Abstract
The human cytomegalovirus 72-kDa immediate-early (IE)1 and 86-kDa IE2 proteins are expressed at the start of infection, and they are believed to exert much of their function through promiscuous transcriptional activation of viral and cellular gene expression. Here, we show that the impaired growth of an IE1-deficient mutant virus in human fibroblasts is efficiently rescued by histone deacetylase (HDAC) inhibitors of three distinct chemical classes. In the absence of IE1 expression, the viral major IE and UL44 early promoters exhibited decreased de novo acetylation of histone H4 during the early phase of infection, and the hypoacetylation correlated with reduced transcription and accumulation of the respective gene products. Consistent with these findings, IE1 interacts specifically with HDAC3 within infected cells. We also demonstrate an interaction between IE2 and HDAC3. We propose that the ability to modify chromatin is fundamental to transcriptional activation by IE1 and, likely, IE2 as well.
Keywords: gene regulation, chromatin remodeling, transcriptional activation
Human cytomegalovirus (HCMV) is a ubiquitous β-herpes virus that can cause life-threatening disease in immunologically immature or compromised individuals, including AIDS patients, transplant recipients, and congenitally infected neonates (1). Infection of permissive cultured cells with HCMV results in the sequential expression of three classes of viral genes termed immediate-early (IE), early, and late (2). Expression of IE genes is a prerequisite for progression into the early phase of the infectious cycle and the subsequent replication of viral DNA, which is, in turn, required for entry into the late phase, culminating in virion assembly (2).
Among the most abundant IE gene products are the UL123-coded 72-kDa IE1 and UL122-coded 86-kDa IE2 proteins. Their mRNAs are derived by differential splicing of a primary transcript, whose synthesis is controlled by the major IE promoter. Both proteins target to the host-cell nucleus and act as promiscuous transcriptional activators. In fact, IE1 and IE2 cooperate to stimulate transcription from various promoters (2, 3). Mutant studies have shown that IE2 is indispensable for viral early gene expression in cultured cells (4, 5). Although IE1-null viruses replicate quite efficiently at a high input multiplicity, the absence of IE1 results in a broad block to HCMV early gene expression at a low multiplicity of infection (6–8). IE1 interacts with constituents of ND10 bodies as well as with chromatin, but it does not appear to bind DNA directly (9–13).
Like the cellular genome, HCMV DNA is associated with histones, at least during certain stages of the infectious cycle (14–16). The expression of cellular genes is regulated by post-translational modification of the amino-terminal tails of core histones by reversible acetylation and subsequent changes in local chromatin structure and composition (reviewed in ref. 17). Steady-state acetylation levels of core histones and other nuclear proteins result from the balance between the opposing activities of two classes of enzymes: histone acetyltransferases and histone deacetylases (HDACs). In general, histone acetyltransferase activity leading to hyperacetylation of histones is associated with transcriptional activation (reviewed in ref. 18), whereas HDAC activity resulting in hypoacetylation is linked to repression (reviewed in ref. 19).
This study demonstrates that HCMV IE1, and probably IE2 as well, promotes viral transcription by antagonizing histone deacetylation.
Materials and Methods
Biological and Chemical Reagents. The plasmids pCGN-IE1, pCGN-IE2, pcDNA-IE1, pGL3-ICP36, and pGL3-MIEP have been described (13, 20, 21). For the construction of pcDNA-IE2, the HCMV IE2-coding region from pCGN-IE2 was inserted into the KpnI site in pcDNA3 (Invitrogen). The plasmid pcDNA-HDAC3-Flag was a gift from E. Verdin (University of California, San Francisco).
Human MRC-5 embryonic lung cells (European Collection of Cell Cultures) and human H1299 lung carcinoma cells were cultured in medium containing 10% FCS. MRC-IE1 cells were generated by transducing MRC-5 fibroblasts with an HCMV IE1-expressing retrovirus that was produced by transfecting ΦNX-Ampho cells (a gift from G. Nolan, Stanford University, Stanford, CA) with a pLXSN-derived plasmid (BD Biosciences Clontech) containing the IE1-coding sequence followed by selection with G418. The IE1-deficient HCMV (CR208) and its parental WT strain (Towne) (7) were provided by E. Mocarski (Stanford University). WT and mutant HCMVs were grown and titered on MRC-5 or MRC-IE1 cells, respectively.
HDAC inhibitors were obtained from Biomol (Helminthosporium carbonum toxin, HC toxin) or Upstate Biotechnology (Lake Placid, NY) [trichostatin A (TSA) and sodium butyrate]. Maximum subtoxic drug concentrations were determined by using a commercial cell-viability assay (CellTiter 96 AQueous One solution cell-proliferation assay, Promega).
