δ::kanMX6 sml1Δ::TRP1 orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, NatMX} |
| YJL5411 | This study | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} rad9Δ::kanMX cdc20::{pMET3-HA3-CDC20, NatMX} |
| YJL5441 | This study | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 rad53::{RAD53-2HA6HIS, TRP1} mrc1Δ::kanMX cdc20::{pMET3-HA3-CDC20, NatMX} |
Genomic DNA from yJK7-2 (Melo et al., 2001) was used as a template to generate a DDC2-GFP, kanMX PCR fragment by using OJL1404 and OJL1405. Genomic DNA from U973 (sml1Δ::TRP1 esr1-1; Rothstein laboratory) was used as a template to generate a sml1Δ::TRP1 polymerase chain reaction (PCR) fragment by using OJL1110 and OJL1111. Genomic DNA from the yeast haploid deletion collection (ResGen; Invitrogen, Carlsbad, CA) was used as a template to generate a rad9Δ::kanMX PCR fragment by using OJL1487 and OJL1488. The entire RAD53 and MRC1 open reading frames were deleted using PCR amplification of the kanMX from pAG25 with tagged primers by using the oligonucleotides indicated in Table 2 (Goldstein and McCusker, 1999).
Table 2.
Oligonucleotides used in this study
| Oligo | Purpose | Sequence |
|---|---|---|
| OJL1404 | DDC2-GFP | AAAGGTACGTGGGACAAGAC |
| OJL1405 | DDC2-GFP | AGACAGCAACACACATCTAG |
| OJL1110 | sml1Δ | ctcgcatcgatAAGGATCACGTTCCTTCTGC |
| OJL1111 | sml1Δ | gcgacctcgagGAAGACATTGCGGGTTCAAG |
| OJL1002 | rad53Δ | GAGAGAATAGTGAGAAAAGATAGTGTTACACAACATCAACcggatccccgggttaattaa |
| OJL1003 | rad53Δ | ctcttaaaaaggggcagcattttctatgggtatttgtcctgaattcgagctcgtttaaac |
| OJL1487 | rad9Δ | GCTCCCCATCAAAATAAGGTC |
| OJL1488 | rad9Δ | TATGTGTCGTCCCAGTACTC |
| OJL1497 | mrc1Δ | AGACAAACAACTAAGGAAGTTCGTTATTCGCTTTTGAACTTATCACCAAATATTTTAGTGcggatccccgggttaattaa |
| OJL1498 | mrc1Δ | CGACTACTTCAAGACAGCTTCTGGAGTTCAATCAACTTCTTCGGAAAAGATAAAAAACCAcatcgatgaattcgagctcg |
Yeast Media
Cells were grown in YEP, synthetic complete (SC), or synthetic (S broth) medium (Guthrie and Fink, 1990) supplemented with 2% dextrose (wt/vol), 2% galactose (wt/vol), 3% raffinose (wt/vol), or 3% raffinose (wt/vol) + 0.05% dextrose (wt/vol). To obtain reproducible induction of rereplication, cells were inoculated from a culture containing 2% dextrose into a culture containing 3% raffinose + 0.05% dextrose and grown for 12-15 h overnight before the experiment commenced.
Cell Proliferation Assay
Yeast cells were diluted in S broth to OD600 measurements of 0.2, and then serially diluted fivefold for six dilutions and spotted onto SDC-Ura or SGalC-Ura plates. For transient pulses of rereplication, cells grown overnight in SRaffC-Ura + 0.05% dextrose were pelleted and resuspended in YEPRaff + 15 μg/ml nocodazole. Once >90% of the cells were arrested as large budded cells, galactose was added to a final concentration of 2%, and samples were removed at various time points, diluted in SD broth, and plated on SDC-Ura plates. Colonies were counted after 72 h at 30°C. All platings were done in triplicate, and two separate experiments were conducted. The mean and SE of the mean are shown. Statistical significance was determined using a Student's t test.
Flow Cytometry Analysis
Cells grown overnight in SRaffC-Met, Ura + 0.05% dextrose were pelleted and resuspended in YEPRaff + 2 mM methionine to arrest cells in metaphase by Cdc20p depletion. Once arrested (>90% large budded cells), nocodazole (15 μg/ml) was added for an additional 30 min. Galactose was then added to a final concentration of 2%, and samples were taken every hour. Cells were fixed and stained with 1 μM Sytox Green (Molecular Probes, Eugene, OR) as described previously (Haase and Lew, 1997). Vertical lines indicate median DNA content after gating from 100 to 1000, which captures all whole, unclumped cells.
DDC2-Green Fluorescent Protein (GFP) Foci
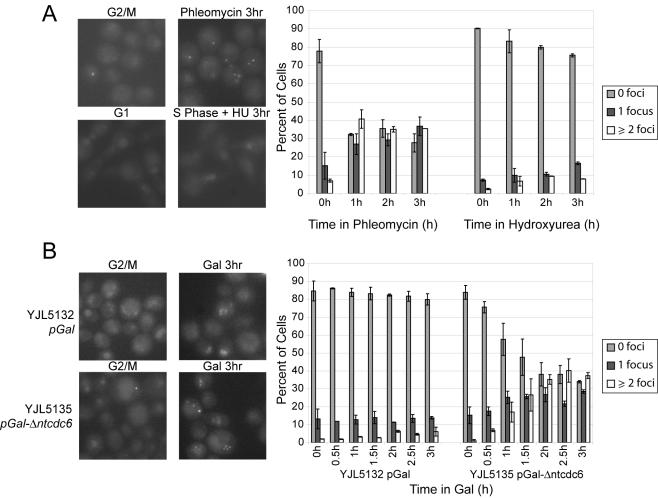
Cells grown overnight in YEPRaff + 0.05% dextrose were pelleted and resuspended in YEPRaff + 15 μg/ml nocodazole. Once >90% of the cells were arrested as large budded cells, galactose was added to 2%, and samples were removed at various time points, washed twice in phosphate-buffered saline (PBS), and visualized using an Olympus BX60 microscope. Pictures were recorded using a Hamamatsu Orca-ER camera and OpenLab3.1.7 software. Fluorescent images were taken in three z sections that bracketed the thickness of the cell, and then projected into one image by using ImageJ's maximum pixel intensity function. Between 60 and 120 cells were scored for zero, one, or two or more foci per cell, for each strain for each time point. To obtain hydroxyurea (HU)-treated cells for the experiment in Figure 3A, cells were grown in YEPD. They were then arrested in G1 (>95% unbudded cells) with 50 ng/ml α factor and released into a HU arrest with the addition of pronase to a final concentration of 100 μg/ml and HU to a final concentration of 0.2 M. Samples were processed for quantification as described above. To obtain phleomycin-treated cells for the experiment in Figure 3A, cells were grown in YEPD, arrested with 15 μg/ml nocodazole (>95% large budded cells), and then treated with phleomycin at a final concentration of 20 μg/ml (Cayla, Toulouse, France). Samples were processed for quantification as described above.
