Abstract
Cortical dysplasia accounts for at least 14% of epilepsy cases, and is mostly seen in children. However, the understanding of molecular mechanisms and pathogenesis underlying cortical dysplasia is limited. The aim of this cross-sectional study is to identify potential key molecules in the mechanisms of cortical dysplasia by screening the proteins expressed in brain tissues of childhood cortical dysplasia patients with epilepsy using isobaric tags for relative and absolute quantitation-based tandem mass spectrometry compared to controls, and several differentially expressed proteins that are not reported to be associated with cortical dysplasia previously were selected for validation using real-time polymerase chain reaction, immunoblotting and immunohistochemistry. 153 out of 3340 proteins were identified differentially expressed between childhood cortical dysplasia patients and controls. And FSCN1, CRMP1, NDRG1, DPYSL5, MAP4, and FABP3 were selected for validation and identified to be increased in childhood cortical dysplasia patients, while PRDX6 and PSAP were identified decreased. This is the first report on differentially expressed proteins in childhood cortical dysplasia. We identified differential expression of FSCN1, CRMP1, NDRG1, DPYSL5, MAP4, FABP3, PRDX6 and PSAP in childhood cortical dysplasia patients, these proteins are involved in various processes and have various function. These results may provide new directions or targets for the research of childhood cortical dysplasia, and may be helpful in revealing molecular mechanisms and pathogenesis and/or pathophysiology of childhood cortical dysplasia if further investigated.
Introduction
Cortical dysplasia is a common cause of epilepsy and accounts for at least 14% of epilepsy cases [1], among whom more than 40% were refractory epilepsy [2]. It happens mostly in childhood [3]. Although previous researches revealed several genetic and acquired causes of childhood cortical dysplasia (CCD) and the mechanisms of its epileptogenesis [4], our understanding of molecular mechanisms and pathogenesis underlying CCD with epilepsy is still limited.
In previous studies, only a few analyzed the proteomics of epilepsy patients using brain tissues or cerebrospinal fluid [5, 6, 7]. However, the proteomics or transcriptomics of CCD with epilepsy has not been analyzed, especially in brain tissues of CCD patients. Isobaric tags for relative and absolute quantitation (iTRAQ) is a comparative proteomic approach that can analyze up to 8 samples in one experiment, and is widely used in proteomic researches in different diseases [8, 9]. Moreover, This study screened the differentially expressed proteins in brain tissues of CCD patients with epilepsy compared to traumatic intracranial hypertension (TIH) patients using iTRAQ-based tandem mass spectrometry and selected several proteins that are differentially expressed or unreported associated with CCD previously for validation using real-time quantitative polymerase chain reaction (qPCR) analysis, immunoblotting and immunohistochemistry. Our result suggests that 153 out of 3340 proteins were diffrentially expressed in patients with CCD compared to controls, and these proteins are mainly involved in mechanisms of catalytic activity, binding, molecule-structuring activity, transporter activity, and enzyme regulation activity. Among these 153 proteins, 8 proteins that have not been associated with CCD, but participate in CCD-related biological processes or have CCD-related molecular functions according to Gene Oncology, including NDRG1, FSCN1, FABP3, DPYSL5, PSAP, MAP4, CRMP1, and PRDX6 were selected and validated.
Materials and methods
Patients and tissue preparation
The study protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University (2013–026), and the study was conducted according to the principle expressed in the Declaration of Helsinki. Written consents were obtained from patients and/or their legal guardians. No author but S. L. and H. Y. had access to information that could identify individual participants during data collection, while no author had access to such information after data collection.