Immunoprecipitation and Western Blot Analyses. For coimmunoprecipitation analyses, H1299 cells were transfected with 10 μg of expression plasmid or empty vector by using calcium phosphate precipitation, or MRC-5 cells were infected with WT or mutant HCMV at a multiplicity of 5 median tissue culture 50% infective dose (TCID50) per cell. Subsequently, cells were harvested and resuspended at 4°C in lysis buffer [20 mM Tris·HCl, pH 8.0/100 mM NaCl/1 mM EDTA/0.1% Nonidet P-40/1 mM DTT/10% glycerol/protease inhibitor mixture (Complete Mini, Roche)], and sonicated on ice, and lysates were clarified by centrifugation at 10,000 × g for 10 min. The supernatants were precleared by using protein A Sepharose (Sigma–Aldrich) and then incubated with appropriate Ab-coupled beads at 4°C for 3 h, followed by four washing steps with wash buffer (50 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% Nonidet P-40). For Western blotting, washed Sepharose beads or whole-cell protein extracts were prepared in lysis buffer [50 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% SDS/1% Nonidet P-40/0.5% sodium deoxycholate/protease inhibitor mixture (Complete Mini, Roche)] and mixed with 2× sample buffer (100 mM Tris·HCl, pH 6.8/4% SDS/0.2% bromophenol blue/20% glycerol/0.2 M 2-mercaptoethanol), followed by heating at 95°C for 5 min. Proteins were then assayed by Western blot analysis as described (13). The following Abs or Ab-agarose conjugates were used in these studies: anti-HCMV IE1, clone 1B12 (20); anti-HCMV IE2, clone 3A9; anti-HCMV pp28, clone 10B4–29 (22), anti-histone H3 (Upstate Biotechnology); anti-acetyl-histone H3 (Upstate Biotechnology); anti-HDAC3, clone 3G6 (Upstate Biotechnology); anti-Flag polyclonal (Sigma–Aldrich); anti-Flag M2 agarose affinity gel (Sigma–Aldrich); and anti-hemagglutinin (HA) agarose, clone HA-7 (Sigma–Aldrich).
Chromatin Immunoprecipitation and RNA Quantification. Chromatin immunoprecipitation assays were performed after the protocol from Upstate Biotechnology with minor modifications. Protein from 5 × 106 MRC-5 cells was immunoprecipitated with a polyclonal rabbit antiserum directed against amino-terminally acetylated histone H4 (Upstate Biotechnology). Quantitative real-time PCR amplifications were carried out in triplicate on 2 μl of purified DNA in the presence of 0.5 μM of each primer and 4 mM Mg2+. The annealing temperature was decreased during the first cycles from 64°C to 56°C at a rate of 0.5°C per cycle. Relative changes in coprecipitated DNA were normalized to input DNA and calculated by using the relative quantification strategy described at www.lightcycler-online.com/lc_support/pdfs/lc_13.pdf. The PCR-amplified regions comprised 284-, 275-, or 230-bp sequences at the 3′ ends of the major IE, UL44, or c-fos promoters, respectively, covering the transcription start sites. The primer sequences were as follows: MIEP, 5′-CTTACGGGACTTTCCTACTTG-3′ (forward) and 5′-CGATCTGACGGTTCACTAA-3′ (reverse); UL44P, 5′-AACCTGAGCGTGTTTGTG-3′ (forward) and 5′-CGTGCAAGTCTCGACTAAG-3′ (reverse); and c-fosP, 5′-GAGACCTCTGAGACAGGAACT-3′ (forward) and 5′-CAGATGCGGTTGGAGTAC-3′ (reverse).
For quantification of viral transcripts, total RNA was isolated from 2.5 × 106 cells with TRIzol (Invitrogen). RNA was further purified by using the RNeasy Mini kit, including a DNase digestion step (Qiagen, Valencia, CA). Purified RNA (5 μg) was converted into cDNA by using an oligo(dT) primer and SuperScript III enzyme (Invitrogen), and quantitative real-time PCR was performed as described above by using 2 μl of 1:10 diluted first-strand cDNA and the following primer pairs: UL44, 5′-GCTGTCGCTCTCCTCTTTCG-3′ (forward) and 5′-TCACGGTCTTTCCTCCAAGG-3′ (reverse); and UL122, 5′-CAAGAGTGGGTTGTCAGCGTG-3′ (forward) and 5′-GGCAGAACTCGGTGACATCC-3′ (reverse).