Figure 3.
Subnuclear Ddc2p foci consistent with DNA damage are formed when rereplication is induced. (A) HU-induced replication stress does not induce subnuclear Ddc2p foci to the same extent as DNA damage. YJL5135 (ddc2:DDC2-GFP pGAL1-Δntcdc6 orc2-cdk6A orc6-cdk4A MCM7-2NLS) growing in medium containing 2% dextrose was arrested in metaphase with 15 μg/ml nocodazole followed by treatment with 20 μg/ml phleomycin to induce DNA damage. A parallel culture was arrested in G1 phase with α factor and released from the arrest into 0.2 M HU to induce replication stress. At hourly intervals after either phleomycin addition or release into HU, cells were scored for subnuclear GFP foci, and the number of cells with zero foci, one focus, or two or more foci was quantified. Representative images at 0 and 3 h are shown. Error bars show SE of the mean from two experiments (n = 60-120 per experiment). (B) Rereplication induces Ddc2p foci. YJL5135 and YJL5132 (ddc2:DDC2-GFP pGAL1 orc2-cdk6A orc6-cdk4A MCM7-2NLS) growing in medium containing 3% raffinose + 0.05% dextrose were arrested in metaphase by the addition of 15 μg/ml nocodazole. Then, 2% galactose was added to induce rereplication in YJL5135 and at 30-min intervals the number of foci per cell was quantified (n = 60-120 per experiment). Representative images and quantification are shown as in A.
Rad53p Immunoblot
Cells grown overnight in SRaffC-Ura + 0.05% dextrose were pelleted and resuspended in YEPRaff + 15 μg/ml nocodazole. Once >90% of the cells were arrested as large budded cells, galactose was added to a final concentration of 2%, and samples were removed at various time points. Cells (8.5 ml) at OD600 0.5-1.0 were pelleted and lysed by vortex mixing and boiling with 300 μl of 0.5-mm glass beads (Biospec Products, Bartlesville, OK) and 300 μl of SDS-PAGE loading buffer [8% glycerol (vol/vol), 100 mM Tris-HCl, pH 6.8, 1.6% SDS (wt/vol), 1.6 × 10-3% bromphenol blue (wt/vol), 100 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride] with protease inhibitors (1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml chymostatin, and 1 mM benzamidine) and phosphatase inhibitors (1 mM Na3VO4, 50 mM NaF, and 50 mM Na β-glycerophosphate). The soluble protein was quantified using a Bradford assay (Bio-Rad, Hercules, CA) with bovine serum albumin as a standard (Sigma-Aldrich, St. Louis, MO). Forty micrograms of each protein sample was electrophoresed on a 7.5% SDS-PAGE gel and transferred to nitrocellulose (Protran BA85; Applied Scientific, San Francisco, CA). The membrane was probed with anti-HA 16B12 (Covance, Berkeley, CA) at 1:1000, followed by sheep anti-mouse horseradish peroxidase (NA931V; Amersham Biosciences, Piscataway, NJ) at 1:2000. Immunoblots were developed with the SuperSignal system (Pierce Chemical, Rockford, IL).
Assaying Induction of a Metaphase Arrest
Cells grown overnight in SRaffC-Met,Ura + 0.05% dextrose were pelleted and resuspended in YEPRaff + 2 mM methionine to arrest cells in metaphase by Cdc20p depletion. Once arrested (>90% large budded cells), galactose was added to a final concentration of 2% for 2 h, and then the cells were filtered and washed with S broth and resuspended in SGalC-Met,Ura + 50 ng/ml α factor. Samples were fixed in 67% ethanol (vol/vol), washed twice with PBS, and resuspended in 50 ng/ml 4′6-diamidino-2-phenylindole (DAPI). Cells were visualized by fluorescence microscopy on an Olympus BX60 microscope and quantified as pre- or postmetaphase based on nuclear morphology. At least 200 cells were scored for each strain for each time point, and the experiment was executed twice. The mean percentage of postmetaphase cells and the SE of the mean from the two experiments are charted.
Pulsed Field Gel Electrophoresis (PFGE)
YJL3244 and YJL3248 cells grown overnight in SRaffC-Met,Ura + 0.05% dextrose were pelleted and resuspended in YEPRaff + 2 mM methionine to arrest cells in metaphase by Cdc20p depletion. Once arrested (>90% large budded cells), nocodazole was added to a final concentration of 15 μg/ml for 30 min, after which galactose was added to a final concentration of 2% at time 0. To obtain HU-treated cells for the experiment in Figure 5, cells were grown in YEPD. They were then arrested in G1 (>95% unbudded cells) with 50 ng/ml α factor and released into a HU arrest with the addition of pronase to a final concentration of 100 μg/ml and HU to a final concentration of 0.2 M. To obtain phleomycin-treated cells for the experiment in Figure 5, cells were grown in YEPD, arrested with 15 μg/ml nocodazole (>95% large budded cells), and then treated with phleomycin at a final concentration of 20 or 200 μg/ml (Cayla).
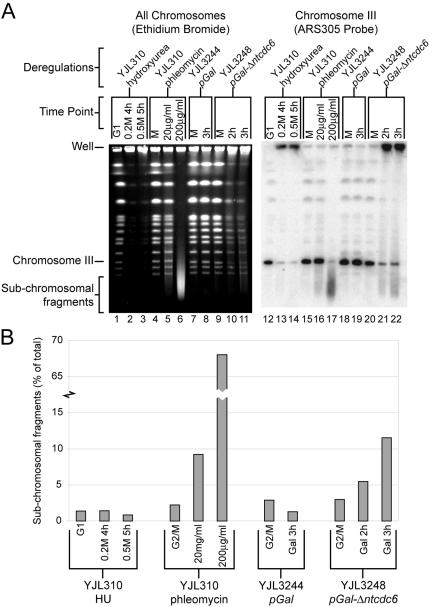
Figure 5.