All the included CCD patients were recruited from 2013 August to 2014 December, and were preoperatively assessed with detailed history, neurological examination, neuropsychological test, ictal and interictal electroencephalography and MRI together with intraoperative electrocorticography for diagnosis and localization of lesions. After surgery, brain tissues from CCD patients were diagnosed by neuropathologists according to consensus of International League Against Epilepsy [10]. All included TIH patients underwent surgery had no history of neurological diseases, and the resected brain tissues were histologically normal. The resected tissues were immediately immersed into liquid nitrogen and strored at -80°C. Brain tissues of sex- and age-matched 8 CCD patients and 8 controls were selected for iTRAQ, brain tissues from the rest 15 CCD patients and 15 controls were used for qPCR, immunoblotting and immunohistochemistry.
Sample preparation and iTRAQ reagents labeling
Total proteins were extracted with iTRAQ lysis buffer. The concentration of proteins was measured using 2-D Quant Kit (Amersham Biosciences, Uppsala, Sweden). Equal amount of proteins from each group were mixed. The pooled samples were subjected to iTRAQ labeling according to the iTRAQ kit protocol (Applied Biosystems, Framingham, MA, USA). Briefly, 2 μl reducing reagent was added to 200 μg protein and centrifuged, then it was incubated at 37°C for 1 hour. 1 μl of Cysteine-Blocking Reagent was added for cysteine blocking. Each protein sample was digested into peptide with 4 μg Trypsin overnight at 37°C. iTRAQ reagents were dissolved in isopropanol, and then mixed with the corresponding sample followed by incubation at room temperature for 3 hours. Samples from CCD patients were labeled with 118 tag and 121 tag, and samples from controls were labeled with 117 tag and 119 tag. All the iTRAQ reagent-labeled samples were then combined. [11, 12]
Peptide fractionation with Isoelectric Focusing (IEF)
The labeled peptides samples were fractionated by IEF on immobilized pH gradient as described previously [13–15]. Briefly, the labeled peptides were dissolved in urea and Pharmalyte solution, applied to IPG strips (pH 3–10), and then focused with an IPGphor system (GE Healthcare Life Sciences Amersham Biosciences, Pittsburg, PA, USA) at 68 kVh. The IPG strips were cut into 36 pieces (0.5cm per piece). Peptides in each pieces was extracted with 0.1% formic acid and 2% acetonitrile and lyophilized and desalted with a C18 Discovery DSC-18 SPE column (Sigma-Aldrich). The desalted peptides were lyophilized again and stored at -20°C for mass spectrometry analysis.
Mass spectrometry and gene oncology analysis
Mass spectrometry was performed with liquid chromatography coupled inline to a QStar mass spectrometer (Applied Biosystems, Framingham. MA, USA). Desalted peptides were reconstituted in a solution containing 0.1% formic acid and 2% acetonitrile, half of which was delivered into a trap column by an online capillary liquid chromatography system (Dionex Ultimate 3000, Amsterdam, The Netherlands). The peptide mixture were automatically separated on a C18-PepMap column (ThermoFisher Dionnex, Sunnyvale, CA, USA) at 0.3 μl/min. The eluent was analysed by OStar Elite Hybrid ESI Quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Framingham. MA, USA) in an information-dependent acquisition mode. Mass spectrometer data acquisition was performed in the positive ion mode, with a selected mass range of 300–1800 m/z. A setting of 2 s was used as the total time for MS/MS events. The two charged peptides which were most abundant, with more than 20 counts, were selected for MS/MS and dynamically excluded for 30 s with ± 50 mDa mass tolerance.
Peptide identification and quantification was performed by ProteinPilot software (Applied Biosystems, Framingham, MA, USA). The search was performed using the International Protein Index (IPI) human database v3.87. Cysteine modification by MMTS was specified as a fixed modification [16].
For protein identification, a generally accepted standard which has been widely used in identifying and quantifying proteins with iTRAQ was taken [17–20]. The protein threshold was set to achieve 95% confidence, False discovery rate (FDR) statistics and 1.3-fold change cut-offs were used to classify the protein expressions as up-regulated (FDR<0.05 and iTRAQ fold-changes above 1.3) or down-regulated (FDR<0.05 iTRAQ and fold-changes below 0.77). For technical variation, while an analysis of repeated iTRAQ experiments established the technical variability to be not more than 30%. The gene oncology of each differentially expressed proteins was searched and classified using PANTHER classification system (www.pantherdb.org). 8 differentially expressed proteins which have not been reported associated with CCD, but may participate in CCD-related biological processes or have CCD-related molecular functions according to previous literature were selected for further validation.