Results
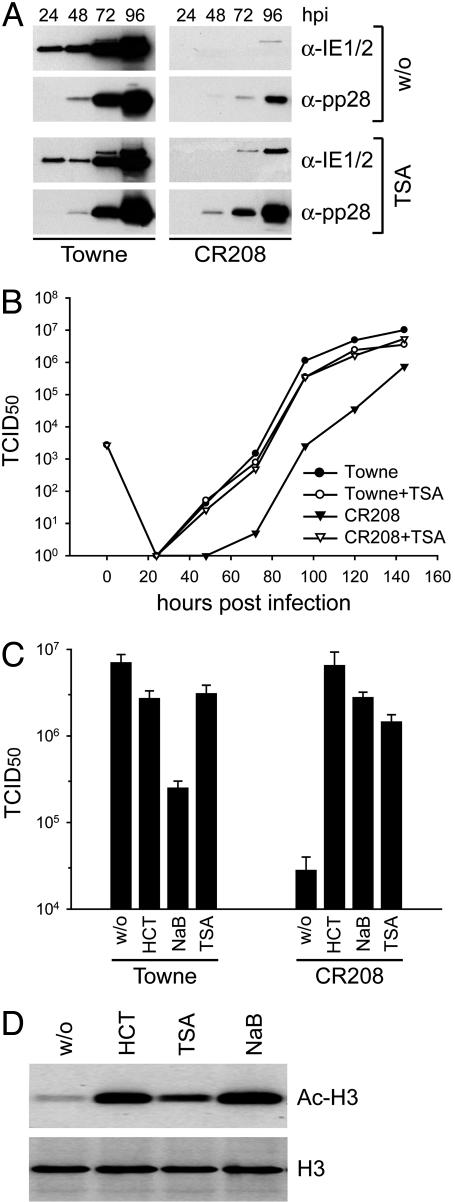
HDAC Inhibitors Rescue the Growth of an IE1-Deficient Mutant Virus. HCMV IE1 is a promiscuous trancriptional activator, and it associates with chromatin (9, 11–13). Also, the murine cytomegalovirus IE1 protein, although it is not related in its sequence to the HCMV IE1 protein, modulates the acetylation of chromatin (23). Consequently, we hypothesized that HCMV IE1 might stimulate gene expression by modifying histone acetylation. As an initial test of this idea, we sought to determine whether the replication of CR208 [an IE1-deficient HCMV mutant whose replication is substantially impaired at low input multiplicities (6, 7)] could be rescued by TSA [a specific inhibitor of class I and II HDACs, which causes global hyperacetylation of core histones (24)]. We infected human MRC-5 fibroblasts with CR208 or its WT parent (Towne) at a multiplicity of 0.1 TCID50 per cell in the continuous presence or absence of TSA. We monitored the accumulation of two virus proteins (Fig. 1A) and the production of infectious progeny (Fig. 1B). As expected, the growth of CR208 was severely delayed when compared with WT, and this impairment was manifest in both reduced viral protein levels and decreased yields of infectious virus. However, in the presence of TSA, the mutant displayed accelerated and enhanced accumulation of viral proteins, and its growth kinetics were similar to the IE1-expressing parental virus. Importantly, the HDAC inhibitor did not stimulate growth of the WT virus, indicating that the drug specifically complemented the lack of the IE1 gene.
Fig. 1.
HDAC inhibitors overcome the growth defect of an IE1-deficient mutant HCMV. (A) Comparison of viral protein accumulation in MRC-5 cells infected with WT (Towne) and mutant (CR208) viruses in the absence or continuous presence (beginning 24 h before infection) of 300 nM TSA. MRC-5 cells were infected at a multiplicity of 0.1 TCID50 per cell, and expression of IE1 and IE2 (IE1/2), as well as the late viral protein pp28, was monitored at 24–96 h postinfection (hpi) by Western blot assay. (B) Multistep growth analysis of WT (Towne, circles) and mutant (CR208, triangles) viruses in the absence (filled symbols) or continuous presence (open symbols) of 300 nM TSA. MRC-5 cells were infected at a multiplicity of 0.1 TCID50 per cell, and virus yields were monitored for 144 h. Symbols identify mean values from two experiments. (C) Titers of WT (Towne) and mutant (CR208) viruses in the absence or continuous presence of the HDAC inhibitors HC toxin (HCT, 100 nM), sodium butyrate (NaB, 1 mM), or TSA (300 nM). MRC-5 cells were infected at a multiplicity of 0.1 TCID50 per cell, and yields were determined 120 h later. Bars represent mean values and SEs from three separate infections. (D) HDAC inhibitors induce histone hyperacetylation. MRC-5 cells were treated with 100 nM HCT, 300 nM TSA, or 1 mM NaB for 24 h, and Western blot analyses were performed by using Abs against acetylated histone H3 (Ac-H3) or the unmodified protein (H3).
To exclude the possibility that TSA, a hydroxamic acid derivative, promotes growth of the mutant virus by means of an effect that is unrelated to inhibition of histone deacetylation, we tested two additional HDAC inhibitors belonging to different chemical classes: sodium butyrate, which is a short-chain fatty acid (25), and HC toxin, which is a cyclic tetrapeptide antibiotic (26). Both drugs were even more effective than TSA in compensating for the loss of IE1 function (CR208, Fig. 1C). HC toxin was particularly potent, raising the yield of CR208 by a factor of >200 to a level that was indistinguishable from the WT virus yield. Also, these results correlated with stronger histone H3 hyperacetylation induced by sodium butyrate and HC toxin compared with TSA (Fig. 1D). Although the continuous presence of TSA or HC toxin had only small inhibitory effects on the growth of the parental Towne virus, sodium butyrate reduced WT yields almost 30-fold (Towne, Fig. 1C) under conditions in which mutant virus yields were still 100-fold enhanced (CR208, Fig. 1C). The inhibitory effects were probably due to adverse consequences of prolonged drug treatment on infected cells because HDAC inhibitors have the potential to induce cell-cycle arrest and apoptotic cell death (e.g., ref. 27). Together, our results show that inhibition of HDAC activity compensates for loss of the IE1 gene during HCMV infection in a highly efficient and specific manner.
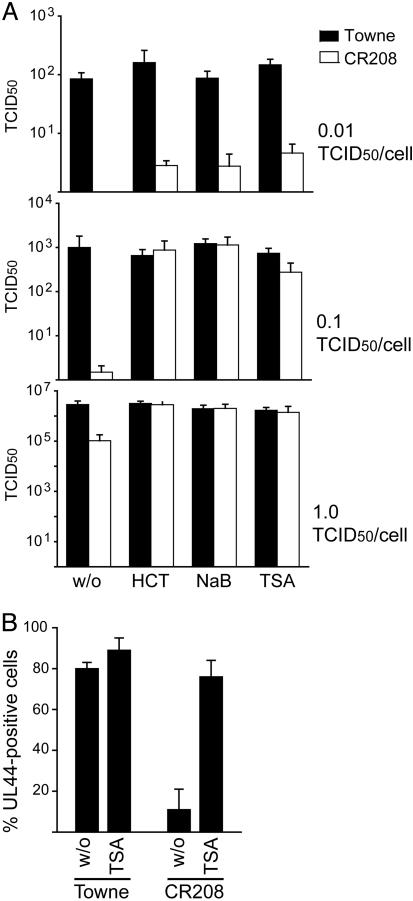
The experiments described so far used continuous drug treatment during multiple rounds of viral replication. Interestingly, in contrast to a relatively low multiplicity of infection (0.01 TCID50 per cell, Fig. 2A), a 24-h pretreatment of cells infected at higher input multiplicities (0.1 or 1 TCID50 per cell, Fig. 2 A) was equally efficient in compensating for the lack of IE1 as a continuous drug treatment throughout the course of infection (Fig. 1). Addition of the drugs at 12 h after infection or later had no stimulatory effect on CR208 (data not shown). Moreover, the percentage of cells initiating early viral gene expression after infection at a multiplicity of ≈1 TCID50 per cell, as determined by immunofluorescent detection of the ppUL44 DNA polymerase accessory protein, was only ≈11% in mutant, compared with ≈80% in WT virus infections (Fig. 2B). This observation is consistent with earlier reports on the phenotype of CR208 (7, 8). Pretreatment with TSA increased the number of pUL44-positive cells in CR208 infections to 76% and had little effect on the WT virus (Fig. 2B). These observations indicate that both IE1 and HDAC inhibitors target very early events in HCMV-infected cells to facilitate HCMV gene expression.
Fig. 2.
Pretreatment of cells with HDAC inhibitors can support normal IE1-mutant virus growth. (A) Effect of pretreatment with HDAC inhibitors on the titers of WT Towne (black bars) and CR208 mutant (white bars) viruses after infection at a multiplicity of 0.01, 0.1, or 1 TCID50 per cell. MRC-5 cells were treated with 100 nM HC toxin (HCT), 2 mM sodium butyrate (NaB), or 500 nM TSA for 24 h before infection, and virus titers were determined at 72 h after infection. Bars represent mean values from three separate infections with SEs. (B) Immunofluorescence analysis showing expression of the ppUL44 early viral protein at 16 h after infection in MRC-5 cells infected at a multiplicity of ≈1 TCID50 per cell with WT (Towne) or mutant (CR208) virus. Cells were pretreated with 500 nM TSA or the respective solvent dimethyl sulfoxide (w/o) for 24 h before infection. Cells in at least 10 fields of view (≈40 cells per field) were counted. Bars represent mean values with SEs presented as the percentage of 4′,6-diamidino-2-phenylindole-stained nuclei that also stained positive for ppUL44.
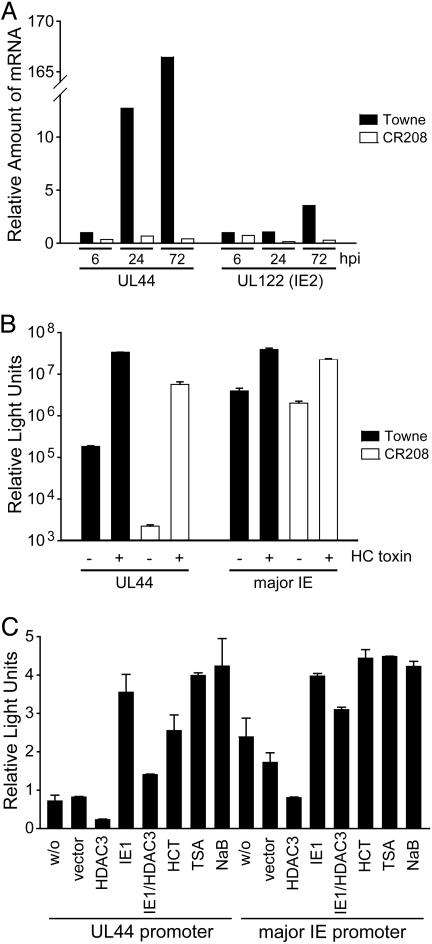
Histone Hypoacetylation and Reduced Transcription in the Absence of IE1. Using real-time RT-PCR, we quantified the accumulation of the major IE-derived IE2 (UL122) and the early UL44 transcripts in WT and mutant virus infections. Both mRNAs accumulated to lower levels in cells infected with CR208 as compared to WT virus (Fig. 3A), and, as has been noted (8), expression of UL44 was more severely affected than IE2. Consistent with these findings, expression from a luciferase reporter controlled by the UL44 promoter was ≈80-fold reduced in mutant as compared to WT virus-infected cells, and HC toxin reversed this effect, activating the UL44 promoter in the absence of IE1 by a factor of >2,500 (Fig. 3B). Similar, but more modest, effects were observed for the major IE promoter (Fig. 3B).
Fig. 3.
The HCMV major IE and UL44 promoters are regulated by histone acetylation and IE1. (A) Quantitative real-time RT-PCR was performed with total RNA isolated from MRC-5 cells at 6–72 h postinfection (hpi) with WT (Towne) or mutant (CR208) virus at a multiplicity of 0.1 TCID50 per cell by using UL44- or UL122 (IE2)-specific primers. (B) Luciferase assays were performed in MRC-5 cells, which were transfected with reporter plasmids pGL3-ICP36 (UL44) or pGL3-MIEP (major IE) and treated with 100 nM HC toxin at 24–40 h after transfection, as indicated. At 24 h after transfection, cells were infected with WT (Towne) or mutant (CR208) viruses at a multiplicity of 1 TCID50 per cell for 16 h. (C) Luciferase assays were performed in which MRC-5 cells were cotransfected with reporter plasmids pGL3-ICP36 (UL44 promoter) or pGL3-MIEP (major IE promoter) and empty vector (w/o) or expression plasmids encoding HDAC3, IE1, or both. As indicated, cells were treated with 100 nM HC toxin (HCT), 2 mM sodium butyrate (NaB), or 500 nM TSA at 24–40 h after transfection. Transfections were performed in triplicate, and mean relative light units with SEs (× 103 for the UL44 promoter and × 106 for the major IE promoter) are presented.
To further confirm that the major IE and UL44 promoters are subject to regulation by histone acetylation/deacetylation, we analyzed the effects of over-expressed HDAC3 on the activity of the luciferase reporter constructs (Fig. 3C). The HDAC significantly inhibited the activity of the major IE and UL44 promoters; IE1 partially overcame the inhibitory effect of HDAC3, and the addition of HDAC inhibitors activated both viral promoters. These results demonstrate that the activity of IE1-responsive HCMV promoters can be modulated by changes in histone acetylation.
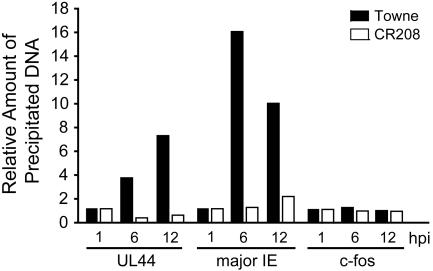
To test directly whether IE1-mediated activation of the major IE and UL44 promoters correlates with the induction of histone acetylation, we performed chromatin immunoprecipitation assays by using an Ab that recognizes acetylated histone H4 (Fig. 4). From 1 to 6 h after infection with WT virus, we observed a 16.1- and 3.8-fold relative increase in H4 acetylation at the major IE or UL44 promoters, respectively. In the same time interval after infection with the IE1-deficient virus, the acetylation levels went up by only 1.3-fold at the major IE and dropped 2.5-fold at the UL44 promoter. At 12 h after infection, H4 acetylation levels decreased at the major IE promoter while still increasing at the UL44 promoter during WT infection. In contrast, there was a slight increase at both promoters after infection with the mutant virus. Nevertheless, at 12 h histone acetylation was still less efficient by a factor of 4.5 (major IE promoter) or 11.6 (UL44 promoter) in mutant vs. WT virus infections. Under the same experimental conditions, both WT and mutant viruses had little effect on H4 acetylation at the cellular fos promoter, suggesting that IE1 might specifically modify the chromatin of viral genes. Similar results were obtained for acetylated histone H3 (data not shown). Our results demonstrate that IE1-mediated transcriptional activation of at least two viral promoters correlates with an increase in histone acetylation.
Fig. 4.
Decreased histone H4 acetylation at the HCMV major IE and UL44 promoters in the absence of IE1. Chromatin immunoprecipitation assays were performed on chromatin from MRC-5 cells infected with WT (Towne) or mutant (CR208) viruses at a multiplicity of 1 TCID50 per cell by using an Ab specific to amino-terminally acetylated histone H4- and UL44-specific, major IE-specific, or c-fos-specific primers for quantitative real-time PCR of coprecipitated DNA. PCRs were performed in triplicate, and mean values normalized to input DNA levels are presented, representing changes in acetylation at 6 and 12 h postinfection (hpi) relative to 1 h postinfection (set at 1).
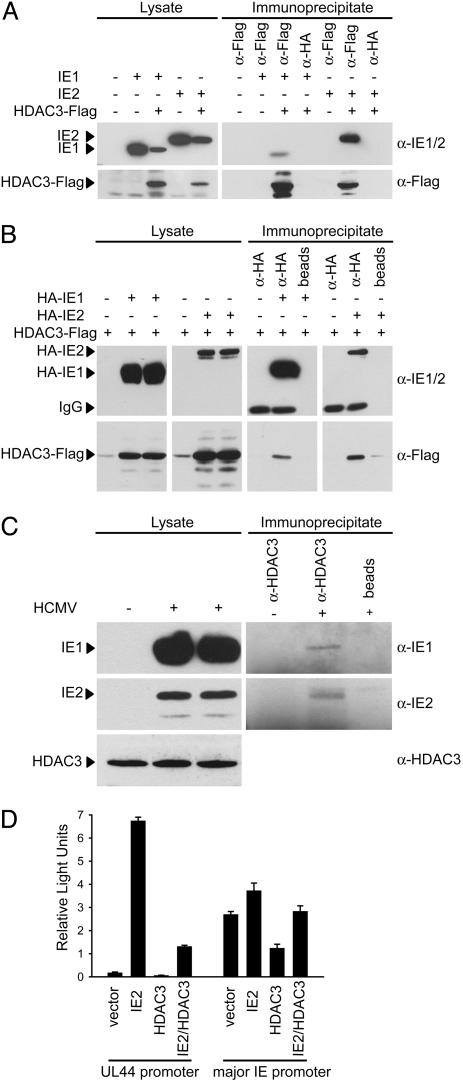
Interaction of IE1 and IE2 with HDAC3. To investigate whether IE1 might antagonize histone deacetylation through complex formation with cellular HDACs, we performed coimmunoprecipitation experiments. As a control, we included the IE2 protein in these assays. We searched for interactions between the viral proteins and HDAC3 because its ectopic expression was previously shown to inhibit HCMV infection (16), and our results demonstrated that it interfered with activity of the major IE and UL44 promoters (Fig. 3C). After cotransfection of expression plasmids encoding IE1 and Flag-tagged HDAC3, the viral protein was coprecipitated from cell extracts with a Flag-specific Ab, but not with a control Ab (Fig. 5A). In the absence of the Flag-HDAC3 protein, IE1 was not detected in Flag-specific immunoprecipitates. IE2 protein was also found to interact with epitope-tagged HDAC3 in this experiment. Relative to the input protein levels (lysate, Fig. 5A), a significantly larger proportion of IE2 was captured (immunoprecipitate, Fig. 5A) with the cellular enzyme as compared to IE1. In similar experiments, cells were cotransfected with plasmids expressing HA-tagged variants of IE1 or IE2 and Flag-tagged HDAC3, and proteins were immunoprecipitated with an HA-specific Ab. The HDAC3 protein was coprecipitated with both HCMV proteins, and again binding to IE2 was more efficient than to IE1 (Fig. 5B, compare lysate to immunoprecipitate). Next, we tested whether the interaction of IE1 and IE2 with endogenous HDAC3 could be detected in extracts of HCMV-infected cells. Ab to HDAC3 coprecipitated both IE1 and IE2, and, again, a higher proportion of IE2 than IE1 in the infected cell extract was immunoprecipitated with the HDAC Ab (Fig. 5C, compare lysate to immunoprecipitate). In sum, our results demonstrate that IE1 and IE2 interact with HDAC3, both in transfected cells where the proteins are over expressed and within HCMV-infected cells.
Fig. 5.
Physical interaction of IE1 and IE2 with HDAC3 in vivo.(A) Coimmunoprecipitation of IE1 or IE2 with epitope-tagged HDAC3 from transfected cells. H1299 cells were transfected with pcDNA-IE1, pcDNA-IE2, pcDNA-HDAC3-Flag, or empty vector as indicated, and immunoprecipitations were performed by using an anti-Flag agarose conjugate or a nonspecific agarose-Ab conjugate. Proteins from immunoprecipitates or whole-cell lysates were detected by Western blotting. (B) Coimmunoprecipitation of epitope-tagged HDAC3 with tagged IE1 or IE2 from transfected cells. H1299 cells were transfected with pCGN-IE1, pCGN-IE2, pcDNA-HDAC3-Flag, or empty vector as indicated, and immunoprecipitations were performed by using an anti-HA agarose conjugate or Sepharose beads without Ab, followed by Western blotting. IgG, Ig heavy chains. (C) Coimmunoprecipitation of IE1 or IE2 with endogenous HDAC3 from infected cells. MRC-5 cells were infected with HCMV (Towne) at a multiplicity of 5 TCID50 per cell, as indicated, and immunoprecipitations were performed by using an anti-HDAC3 mAb or empty beads. Proteins from immunoprecipitates or whole-cell lysates were detected by Western blotting. (D) Luciferase assays from cells cotransfected with pGL3-ICP36 (UL44) or pGL3-MIEP (major IE) and empty vector, pcDNA-IE2, pcDNA-HDAC3-Flag, or combinations thereof, as indicated. Transfections were performed in triplicate, and bars represent mean relative light units with SEs (× 103 for the UL44 promoter and × 106 for the major IE promoter).
Finally, given the association of IE2 with the HDAC, we tested whether IE2 could activate HDAC3-repressed UL44 and major IE promoters (Fig. 5D). Both repressed promoters were activated by IE2, consistent with the view that IE2 can bind and block the activity of HDAC3.
Discussion
IE1 promotes viral transcription by antagonizing histone deacetylation. This conclusion rests on three observations. First, HDAC inhibitors complemented the growth of an IE1-deficient mutant HCMV, but did not enhance the replication of WT virus (Figs. 1 and 2). Second, as reported (7, 8), the expression of viral mRNAs was reduced in mutant virus-infected cells (Fig. 3A), and, consistent with the effect of HDAC inhibitors, the major IE and UL44 promoters were found to be hypoacetylated in IE1-deficient mutant as compared to WT virus-infected cells (Fig. 4). Third, IE1 and HDAC3 can be coimmunoprecipitated from extracts of transfected (Fig. 5 A and B) and virus-infected cells (Fig. 5C). Presumably, IE1 interacts with HDAC3 in such a way as to inhibit its activity. A role for IE1 in the regulation of HDAC activity fits well with its localization to cellular chromatin (9, 11–13). In addition to the 72-kDa IE1 protein, mRNA splice variants give rise to three smaller IE1 proteins comprised of portions of the 72-kDa protein, and two of these variants inhibit HCMV gene expression, when ectopically produced from transfected plasmids (28). It is conceivable that this inhibition acts from a dominant-negative effect of the smaller IE1 proteins, antagonizing the ability of the 72-kDa IE1 protein to block HDAC function.
Although the case is not as strong as for IE1, it is very likely that IE2 also antagonizes histone deacetylation. IE2 and HDAC3 can be coimmunoprecipitated from extracts of transfected and infected cells (Fig. 5 A–C) and, consistent with the interaction, IE2 can overcome HDAC3-mediated inhibition of the major IE and UL44 promoters in reporter assays (Fig. 5D).
The IE1 protein of murine cytomegalovirus has been previously shown to interact with HDAC2 (23). Like HCMV IE1, the murine virus IE1 protein is an IE transcriptional activator protein, but the two proteins are not related in their sequence. We tested whether HCMV IE1 and HDAC2 can be coimmunoprecipitated, and we could not detect an interaction (data not shown). Consequently, although the two viruses both target HDACs, the IE1 proteins from the human and murine viruses appear to differ in the details of their activities. The replication of other herpes viruses also has been shown to be controlled by histone acetylation. For example, HDAC inhibitors accelerate viral mRNA accumulation in ICP0-deficient herpes simplex virus type 1 (29), and ICP0 binds to HDAC4, -5, and -7 (30).
We do not yet know whether IE1 and IE2 interact with HDACs other than HDAC3 or whether they target the same or different subsets of HDACs. IE1 and IE2 mRNAs are derived from the same transcription unit by differential splicing, and the two proteins share 85 amino-terminal amino acids encoded by two common exons. It is possible that this common region contains the domain responsible for interaction with HDACs, making it likely that the two proteins would target the same set of HDACs. We also do not know whether IE1 and IE2 interact directly or indirectly with HDAC3. Both proteins contain zinc-finger-like motifs located outside of their common domains, and these motifs might sponsor direct HDAC interactions because zinc finger domains are responsible for the interaction of nuclear receptors with HDAC3 (31). IE1 binds the PML protein (11) and PML binds several HDACs, including HDAC2 and HDAC3 (32), providing a potential mechanism for an indirect IE1-HDAC3 interaction. However, Tang and Maul (23) have ruled out this possibility for murine cytomegalovirus IE1 by showing that the HDAC2 interaction occurs in PML–/–cells.
HC toxin restored CR208 replication to WT levels (Fig. 1C). Because an IE1-null mutant can be fully complemented by an HDAC inhibitor, it is possible that modulation of HDAC function might be the sole, primary biochemical function of IE1. However, IE1 has been reported to physically interact with constituents of the basal transcription factor TFIID and transcription factors E2F, CTF-1, and Sp1 (33–36). Perhaps, these interactions serve to target IE1's HDAC inhibitory activity to the vicinity of the cellular transcription factors, and this localization, in turn, influences function of the targeted factor. Alternatively, HC toxin might be a more potent HDAC inhibitor than IE1, and its global effect on histone acetylation might compensate for the loss of additional IE1 functions.
The IE2 protein has been shown to bind the p300- and CBP-associated factor (P/CAF) histone acetyltransferase, potentiating its ability to activate transcription (37). Here, we demonstrate that IE1, and likely IE2, interacts with and inhibits HDAC3. Chromatin remodeling is a major mechanism by which HCMV IE proteins activate transcription.
Acknowledgments
We thank R. Greaves (Imperial College, London), E. Mocarski, G. Nolan, and E. Verdin for generous gifts of reagents, H. H. Niller for critical comments, S. Krauss and J. Kaps for experimental help, and H. Wolf for continuous support. This work was supported by National Institutes of Health Grant CA85786 (to T.S.) and a Deutsche Forschungsgemeinschaft Emmy Noether Fellowship NE 791/1-1 (to M.N.).
Author contributions: M.N., C.P., and T.S. designed research; M.N. and C.P. performed research; M.N. and C.P. analyzed data; and M.N., C.P., and T.S. wrote the paper.
Abbreviations: HCMV, human cytomegalovirus; IE, immediate-early; HDAC, histone deacetylase; HC toxin, Helminthosporium carbonum toxin; TSA, trichostatin A; TCID50, tissue culture 50% infective dose; HA, hemagglutinin.
References
- 1.Pass, R. F. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Strauss, S. E. (Lippincott, Philadelphia), Vol. 2, pp. 2675–2705. [Google Scholar]
- 2.Mocarski, E. S. & Courcelle, C. T. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Strauss, S. E. (Lippincott, Philadelphia), Vol. 2, pp. 2629–2673. [Google Scholar]
- 3.Castillo, J. P. & Kowalik, T. F. (2002) Gene 290, 19–34. [DOI] [PubMed] [Google Scholar]
- 4.Marchini, A., Liu, H. & Zhu, H. (2001) J. Virol. 75, 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heider, J. A., Bresnahan, W. A. & Shenk, T. E. (2002) Proc. Natl. Acad. Sci. USA 99, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mocarski, E. S., Kemble, G. W., Lyle, J. M. & Greaves, R. F. (1996) Proc. Natl. Acad. Sci. USA 93, 11321–11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greaves, R. F. & Mocarski, E. S. (1998) J. Virol. 72, 366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gawn, J. M. & Greaves, R. F. (2002) J. Virol. 76, 4441–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafemina, R. L., Pizzorno, M. C., Mosca, J. D. & Hayward, G. S. (1989) Virology 172, 584–600. [DOI] [PubMed] [Google Scholar]
- 10.Korioth, F., Maul, G. G., Plachter, B., Stamminger, T. & Frey, J. (1996) Exp. Cell Res. 229, 155–158. [DOI] [PubMed] [Google Scholar]
- 11.Ahn, J. H., Brignole, E. J., III, & Hayward, G. S. (1998) Mol. Cell. Biol. 18, 4899–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson, G. W., Kelly, C., Sinclair, J. H. & Rickards, C. (1998) J. Gen. Virol. 79, 1233–1245. [DOI] [PubMed] [Google Scholar]
- 13.Nevels, M., Brune, W. & Shenk, T. (2004) J. Virol. 78, 7803–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kierszenbaum, A. L. & Huang, E. S. (1978) J. Virol. 28, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St. Jeor, S., Hall, C., McGaw, C. & Hall, M. (1982) J. Virol. 41, 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, J. C., Fischle, W., Verdin, E. & Sinclair, J. H. (2002) EMBO J. 21, 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizuka, M. & Smith, M. M. (2003) Curr. Opin. Genet. Dev. 13, 154–160. [DOI] [PubMed] [Google Scholar]
- 18.Carrozza, M. J., Utley, R. T., Workman, J. L. & Cote, J. (2003) Trends Genet. 19, 321–329. [DOI] [PubMed] [Google Scholar]
- 19.Thiagalingam, S., Cheng, K. H., Lee, H. J., Mineva, N., Thiagalingam, A. & Ponte, J. F. (2003) Ann. N.Y. Acad. Sci. 983, 84–100. [DOI] [PubMed] [Google Scholar]
- 20.Zhu, H., Shen, Y. & Shenk, T. (1995) J. Virol. 69, 7960–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romanowski, M. J. & Shenk, T. (1997) J. Virol. 71, 1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva, M. C., Yu, Q. C., Enquist, L. & Shenk, T. (2003) J. Virol. 77, 10594–10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang, Q. & Maul, G. G. (2003) J. Virol. 77, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida, M., Kijima, M., Akita, M. & Beppu, T. (1990) J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]
- 25.Davie, J. R. (2003) J. Nutr. 133, 2485S–2493S. [DOI] [PubMed] [Google Scholar]
- 26.Brosch, G., Ransom, R., Lechner, T., Walton, J. D. & Loidl, P. (1995) Plant Cell 7, 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks, P. A., Richon, V. M. & Rifkind, R. A. (2000) J. Natl. Cancer Inst. 92, 1210–1216. [DOI] [PubMed] [Google Scholar]
- 28.Awasthi, S., Isler, J. A. & Alwine, J. C. (2004) J. Virol. 78, 8191–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon, A. P., Liang, Y. & Roizman, B. (2003) J. Virol. 77, 12671–12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomonte, P., Thomas, J., Texier, P., Caron, C., Khochbin, S. & Epstein, A. L. (2004) J. Virol. 78, 6744–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franco, P. J., Li, G. & Wei, L. N. (2003) Mol. Cell. Endocrinol. 206, 1–12. [DOI] [PubMed] [Google Scholar]
- 32.Wu, W. S., Vallian, S., Seto, E., Yang, W. M., Edmondson, D., Roth, S. & Chang, K. S. (2001) Mol. Cell. Biol. 21, 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayhurst, G. P., Bryant, L. A., Caswell, R. C., Walker, S. M. & Sinclair, J. H. (1995) J. Virol. 69, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolis, M. J., Pajovic, S., Wong, E. L., Wade, M., Jupp, R., Nelson, J. A. & Azizkhan, J. C. (1995) J. Virol. 69, 7759–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukac, D. M., Harel, N. Y., Tanese, N. & Alwine, J. C. (1997) J. Virol. 71, 7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yurochko, A. D., Mayo, M. W., Poma, E. E., Baldwin, A. S., Jr., & Huang, E. S. (1997) J. Virol. 71, 4638–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryant, L. A., Mixon, P., Davidson, M., Bannister, A. J., Kouzarides, T. & Sinclair, J. H. (2000) J. Virol. 74, 7230–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]