Rereplication induces double-stranded breaks. (A) Rereplication generates subchromosomal fragments. (Lanes 1-11) PFGE stained with ethidium bromide. (Lanes 12-22) Southern blot of PFGE probed with ARS305 fragment to detect chromosome III. (Lanes 1-3, 12-14) YJL310 (CDC6 ORC2 ORC6 MCM7) was arrested in G1 phase with α factor and then released from the arrest into the indicated amounts of HU for the indicated times. (Lanes 4-6, 15-17) YJL310 was arrested in metaphase with 15 μg/ml nocodazole and then treated for 2 h with the indicated amount of phleomycin. (Lanes 7-8, 18-19) YJL3244 (pGAL1 orc2-cdk6A orc6-cdk4A MCM7-2NLS pMET3-HA3-CDC20) was arrested in metaphase in medium containing 3% raffinose and 2 mM methionine. Once arrested, galactose was added to 2%. (Lanes 9-11, 20-22) YJL3248 (pGAL1-Δntcdc6 orc2-cdk6A orc6-cdk4A MCM7-2NLS pMET3-HA3-CDC20) was treated as for YJL3244. DNA from equivalent numbers of cells were loaded in each lane, except twice as many cells were loaded in lanes 1-3 and 12-14 to compensate for their G1 or nearly G1 DNA content. (B) Quantification of subchromosomal fragment from Southern blot in A. The intensity of subchromosomal signal is shown as a percentage of the total signal for each lane.
To make plugs for PFGE, 6 × 108 cells were washed twice with ice-cold 50 mM EDTA and resuspended to 500 μl with 50°C SCE (1 M sorbitol, 0.1 M Na citrate, and 10 mM EDTA). Lyticase was added to a final concentration of 150 U/ml, and 250 μl of the sample was mixed with 250 μl of molten, 50°C 1% SeaPlaque GTG LMP agarose (FMC Bioproducts, Rockland, ME), and then aliquoted into disposal plug molds (170-3713; Bio-Rad). The plug molds were allowed to solidify at 4°C, and then placed in SCEM + lyticase [1 M sorbitol, 0.1 M Na citrate, 10 mM EDTA, 5% β-mercaptoethanol (vol/vol), and 160 U/ml lyticase] for 24 h at 37°C. Plugs were then washed three times in T10E1 (10 mM Tris, pH 8.0, and 1 mM EDTA) for 15 min each wash and resuspended in proteinase K solution [1% sarcosyl (wt/vol), 0.5 M EDTA, and 2 mg/ml proteinase K] for 48 h at 55°C. Finally, plugs were washed three times in T10E1 for 15 min each wash and left overnight at 37°C in T10E1, which removes background fluorescence during ethidium bromide visualization of the gel.
Plugs were cut in half and loaded on a 1% SeaKem LE agarose (wt/vol) gel in 0.5× TBE (45 mM Tris, 45 mM borate, and 1 mM EDTA). The gel was electrophoresed in 14°C 0.5× TBE on a CHEF DR-III system with initial switch time of 50 s, final switch time of 90 s, run time of 22 h, voltage of 6 V, and angle of 120°. The gel was stained with 0.5 μg/ml ethidium bromide in 0.5× TBE for 1.5 h, destained in deionized water for 2 h, and imaged with an AlphaImager. The DNA was then nicked in 0.5 M HCl for 1 h, denatured in 1.5 M NaCl, 0.5 M NaOH for 40 min, and neutralized in 3M NaCl, 55 mM Tris base, 455 mM Tris-HCl for 40 min. The DNA was then transferred to a GeneScreen Plus nylon membrane and cross-linked with 0.12 J of UV light in a UV Stratalinker 1800 (Stratagene, La Jolla, CA). The membrane was probed with an ARS305 fragment (Nguyen et al., 2001) and imaged and quantified with a Storm 840 (Amersham Biosciences).
RESULTS
Rereplication Rapidly Blocks Cell Proliferation
Previous work in our laboratory established yeast strains in which rereplication can be induced in metaphase-arrested cells (Nguyen et al., 2001). These yeast strains contain genetic alterations that make three replication initiation proteins refractory to the inhibitory effect of the CDK Cdc28p. The CDK phosphorylation of two subunits of the origin recognition complex, Orc2p and Orc6p, was blocked by mutating their CDK consensus phoshorylation sites (orc2-6A, orc6-4A). Cdc28p-directed nuclear exclusion of the Mcm2-7p complex (Labib et al., 1999; Nguyen et al., 2000) was prevented by fusing two tandem copies of the simian virus 40 nuclear localization signal to Mcm7p (MCM7-2NLS). Finally, CDK regulation of Cdc6p was disrupted by integrating pGAL1-Δntcdc6, which expresses an N-terminally truncated and slightly stabilized Cdc6p (Δntcdc6p), under the control of the galactose-inducible GAL1 promoter (Drury et al., 1997). In this rereplicating strain, rereplication is detectable only after Δntcdc6p is induced by growth in galactose-containing medium. A parallel strain, containing pGAL1 instead of pGAL1-Δntcdc6, does not rereplicate and serves as a negative control strain (Figure 1A).
Figure 1.
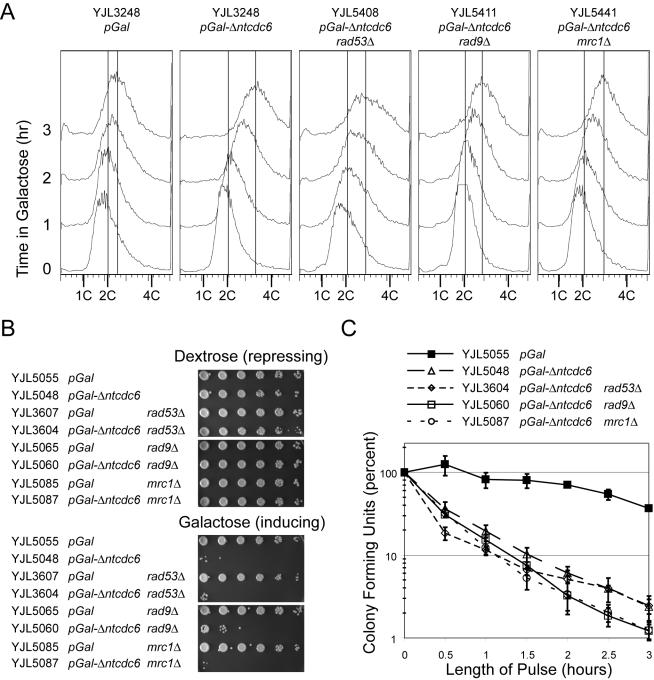
Induction of rereplication rapidly blocks cell proliferation. (A) Checkpoint-deficient strains are capable of rereplicating. Cells with the indicated genotypes plus pMET3-HA3-CDC20 orc2-cdk6A orc6-cdk4A MCM7-2NLS were grown in medium containing 3% raffinose + 0.05% dextrose. Metaphase arrest was induced by adding 2 mM methionine, to transcriptionally deplete Cdc20p, and 15 μg/ml nocodazole. Then, 2% galactose was added, and samples were taken hourly for flow cytometry. Vertical lines indicate the median DNA content for the 0- and 3-h time points. (B) Constitutive induction of rereplication prevents cell proliferation. Cells with the indicated genotypes plus orc2-cdk6A orc6-cdk4A MCM7-2NLS were grown on plates containing 2% dextrose and serially diluted into S broth with fivefold dilutions. The dilutions were plated on medium containing either 2% dextrose, which represses rereplication, or 2% galactose, which induces rereplication in strains containing pGAL1-Δntcdc6. (C) Transient induction of rereplication rapidly inhibits colony forming potential. Cells with the indicated genotypes plus orc2-cdk6A orc6-cdk4A MCM7-2NLS were grown in medium containing 3% raffinose plus 0.05% dextrose and arrested in metaphase with addition of 15 μg/ml nocodazole. Galactose (2%) was added for the indicated number of hours to allow for transient induction of rereplication and cells were then plated on medium containing 2% dextrose to score colony-forming units (CFU). For each strain, the CFU is expressed as a percentage of the CFU present at time 0 h. Error bars show SE of the mean from two experiments.