Real-time qPCR analysis
Total RNA was extracted using Trizol (Thermofisher, Waltham, USA). Extracted RNA was reverse transcribed into cDNA by A3500 Reverse Transcription System (Promega, Madison, WI, USA). qPCR was performed using TaqMan GeneExpression Kit in ABI 7900HT system. The sequences of primers (OriGene Technologies, Inc. Rockville, USA) were NDRG1 (HP209104), FSCN1 (HP206673), FABP3 (HP207465), DPYSL5 (HP213501), CRMP1 (HP232913), PRDX6 (HP208150), PSAP (HP231407), MAP4 (HP206072), and β-Actin (HP204660). The mRNA expression level were analyzed using ΔΔCt method.
Immunoblotting analysis
Total proteins were extracted with RIPA Lysis Buffer and the concentrations were determined with BCA Kit (Beyotime, Haimen, China). Protein samples were loaded to 10% SDS-PAGE gel for electrophoresis and transferred to PVDF membranes. The membranes were incubated in 0.4% gelatin for 1 hour at room temperature and then incubated in primary antibodies (CRMP1, DPYSL5, FSCN1, NDRG1, PRDX6) (1:1000–1:10000 dilution, Abcam, Cambridge, UK) at 4°C overnight. After washed with TBST buffer, the membranes were incubated in HRP-conjugated secondary antibody (1:5000 dilution) for 1h at room temperature. The protein bands were visualized using ECL detection system (Millipore, Germany) and analyzed using Quantity One software (Bio-Rad Laboratories, Hercules, CA, USA).
Immunohistochemistry
Brain tissues from CCD patients and controls were fixed with 4% paraformaldehyde overnight at 4°C, then routinely embedded in paraffin and sectioned. After dewaxing and rehydration, the sections were boiled in citrate buffer (pH 6.0) in microwave oven for 20 min for antigen retrieval. Endogenous peroxidase activity was quenched by treatment with 3% H2O2 for 10 min. The sections were blocked with goat serum for 30 min and incubated at 4oC overnight with primary antibodies (CRMP1 1:250, DPYSL5 1:100 and FSCN1 1:250). Then sections were incubated in goat-anti-rabbit secondary antibodies (30 min, 37°C) and visualized using DAB (ZSGB-Bio, Beijing, China). After counterstain with hematoxylin and dehydrated, sections were evaluated under microscope. A semi-quantitative score was applied to the images obtained from the microscope inspection. The staining intensity ranged from 1 to 3 and the percentage of positive cells was measured manually in a range from 0 to 100%. Staining intensity (1–3) was multiplied by the percentage of positive cells (0–100) to obtain a final score ranging from 0 to 300 [21]. This method was repeated three times for the immunohistochemistry of each antibody in the brain tissue samples.
Statistical analysis
Data was expressed as mean±SD, FDR statistics was used to identified differentially expressed proteins, FDR<0.05 was considered statistically significant. Intergroup differences in immunoblotting and immunohistochemistry between the CCD group and the control group were analyzed using t test or rank sum test. p<0.05 was considered statistically significant.
Results
Demographics and clinical characters of patients
23 CCD patients (11 female, age 6.96±3.70, disease course 2.76±1.61 years) and 23 TIH patients (8 Female, age 7.22±3.10 years) who underwent surgery were included in this study. (Table 1)
Table 1. Patient demographics and clinical characteristics.
| Characteristic | CCD group | Controls |
|---|---|---|
| Age (year), mean ± SD | 6.96±3.70 | 7.22±3.10 |
| Sex (male/female) | 12/11 | 15/8 |
| Diagnosis | CCD with epilepsy | Trauma |
| Tissue pathology | Cortical dysplasia | Normal |
| Resection tissue | Neocortex | Neocortex |
CCD, Childhood cortical dysplasia.
Differentially expressed proteins revealed by iTRAQ analysis
Brain tissues of 16 randomly selected patients (n = 8 for each group) were analyzed with iTRAQ. In total, 3440 proteins were found with 95% confidence, among which 153 proteins were differentially expressed (FDR<0.05), including 64 up-regulated and 89 down-regulated proteins (Tables 2 & 3).
Table 2. 64 up-regulated proteins in childhood cortical dysplasia patients with epilepsy compared to controls by iTRAQ.
| Accession | Gene Name | Protein | 118:117 | 121:119 |
|---|---|---|---|---|
| IPI:IPI00478003.3 | A2M | Alpha-2-macroglobulin | 6.607 | 6.546 |
| IPI:IPI00335509.3 | DPYSL5 | Dihydropyrimidinase-related protein 5 | 5.152 | 4.966 |
| IPI:IPI00215801.1 | RBM39 | Isoform 2 of RNA-binding protein 39 | 4.966 | 4.246 |
| IPI:IPI00291932.1 | ACAN | Isoform 3 of Aggrecan core protein | 3.311 | 3.698 |
| IPI:IPI00923597.2 | NDRG1 | cDNA FLJ39243 fis, clone OCBBF2008283, highly similar to Protein NDRG1 | 3.631 | 3.597 |
| IPI:IPI00647915.1 | TAGLN2 | TAGLN2 24 kDa protein | 2.965 | 3.597 |
| IPI:IPI00218993.1 | HSPH1 | Isoform Beta of Heat shock protein 105 kDa | 3.436 | 3.532 |
| IPI:IPI00220213.2 | TNC | Isoform 4 of Tenascin | 3.342 | 3.404 |
| IPI:IPI00744780.2 | BCAS1 | Isoform 2 of Breast carcinoma-amplified sequence 1 | 3.767 | 3.311 |
| IPI:IPI00026237.1 | MAG | Myelin-associated glycoprotein | 3.467 | 3.251 |
| IPI:IPI00640953.1 | SIRT2 | Sirtuin-2 | 3.221 | 2.992 |
| IPI:IPI00219684.3 | FABP3 | Fatty acid-binding protein, heart | 2.992 | 2.831 |
| IPI:IPI00641181.5 | MARCKSL1 | MARCKS-related protein | 2.704 | 2.831 |
| IPI:IPI00415014.3 | MAP1LC3A | Isoform 1 of Microtubule-associated proteins 1A/1B light chain 3A | 3.221 | 2.729 |
| IPI:IPI00553211.1 | ERMN | Isoform 2 of Ermin | 2.729 | 2.704 |
| IPI:IPI00032958.3 | ANLN | Isoform 2 of Actin-binding protein anillin | 2.754 | 2.582 |
| IPI:IPI00298497.3 | FGB | Fibrinogen beta chain | 2.630 | 2.559 |
| IPI:IPI00295777.6 | GPD1 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic | 2.805 | 2.489 |
| IPI:IPI00556376.2 | CRMP1 | dihydropyrimidinase-related protein 1 isoform 1 | 2.630 | 2.489 |
| IPI:IPI00295469.5 | CPNE6 | cDNA FLJ55997, highly similar to Copine-6 | 2.399 | 2.270 |
| IPI:IPI00854567.3 | KIAA1598 | Isoform 2 of Shootin-1 | 2.312 | 2.249 |
| IPI:IPI00022463.1 | TF | Serotransferrin | 2.270 | 2.249 |
| IPI:IPI00173346.3 | PGM2L1 | Glucose 1,6-bisphosphate synthase | 1.803 | 2.188 |
| IPI:IPI00059135.1 | PPP1R14A | Isoform 1 of Protein phosphatase 1 regulatory subunit 14A | 2.606 | 2.148 |
| IPI:IPI00157414.3 | ENPP6 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 6 | 2.312 | 2.109 |
| IPI:IPI00396130.5 | SRCIN1 | Isoform 4 of SRC kinase signaling inhibitor 1 | 2.128 | 2.089 |
| IPI:IPI00856045.1 | AHNAK2 | Isoform 1 of Protein AHNAK2 | 2.089 | 2.089 |
| IPI:IPI00329719.1 | MYO1D | Myosin-Id | 2.291 | 2.070 |
| IPI:IPI00007702.1 | HSPA2 | Heat shock-related 70 kDa protein 2 | 2.070 | 2.070 |
| IPI:IPI00027223.2 | IDH1 | Isocitrate dehydrogenase [NADP] cytoplasmic | 2.070 | 2.051 |
| IPI:IPI00747810.2 | FSCN1 | FSCN1 protein (Fragment) | 2.270 | 1.977 |
| IPI:IPI00940816.2 | ARHGEF2 | Isoform 3 of Rho guanine nucleotide exchange factor 2 | 2.070 | 1.977 |
| IPI:IPI00021841.1 | APOA1 | Apolipoprotein A-I | 2.089 | 1.905 |
| IPI:IPI00878314.1 | MAP4 | 110 kDa protein | 2.070 | 1.905 |
| IPI:IPI00553177.1 | SERPINA1 | Isoform 1 of Alpha-1-antitrypsin | 2.148 | 1.888 |
| IPI:IPI00029111.3 | DPYSL3 | Collapsin response mediator protein 4 long variant | 1.941 | 1.871 |
| IPI:IPI00873622.3 | WDR1 | Putative uncharacterized protein WDR1 | 1.786 | 1.837 |
| IPI:IPI00045051.3 | PURB | Transcriptional activator protein Pur-beta | 1.941 | 1.803 |
| IPI:IPI00760925.2 | MYO18A | Isoform 3 of Myosin-XVIIIa | 1.820 | 1.786 |
| IPI:IPI00554737.3 | PPP2R1A | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | 1.500 | 1.770 |
| IPI:IPI00004560.1 | DCLK1 | Isoform 2 of Serine/threonine-protein kinase DCLK1 | 1.854 | 1.706 |
| IPI:IPI00022388.2 | DPYSL4 | Dihydropyrimidinase-related protein 4 | 1.837 | 1.706 |
| IPI:IPI00304409.3 | CARHSP1 | Calcium-regulated heat stable protein 1 | 1.706 | 1.690 |
| IPI:IPI00926256.1 | SLC4A1 | Band 3 anion transport protein | 1.629 | 1.690 |
| IPI:IPI00010133.3 | CORO1A | Coronin-1A | 1.820 | 1.660 |
| IPI:IPI00021766.5 | RTN4 | Isoform 1 of Reticulon-4 | 1.871 | 1.629 |
| IPI:IPI00218414.5 | CA2 | Carbonic anhydrase 2 | 1.871 | 1.629 |
| IPI:IPI00916847.1 | OLA1 | OLA1 47 kDa protein | 2.109 | 1.614 |
| IPI:IPI00186966.3 | BIN1 | Isoform IIA of Myc box-dependent-interacting protein 1 | 1.803 | 1.585 |
| IPI:IPI00965262.1 | CLASP2 | 166 kDa protein | 1.754 | 1.585 |
| IPI:IPI00185159.7 | BAIAP2 | Isoform 4 of Brain-specific angiogenesis inhibitor 1-associated protein 2 | 1.614 | 1.528 |
| IPI:IPI00940222.1 | AKAP12 | Isoform 3 of A-kinase anchor protein 12 | 1.600 | 1.500 |
| IPI:IPI00479514.2 | CACNA2D1 | Isoform 2 of Voltage-dependent calcium channel subunit alpha-2/delta-1 | 1.675 | 1.486 |
| IPI:IPI00017895.3 | GPD2 | Isoform 1 of Glycerol-3-phosphate dehydrogenase, mitochondrial | 1.600 | 1.486 |
| IPI:IPI00385612.2 | SLC8A2 | Putative uncharacterized protein DKFZp761D171 | 1.514 | 1.486 |
| IPI:IPI00294187.1 | PADI2 | Protein-arginine deiminase type-2 | 1.459 | 1.486 |
| IPI:IPI00455620.3 | RUFY3 | protein RUFY3 isoform 1 | 1.445 | 1.486 |
| IPI:IPI00942902.1 | GDA | Guanine deaminase | 1.472 | 1.472 |
| IPI:IPI00022774.3 | VCP | Transitional endoplasmic reticulum ATPase | 2.965 | 1.459 |
| IPI:IPI00910602.1 | NEFH | Isoform 1 of Neurofilament heavy polypeptide | 1.614 | 1.445 |
| IPI:IPI00465436.4 | CAT | Catalase | 1.644 | 1.380 |
| IPI:IPI00159927.2 | NCAN | Neurocan core protein | 1.675 | 1.330 |
| IPI:IPI00306667.5 | CNP | Isoform CNPII of 2',3'-cyclic-nucleotide 3'-phosphodiesterase | 1.393 | 1.318 |
| IPI:IPI00456623.2 | BCAN | Isoform 1 of Brevican core protein | 1.432 | 1.306 |
Table 3. 89 down-regulated proteins in childhood cortical dysplasia patients with epilepsy compared to controls by iTRAQ.
| Accession | Gene Name | Protein | 118:117 | 121:119 |
|---|---|---|---|---|
| IPI:IPI00480085.6 | DNM3 | Putative uncharacterized protein DNM3 | 0.711 | 0.738 |
| IPI:IPI00909720.1 | PSD3 | cDNA FLJ54694, highly similar to Pleckstrin and Sec7 domain-containing protein3 | 0.679 | 0.738 |
| IPI:IPI00219446.5 | PEBP1 | Phosphatidylethanolamine-binding protein 1 | 0.679 | 0.731 |
| IPI:IPI00418471.6 | VIM | Vimentin | 0.738 | 0.718 |
| IPI:IPI00789794.1 | DLG4 | disks large homolog 4 isoform 2 | 0.731 | 0.711 |
| IPI:IPI00024990.6 | ALDH6A1 | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | 0.745 | 0.705 |
| IPI:IPI00011515.1 | PACSIN1 | Protein kinase C and casein kinase substrate in neurons protein 1 | 0.745 | 0.698 |
| IPI:IPI00926312.1 | OGDH | oxoglutarate dehydrogenase isoform 3 precursor | 0.745 | 0.685 |
| IPI:IPI00031804.1 | VDAC3 | Isoform 1 of Voltage-dependent anion-selective channel protein 3 | 0.698 | 0.685 |
| IPI:IPI00007682.2 | ATP6V1A | V-type proton ATPase catalytic subunit A | 0.766 | 0.673 |
| IPI:IPI00003968.1 | NDUFA9 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | 0.649 | 0.673 |
| IPI:IPI00217871.4 | ALDH4A1 | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | 0.759 | 0.667 |
| IPI:IPI00954954.1 | CLU | clusterin isoform 3 | 0.752 | 0.667 |
| IPI:IPI00299402.1 | PC | Pyruvate carboxylase, mitochondrial | 0.679 | 0.667 |
| IPI:IPI00007812.1 | ATP6V1B2 | V-type proton ATPase subunit B, brain isoform | 0.711 | 0.661 |
| IPI:IPI00026216.4 | NPEPPS | Puromycin-sensitive aminopeptidase | 0.673 | 0.655 |
| IPI:IPI00386271.4 | SLC25A12 | Calcium-binding mitochondrial carrier protein Aralar1 | 0.698 | 0.649 |
| IPI:IPI00167215.6 | HEPACAM | Isoform 1 of Hepatocyte cell adhesion molecule | 0.619 | 0.637 |
| IPI:IPI00940744.1 | NDUFS1 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 0.608 | 0.637 |
| IPI:IPI00009439.1 | SYT1 | Synaptotagmin-1 | 0.738 | 0.625 |
| IPI:IPI00300568.4 | SYN1 | Isoform IA of Synapsin-1 | 0.614 | 0.625 |
| IPI:IPI00017855.1 | ACO2 | Aconitate hydratase, mitochondrial | 0.711 | 0.608 |
| IPI:IPI00219078.5 | ATP2A2 | Isoform 1 of Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 0.631 | 0.592 |
| IPI:IPI00847322.1 | SOD2 | superoxide dismutase 2, mitochondrial isoform A precursor | 0.766 | 0.586 |
| IPI:IPI00018342.5 | AK1 | Adenylate kinase isoenzyme 1 | 0.679 | 0.575 |
| IPI:IPI00009532.5 | ABAT | cDNA FLJ56034, highly similar to 4-aminobutyrate aminotransferase, mitochondrial | 0.597 | 0.570 |
| IPI:IPI00218660.3 | ITPR1 | Isoform 4 of Inositol 1,4,5-trisphosphate receptor type 1 | 0.470 | 0.570 |
| IPI:IPI00873201.1 | PSAP | Isoform Sap-mu-6 of Proactivator polypeptide | 0.457 | 0.565 |
| IPI:IPI00328156.9 | MAOB | Amine oxidase [flavin-containing] B | 0.619 | 0.555 |
| IPI:IPI00219219.3 | LGALS1 | Galectin-1 | 0.586 | 0.550 |
| IPI:IPI00746777.3 | ADH5 | Alcohol dehydrogenase class-3 | 0.515 | 0.550 |
| IPI:IPI00016801.1 | GLUD1 | Glutamate dehydrogenase 1, mitochondrial | 0.679 | 0.545 |
| IPI:IPI00941244.1 | AQP4 | 33 kDa protein | 0.673 | 0.545 |
| IPI:IPI00006663.1 | ALDH2 | Aldehyde dehydrogenase, mitochondrial | 0.643 | 0.545 |
| IPI:IPI00028520.2 | NDUFV1 | Isoform 1 of NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | 0.565 | 0.545 |
| IPI:IPI00946334.1 | NDUFS2 | dehydrogenase (ubiquinone) Fe-S protein 2 isoform 2 precursor | 0.530 | 0.545 |
| IPI:IPI00643720.3 | OGDHL | 2-oxoglutarate dehydrogenase-like, mitochondrial | 0.614 | 0.540 |
| IPI:IPI00023591.1 | PURA | Transcriptional activator protein Pur-alpha | 0.718 | 0.535 |
| IPI:IPI00009771.6 | LMNB2 | Lamin-B2 | 0.625 | 0.530 |
| IPI:IPI00015602.1 | TOMM70A | Mitochondrial import receptor subunit TOM70 | 0.597 | 0.530 |
| IPI:IPI00011229.1 | CTSD | Cathepsin D | 0.575 | 0.530 |
| IPI:IPI00013508.5 | ACTN1 | Alpha-actinin-1 | 0.530 | 0.530 |
| IPI:IPI00021088.1 | KCNAB2 | Isoform 1 of Voltage-gated potassium channel subunit beta-2 | 0.373 | 0.530 |
| IPI:IPI00383807.1 | SLC4A4 | Electrogenic Na+ bicarbonate cotransporter (Fragment) | 0.631 | 0.525 |
| IPI:IPI00479877.4 | ALDH9A1 | 4-trimethylaminobutyraldehyde dehydrogenase | 0.488 | 0.520 |
| IPI:IPI00413060.1 | SYNPO | Isoform 3 of Synaptopodin | 0.461 | 0.520 |
| IPI:IPI00004358.4 | PYGB | Glycogen phosphorylase, brain form | 0.685 | 0.515 |
| IPI:IPI00007087.4 | FBXO2 | F-box only protein 2 | 0.501 | 0.511 |
| IPI:IPI00008485.1 | ACO1 | Cytoplasmic aconitate hydratase | 0.711 | 0.506 |
| IPI:IPI00411706.1 | ESD | S-formylglutathione hydrolase | 0.479 | 0.501 |
| IPI:IPI00017704.3 | COTL1 | Coactosin-like protein | 0.466 | 0.501 |
| IPI:IPI00291175.7 | VCL | Isoform 1 of Vinculin | 0.457 | 0.501 |
| IPI:IPI00025796.3 | NDUFS3 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | 0.540 | 0.497 |
| IPI:IPI00657774.1 | STX1B | Syntaxin 1B alternative isoform deltaTM | 0.631 | 0.492 |
| IPI:IPI00220271.3 | AKR1A1 | Alcohol dehydrogenase [NADP+] | 0.555 | 0.492 |
| IPI:IPI00006579.1 | COX4I1 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | 0.511 | 0.488 |
| IPI:IPI00021812.2 | AHNAK | Neuroblast differentiation-associated protein AHNAK | 0.565 | 0.483 |
| IPI:IPI00418169.3 | ANXA2 | Isoform 2 of Annexin A2 | 0.413 | 0.453 |
| IPI:IPI00016077.1 | GBAS | Protein NipSnap homolog 2 | 0.550 | 0.441 |
| IPI:IPI00645031.1 | CRYL1 | Isoform 2 of Lambda-crystallin homolog | 0.483 | 0.441 |
| IPI:IPI00301180.4 | SLC12A5 | Isoform 2 of Solute carrier family 12 member 5 | 0.479 | 0.441 |
| IPI:IPI00027497.5 | GPI | Glucose-6-phosphate isomerase | 0.705 | 0.433 |
| IPI:IPI00872379.1 | ANXA5 | 36 kDa protein | 0.373 | 0.421 |
| IPI:IPI00010130.3 | GLUL | Glutamine synthetase | 0.649 | 0.417 |
| IPI:IPI00514285.2 | PTGDS | Prostaglandin D2 synthase 21kDa | 0.497 | 0.413 |
| IPI:IPI00946099.1 | SRI | Putative uncharacterized protein SRI | 0.328 | 0.409 |
| IPI:IPI00216138.6 | TAGLN | Transgelin | 0.302 | 0.409 |
| IPI:IPI00013043.1 | TPPP | Tubulin polymerization-promoting protein | 0.685 | 0.394 |
| IPI:IPI00219067.4 | GSTM2 | Glutathione S-transferase Mu 2 | 0.497 | 0.391 |
| IPI:IPI00302592.2 | FLNA | Isoform 2 of Filamin-A | 0.350 | 0.391 |
| IPI:IPI00005038.1 | HRSP12 | Ribonuclease UK114 | 0.406 | 0.387 |
| IPI:IPI00514424.4 | PPT1 | Palmitoyl-protein thioesterase 1 | 0.394 | 0.377 |
| IPI:IPI00641737.1 | HP | Haptoglobin | 0.429 | 0.356 |
| IPI:IPI00303568.3 | PTGES2 | Prostaglandin E synthase 2 | 0.429 | 0.356 |
| IPI:IPI00013698.3 | ASAH1 | N-acylsphingosine amidohydrolase (Acid ceramidase) 1, isoform CRA_c | 0.433 | 0.353 |
| IPI:IPI00604710.2 | SLC3A2 | Isoform 1 of 4F2 cell-surface antigen heavy chain | 0.488 | 0.347 |
| IPI:IPI00021828.1 | CSTB | