Abstract
MicroRNAs (miRNAs) impinge on the translation and stability of their target mRNAs, and play key roles in development, homeostasis and disease. The gene regulation mechanisms they instigate are largely mediated through the CCR4–NOT deadenylase complex, but the molecular events that occur on target mRNAs are poorly resolved. We observed a broad convergence of interactions of germ granule and P body mRNP components on AIN-1/GW182 and NTL-1/CNOT1 in Caenorhabditis elegans embryos. We show that the miRISC progressively matures on the target mRNA from a scanning form into an effector mRNP particle by sequentially recruiting the CCR4–NOT complex, decapping and decay, or germ granule proteins. Finally, we implicate intrinsically disordered proteins, key components in mRNP architectures, in the embryonic function of lsy-6 miRNA. Our findings define dynamic steps of effector mRNP assembly in miRNA-mediated silencing, and identify a functional continuum between germ granules and P bodies in the C. elegans embryo.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22 nucleotide (nt)-long RNAs that impinge on gene expression to regulate a broad variety of biological processes (1). miRNAs direct silencing from within the miRNA-induced silencing complex (miRISC), an assembly of an Argonaute (ALG-1 and -2 in Caenorhabditis elegans) and GW182 proteins (AIN-1 and -2) (2,3). The miRISC typically recognizes 3΄ un-translated region (3΄UTR) sequences of target messenger RNAs (mRNAs) through imperfect base-pairing with miRNAs (1). Cognate interactions instigate a series of gene-silencing mechanisms, which include mRNA translation repression, deadenylation, decapping and decay (4–9). The relative contribution of each of these events is still a matter of debate, and likely depends on cellular context. The multi-subunit CCR4–NOT deadenylase complex is a key effector in the several mechanistic aspects of miRNA-mediated silencing (5,10,11). The scaffolding subunit CNOT1 (NOT-like 1, or NTL-1 in C. elegans) directly interacts with GW182 in vitro (12–14), and either alone or in combination with other CCR4–NOT subunits (15,16), further tethers other effector components such as the RNA helicase DDX6 (15–17) or the distinct PAN2/3 deadenylase complex (18).
A significant fraction of the Argonaute and GW182 proteins localize to processing bodies (P bodies), which are dynamic assemblies of RNA and proteins observed as distinctively large foci throughout the cell cytoplasm (5,19–25). Their full composition is unknown, but numerous other factors implicated in mRNA processing, such as decapping enzymes (Dcp1/2) and activators (Pat1 and the Lsm1–7 complex) and the 5΄→3΄ exonuclease Xrn1, co-localize in P bodies (26–29). While they do concentrate several key miRNA co-factors, detectable P bodies as distinct cytoplasmic foci are not required for miRNA-mediated silencing. Genetic depletion of components often results in their reduction in size or abundance without impairing miRNA-mediated silencing (30–32).
P bodies belong to a broad and functionally diverse group of electron-dense and membrane-less cellular foci referred to as messenger ribonucleoprotein (mRNP) granules. mRNPs include stress granules, transport granules, chromatoid bodies in male germ cells and germ granules in oocytes and embryos (33–35). mRNP functions have been largely inferred based on co-localization of proteins, enzymatic functions attributed to resident proteins and interactions in vitro. Germ granules are thought to be sites of mRNA storage for germ cell lineage functions (36–40), whereas P bodies are instead being primarily associated with mRNA processing and decay (26). What determines the structural and functional frontiers or the interactions between the distinct mRNP subtypes is not well defined. High-resolution, live imaging studies in C. elegans embryos revealed that germ granules exhibit liquid droplet-like behavior, which allows rapid phase transitions of dissolution and condensation (41). Such a behavior is consistent with a dynamic molecular scaffold of multivalent protein–RNA complexes, lending grounds to a model explaining assemblies of large cytoplasmic mRNPs like P bodies and germ granules (42). Recent studies uncovered a key contribution for intrinsically disordered regions, often encoded in RNA-binding proteins, in mRNP granule architecture and dynamics (43–48).
Through a combination of proteomics, genetics and novel cell-free assays in C. elegans, we delineate the molecular events leading to and occurring during embryonic miRNA-mediated silencing. We identify a striking convergence of interactions between germ granule and P body components with AIN-1 and NTL-1. We show that scanning miRISC and mRNP components assemble sequentially on mRNA targets. Furthermore, we show that miRISC components co-fractionate and associate with intrinsically disordered germ granule proteins and implicate MEG-2 in the embryonic function of the lsy-6 miRNA. We thus identify new molecular events underlying embryonic miRNA functions, and suggest a role for mRNP granule components in specializing their silencing mechanisms.
MATERIALS AND METHODS
Caenorhabditis elegans strains and RNAi
Caenorhabditis elegans were cultured using standard techniques as described (49). RNAi was performed as in Fire et al. (50) and Timmons et al. (51) on L4 animals and progeny (embryos) were harvested. Worm strains used: N2 Bristol (wild-type: wt), MJS26 (ALG-2::GFP, described in 52), FD21 (AIN-1-LAP, unc-119(ed3); tagIs1271), EV465 (NTL-1::LAP, described in 53), meg-1(vr10), meg-2(ok1937), MH2636 (otIs114(plim-6::GFP, rol-6, lsy-6(ot150)), FD14 (meg-1(vr10); otIs114; lsy-6(ot150), rol-6), FD15 (meg-1(vr10); otIs114, rol-6), FD16 (meg-2(ok1937); otIs114; lsy-6(ot150), rol-6), FD17 (meg-2(ok1937); otIs114, rol-6), JH3251 (PGL-1-3xFLAG) and JH3292 (MEG-1-1xFLAG) are C-terminal FLAG insertions in the genomic pgl-1 and meg-1 loci and were generated using CRISPR/Cas9 system by the laboratory of Geraldine Seydoux, FD22 (3xFLAG-MEG-2) is a N-terminal 3xFLAG insertion in genomic meg-2 locus, generated using CRISPR/Cas9 system. All strains were grown at 22°C, except strains used in assessing meg-1 and meg-2 genetic interactions with lsy-6, which were maintained at 16 or 19°C, as indicated.
Plasmids
The Renilla reniformis luciferase (RL) constructs containing miR-35 wt or mutated sites have been described in Wu et al. (54).
Preparation of C. elegans embryonic extract for translation assays, deadenylation assays, deadenylated RNA immunoprecipitation (DRIP)
Caenorhabditis elegans embryo extracts were prepared as described in Wu and Duchaine (55), except that calf-liver tRNA was omitted from the extract.
In vitro transcription and deadenylation assays
Assays were setup and performed as described in Wu et al. (54). The RL constructs encoding RL 6x pA86, RL 6xmut pA86, RL 6x pA0 and RL 6xmut pA0 RNAs were linearized with MfeI, and in vitro transcribed using MAXIscript® T7 Transcription Kit (Ambion). These RNAs were labeled with [α-32P] UTP and capped with anti-reverse cap analog (ARCA, Ambion).
Deadenylated RNA immunoprecipitation (DRIP)
Deadenylation assay was conducted as described above with the following modification: Prior to deadenylation reaction, C. elegans embryonic extract was pre-cleared with pre-equilibrated Dynabeads® Protein G (Life Technologies) for 1 h at 4°C with rotation in DRIP buffer (24 mM HEPES-KOH pH 7.4, 25 mM KOAc, 1.28 mM Mg(OAc)2, 0.1 U/μl Ribolock RNase inhibitor (Fermentas), 1 mM dithiothreitol (DTT)). A total of 50 μl-deadenylation reaction mixture was setup per time point. Deadenylation assay was then conducted over a 3-h time-course. During incubation, mouse anti-GFP antibody (Roche) was added to Dynabeads® Protein G and incubated for 1 h at 4°C with rotation. The reaction mixture was then incubated with 50 μl of a 1:1 suspension of anti-GFP-Dynabeads® Protein G for 30 min at 4°C with rotation. After the immunoprecipitation step, beads were washed four times with DRIP buffer. Washes were performed at 4°C with rotation. The beads were then transferred into two tubes, one for western blot analysis, and the other for Proteinase K treatment and RNA extraction. Proteinase K treatment was performed by resuspending the beads in 90 μl Proteinase K buffer (200 mM Tris–HCl pH 8, 25 mM ethylenediaminetetraacetic acid (EDTA) pH 8, 30 mM NaCl, 2% sodium dodecyl sulphate (SDS)) and 10 μl Proteinase K (10 μg/μl) for 10 min at room temperature. The eluted RNA was purified using phenol/chloroform and ethanol precipitation, followed by separation on a 4% polyacrylamide/urea gel and autoradiography. The percentage of RNA brought down by IP was calculated based on ImageJ measurements of the intensity of the bands corresponding to full length and deadenylated RNAs in bound and unbound fractions.
Extract preparation and multidimensional protein identification (MuDPIT)
Embryo pellets were homogenized in lysis buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 with Complete EDTA-free protease inhibitors (Roche)) and cleared by 16 100 × g centrifugation. FLAG-tagged proteins were purified using ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich A2220) following extract preparation. With our culture methods, embryonic staging may differ within individual preparations and from batch to batch. Prior to adding the matrices, the clarified lysate was quantified and diluted to 5 mg/ml concentration in lysis buffer (50 μl bead slurry was added for 1 ml IP volume). Immunoprecipitations were carried out at 4°C for 2 h, and beads were then washed four times in the lysis buffer. Bound proteins were eluted using the 3xFLAG peptide (Sigma-Aldrich F4799). A fraction of the eluate (1/10th for AIN-1-LAP and 1/3rd for NTL-1-LAP) was monitored by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by western blot analysis. Non-transgenic N2 embryos were used as controls for the purifications. MuDPIT was performed as described in Duchaine et al. (56).
Assessment for siRNA or miRNA pathway proteins
The studies integrated for the analysis of factors implicated in miRNA and other RNAi-related pathways are as follows: let-7 phenotype (WormBase (WS220), 57), let-7 sensitized (58), Drosophila miRNA and siRNA (59), DCR-1 Co-IP (56), ERI-1 Co-IP (60), AIN-2 Co-IP (3), suppression of transgene silencing in eri-1 and dsGFP RNAi (61), germline co-suppression defect (62), SynMuv suppression (63). The generation and analysis of the 11 screens was previously described in Tabach et al. (57). The hyper-geometric P-values were calculated from a population of 17 000 genes.
Gene ontology (GO) term analysis
Gene ontology (GO) term over-representation test (release 20150430) for GO cellular component annotations was determined using the PANTHER Classification System (http://www.pantherdb.org) (64,65). Identified proteins listed in Supplementary Table S1 (AIN-1 interactors) and S2 (NTL-1 interactors) served as the analyzed list, and the C. elegans genes in the PANTHER database served as the reference list.
Nuclease sensitivity assay
At the end of a 3-h deadenylation reaction, 0.005 U/μl of micrococcal nuclease (MNase, Roche) and 1 mM CaCl2 was added to the reaction mixture. Aliquots of the MNase-treated reaction mixture were withdrawn over a 15-min treatment and the MNase treatment was stopped by the addition of 20 mM glycol ether diamine tetraacetic acid (EGTA). RNA was extracted and analyzed by autoradiography as described in Wu et al. (54).
Biotinylated isoxazole (b-isox)-mediated precipitation
Caenorhabditis elegans embryonic pellets were resuspended in a lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 1 μM DTT) supplemented with protease, phosphatase and RNase inhibitors. The extracts were homogenized with a pre-chilled Kontes dounce homogenizer and then centrifuged three times at 14 000 rpm for 10 min at 4°C. The samples were exposed to 100 μM of the b-isox chemical (Sigma Aldrich) and rotated at 4°C for 90 min. The incubated reaction was then centrifuged at 10 000 × g for 10 min to pellet the precipitates. The pellet was washed twice in the lysis buffer and resuspended in 1× SDS loading buffer for protein analysis. Proteins in the supernatant fractions were precipitated by addition of four volumes of cold acetone, incubated for 1 h at −20°C and centrifuged at 15 000 × g for 10 min to pellet the precipitates.
Western blotting
Protein samples from DRIP were separated on a 6% SDS-PAGE and analyzed by western blot. Protein samples for CCR-4 and CCF-1 western blot analysis were separated on a 10% SDS-PAGE. Samples from b-isox-mediated precipitation were resolved on NuPAGE 4–12% Tris-Glycine gradient gels (Invitrogen). Antibodies used were: mouse monoclonals against GFP (Roche), alpha tubulin (Abcam), FLAG (Sigma-Aldrich); rabbit polyclonals against PAB-1/2 (66), DCR-1, ALG-1/2, RDE-4 (56), CGH-1 (67), GLH-1 (68), PAN-1 (69), IFE-1, IFE-2 (70) and AIN-1 (gift from Dr Martin Simard); rat polyclonal against DCAP-2 (71). HRP-conjugated goat anti-rabbit, anti-mouse and anti-rat (Sigma-Aldrich) and mouse TrueBlot® (eBioscience) were used as secondary antibodies. For CCF-1 and CCR-4, rabbit polyclonal antisera were raised against the following peptides at Capralogics: KGGLQEVADQLDVKRQGVR (CCF-1, 3755) and VHRVLTEDEIASGRSTRWTELE (CCR-4, 3756). For NTL-1, the region corresponding to amino acid position 650–950 was amplified from cDNA using forward primer: 5΄-ATAATAGGATCCAGGTAATGAAAGAGAACTCGG-3΄ (Tdo1707); and reverse primer: 5΄-TATTATGGATCCCAAATTTTCCACTGACATCGC-3΄ (Tdo1708) and cloned into pCAL-KC via BamHI/BamHI. This construct was used as a template for generating the antigen for mouse polyclonal against NTL-1 (4162, Capralogics Inc.). Sera for CCF-1, CCR-4 and NTL-1 were used at 1:1000 dilution in 5% non-fat dry milk in 0.1% Tween-phosphate buffered saline overnight at 4°C.
RESULTS
Germ granule and P body proteins are enriched among miRISC interactions
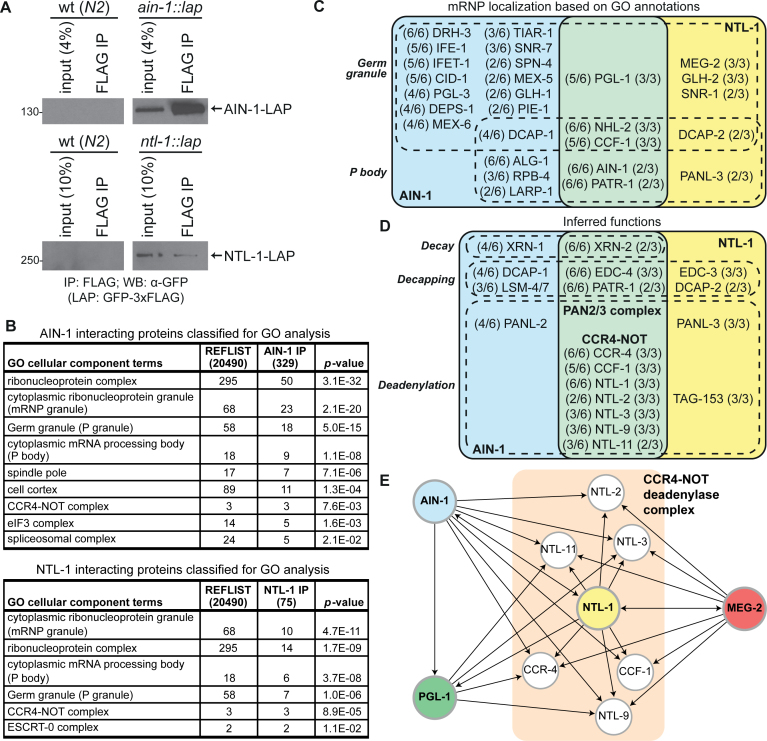
mRNA deadenylation is a prevalent outcome for miRNA targets in diverse systems and this activity has been largely attributed to the CCR4–NOT deadenylase complex (72). The molecular interactions of miRISC with mRNA processing machineries in the embryo are still unknown. To capture the physical interactions between miRISC and its effectors in the C. elegans embryo, we performed immunoprecipitation (IP)-MuDPIT proteomics on the miRISC protein AIN-1, a C. elegans ortholog to GW182 and on the CCR4–NOT complex scaffold NTL-1, the ortholog of CNOT1. LAP (GFP-3xFLAG)-tagged AIN-1 and NTL-1 proteins were immuno-purified from C. elegans transgenic embryos expressing tag fusions at endogenous levels (Figure 1A, and see ‘Materials and Methods’ section). Recovered fractions were analyzed using multi-dimensional protein identification technology (MuDPIT) (73–75). Six independent biological replicates were analyzed for AIN-1, and three were analyzed for NTL-1. Only candidate interactions detected in at least two independent biological experiments were retained, and proteins also found in negative control samples (non-transgenic strains) were disqualified. A total of 340 proteins were detected in at least two samples for AIN-1 purifications (Supplementary Table S1), while 78 candidate interactions were identified from NTL-1 sample analyses (Supplementary Table S2).
Figure 1.
Germ granule and P body proteins are enriched among miRISC interactions. (A) Western blots of embryo lysates and FLAG immunoprecipitations (IP) from wild-type (wt) non-transgenic (N2, left panels) and transgenic animals expressing LAP-tagged AIN-1 (top right panel) or NTL-1 (bottom right panel). (B) Gene Ontology (GO) analysis of cellular component terms on AIN-1 and NTL-1 proteins detected by MuDPIT. Among the proteins retained from at least two biological replicates, only 329/340 AIN-1 interactors and 75/78 NTL-1 interactors were classified for GO analysis. (C) Venn diagram of proteins with GO annotations to cellular component terms related to germ granules and P bodies. (D) Venn diagram of a subset of proteins with inferred functions in deadenylation, decapping and RNA decay. Fractions in the Venn diagrams indicate the number of times the corresponding protein was detected in each independent IP (out of 6 for AIN-1, and out of 3 for NTL-1). (E) Network of proteins converging on the CCR4–NOT deadenylase complex, as detected by MuDPIT analyses in Caenorhabditis elegans embryonic extracts. Target proteins (using FLAG immunoprecipitation) are shown in bold and in colored/shaded circles. Arrowheads indicate detected interactions.
AIN-1 and NTL-1 interaction datasets significantly overlapped with previous phylogenetic profiling (co-evolution), genome-wide RNAi screens and proteomic analyses that identified genes of the RNAi and miRNA pathways (71/340 for AIN-1, P-value: 1.14 × 10−37; 18/78 for NTL-1, P-value: 6.33 × 10−12; (Supplementary Tables S3–5, and see ‘Materials and Methods’ section) (57). Genes encoding 25 of the 71 proteins shared with AIN-1 proteomics were identified in an RNAi screen for enhancement of the let-7 phenotypes in a sensitized background (P-value: 6.6 × 10−9) (58), and 29/71 displayed the same phenotype in other independent RNAi experiments (P-value: 1.7 × 10−12). Extensive and significant overlap is also observed between AIN-1 datasets and results of a screen for miRNA factors in Drosophila (17/71, P-value: 3.4 × 10−14). NTL-1 datasets significantly overlap with let-7 phenotype screen (6/18; P-value: 0.002). Finally, both AIN-1 and NTL-1 interactions further overlap with proteomic and genetic screens for RNAi pathway factors (Supplementary Table S3). These results indicate that both AIN-1 and NTL-1 interactions are functionally relevant to the miRNA and RNAi pathways in a diverse variety of cellular and species contexts.
GO classification using the PANTHER system (64,65) revealed a strong enrichment for annotations to cytoplasmic RiboNucleoProtein granules (mRNP granules) (Figure 1B). A total of 23 out of 329 AIN-1 interactions (P-value: 2.1 × 10−20), among 195/329 proteins with classified terms and 10 out of 75 NTL-1 interactions (49/75 classified; P-value: 4.7 × 10−11) were annotated as cytoplasmic mRNP granules. More specifically, P body components were enriched among AIN-1 and NTL-1 interactions. P body components were annotated to 9 interactions with AIN-1 (P-value: 1.1 × 10−8), and 6 NTL-1 interactions (P-value: 3.7 × 10−8). Detected P body proteins among the interactions included several of the CCR4–NOT complex subunits, the PAN2/3 deadenylase complex, the decapping enzymes DCAP-1/2 and the decapping activator PATR-1 (Figure 1C and D). Finally NHL-2, a member of the TRIM-NHL family of proteins and a miRISC cofactor (76) which localizes to P bodies and germ granules based on GO annotations (76,77), was among the most consistently detected interactions in both NTL-1 and AIN-1 purifications.
Surprisingly, germ granule (also known as P granules in C. elegans) proteins were strongly enriched among AIN-1 interactions (18 interactions, P-value: 5.0 × 10−15) and in NTL-1 interaction datasets (7 interactions, P-value: 1.0 × 10−6) (Figure 1C). Interactions detected with AIN-1 include proteins known to play critical roles in germline determination and functions including PGL-1, PGL-3 (78,79), CCCH Zinc finger proteins PIE-1 (80), MEX-5 and MEX-6 (81), the snRNP spliceosome component SNR-7 (82), DEAD-box RNA helicases DRH-3 (83) and GLH-1 (68), a close C. elegans homolog to Drosophila VASA. eIF4E homolog IFE-1 and 4E transporter and translation regulator IFET-1, both known residents of germ granules in C. elegans (84,85), were detected among interactions with AIN-1.
Whereas some of the detected proteins reside and/or function within germ granules, others are known for their structural function in mRNP assembly itself. This is the case for MEG-2 protein, detected in 3/3 NTL-1 purifications (Figure 1C and Supplementary Table S2) and its paralog MEG-1, which was detected with lesser consistency and at lower peptide coverage (not shown). MEG-2 and MEG-1 lack any recognizable domains, are constituted of inherently disordered regions rich in serine and localize to germ granules (48,86). Both proteins act at least in part redundantly in germline development and germ granule architecture dynamics. Interestingly, MEG-1 was recently shown to be a target of the MBK-2(DYRK) kinase and of the PPTR-1/2(PP2A) phosphatase, with activities that modulate germ granule assembly (48). PPTR-1 is also detected among NTL-1 interactions, in 2/3 biological replicates (Supplementary Table S2) and MBK-2 was detected with poorer consistency, in one out of three NTL-1 purification samples (not shown).
Overall, our comparative proteomic analyses reveal the physical linkage of the miRISC core component AIN-1 and its effector complex scaffold protein NTL-1 with mRNPs. It further identifies previously unrecognized interactions with key germ granule components.
Coupled expression and function of the CCR4–NOT complex subunits in embryonic miRNA-mediated deadenylation
Intersect of the datasets revealed an extensive overlap of the interactions with the CCR4–NOT complex and AIN-1. About 48% of the detected NTL-1 interactions were also detected in the AIN-1 IP (Table 1), and the enrichment of CCR4–NOT complex components is highlighted through GO analysis on the AIN-1 associated proteins (Figure 1B; P-value: 7.6 × 10−3).
Table 1. Comparative proteomics of AIN-1- and NTL-1-interacting proteins.
| NTL-1::LAP | AIN-1::LAP | ||||||
|---|---|---|---|---|---|---|---|
| Sequence name | Protein | # datasets detected | coverage (peptide counts) | # datasets detected | coverage (peptide counts) | Homology/Domain | Description |
| ZC518.3 | CCR-4 | 3/3 | 61% (40) | 6/6 | 20% (6) | Ccr4/CNOT6, CNOT6L | CCR4–NOT subunit |
| F57B9.2 | LET-711 | 3/3 | 43% (121) | 6/6 | 10% (17) | CNOT1 | CCR4–NOT subunit |
| B0513.1 | LIN-66 | 3/3 | 10% (4) | 6/6 | 17% (6) | none detected | translational regulation |
| Y44E3A.6 | Y44E3A.6 | 3/3 | 12% (7) | 6/6 | 13% (7) | EDC4 | decapping activator |
| C07G1.5 | HGRS-1 | 3/3 | 6% (3) | 6/6 | 12% (6) | Vps27p,FYVE Zn finger | ESCRT-0 component |
| F26F4.7 | NHL-2 | 3/3 | 8% (6) | 6/6 | 10% (6) | TRIM-NHL | miRISC component |
| F31E3.3 | RFC-4 | 3/3 | 12% (3) | 5/6 | 13% (3) | RFC4 | DNA replication |
| ZK381.4 | PGL-1 | 3/3 | 4% (3) | 5/6 | 5% (2) | none detected | RGG box motif, P granules |
| C18H9.3 | C18H9.3 | 3/3 | 4% (3) | 5/6 | 4% (2) | GIGYF1/2 | GYF domain protein |
| Y56A3A.20 | CCF-1 | 3/3 | 39% (19) | 5/6 | 18% (4) | Caf1/CNOT7 | CCR4–NOT subunit |
| T01B7.6 | TRCS-2 | 3/3 | 4% (3) | 5/6 | 7% (3) | none detected | uncharacterized |
| H28G03.1 | H28G03.1 | 3/3 | 13% (3) | 4/6 | 12% (2) | RNA-binding | uncharacterized |
| Y56A3A.1 | NTL-3 | 3/3 | 58% (44) | 3/6 | 8% (4) | CNOT3 | CCR4–NOT component |
| F13D12.2 | LDH-1 | 3/3 | 12% (3) | 3/6 | 15% (4) | LDHB | lactate dehydrogenase |
| K10B3.8 | GPD-2 | 3/3 | 14% (3) | 3/6 | 11% (3) | GAPDH | glycolysis |
| K10B3.7 | GPD-3 | 3/3 | 14% (3) | 3/6 | 11% (3) | GAPDH | glycolysis |
| R11A8.7 | R11A8.7 | 3/3 | 5% (9) | 3/6 | 2% (3) | Q/N-rich domain | uncharacterized |
| F56A3.4 | SPD-5 | 3/3 | 4% (3) | 3/6 | 4% (3) | coiled coil domain | cell division |
| C26E6.3 | NTL-9 | 3/3 | 45% (27) | 3/6 | 10% (2) | RQCD1 | CCR4–NOT component |
| B0286.4 | NTL-2 | 3/3 | 42% (14) | 2/6 | 10% (2) | CNOT2 | CCR4–NOT component |
| C06G1.4 | AIN-1 | 2/3 | 6% (3) | 6/6 | 53% (37) | GW182/TNRC6 | miRISC component |
| F43G6.9 | PATR-1 | 2/3 | 7% (4) | 6/6 | 10% (5) | PAT1 | mRNA decay |
| Y116A8C.35 | UAF-2 | 2/3 | 10% (2) | 6/6 | 23% (5) | U2AF35, RRM | splicing |
| Y48B6A.3 | XRN-2 | 2/3 | 4% (3) | 6/6 | 13% (7) | XRN2 | 5΄-3΄ exoribonuclease |
| F31D4.3 | FKB-6 | 2/3 | 12% (4) | 5/6 | 8% (2) | TPR repeat | protein folding |
| C34G6.7 | STAM-1 | 2/3 | 12% (4) | 5/6 | 16% (4) | Q/N-rich domain, SH3 | protein transport |
| R05D3.7 | UNC-116 | 2/3 | 11% (7) | 5/6 | 7% (4) | kinesin-1 heavy chain | intracellular transport |
| T25G12.5 | ACDH-7 | 2/3 | 7% (2) | 5/6 | 10% (3) | ACADM | acyl-CoA dehydrogenase |
| Y34D9A.10 | VPS-4 | 2/3 | 7% (3) | 5/6 | 11% (3) | VPS4B, VPS4A | vacuolar protein sorting |
| W01B11.3 | NOL-5 | 2/3 | 6% (3) | 5/6 | 15% (5) | NOP58 | nucleolar RNP |
| Y74C10AR.1 | EIF-3.i | 2/3 | 18% (5) | 4/6 | 20% (5) | EIF3I | translation initiation |
| T12E12.4 | DRP-1 | 2/3 | 11% (6) | 4/6 | 5% (3) | DRP1 | dynamin-related protein |
| Y73F8A.25 | NTL-11 | 2/3 | 5% (3) | 3/6 | 8% (3) | CNOT11 | CCR4–NOT component |
| Y54G9A.6 | BUB-3 | 2/3 | 10% (2) | 3/6 | 12% (3) | BUB3 | mitotic checkpoint |
| F35G12.2 | IDHG-1 | 2/3 | 8% (2) | 3/6 | 10% (2) | isocitrate dehydrogenase | tricarboxylic acid cycle |
| Y59A8B.6 | PRP-6 | 2/3 | 3% (2) | 3/6 | 5% (3) | PRPF6 | pre-mRNA processing |
| T23B5.1 | PRMT-3 | 2/3 | 3% (2) | 3/6 | 5% (3) | PRMT9 | methyltransferase |
| ZK1053.4 | ZK1053.4 | 2/3 | 4% (2) | 3/6 | 3% (2) | coiled-coil domain | SEPA-1 family, autophagy |
A list of 38 proteins detected in both AIN-1 and NTL-1 immunoprecipitations. Proteins that were detected only once in each immunoprecipitation and found in the negative control (non-transgenic wt N2 background) were excluded. Homology data and description for each protein were obtained from Wormbase WS250 and UniProt database.
Among shared interactions, the CCR4–NOT catalytic subunit CCR-4 (CCR-4a/b; orthologous to CNOT6/6L) scored among the very highest in percentage of peptide coverage and in the number of detected peptides, and was detected in all samples analyzed. CCF-1 (CAF1), the other deadenylase catalytic subunit of the complex, was detected in 5/6 AIN-1 samples and in 3/3 NTL-1 samples. Together with CCF-1, CCR-4 and NTL-1, a total of 7 known subunits of the CCR4–NOT complex were common to both AIN-1 and NTL-1 purifications, including NTL-2, NTL-3, NTL-9 and NTL-11 (Figure 1D). Decapping co-factors PATR-1 and EDC-4 (named based on homology with human Edc4) and the mRNA decay enzyme 5΄→3΄ exonuclease XRN-2 were also detected in both groups of datasets (Figure 1D and Table 1). Finally, and in spite of extensive overlap, some of the best-detected proteins in NTL-1 purifications were absent from any AIN-1 interaction datasets. In particular, TAG-153 is an uncharacterized paralog of NTL-2 (Supplementary Figure S1), a member of the NOT2/3/5 family and was among the proteins most consistently detected in NTL-1 purifications (Supplementary Table S2). While NTL-2 is consistently detected in 3/3 NTL-1 IP, TAG-153 is absent from all six AIN-1 interaction replicates. This may suggest specialization of distinct and functionally non-redundant CCR4–NOT complexes in miRNA-mediated silencing.
The extent of CCR4–NOT interactions with intrinsically disordered protein MEG-2 and germ granule component PGL-1 was further validated by reciprocal IP-MuDPIT (Figure 1E). All 7 subunits of CCR4–NOT complex were consistently detected in 3/3 MEG-2 samples, and CCR-4, NTL-3, NTL-9 and NTL-11 were detected in 2/3 PGL-1 samples (Supplementary Table S6).
Collectively, these data reveal that embryonic miRISC physically interacts with mRNA deadenylation and decay machineries, and position i-AIN-1 as a bridge between the miRNA-dedicated ALG-1/2 Argonaute proteins and their gene-silencing effectors, and ii-CCR4–NOT deadenylase complex as a hub in the network of mRNPs in C. elegans embryos.
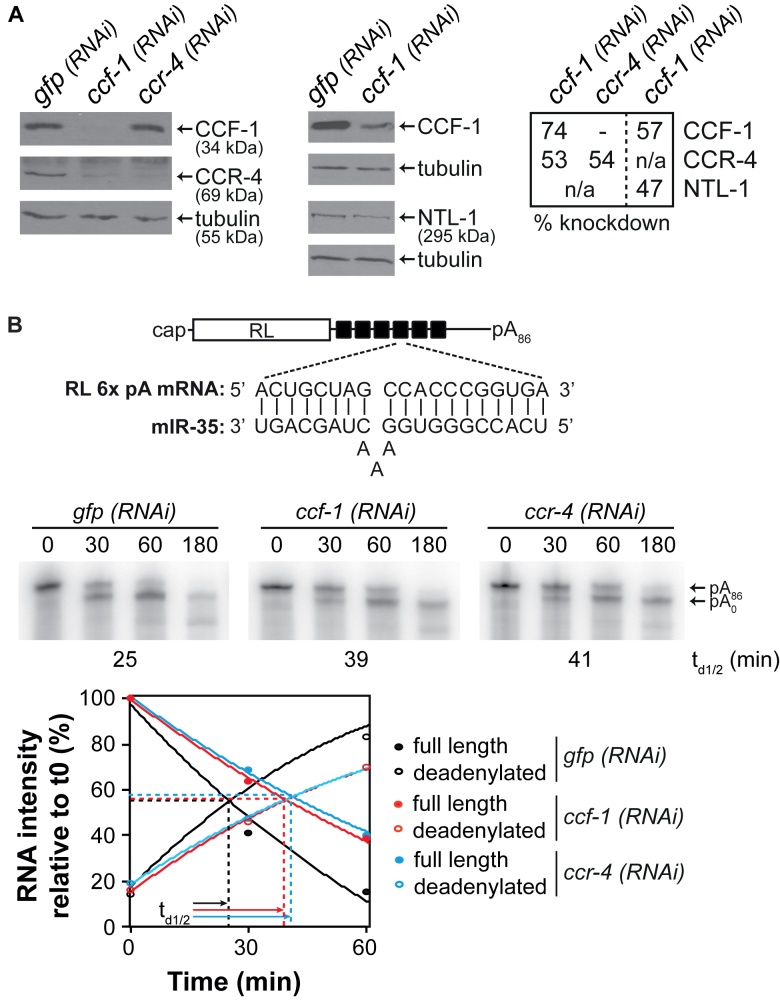
The CCR4–NOT complex had never been functionally linked to miRNA-mediated silencing mechanisms in C. elegans embryos. To formally test the implications of CCF-1 and CCR-4 in embryonic miRNA-mediated deadenylation, we exploited an in vitro embryonic extract previously developed in our lab (54,55) and proficient for miRNA-mediated silencing and deadenylation. For this, an in vitro transcribed, radiolabeled polyadenylated RL reporter RNA bearing six miR-35 binding sites (RL 6x pA, Figure 2B) was incubated in wt or genetically-depleted extracts over a time-course of 3 h. RNA was extracted, and deadenylation was monitored and quantified using denaturing electrophoresis and autoradiography. Because strong genetic depletion of ccf-1 and ccr-4 results in pleiotropic defects including sterility and, in the case of ccf-1 mutants, embryonic and larval lethality (53,87), null alleles or strong RNAi depletions could not be used in extract preparation. Instead, we generated cell-free embryonic extracts wherein ccf-1 and ccr-4 expression was mildly reduced by RNAi (Figure 2A and Supplementary Figure S2, see ‘Materials and Methods’ section). In extracts derived from wt embryos subjected to mock (gfp) RNAi, the RL 6x pA mRNA reached half-deadenylation time (td1/2) at 25 min (Figure 2B). In contrast, deadenylation of the reporter was significantly delayed under mild ccf-1 (RNAi) (74% knockdown, Figure 2A) and in ccr-4 (RNAi) depletions (54% knockdown, Figure 2A), delaying half-deadenylation times to 39 and 41 min, respectively (Figure 2B). A similar effect was observed when a reporter bearing three miR-35 binding sites was examined (Supplementary Figure S2). Interestingly, while examining knockdowns of ccf-1 and ccr-4 by western blot, we observed a decrease in CCR-4 protein expression under ccf-1 (RNAi) depletion, while ccr-4 (RNAi) did not significantly impact CCF-1 protein expression (Figure 2A). Furthermore, NTL-1 expression was significantly decreased (47% reduction) even under mild (57%) ccf-1(RNAi) knockdown.
Figure 2.
Coupled expression and function of the CCR4–NOT complex subunits in embryonic miRNA-mediated deadenylation. (A) Western blot analyses of embryonic extracts exposed to mock (gfp) RNAi, ccf-1 (RNAi) or ccr-4 (RNAi) probed with anti-CCF-1, anti-CCR-4, anti-NTL-1 and anti-tubulin (loading control) antibodies. Percentage of knockdown was quantified using ImageJ on western blots. (B) Deadenylation time course of RL 6x pA in wt embryonic extracts exposed to mock (gfp) RNAi, ccf-1 RNAi or ccr-4 RNAi. The relative intensity of the bands corresponding to full length and deadenylated RNAs was measured using ImageJ. A second-order polynomial regression was used, and the time of half-deadenylation (td1/2, intersection point between the full length and deadenylated RNA) was calculated using the quadratic formula. Schematic representation of the RL 6x reporter RNA used and the sequences of miR-35 and miR-35 binding site are also shown. Images are representative of at least three independent experiments.
These results are reminiscent of the coupled stability of the CCR4–NOT complex subunits in diverse species (53,88,89). We note that such results make it difficult to genetically disambiguate the relative or redundant contributions of the catalytic subunits in miRNA-mediated silencing. Nonetheless, these results show that CCR-4 and/or CCF-1 contribute to miRNA-mediated deadenylation in C. elegans embryos.
Step-wise assembly of miRISC effector complexes on mRNA targets
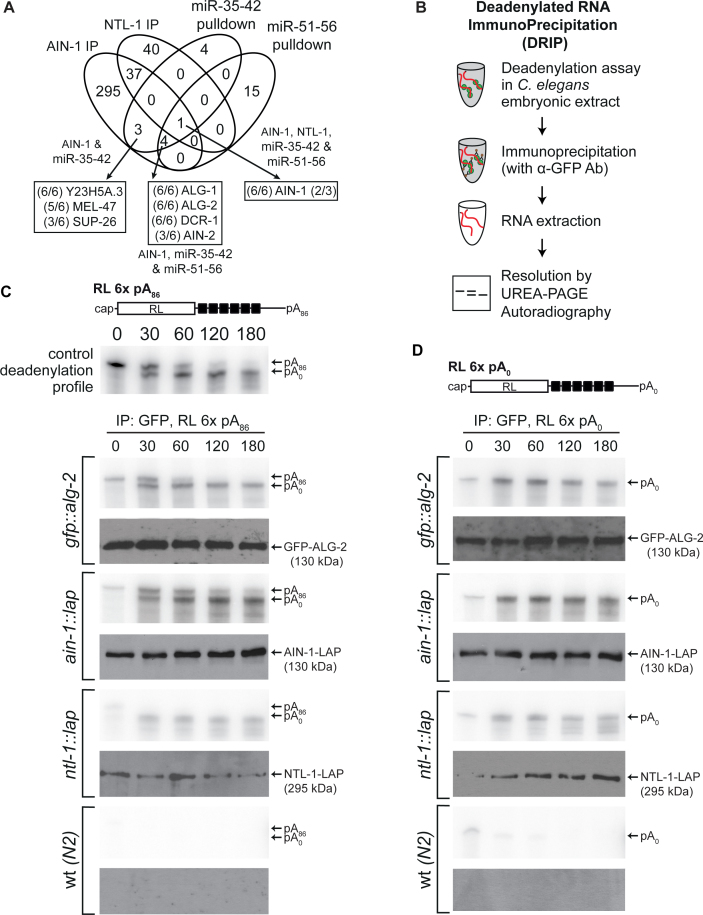
We had previously performed MuDPIT proteomic analyses on miRISC captures using miRNA target analogs (2΄-O-Me modified and biotinylated oligonucleotides) encoding binding sites for the maternal miR-35-42 and the zygotic miR-51-56 embryonic miRNA families (54,90,91). Instead of being based upon relatively stable protein-protein interactions like IP, this strategy of miRISC capture relies solely on its ability to specifically find and bind miRNA target sequences in a single step purification from a complex lysate mixture. When target analog capture and AIN-1 interactions were compared, only eight proteins were detected in both datasets (Figure 3A and Supplementary Table S7), which primarily reflect the known central components of miRISC. AIN-1, AIN-2, ALG-1, ALG-2 and DCR-1 were among the best-detected proteins in overlaps between AIN-1 IP, miR-35-42 and miR-51-56 target analog captures. The overlap also revealed factors of unknown or poorly characterized purpose in miRNA functions, which were detected at lower peptide coverage and in fewer replicates (SUP-26, Y23H5A.3, MEL-47). In stark contrast with AIN-1 IP datasets, none of the detected proteins in target analog captures are known components of the CCR4–NOT complex, any of the mRNP granules, or known mRNA decay machineries. Furthermore, while ALG-1 or ALG-2 were the best-detected interactions in AIN-1 IP based on coverage percentage or peptide counts (ALG-2: 68% coverage, 72 peptides; ALG-1: 63%, 76 peptides), neither was detected among interactions with NTL-1. Such a discontinuity between miRISC in its target recognition form, as captured using analog pull-down and the deadenylation and decay machineries interaction with AIN-1, lies at odds with the rapid and processive deadenylation of miRNA targets, which pervades in C. elegans embryonic cell-free systems (54). Such disparities in capturing the miRISC–CCR4–NOT interactions were noticeable in other systems where miRISC target analog capture was used (11), but also with distinct methods of Ago-complexes purification (92,93). We note that since target analog capture identifies endogenous miRISC components on the basis of its scanning activity, absence of deadenylation and decay machineries cannot be due to protein tagging artifacts.
Figure 3.
Step-wise assembly of miRISC effector complexes on mRNA targets. (A) Venn diagram of proteins interacting with AIN-1, NTL-1 and 2΄-O-Me captured miRISC. Fractions indicate the number of times the corresponding protein was detected in each independent IP (out of 6 for AIN-1, and out of 3 for NTL-1). (B) Flow chart of the procedure for Deadenylated RNA ImmunoPrecipitation (DRIP) assay. (C and D) DRIP profiles of RL 6x pA86 (C) and RL 6x pA0 (D) that represent target RNAs associated to immunoprecipitated proteins at each time point, as determined by autoradiography. Top panel in (C) is representative of a deadenylation assay time course carried out in wt (N2) extract prior to the IP step. Western blots on GFP IPs of embryo-stage transgenic animals carrying GFP-ALG-2, AIN-1-LAP or NTL-1-LAP using anti-GFP are shown below each autoradiograph. (C and D) are representative of at least three independent experiments.
We reasoned that the interactions detected with AIN-1 and NTL-1 may represent biochemically distinct form(s) of miRISC, involved in the effector step(s) of miRNA-mediated silencing, in contrast to, and perhaps downstream of, target recognition or scanning miRISC. To test this hypothesis, we developed an in vitro assay to detect interactions of miRISC components with targeted mRNAs prior to and during the course of deadenylation. The Deadenylated RNA-ImmunoPrecipitation (DRIP, Figure 3B) assay combines the C. elegans embryonic cell-free extract capable of miRNA-mediated silencing and deadenylation, with RNA immunoprecipitation. Radiolabeled RL 6x pA reporter was incubated in wt extract as above, or in extracts derived from transgenic animals expressing GFP-tagged miRISC proteins, over a course of 3 h. The reporter was deadenylated with similar kinetics in extracts derived from transgenic animals (Supplementary Figure S3A). Time points were chosen to reflect the state of the target mRNA prior to (Figure 3C, top panel; 0 min), during (30, 60 min) and after deadenylation (120, 180 min). IP was performed at each time point on core miRISC components, the Argonaute ALG-2, the GW182 ortholog AIN-1 and on the scaffolding subunit of the CCR4–NOT deadenylase complex, NTL-1. RNA was then extracted and resolved by urea-polyacrylamide gel electrophoresis and autoradiography. Importantly, the same monoclonal antibody directed against GFP was used for IP and exhibited minimal background when no fusion was present in the extract (Figure 3C, wt (N2) panel). When GFP-ALG-2 was recovered by IP, both full-length RL 6x pA86 and its deadenylated form were detected. Full-length RL 6x pA86 was detected at 0, 30 and 60 min, while the deadenylated species was detected at 30 min and at all later time points of the 3-h course. A similar profile was observed with AIN-1-LAP IP; AIN-1 associated with both the polyadenylated reporter and the deadenylated RNA species and remained stably associated post-deadenylation. In contrast, only the deadenylated species of RL 6x was detected in the NTL-1 IP during the time course (Figure 3C, NTL-1 panel). This observation indicates that its association with mRNA targets occurs on the mRNA and later then the initial recognition by scanning miRISC. Furthermore, it is consistent with a highly processive activity of the CCR4–NOT complex. These interactions were maintained in a poly(A) tail-independent manner; ALG-2, AIN-1 and NTL-1 remained stably associated with the target mRNAs long after completion of deadenylation. In line with this conclusion, DRIP profiles of 6x transcripts lacking a poly(A) tail (RL 6x pA0) closely mirrored the profiles of RL 6x pA86 (Figure 3D). Targeted RL 6x reporters were strongly enriched in ALG-2 and AIN-1 IP in comparison with reporters bearing mutations in seed-binding sequences, which remained fully polyadenylated (RL 6xmut pA; Supplementary Figure S3B) (54). We noticed however that although weakly, RL 6xmut pA86 reporter co-immunoprecipitated with ALG-2 and AIN-1 significantly above background, but not with NTL-1. This may be indicative of non-cognate low-affinity miRISC scanning interactions.
Taken together, these results show that the interaction with scanning miRISC precedes the recruitment of CCR4–NOT complex scaffolded by NTL-1 on the target mRNA. It further indicates that their interactions do not depend on the presence of poly(A) tail, and persist long after deadenylation. We note that although stable, this complex might reflect an intermediate in a de-capping and decay pathway that is not observed in this extract.
miRISC interactions seclude target mRNAs in nuclease-refractory mRNPs
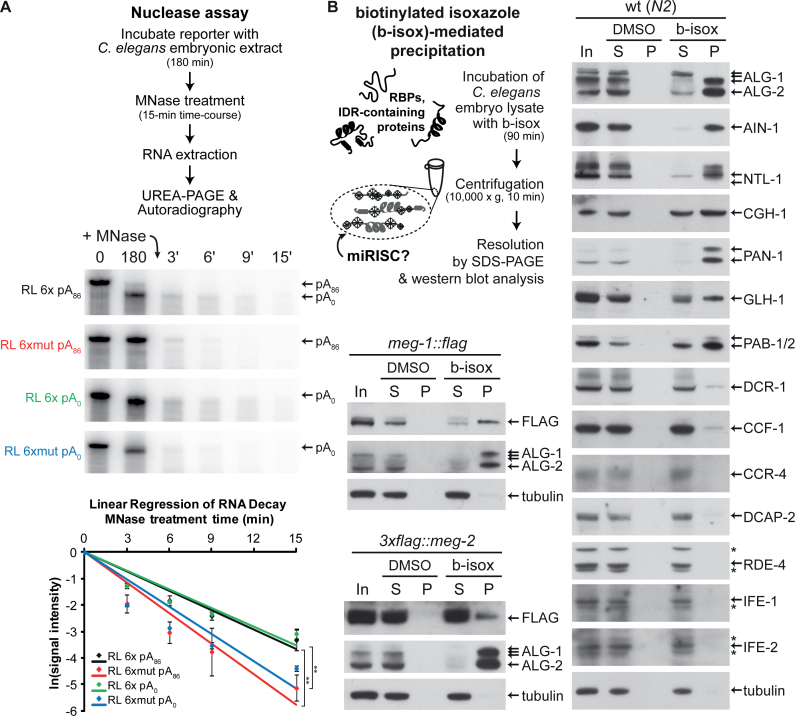
Considering the breadth of interactions of miRISC with its effector machinery on target mRNAs demonstrated by the above proteomic analyses and DRIP profiles, we reasoned that assembly of mRNP granules could sequester mRNA targets. To test this idea in vitro, we subjected the assembled complexes to a nuclease-resistance assay (Figure 4A). Radiolabeled polyadenylated RL 6x pA was incubated in cell-free extract until its complete deadenylation (180 min), and then challenged with serial dilutions of hindrance-sensitive micrococcal nuclease (MNase) over a 15-min time-course (Supplementary Figure S4A). RL 6xmut pA was used as control. Both targeted and untargeted reporters decayed as a result of the MNase treatment, but RL 6x reporters resisted significantly better than the non-targeted RL 6xmut reporter (Figure 4A). Full-length RL 6x reporter remained visible at the 6- and 9-min MNase treatment time points, when the RL 6xmut reporter was entirely degraded. We note that the entire 6x reporter mRNA, and not only the sequence encoding and surrounding miRNA-binding sites are protected in this manner, suggesting that the entire reporter is sequestered by miRISC and co-factors in the extract. Quantitation of independent replicates confirmed that the targeted RL 6x pA reporter was significantly less sensitive to MNase treatment than a non-targeted RL 6xmut pA reporter (Figure 4A, graphical panel). When the nuclease assay was conducted on un-adenylated transcripts (RL 6x pA0 and RL 6xmut pA0), the same outcome was observed with no significant difference in progression, indicating that mRNP assembly is independent of poly(A) tail presence. In the absence of extract, full-length 6x and 6xmut reporters were completely and rapidly degraded upon exposure to MNase, even at greatest dilutions (Supplementary Figure S4B), indicating that refraction to MNase is conferred by interactions between reporter sequences and the extract. Overall, these results imply that miRNP assembly secludes miRNA-targeted mRNA, and raise the possibility that mRNP assembly on target mRNA may contribute to silencing.
Figure 4.
miRISC secludes target mRNAs in nuclease-refractory mRNPs, and are selectively precipitated by biotinylated isoxazole. (A) Top panel: schematic diagram of nuclease assay. Middle panel: PAGE-autoradiography of reporter mRNAs (RL 6x and mut, +/− poly(A) tail). RNAs at 0 and 180 min are on the left to show their integrity at the start and end of the 3-h incubation in the embryonic extract, prior to nuclease treatment. MNase was then added to the reaction after 180 min and RNA integrity was monitored over a 15-min (denoted by t') MNase treatment. Bottom panel: the intensity of the RNAs following MNase treatment was quantified using ImageJ from triplicate experiments conducted using the same extract preparation. A logarithmic regression using a linear model was used to analyze the rate of RNA reporter decay. Autoradiograph from one replicate is presented. Experiment was reproduced at least twice in independent extract preparations. Error bars indicate standard deviation. Statistical significance was calculated using one-tailed t-test (**P < 0.01). (B) Schematic representation of the biotinylated isoxazole (b-isox)-mediated precipitation and western blots on Caenorhabditis elegans embryonic lysates (50 μg total protein) following exposure to b-isox (100 μM final). Lysates were derived from wt (N2), and FLAG-tagged meg-1 and meg-2 strains. DMSO, used as the solubilizing agent for b-isox, served as a mock control. In indicates input, S indicates soluble content and P indicates precipitate. Asterisks (*) indicate non-specific bands.
Selective precipitation of miRISC by biotinylated isoxazole
We elected to further characterize the association of miRISC with mRNPs using biotinylated isoxazole (b-isox), a compound causing aggregation of proteins rich in intrinsically disordered regions that are key determinants for mRNP assembly (44,94–96). Precipitation using this reagent selectively enriches constituents of mRNPs, and at least some of their associated proteins (44,95). Selective co-precipitation with b-isox from C. elegans embryonic lysates was assessed by western blotting for a panel of proteins related to miRNA function, RNAi, translation, mRNA processing, P bodies and germ granules (Figure 4B). Strikingly, miRISC components ALG-1/2 and AIN-1 were strongly enriched in the b-isox precipitate. The AIN-1-LAP fusion fractionated in a similar manner (Supplementary Figure S5). NTL-1, the poly(A) binding proteins PAB-1/2, the C. elegans DDX6 ortholog CGH-1 and the germ granule constituents (PAN-1, GLH-1 and MEG-1) were all preferentially co-precipitated with b-isox. Curiously, while MEG-1 and MEG-2 paralogs are rich in intrinsically disordered regions, the two FLAG-tagged fusion proteins behave differently with regards to b-isox precipitation. MEG-1 is strongly enriched in the precipitate, while a more limited portion of MEG-2 is selectively precipitated. Interestingly, unlike the CCR4–NOT complex scaffold NTL-1, its catalytic subunits CCR-4 and CCF-1 co-precipitate only in limited amounts. A minor fraction of DCR-1 was also detected in the precipitate fraction. Finally, b-isox precipitation was highly selective; the dsRNA-binding protein RDE-4, the cap-binding proteins IFE-1 and IFE-2, and tubulin were not recovered in the pellet fraction.
With these results, and in line with initial work on b-isox by Steven McKnight's group (44,95), the selective precipitation of mRNP proteins with b-isox has now been well characterized. However, we still do not rule out that part of the selectivity of b-isox precipitation may be due to its inherent compatibility or incompatibility with individual proteins. Notwithstanding this reservation, the strong selective enrichment of ALG-1/2, AIN-1 and NTL-1 proteins in b-isox precipitates lends further support to their association with mRNPs in vivo.
Inherently disordered proteins participate in the regulation of cog-1 mRNA by lsy-6 miRNA
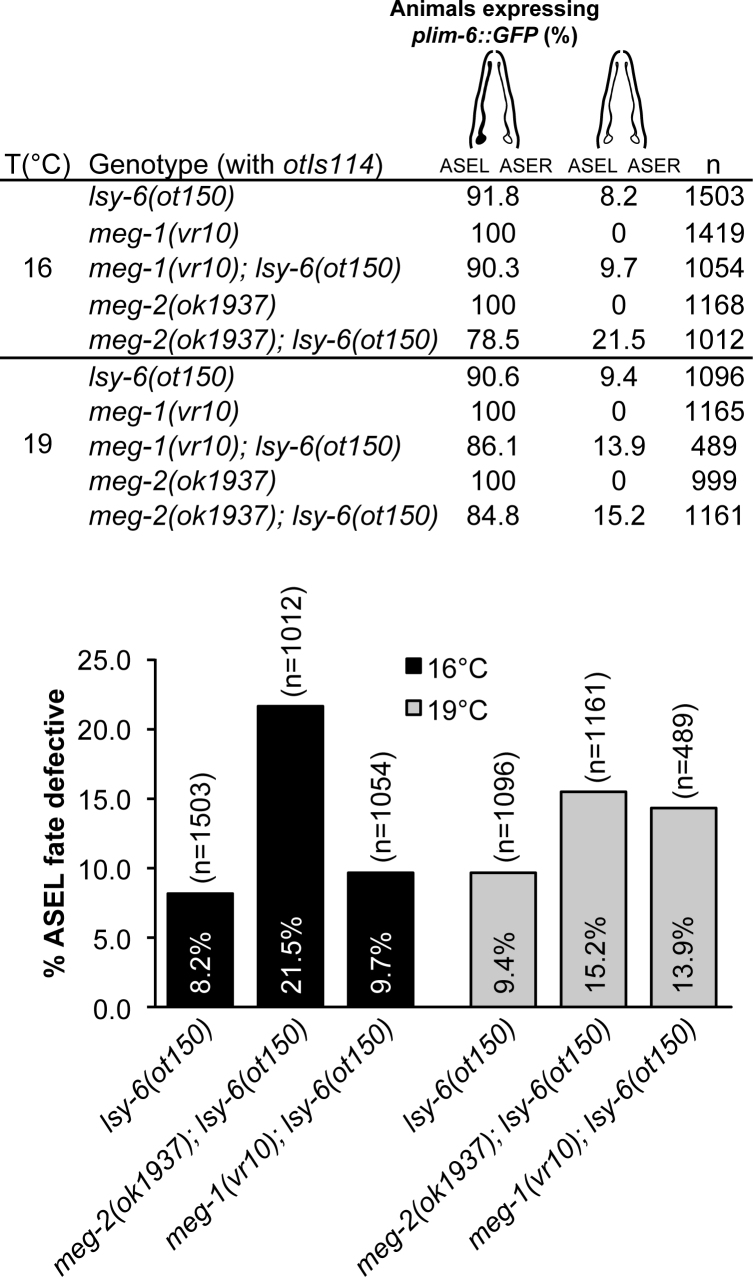
The structural role of MEG proteins in germ granule assembly and disassembly has recently been described (48,86). Consistent and extensive detection of MEG-2 and CCR4–NOT complex interactions (Figure 1) and architectural implications of intrinsically disordered proteins in mRNPs suggest that MEG proteins may contribute in miRNA-mediated silencing. To determine whether intrinsically disordered MEG proteins are implicated in embryonic miRNA function, we first tested the effects of meg-2 loss on the activity of the lsy-6 miRNA (Figure 5). lsy-6 functions during embryogenesis in the developmental specification of two bilaterally asymmetric neurons, ASEL and ASER, by downregulating its target, cog-1 (97). Animals lacking lsy-6 expression fail to downregulate cog-1 in the ASEL, resulting in the ASEL neuron adopting the ASER fate. The hypomorphic lsy-6(ot150) allele encodes a mutation in the conserved regulatory element in the lsy-6 promoter that leads to the reduction of lsy-6, but does not eliminate its function, resulting in a partially penetrant ASEL fate specification phenotype (98). This sensitized background has been extensively used to look at genetic interactions with the miRNA pathway (66,76,99–102). ASEL fate was assayed by scoring for the expression of the ASEL-specific plim-6::GFP, a transcriptional reporter that serves as an indicator for successful cog-1 silencing by lsy-6. Loss of meg-2 in lsy-6(ot150) significantly enhanced the ASEL fate specification phenotype, with the absence of reporter expression in ASEL detected at 21.5%, compared to 8.2% in lsy-6(ot150) animals, thus more than doubling the penetrance of the phenotype (Figure 5). This effect was modulated by temperature and the exacerbated lsy-6 phenotype was more prominent when animals were grown at 16°C than at 19°C (21.5% at 16°C compared to 15.2% at 19°C). In meg-2 mutants with wt lsy-6 expression, the reporter was expressed in the ASEL of every animal, indicating that removal of meg-2 activity on its own did not affect ASEL fate specification. meg-1 mutants had no effect on lsy-6 mutants at 16°C, and a mild incidence on ASEL specification when animals were grown at 19°C (from 9.7 to 13.9%).
Figure 5.
Loss of intrinsically disordered meg-2 impinges on cog-1 mRNA regulation by lsy-6 miRNA. The plim-6::GFP expression (denoted in black in schematic diagram) indicates ASEL neuronal cell fate. plim-6::GFP mis-expression phenotypes were quantified in lsy-6, meg-2 and meg-1 single mutants and lsy-6; meg-2 and lsy-6; meg-1 compound mutants. n = animals scored for each genotype.
These results suggest that meg-2 directly or indirectly participates in the function of lsy-6 miRNA in silencing cog-1 expression during embryogenesis, while its paralog meg-1 may have a more limited contribution.
DISCUSSION
Through concerted proteomics and interaction analyses, cell-free assays and genetics, we resolved temporal events leading to silencing by miRISC and identified a role for mRNPs and intrinsically disordered proteins in the functions of embryonic miRNAs. Our results support a model wherein progressive mRNP assembly on target mRNA is an integral part of the mechanism of miRNA-mediated silencing in the embryo (Figure 6). This model improves the previous static view on miRISC interactions, and opens up new possibilities into how developmental contexts modulate silencing mechanisms dictated by miRNAs.
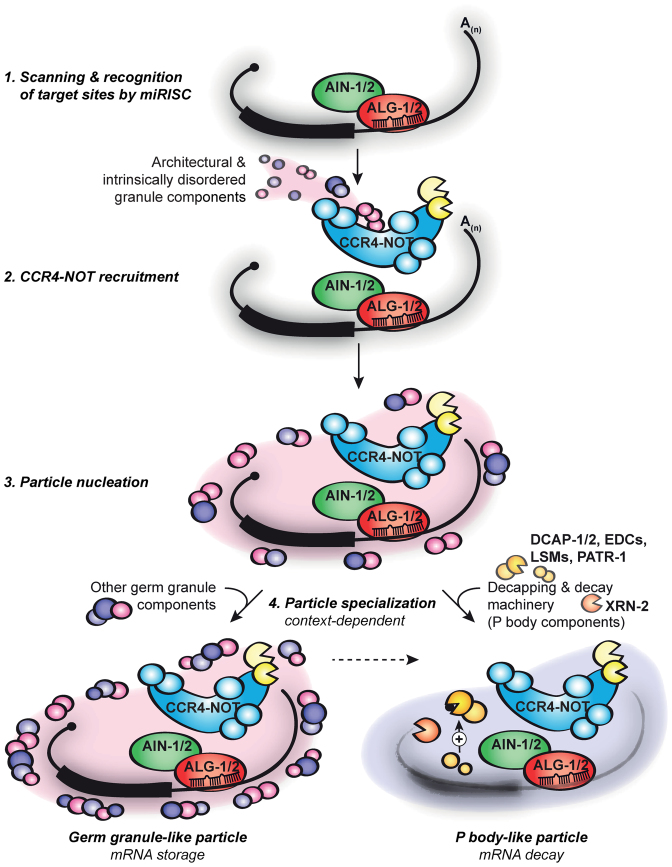
Figure 6.
Model: assembly and specialization of mRNPs on mRNAs in embryonic miRNA-mediated silencing. The miRISC (ALG-1/2 and AIN-1/2, and other accessory proteins) scans and recognizes mRNA targets (1). The CCR4–NOT deadenylase complex, along with tethered architectural and intrinsically disordered granule constituents (MEG-1 and MEG-2, and PGL proteins), is sequentially recruited to target mRNAs (2) and nucleates an mRNP particle (3). Proteomics analyses on AIN-1 and NTL-1 reveal a convolution of germ granule and P body-like mRNP complexes. Such diversity indicates the possibility of particle specialization that depends on cellular and developmental context for modulating the miRISC output on target silencing by storing the mRNA or subjecting it to decay (see ‘Discussion’ section).
Scanning miRISC and effector miRISC are distinct
We provide three distinct lines of experimental evidence supporting the view that miRISC biochemically matures from a ‘free’ scanning miRISC, to a mRNA-bound form which tethers effector components of miRNA-mediated silencing. Firstly, interaction datasets generated with AIN-1 IP contrast with miRISC-associated components captured through 2΄-O-methyl target analog affinity. Whereas in both cases the Argonautes ALG-1 and ALG-2 were the best-detected interactions by far, the mRNA deadenylase, the processing machineries or germ granule components were not detected in target analog captures. Secondly, while NTL-1 could be specifically recruited to miRISC-bound reporters in DRIP assays and AIN-1 was consistently detected among NTL-1 interactions, neither ALG-1 nor ALG-2 Argonautes could be detected in NTL-1 IPs. Thirdly and most decisively, scanning and effector miRISC could be resolved in time; DRIP results indicate that ALG-2 and AIN-1 association on the polyadenylated form of the 6xmiR-35 reporter precedes association with NTL-1 or the consequent mRNA deadenylation.
These findings are in logical line with previous conclusions drawn from Drosophila and human cells, which biochemically resolved the ‘miRISC-loading complex’ or RLC from mature miRISC. RLC complexes lack GW182, but contain Dicer and exhibit pre-miRNA processing activity, while ‘mature miRISC’ contains GW182 but lacks Dicer and pre-miRNA-processing activity (103,104). Hence, a tentative integrated view on data obtained across species and systems is that Argonaute-containing complexes are progressively remodeled from loading, to scanning, to the several steps of target silencing, to recycling (105–107).
We note that significant circumstantial evidence supports the possibility that multiple alternative miRISC maturation pathways may co-occur. A previous report examined AIN-2 interactions and mainly revealed interactions with components of the translation initiation machinery, but did not detect deadenylase, decapping, decay or mRNP components (3) that are pre-eminent with AIN-1. The fact that AIN-2 was detected in our AIN-1 proteomic analyses indicates that such pathways may not be mutually exclusive.
CCR4–NOT/AIN-1 interactions nucleate mRNPs on miRNA targets
Our work provides a unique glimpse on the intricate interactions that prevail in embryonic miRISC mRNPs and on their biological significance. The above-described sequential recruitment of the CCR4–NOT scaffold NTL-1 on miRNA targets, the breadth of the interactions of AIN-1 and NTL-1 with P body and germ granule proteins, the refraction of miRNA reporters to MNase challenge and the selective precipitation of miRISC with biotinylated isoxazole support the assembly of a mRNP microenvironment on miRNA targets. We furthermore note that some of the detected interactions are independently corroborated in a recent protein–protein interaction network involved in embryonic polarity and germ granule assembly (108). AIN-1 was detected in PIE-1 and CAR-1 IP proteomics, and NTL-1, CCF-1, CCR-4 and MEG-2 were detected in MBK-2 IPs.
It has long been noticed that a fraction of the miRISC components, such as GW182 homologs, Argonautes and small RNAs, localize to P body and/or P body like mRNPs (5,19,21–25). A key question is how the mechanisms at work in the assembly of organelle-scale P bodies or germ granules relate to miRISC functions and dynamics. Important insight can be gained by considering a closely related paradigm. The Gavis group used quantitative single-molecule imaging to examine assembly of mRNP into germ granules in the Drosophila oocyte (109). Detailed examination of stoichiometry and mRNP dynamics revealed that localized mRNAs are assembled and transported as single-mRNA RNP complexes into the oocyte, and are later merged as germ granules in the germ plasm. Build-up into germ granules is preferential for mRNPs that contain the same mRNA species, and mimics a positive-feedback dynamic, which could play a role in precipitating high-scale germ granule mRNPs. Altogether, this suggests that the content and assembly processes of single-mRNA and greater-scale mRNPs can be distinctly controlled, and progress along defined spatio-temporal steps (109). If one projects this concept of mRNP reorganization into a miRNA-mediated silencing analogy, progression from single-mRNA-bound miRISC to greater scaffolds may be a consequence of the recruitment of CCR4–NOT and its associated proteins (Figure 6). Specifically, tethering intrinsically disordered proteins to miRISC through NTL-1 interactions, or their combination with determinants of GW182 homologs (110), could trigger phase transition to larger dynamic mRNP granules and thus provide an enhanced microenvironment for mRNA seclusion, storage or for decapping and decay.
Context and miRNP function: to decay or not to decay?
De-repression of lsy-6 reporters in vivo under depletion of intrinsically disordered proteins, and DRIP profiles of RL 6x reporters in vitro suggest that miRISC-instigated mRNP assembly contributes to miRNA target silencing in the C. elegans embryo. At first glance, this result may stand at odds with experiments in Drosophila S2 cells, wherein impairment of P body formation by knockdown of the decapping factors (Lsm1 and Lsm3) did not prevent miRNA reporter silencing (31). This observation led to the interpretation that P bodies arise as a consequence of miRNA-mediated silencing rather than being a cause (31). Such results, however, could not rule out the possibility that putative P body functions are redundant with other aspects of miRNA-mediated silencing in S2 cells or that a sufficient function for a lesser-scale miRISC mRNP scaffold on target mRNAs. In addition, substantial evidence supports the idea that developmental context defines the composition and functions of P bodies and mRNPs in general. Work by the Evans and Schisa groups in C. elegans has already highlighted the diversity of mRNPs during oocyte maturation (111,112) and in early embryo (111). mRNPs that contain components such as CAR-1 and CGH-1 have distinct functions in maternal mRNA translation repression and degradation (111). This work and the results from the Seydoux group further indicate that the composition and function of mRNPs rapidly progress during early development (40,111). This diversity indicates that interactions detected here with NTL-1 and AIN-1 reflects a convolution of functionally distinct germ granule and P body-like particles that occur in the different cell lineages, merged in our embryonic lysate preparations. The interactors PGL-1, GLH-1 and MEG-1 are distinctly detectable in P lineage blastomeres during C. elegans embryogenesis, where they are important for germ granule architecture and stability (68,78,79,86,113,114). MEG-2, while partially functionally redundant with MEG-1 in the germline, is more broadly expressed and extends to somatic blastomeres (86). De-capping factors DCAP-1 and DCAP-2 co-localize with PGL-1 in P1 germline blastomeres in germ granules, but are also closely associated with P bodies throughout C. elegans lifespan (40,71). The PATR-1 decapping co-factor is also detected both in germline and somatic P bodies, but progressively accumulates in somatic blastomeres (40). The C. elegans homolog of the eIF4E-transporter, IFET-1 is partitioned into germ cells after the 4-cell stage, where it functions as a translational repressor of germ granule localized RNAs (85). Finally, the Sm-like (LSM) proteins, thought to couple deadenylation with mRNA decay (115), are enriched in somatic P bodies from the 3/4-cell stages, a window that coincides with maternal mRNA decay and such a localization requires NTL-1 (40,116).
The relative contributions of the translation repression, and deadenylation and decay components of miRNA-mediated silencing is still a matter of debate and remain under scrutiny in various systems and cell types. Our findings support the possibility that specialization of mRNPs can modulate miRISC output. It is thus reasonable that the extent and nature of functions of mRNPs in miRNA-mediated silencing mechanisms should be systematically considered in specific developmental and cellular contexts.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Martin J. Simard for providing the gfp::alg-2 strain and AIN-1 antibody, Dr Christian Eckmann and the C. elegans TransgeneOme platform for the ain-1::lap and ntl-1::lap strains, and Dr Valerie Reinke and the Caenorhabditis Genetics Center (CGC) for the meg-1 and meg-2 strains used in this study. We also thank Dr Geraldine Seydoux and Dr Deepika Calidas for the meg-1::1xflag and pgl-1::3xflag strains and comments on the manuscript.
Author contributions: E.W. analyzed the MuDPIT data and conducted all the experiments, except for the following: A.A.V. and J.W. carried out the MuDPIT on AIN-1, NTL-1, MEG-2, and PGL-1 IP samples; C.C. from N.S. laboratory suggested and performed the b-isox-mediated precipitation; M.F. from T.F.D. laboratory assisted with the NTL-1 and PAB-1/2 antibody production and with the data analysis from the MNase sensitivity assay using R software; E.C. from Y.T. laboratory conducted the computational assessment for siRNA and miRNA pathway proteins. M.S. generated the ain-1::lap and ntl-1::lap strains used for immunoprecipitation, DRIP analysis and b-isox-mediated precipitation. E.W. and T.F.D. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) [MOP 123352 to T.F.D.]; Fonds de la Recherche en Santé du Québec (FRQS); Chercheur-Boursier Senior Salary Award (to T.F.D.); Canadian Institutes of Health Research Frederick Banting and Charles Best Doctoral Research Award (to E.W.). Funding for open access charge: CIHR [MOP 123352].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C.. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001; 106:23–34. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L., Ding L., Cheung T.H., Dong M.Q., Chen J., Sewell A.K., Liu X., Yates J.R. 3rd, Han M.. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007; 28:598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E.. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005; 122:553–563. [DOI] [PubMed] [Google Scholar]
- 5.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E.. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006; 20:1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S.F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E.. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007; 21:2558–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F.. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006; 312:75–79. [DOI] [PubMed] [Google Scholar]
- 8.Mathonnet G., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S. et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007; 317:1764–1767. [DOI] [PubMed] [Google Scholar]
- 9.Wu L., Fan J., Belasco J.G.. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:4034–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E.. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009; 15:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A. et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009; 35:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun J.E., Huntzinger E., Fauser M., Izaurralde E.. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011; 44:120–133. [DOI] [PubMed] [Google Scholar]
- 13.Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W.. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011; 18:1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F. et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011; 18:1211–1217. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Boland A., Kuzuoglu-Ozturk D., Bawankar P., Loh B., Chang C.T., Weichenrieder O., Izaurralde E.. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell. 2014; 54:737–750. [DOI] [PubMed] [Google Scholar]
- 16.Mathys H., Basquin J., Ozgur S., Czarnocki-Cieciura M., Bonneau F., Aartse A., Dziembowski A., Nowotny M., Conti E., Filipowicz W.. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell. 2014; 54:751–765. [DOI] [PubMed] [Google Scholar]
- 17.Rouya C., Siddiqui N., Morita M., Duchaine T.F., Fabian M.R., Sonenberg N.. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA. 2014; 20:1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng D., Ezzeddine N., Chen C.Y., Zhu W., He X., Shyu A.B.. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008; 182:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding L., Spencer A., Morita K., Han M.. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005; 19:437–447. [DOI] [PubMed] [Google Scholar]
- 20.Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J.. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002; 13:1338–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K.. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005; 7:1267–1274. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R.. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005; 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T.. Identification of novel argonaute-associated proteins. Curr. Biol. 2005; 15:2149–2155. [DOI] [PubMed] [Google Scholar]
- 24.Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W.. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005; 309:1573–1576. [DOI] [PubMed] [Google Scholar]
- 25.Sen G.L., Blau H.M.. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005; 7:633–636. [DOI] [PubMed] [Google Scholar]
- 26.Sheth U., Parker R.. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003; 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingelfinger D., Arndt-Jovin D.J., Luhrmann R., Achsel T.. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002; 8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 28.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002; 22:8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B.. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002; 21:6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cougot N., Babajko S., Seraphin B.. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004; 165:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E.. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007; 27:3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J.H., Yang W.H., Gulick T., Bloch K.D., Bloch D.B.. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005; 11:1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchan J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014; 11:1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decker C.J., Parker R.. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012; 4:a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voronina E., Seydoux G., Sassone-Corsi P., Nagamori I.. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 2011; 3:a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boag P.R., Atalay A., Robida S., Reinke V., Blackwell T.K.. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 2008; 182:543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen Chi M., Chalmel F., Agius E., Vanzo N., Khabar K.S., Jegou B., Morello D.. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One. 2009; 4:e4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noble S.L., Allen B.L., Goh L.K., Nordick K., Evans T.C.. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 2008; 182:559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soderstrom K.O., Parvinen M.. Incorporation of (3H)uridine by the chromatoid body during rat spermatogenesis. J. Cell Biol. 1976; 70:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallo C.M., Munro E., Rasoloson D., Merritt C., Seydoux G.. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 2008; 323:76–87. [DOI] [PubMed] [Google Scholar]
- 41.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A.. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009; 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- 42.Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012; 483:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C., Eckmann C.R., Myong S., Brangwynne C.P.. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y., Protter D.S., Rosen M.K., Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P.. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015; 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015; 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., Seydoux G.. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014; 3:e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 51.Timmons L., Court D.L., Fire A.. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001; 263:103–112. [DOI] [PubMed] [Google Scholar]
- 52.Vasquez-Rifo A., Jannot G., Armisen J., Labouesse M., Bukhari S.I., Rondeau E.L., Miska E.A., Simard M.J.. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One. 2012; 7:e33750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nousch M., Techritz N., Hampel D., Millonigg S., Eckmann C.R.. The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 2013; 126:4274–4285. [DOI] [PubMed] [Google Scholar]
- 54.Wu E., Thivierge C., Flamand M., Mathonnet G., Vashisht A.A., Wohlschlegel J., Fabian M.R., Sonenberg N., Duchaine T.F.. Pervasive and cooperative deadenylation of 3΄UTRs by embryonic microRNA families. Mol. Cell. 2010; 40:558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu E., Duchaine T.F.. Cell-free microRNA-mediated translation repression in Caenorhabditis elegans. Methods Mol. Biol. 2011; 725:219–232. [DOI] [PubMed] [Google Scholar]
- 56.Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D. Jr, Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G. et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006; 124:343–354. [DOI] [PubMed] [Google Scholar]
- 57.Tabach Y., Billi A.C., Hayes G.D., Newman M.A., Zuk O., Gabel H., Kamath R., Yacoby K., Chapman B., Garcia S.M. et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013; 493:694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parry D.H., Xu J., Ruvkun G.. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr. Biol. 2007; 17:2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou R., Hotta I., Denli A.M., Hong P., Perrimon N., Hannon G.J.. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell. 2008; 32:592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thivierge C., Makil N., Flamand M., Vasale J.J., Mello C.C., Wohlschlegel J., Conte D. Jr, Duchaine T.F.. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat. Struct. Mol. Biol. 2012; 19:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J.K., Gabel H.W., Kamath R.S., Tewari M., Pasquinelli A., Rual J.F., Kennedy S., Dybbs M., Bertin N., Kaplan J.M. et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005; 308:1164–1167. [DOI] [PubMed] [Google Scholar]
- 62.Robert V.J., Sijen T., van Wolfswinkel J., Plasterk R.H.. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005; 19:782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui M., Kim E.B., Han M.. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2006; 2:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mi H., Muruganujan A., Thomas P.D.. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013; 41:D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas P.D., Kejariwal A., Campbell M.J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S. et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003; 31:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flamand M.N., Wu E., Vashisht A., Jannot G., Keiper B.D., Simard M.J., Wohlschlegel J., Duchaine T.F.. Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Res. 2016; 44:5924–5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boag P.R., Nakamura A., Blackwell T.K.. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005; 132:4975–4986. [DOI] [PubMed] [Google Scholar]
- 68.Gruidl M.E., Smith P.A., Kuznicki K.A., McCrone J.S., Kirchner J., Roussell D.L., Strome S., Bennett K.L.. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13837–13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao G., Deeb F., Mercurio J.M., Parfenova A., Smith P.A., Bennett K.L.. PAN-1, a P-granule component important for C. elegans fertility, has dual roles in the germline and soma. Dev. Biol. 2012; 364:202–213. [DOI] [PubMed] [Google Scholar]
- 70.Jankowska-Anyszka M., Lamphear B.J., Aamodt E.J., Harrington T., Darzynkiewicz E., Stolarski R., Rhoads R.E.. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem. 1998; 273:10538–10542. [DOI] [PubMed] [Google Scholar]
- 71.Lall S., Piano F., Davis R.E.. Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell. 2005; 16:5880–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jonas S., Izaurralde E.. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015; 16:421–433. [DOI] [PubMed] [Google Scholar]
- 73.MacCoss M.J., McDonald W.H., Saraf A., Sadygov R., Clark J.M., Tasto J.J., Gould K.L., Wolters D., Washburn M., Weiss A. et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Washburn M.P., Wolters D., Yates J.R. 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001; 19:242–247. [DOI] [PubMed] [Google Scholar]
- 75.Wolters D.A., Washburn M.P., Yates J.R. 3rd. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001; 73:5683–5690. [DOI] [PubMed] [Google Scholar]
- 76.Hammell C.M., Lubin I., Boag P.R., Blackwell T.K., Ambros V.. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell. 2009; 136:926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hyenne V., Desrosiers M., Labbe J.C.. C. elegans Brat homologs regulate PAR protein-dependent polarity and asymmetric cell division. Dev. Biol. 2008; 321:368–378. [DOI] [PubMed] [Google Scholar]
- 78.Kawasaki I., Amiri A., Fan Y., Meyer N., Dunkelbarger S., Motohashi T., Karashima T., Bossinger O., Strome S.. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004; 167:645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S.. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998; 94:635–645. [DOI] [PubMed] [Google Scholar]
- 80.Mello C.C., Schubert C., Draper B., Zhang W., Lobel R., Priess J.R.. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996; 382:710–712. [DOI] [PubMed] [Google Scholar]
- 81.Schubert C.M., Lin R., de Vries C.J., Plasterk R.H., Priess J.R.. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell. 2000; 5:671–682. [DOI] [PubMed] [Google Scholar]
- 82.Barbee S.A., Lublin A.L., Evans T.C.. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr. Biol. 2002; 12:1502–1506. [DOI] [PubMed] [Google Scholar]
- 83.Nakamura M., Ando R., Nakazawa T., Yudazono T., Tsutsumi N., Hatanaka N., Ohgake T., Hanaoka F., Eki T.. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells. 2007; 12:997–1010. [DOI] [PubMed] [Google Scholar]
- 84.Amiri A., Keiper B.D., Kawasaki I., Fan Y., Kohara Y., Rhoads R.E., Strome S.. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001; 128:3899–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sengupta M.S., Low W.Y., Patterson J.R., Kim H.M., Traven A., Beilharz T.H., Colaiacovo M.P., Schisa J.A., Boag P.R.. ifet-1 is a broad-scale translational repressor required for normal P granule formation in C. elegans. J. Cell Sci. 2013; 126:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leacock S.W., Reinke V.. MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics. 2008; 178:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molin L., Puisieux A.. C. elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4-NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene. 2005; 358:73–81. [DOI] [PubMed] [Google Scholar]
- 88.Boland A., Chen Y., Raisch T., Jonas S., Kuzuoglu-Ozturk D., Wohlbold L., Weichenrieder O., Izaurralde E.. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat. Struct. Mol. Biol. 2013; 20:1289–1297. [DOI] [PubMed] [Google Scholar]
- 89.Temme C., Zhang L., Kremmer E., Ihling C., Chartier A., Sinz A., Simonelig M., Wahle E.. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010; 16:1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P.. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294:858–862. [DOI] [PubMed] [Google Scholar]
- 91.Stoeckius M., Maaskola J., Colombo T., Rahn H.P., Friedlander M.R., Li N., Chen W., Piano F., Rajewsky N.. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat. Methods. 2009; 6:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hock J., Weinmann L., Ender C., Rudel S., Kremmer E., Raabe M., Urlaub H., Meister G.. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007; 8:1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Landthaler M., Gaidatzis D., Rothballer A., Chen P.Y., Soll S.J., Dinic L., Ojo T., Hafner M., Zavolan M., Tuschl T.. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008; 14:2580–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Decker C.J., Teixeira D., Parker R.. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007; 179:437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012; 149:768–779. [DOI] [PubMed] [Google Scholar]
- 96.Reijns M.A., Alexander R.D., Spiller M.P., Beggs J.D.. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008; 121:2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnston R.J., Hobert O.. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003; 426:845–849. [DOI] [PubMed] [Google Scholar]
- 98.Sarin S., O'Meara M.M., Flowers E.B., Antonio C., Poole R.J., Didiano D., Johnston R.J. Jr, Chang S., Narula S., Hobert O.. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics. 2007; 176:2109–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vasquez-Rifo A., Bosse G.D., Rondeau E.L., Jannot G., Dallaire A., Simard M.J.. A new role for the GARP complex in microRNA-mediated gene regulation. PLoS Genet. 2013; 9:e1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang P., Zhang H.. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 2013; 14:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zinovyeva A.Y., Bouasker S., Simard M.J., Hammell C.M., Ambros V.. Mutations in conserved residues of the C. elegans microRNA Argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS Genet. 2014; 10:e1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ren Z., Veksler-Lublinsky I., Morrissey D., Ambros V.. Staufen negatively modulates microRNA activity in Caenorhabditis elegans. G3 (Bethesda). 2016; 6:1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fukaya T., Tomari Y.. MicroRNAs mediate gene silencing via multiple different pathways in drosophila. Mol. Cell. 2012; 48:825–836. [DOI] [PubMed] [Google Scholar]
- 104.Miyoshi K., Okada T.N., Siomi H., Siomi M.C.. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009; 15:1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O.. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009; 11:1143–1149. [DOI] [PubMed] [Google Scholar]
- 106.Lee Y.S., Pressman S., Andress A.P., Kim K., White J.L., Cassidy J.J., Li X., Lubell K., Lim do H., Cho I.S. et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009; 11:1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu P.H., Isaji M., Carthew R.W.. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol. Cell. 2013; 52:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen J.X., Cipriani P.G., Mecenas D., Polanowska J., Piano F., Gunsalus K.C., Selbach M.. In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into p granule dynamics. Mol. Cell Proteomics. 2016; 15:1642–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Little S.C., Sinsimer K.S., Lee J.J., Wieschaus E.F., Gavis E.R.. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 2015; 17:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang K.L., Chadee A.B., Chen C.Y., Zhang Y., Shyu A.B.. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013; 19:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hubstenberger A., Cameron C., Noble S.L., Keenan S., Evans T.C.. Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J. Cell Biol. 2015; 211:703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wood M.P., Hollis A., Severance A.L., Karrick M.L., Schisa J.A.. RNAi screen identifies novel regulators of RNP granules in the Caenorhabditis elegans germ line. G3 (Bethesda). 2016; 6:2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuznicki K.A., Smith P.A., Leung-Chiu W.M., Estevez A.O., Scott H.C., Bennett K.L.. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 2000; 127:2907–2916. [DOI] [PubMed] [Google Scholar]
- 114.Spike C., Meyer N., Racen E., Orsborn A., Kirchner J., Kuznicki K., Yee C., Bennett K., Strome S.. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics. 2008; 178:1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tharun S., Parker R.. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001; 8:1075–1083. [DOI] [PubMed] [Google Scholar]
- 116.Seydoux G., Fire A.. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994; 120:2823–2834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.