Further characterization of these strains initially revealed that sustained rereplication leads to a dramatic decrease plating efficiency (Figure 1B). Both the pGAL1-Δntcdc6 rereplicating strain and pGAL1 control strain grew with similar efficiency when plated on medium containing dextrose, which represses the pGAL1 promoter. However, when cells were plated on medium containing galactose, the pGAL1- Δntcdc6 rereplicating strain showed a decrease in plating efficiency by at least three orders of magnitude. In the absence of perturbations of ORC and MCM, expression of Δntcdc6p had no effect on cell growth as assayed by colony size or plating efficiency on galactose-containing medium (our unpublished data).
Significant inhibition of cell proliferation also could be seen with transient induction of rereplication (Figure 1C). Both the pGAL1-Δntcdc6 rereplicating strain and pGAL1 control strain were arrested in metaphase with nocodazole then exposed to galactose to induce rereplication. After varying amounts of time in galactose, cells were plated on dextrose-containing medium to assess the number of cells that could give rise to viable colonies (colony-forming units). Because Δntcdc6p becomes undetectable within 30 min after galactose-induced cells are repressed by the addition of dextrose (Nguyen et al., 2001), we expected reinitiation to end after cell plating. The pGAL1 control strain showed only a slight decrease in colony-forming units after 3 h in galactose. In contrast, the pGAL1-Δntcdc6 rereplicating strain showed a fivefold decrease in colony-forming units after only 30 min in galactose and a nearly 50-fold decrease after 3 h, a statistically significant difference (p < 0.002).
Rereplication Induces a RAD53-dependent Metaphase Checkpoint Arrest
To determine how rapidly rereplicating cells cease dividing, we examined cells microscopically 2 d after transient exposure to galactose. Most rereplicating cells that did not give rise to colonies also did not rebud (our unpublished data), indicating that the cells could not progress beyond the G1 commitment point of the next cell cycle. To pinpoint where in the cell cycle these cells were blocked, we arrested cells in metaphase by depleting them of Cdc20p, which is required for the metaphase-anaphase transition (Schwab et al., 1997; Visintin et al., 1997), induced rereplication with galactose for 2 h, and then restored Cdc20p expression to remove the original metaphase block. α Factor was added to trap any cells that progressed into G1 phase of the next cell cycle (Figure 2A). Cell and nuclear morphology were used to distinguish between cells that were in metaphase and cells that were postmetaphase (anaphase/telophase or G1 phase). More than 90% of the pGAL1-negative control cells proceeded past metaphase and accumulated in G1 phase. In contrast, <20% of the pGAL1-Δntcdc6-rereplicating cells had exited metaphase 5 h after Cdc20p expression was restored. Similar results were obtained when these cells were monitored after rereplication was induced for only 1 h instead of 2 h (our unpublished data). Because rereplication was barely detectable by flow cytometry after 1 h of induction (Figure 1A), these data suggest that even limited rereplication induces a metaphase arrest.
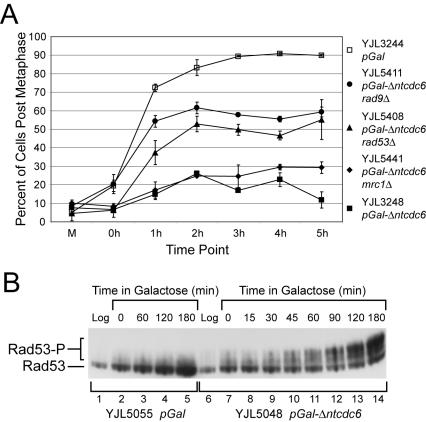
Figure 2.
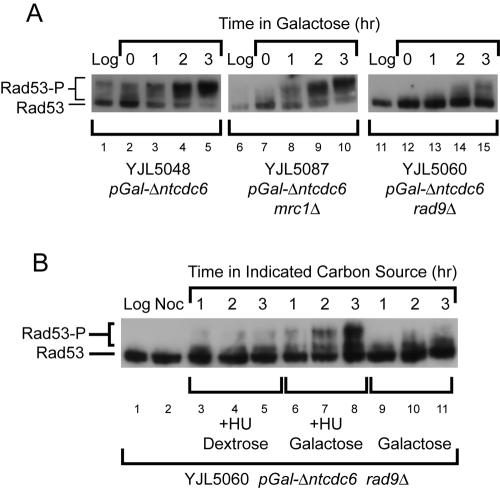
Rereplication induces a RAD53-dependent checkpoint response. (A) Rereplication induces a metaphase arrest that is dependent in part on RAD53 and RAD9. Cells with the indicated genotypes plus pMET3-HA3-CDC20 orc2-cdk6A orc6-cdk4A MCM7-2NLS were arrested in metaphase by transcriptional depletion of Cdc20p in medium containing 3% raffinose and 2 mM methionine (M). Then, 2% galactose was added for 2 h to allow the induction of rereplication followed, at time 0 h, by release from the Cdc20p depletion arrest by transfer of cells to medium lacking methionine but containing 2% galactose and α factor. At hourly intervals after the release, DAPI-stained cells were scored (n = 300) as pre- or postmetaphase. The percentage of postmetaphase cells is shown for each strain, along with the SE of the mean. (B) Rereplication induces phosphorylation of Rad53p. Cells containing the indicated genotypes plus RAD53-HA orc2-cdk6A orc6-cdk4A MCM7-2NLS were grown in medium containing 3% raffinose + 0.05% dextrose and arrested in metaphase with 15 μg/ml nocodazole. Then, 2% galactose was added to allow the induction of rereplication, and at the indicated times samples were harvested for immunoblot analysis of Rad53p-HA. The hypophosphorylated protein is indicated by Rad53 and the hyperphosphorylated protein is indicated by Rad53-P.
In budding yeast, genotoxic stresses such as replication fork stalls or DNA damage induce a metaphase arrest that requires activation of the checkpoint kinase Rad53p (Allen et al., 1994; Weinert et al., 1994; Sanchez et al., 1996; Sun et al., 1996), the homolog of Chk2 in mammalian cells and Cds1 in Schizosaccharomyces pombe. To determine whether rereplication might activate these pathways, we induced rereplication in a rad53Δ mutant background and monitored the ability of these cells to progress past metaphase. Flow cytometry demonstrated that rereplication was still induced in the presence of the rad53Δ mutation (Figure 1A), and vital staining with phloxine B showed that most of the cells remained metabolically alive after 3 h of induction (our unpublished data). The percentage of cells that could complete metaphase, however, increased from <20% to nearly 50%. This result suggests that a significant portion of the checkpoint-proficient rereplicating cells were arrested solely in response to a RAD53-dependent checkpoint. The remaining 50% of the cells also seemed to activate this checkpoint (see below) but presumably stayed arrested because they were subjected to an additional RAD53-independent metaphase block (see Discussion).
Additional evidence that rereplication activates a RAD53-dependent checkpoint response was obtained by examining Rad53p directly. Activation of Rad53p protein kinase is tightly correlated with its hyperphosphorylation (Allen et al., 1994; Weinert et al., 1994; Sanchez et al., 1996; Sun et al., 1996), a modification that retards Rad53p mobility during gel electrophoresis. After inducing rereplication with galactose in metaphase-arrested cells, we monitored the phosphorylation state of Rad53p by immunoblotting total cell lysates (Figure 2B). In the pGAL1 control strain, Rad53p remained hypophosphorylated for the duration of the galactose induction, consistent with the absence of any checkpoint arrest of the cell cycle. In the pGAL1-Δntcdc6-rereplicating strain, however, Rad53p hyperphosphorylation was detected within 45 min of induction, and the majority of the protein became hyperphosphorylated by 120 min. Together, the metaphase arrest and Rad53p hyperphosphorylation indicate that Rad53p is activated as part of a checkpoint response triggered by rereplication. The nearly complete conversion of Rad53p to the hyperphosphorylated form (Figures 2B and 4A) further suggests that this response was activated in almost all rereplicating cells.
Figure 4.
The checkpoint response induced by rereplication is dependent on Rad9p and not Mrc1p. (A) Cells with the indicated genotypes plus orc2-cdk6A orc6-cdk4A MCM7-2NLS were grown in 3% raffinose + 0.05% dextrose and arrested in metaphase by the addition of 15 μg/ml nocodazole. Then, 2% galactose was added, and at the indicated times samples were harvested for immunoblot analysis of Rad53p-HA. The hypophosphorylated protein is indicated by Rad53 and the hyperphosphorylated protein is indicated by Rad53-P. (B) The rad9Δ cells are capable of responding to stalled rereplication forks. YJL5060 (rad9Δ pGAL1-Δntcdc6 orc2-cdk6A orc6-cdk4A MCM7-2NLS) grown in medium containing 3% raffinose + 0.05% dextrose was arrested at metaphase with 15 μg/ml nocodazole and split into three cultures: 0.2 M HU and 2% dextrose were added to the first culture; 0.2 M HU and 2% galactose were added to the second; and 2% galactose was added to the third. Immunoblot analysis was performed as described in A.
Rereplication Induces Formation of Ddc2-GFP Foci
Because the genome is only partial rereplicated in our strains, many rereplication forks cannot be properly terminating with a converging fork from the adjacent replicon. This suggests that many of the rereplication forks must be stalled or disrupted, potentially signaling replication stress, DNA damage, or both. Analysis of the Ddc2p response to rereplication provided an initial hint that rereplication elicits a checkpoint response to DNA damage. Like Rad53p, Ddc2p is required for the response to both DNA damage and replication stress. Ddc2p in complex with Mec1p is recruited to both sites of double-strand breaks (Kondo et al., 2001; Melo et al., 2001) and stalled replication forks (Katou et al., 2003; Osborn and Elledge, 2003) as part of the sensing of these lesions by the checkpoint pathways. Previous studies established that Ddc2p relocalizes from a diffuse nuclear distribution to punctate subnuclear foci in response to DNA damage (Melo et al., 2001). We observed that similar foci are not generated in response to HU in our strains, thereby providing a possible way to distinguish between the two responses (Figure 3A).
This distinction was demonstrated in a pGAL1-Δntcdc6 rereplicating strain where DDC2 was replaced by DDC2-GFP. Initial experiments were performed in dextrose-containing medium to ensure tight repression of pGAL1- Δntcdc6. The rereplicating strain was arrested in metaphase with nocodazole, exposed to 20 μg/ml of the DNA damaging agent phleomycin, and examined by fluorescence microscopy. Within 1 h, one or more subnuclear foci of Ddc2p-GFP were observed in most cells (Figure 3A), consistent with previously published observations. In contrast, when these cells were released from a G1 arrest into S phase in the presence of 0.2 M HU, there was little induction of Ddc2p-GFP subnuclear foci even 3 h after imposition of the replication block (Figure 3A). If phleomycin is added to these cells, subnuclear Ddc2p-GFP foci occur within an hour, indicating that damage-induced foci are observable in HU-arrested cells (our unpublished data). Similar results were observed in wild-type cells not containing any perturbations of ORC, Mcm2-7, or Cdc6.
To examine the localization of Ddc2p after rereplication, the pGAL1-Δntcdc6 rereplicating and pGAL1 control strains containing DDC2-GFP were arrested in metaphase, induced with galactose, and examined at 30-min intervals by fluorescence microscopy. In the pGAL1-Δntcdc6 strain, within 1 h of induction of rereplication, there was a significant increase in Ddc2p-GFP subnuclear foci (Figure 3B). Within 2 h, the number of cells with foci and the number of foci per cell were quantitatively similar to the response observed with the addition of the DNA damaging agent phleomycin. Little increase in Ddc2p-GFP foci was observed in the pGAL1 control strain. Thus, these findings suggest that rereplication induces a DNA damage checkpoint.
Rereplication Induces a DNA Damage Response
For a more definitive examination of whether rereplication was triggering a DNA damage response, a replication stress response, or both, we took advantage of the genetic distinction between these two checkpoint pathways in budding yeast. Both pathways converge on RAD53 and induce a metaphase arrest. However, upstream of RAD53, the DNA damage response is predominantly dependent on RAD9, whereas the replication stress response is predominantly dependent on MRC1. We individually deleted each gene in the pGAL1-Δntcdc6 rereplicating strain and the pGAL1 control strain and investigated whether the metaphase arrest and Rad53p hyperphosphorylation induced by rereplication was dependent on either gene. Initial experiments established that rereplication was still induced on all chromosomes in the mrc1Δ and rad9Δ mutants (Figure 1A; our unpublished data).
As described above, the proportion of cells arrested in metaphase due to rereplication was approximately halved when RAD53 was deleted. A slightly higher reduction was observed when RAD9 was deleted, whereas a much smaller reduction was observed upon deletion of MRC1 (Figure 2A). Thus, nearly half of the rereplicating cells that are arrested in metaphase are solely held at that arrest by a RAD9-dependent pathway. The remainder, as discussed previously, seem to be subjected to an additional metaphase block. The hyperphosphorylation of Rad53p induced during rereplication (Figure 4A, lanes 1-5) also was dramatically reduced in a rad9Δ mutant background (Figure 4A, lanes 11-15). The simplest interpretation of these results is that the Rad53p phosphorylation and RAD53-dependent metaphase arrest induced by rereplication is primarily triggered through the RAD9-dependent DNA damage response pathway.
The virtually complete dependence of Rad53p hyperphosphorylation on RAD9 suggests that rereplication generates little or no MRC1-dependent signaling of replication stress. Alternatively, one could hypothesize that the rad9Δ mutation, the metaphase state of the cell, or an insufficient number of rereplicating forks, somehow prevents the detection of replication stress in our rereplicating cells. For example, if Mrc1p did not properly assemble onto rereplication forks during reinitiation as it normally does at replication forks during normal initiation, the rad9Δ cells would be unable to signal the presence of stalled forks.
To demonstrate that we can indeed detect replication stress during rereplication in a rad9Δ mutant, the mutant strain was arrested in metaphase, split into three separate culture conditions, and each harvested for immunoblot analysis of Rad53p. Galactose was added to one culture to induce rereplication. As described above, there was little Rad53p hyperphosphorylation because of the rad9Δ mutation (Figure 4B, lanes 9-11). Galactose and HU were added to a second culture to induce replication stress during re-replication. In these cells, robust Rad53p hyperphosphorylation could now be observed (Figure 4B, lanes 6-8), presumably through activation of the MRC1-dependent replication stress response pathway. Finally, dextrose and HU were added to the third culture. Dextrose represses the pGAL1 promoter and stifles any induction of rereplication. No Rad53p hyperphosphorylation was observed in this culture (Figure 4B, lanes 3-5), confirming that rereplication forks were generating the HU-induced replication stress response observed in the second culture. Thus, the MRC1-dependent replication stress response pathway is capable of sensing stalled rereplication forks during a metaphase arrest in a rad9Δ background. The lack of any significant activation of this pathway in the absence of HU suggests that stalled rereplication forks are not triggering the checkpoint response observed in rereplicating cells. Consistent with this conclusion is the observation that the extent and kinetics of Rad53p hyperphosphorylation induced by rereplication are unchanged by deletion of MRC1 (Figure 4A, lanes 6-10). Together, our data suggest that DNA damage, and not replication stress, is the predominant genotoxic insult accumulating as a consequence of rereplication.
Rereplication Induces Double-stranded Breaks
Given the induction of a DNA damage response, we looked for direct evidence of DNA damage induced by rereplication. We assayed whether rereplication results in double-stranded breaks by monitoring the appearance of subchromosomal fragments by PFGE. To verify that PFGE can detect chromosome fragmentation, we examined yeast chromosomes from metaphase-arrested cells treated with phleomycin, which generates double-stranded breaks. At high doses of phleomycin, all chromosomes were converted to a heterogeneous pool of subchromosomal fragments (Figure 5A, lanes 4-6). These results were confirmed by Southern blot analysis of these gels, by using ARS305 to probe for chromosome III (Figure 5A, lanes 15-17).
Similar chromosome fragmentation was not observed in cells arrested in S phase with HU (Figure 5A, lanes 1-3 and 12-14). Replicating structures, such as replication bubbles and forks, are thought to significantly retard DNA mobility during PFGE, and whole chromosomes with many replicating structures are retained in gel loading wells (Hennessy et al., 1990). Nonetheless, the absence of any significant subchromosomal fragments even after prolonged HU arrest suggests that there is no rapid or widespread degeneration of stressed replication forks to double-stranded breaks.
Like HU treatment, rereplication caused the majority of each chromosome to be retained in the wells. However, rereplication also generated subchromosomal fragments, which looked like a smear of DNA migrating from below the smallest chromosome up toward the well (Figure 5A, lanes 9-11). This could be seen more clearly by Southern blot analysis, which showed an accumulation of chromosome III fragments migrating faster than the smallest full-length chromosome (Figure 5A, lanes 20-22) in amounts comparable with those generated by 20 μg/ml phleomycin (Figure 5B). This induction of subchromosomal fragments was specific to rereplicating cells, because no such induction was seen in the control strain (Figure 5A, lanes 18-19). Similar subchromosomal fragments were observed when the Southern blots were probed for chromosome 4 and 7 (our unpublished data). Thus, rereplication, but not replication stressed by HU, generates double-stranded DNA breaks.
Checkpoint Responses Do Not Reduce the Lethality Induced by Rereplication
By mobilizing a corrective response and delaying the cell cycle, checkpoint pathways help to protect cells from insults that would disrupt the proper transmission of genetic information. In some cases, however, recovery from the insult may not be possible despite the activation of a checkpoint. For example, degradation of Mcm proteins in the middle of S phase disrupts active replication forks and seems to activate the replication stress response: Rad53p is hyperphosphorylated and cells experience a RAD9-independent metaphase arrest (Labib et al., 2001). Despite the activation of this checkpoint, cells are unable to recover their ability to replicate after Mcm proteins are restored (Labib et al., 2001), presumably because Mcm proteins cannot be reloaded onto the disrupted replication forks. To determine whether the DNA damage response is able to protect cells from the amount and type of DNA lesions generated by rereplication, we examined the viability of rereplicating cells that harbor deletions in RAD53, RAD9, or MRC1. Strains deleted for any of these genes showed similar decreases in viability as checkpoint-proficient strains when subjected to constitutive or transient (p > 0.35 at 3 h) rereplication (Figure 1, B and C). This suggests that the extent of rereplication in these cells generates an amount or type of lethal genotoxic stress that is irreparable.
DISCUSSION
Eukaryotic cells use multiple overlapping mechanisms to prohibit reinitiation of DNA replication within a single cell cycle. An obvious reason why cells might impose such extensive and layered safeguards is that even a low frequency and amount of extra DNA synthesis could eventually alter genome content. We report here that rereplication can induce an immediate and severe threat to the cell. Rereplicating cells rapidly and permanently cease cell division. They phosphorylate Rad53p in a RAD9-dependent manner and arrest in metaphase. This checkpoint response is unlikely to be a novel “rereplication checkpoint.” Rather, we infer from the stereotypical DNA damage response that rereplication rapidly generates DNA lesions that are recognized by the cell as DNA damage. Thus, the use of multiple mechanisms to prevent rereplication not only preserves genome content in the long-term but also protects cells from lethal genomic insults in the short-term.
Surprisingly, we have been able to demonstrate that re-replication triggers little or no replication stress response, even though rereplication forks fail to complete a full round of replication. The Rad53p phosphorylation observed during rereplication was almost exclusively dependent on RAD9, which signals DNA damage, and was independent of MRC1, which signals replication stress. Similarly, the metaphase arrest induced by rereplication was more dependent on RAD9 than on MRC1. Importantly, the absence of a replication stress response was not due to an inability to respond to replication stress. In a rad9Δ mutant background, where rereplication by itself failed to induce Rad53p phosphorylation, the addition of HU to stress the rereplicating forks leads to robust and persistent Rad53p phosphorylation. The simplest interpretation of these data is that rereplicating forks fail to complete a full round of replication, not because they eventually stall, but because they somehow degenerate into DNA lesions that are recognized as DNA damage. These results contrast with those obtained in human cells depleted of geminin, where the resulting rereplication can be associated with the replication stress response (Melixetian et al., 2004; Zhu et al., 2004). Whether these contrasting results reflect differences in species or protocol for inducing rereplication remains to be addressed in the future.
A key question raised by these findings is how rereplication generates DNA lesions without inducing a stalled fork response. Because a prompt DNA damage response is observed in almost all cells in the presence of the microtubule depolymerizing agent nocodazole, the lesions are unlikely to be a consequence of spindle tension on partially replicated chromosomes. Consistent with this, we can induce rereplication and observe the attendant DNA damage response during S phase (our unpublished data), suggesting that a mitotic state is not required to generate the lesion. Moreover, preliminary evidence suggests that elongation is restrained during rereplication (our unpublished data), raising the possibility that rereplicating replisomes encounter problems that could lead to DNA lesions. We therefore suspect that the lesions are generated by the act of rereplication itself.
Any molecular model for how these lesions are generated must explain why they are generated during rereplication and not during normal replication. One possible explanation is that the first round of replication structurally alters chromosomes in a manner that interferes with their rereplication within the same cell cycle; sister chromatid cohesion, which is established during DNA replication, provides precedence for such a replication-coupled change in chromosome state (reviewed in Nasmyth, 2001). Other possible explanations include hypothetical problems specific to rereplication such as poor coordination of histone synthesis and/or nucleosome assembly with rereplication (Verreault, 2003), rereplicating forks from later rounds of rereplication overtaking rereplicating forks from earlier rounds, or defective assembly of replisomes during reinitiation.
An important approach to understanding how rereplication generates DNA damage is to characterize the molecular structure of the primary lesions that are induced. Importantly, these primary lesions may not be the chromosomal breaks that we observed by PFGE. Other abnormal DNA structures that could trigger the DNA damage response might be generated earlier before degenerating into chromosomal breaks. Fork collapse, for example, can generate “chicken feet” structures (Sogo et al., 2002), which expose free double-stranded DNA ends without cleaving the chromosome. Further analysis of rereplicating DNA will hopefully yield more insight into the structure of these primary lesions and the molecular mechanisms by which they are generated.
Although rereplication induces a RAD9-dependent checkpoint response, this response offers little protection against the lethal consequences of rereplication (Figure 1B). This lack of protection is reminiscent of the futile induction of a RAD9-independent checkpoint response after complete Mcm degradation in S phase (Labib et al., 2001). Loss of Mcm proteins from replication forks is apparently irreparable even after resynthesis of the proteins, because there is no efficient mechanism to reload Mcm proteins at forks. Similarly, in our rereplicating cells the damage induced by re-replication may be irreparable and overwhelm any possible protective effect of the DNA damage response. Additionally, other lethal problems may arise from rereplication that are not dependent on DNA damage and cannot be corrected by the DNA damage response. Such additional problems might account for the partial persistence of metaphase-arrested cells when rereplication is induced in the absence of RAD53 or RAD9 (Figure 2A). Fully understanding the lethal consequences of rereplication will require further molecular characterization of the terminal phenotype of rereplicating cells.
The extra copies of genes that are generated by rereplication have long been considered a possible source of genomic instability. Our observation that DNA damage is generated during rereplication suggests an additional way by which rereplication might generate genomic changes. Interestingly, in mammalian cells, overexpression of a single replication initiation protein Cdt1 can induce subtle rereplication (Vaziri et al., 2003) and has been implicated in tumorigenesis (Arentson et al., 2002). Thus, rereplication may be another potential source for the genomic instability associated with tumorigenesis.
Acknowledgments
We thank Anita Sil, David Toczyski, David Morgan, Hiten Madhani, Carol Gross, and Alexander Johnson for helpful discussions and comments on the manuscript. We thank Alexander Johnson for use of the CHEF gel apparatus. We also thank Erin Quan and Emily Wang for help making initial observations of the checkpoint response. This work was supported by grants to J. L. from the American Cancer Society (RPG-99-169-01-CCG) and the National Institutes of Health (R01 GM59704). B. G. was supported by a National Science Foundation Predoctoral Fellowship (DGE-0202754) and a Department of Defense Breast Cancer Predoctoral Fellowship (W81 × WH-04-1-0409).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-09-0833. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0833.
References
- Alcasabas, A. A., Osborn, A. J., Bachant, J., Hu, F., Werler, P. J., Bousset, K., Furuya, K., Diffley, J. F., Carr, A. M., and Elledge, S. J. (2001). Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- Allen, J. B., Zhou, Z., Siede, W., Friedberg, E. C., and Elledge, S. J. (1994). The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8, 2401-2415. [DOI] [PubMed] [Google Scholar]
- Arentson, E., Faloon, P., Seo, J., Moon, E., Studts, J. M., Fremont, D. H., and Choi, K. (2002). Oncogenic potential of the DNA replication licensing protein CDT1. Oncogene 21, 1150-1158. [DOI] [PubMed] [Google Scholar]
- Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- Broek, D., Bartlett, R., Crawford, K., and Nurse, P. (1991). Involvement of p34cdc2 in establishing the dependency of S phase on mitosis [see comments]. Nature 349, 388-393. [DOI] [PubMed] [Google Scholar]
- Brown, E. J., and Baltimore, D. (2003). Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17, 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann, C., Diffley, J. F., and Nasmyth, K. A. (1995). S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 5, 1257-1269. [DOI] [PubMed] [Google Scholar]
- Detweiler, C. S., and Li, J. J. (1998). Ectopic induction of Clb2 in early G1 phase is sufficient to block prereplicative complex formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95, 2384-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L. S., Perkins, G., and Diffley, J. F. (1997). The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16, 5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L. S., Perkins, G., and Diffley, J. F. (2000). The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10, 231-240. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Chi, Y., Yang, P., and Campbell, J. L. (1999). Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10, 3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili, A. (1998). MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2, 183-189. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and McCusker, J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan, V., Simancek, P., Houchens, C., Snaith, H. A., Frattini, M. G., Sazer, S., and Kelly, T. J. (2001). Redundant control of rereplication in fission yeast. Proc. Natl. Acad. Sci. USA 98, 13114-13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottifredi, V., Shieh, S., Taya, Y., and Prives, C. (2001). p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl. Acad. Sci. USA 98, 1036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. (eds.) (1990). Guide to Yeast Genetics and Molecular Biology, New York: Academic Press.
- Haaf, T., Golub, E. I., Reddy, G., Radding, C. M., and Ward, D. C. (1995). Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92, 2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, S. B., and Lew, D. J. (1997). Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 283, 322-332. [DOI] [PubMed] [Google Scholar]
- Hennessy, K. M., Clark, C. D., and Botstein, D. (1990). Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 4, 2252-2263. [DOI] [PubMed] [Google Scholar]
- Hua, X. H., Yan, H., and Newport, J. (1997). A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J. Cell Biol. 137, 183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli, P. V., Brown, G. W., Muzi-Falconi, M., Tien, D., and Kelly, T. J. (1997). Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 11, 2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. (2003). S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078-1083. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. (2001). Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294, 867-870. [DOI] [PubMed] [Google Scholar]
- Labib, K., Diffley, J. F., and Kearsey, S. E. (1999). G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1, 415-422. [DOI] [PubMed] [Google Scholar]
- Labib, K., Kearsey, S. E., and Diffley, J. F. (2001). MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell 12, 3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Hong, B., Choi, J. M., Kim, Y., Watanabe, S., Ishimi, Y., Enomoto, T., Tada, S., and Cho, Y. (2004). Structural basis for inhibition of the replication licensing factor Cdt1 by Geminin. Nature 430, 913-917. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., Mondesert, O., Leatherwood, J., and Russell, P. (1998). Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol. Biol. Cell 9, 63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, T. J., and Kirschner, M. W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043-1053. [DOI] [PubMed] [Google Scholar]
- Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004). Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165, 473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, J., and Toczyski, D. (2002). A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14, 237-245. [DOI] [PubMed] [Google Scholar]
- Melo, J. A., Cohen, J., and Toczyski, D. P. (2001). Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15, 2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylov, I. S., Kondo, T., Jones, L., Ryzhikov, S., Tanaka, J., Zheng, J., Higa, L. A., Minamino, N., Cooley, L., and Zhang, H. (2002). Control of DNA replication and chromosome ploidy by Geminin and Cyclin A. Mol. Cell. Biol. 22, 1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. (1991). The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66, 743-758. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2001). Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673-745. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., Irie, K., and Li, J. J. (2000). Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10, 195-205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., and Li, J. J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068-1073. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., Lygerou, Z., Nishimoto, T., and Nurse, P. (2000). The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404, 625-628. [DOI] [PubMed] [Google Scholar]
- Nyberg, K. A., Michelson, R. J., Putnam, C. W., and Weinert, T. A. (2002). Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617-656. [DOI] [PubMed] [Google Scholar]
- Osborn, A. J., and Elledge, S. J. (2003). Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17, 1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, L. M., Herr, A., McGarry, T. J., and Richardson, H. (2001). The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 15, 2741-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny, Y., Delacote, F., Vares, G., Petitot, F., Lambert, S., Averbeck, D., and Lopez, B. S. (2001). Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20, 3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, Y., Desany, B. A., Jones, W. J., Liu, Q., Wang, B., and Elledge, S. J. (1996). Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357-360. [DOI] [PubMed] [Google Scholar]
- Sauer, K., Knoblich, J. A., Richardson, H., and Lehner, C. F. (1995). Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 9, 1327-1339. [DOI] [PubMed] [Google Scholar]
- Saxena, S., Yuan, P., Dhar, S. K., Senga, T., Takeda, D., Robinson, H., Kornbluth, S., Swaminathan, K., and Dutta, A. (2004). A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol. Cell 15, 245-258. [DOI] [PubMed] [Google Scholar]
- Schwab, M., Lutum, A. S., and Seufert, W. (1997). Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90, 683-693. [DOI] [PubMed] [Google Scholar]
- Sogo, J. M., Lopes, M., and Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599-602. [DOI] [PubMed] [Google Scholar]
- Sun, Z., Fay, D. S., Marini, F., Foiani, M., and Stern, D. F. (1996). Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10, 395-406. [DOI] [PubMed] [Google Scholar]
- Tada, S., Li, A., Maiorano, D., Mechali, M., and Blow, J. J. (2001). Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3, 107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, G. W., and Lowndes, N. F. (2003). Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 31, 242-246. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., Wernic, D., Poupart, M. A., Koonin, E. V., and Nasmyth, K. (2000). Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375-386. [DOI] [PubMed] [Google Scholar]
- Vas, A., Mok, W., and Leatherwood, J. (2001). Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell. Biol. 21, 5767-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri, C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., Wagle, N., Hwang, D. S., and Dutta, A. (2003). A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11, 997-1008. [DOI] [PubMed] [Google Scholar]
- Verreault, A. (2003). Histone deposition at the replication fork: a matter of urgency. Mol. Cell 11, 283-284. [DOI] [PubMed] [Google Scholar]
- Visintin, R., Prinz, S., and Amon, A. (1997). CDC20 and CDH 1, a family of substrate-specific activators of APC-dependent proteolysis. Science 278, 460-463. [DOI] [PubMed] [Google Scholar]
- Ward, I. M., and Chen, J. (2001). Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759-47762. [DOI] [PubMed] [Google Scholar]
- Weinert, T. A., Kiser, G. L., and Hartwell, L. H. (1994). Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8, 652-665. [DOI] [PubMed] [Google Scholar]
- Wilmes, G. M., Archambault, V., Austin, R. J., Jacobson, M. D., Bell, S. P., and Cross, F. R. (2004). Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18, 981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel, J. A., Kutok, J. L., Weng, A. P., and Dutta, A. (2002). Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am. J. Pathol. 161, 267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin, J., Buck, V., Nurse, P., and Millar, J. B. (2002). Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111, 419-431. [DOI] [PubMed] [Google Scholar]
- Yanow, S. K., Lygerou, Z., and Nurse, P. (2001). Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 20, 4648-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, W., Feng, H., Santiago, F. E., and Kipreos, E. T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885-889. [DOI] [PubMed] [Google Scholar]
- Zhu, W., Chen, Y., and Dutta, A. (2004). Rereplication by depletion of Geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24, 7140-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]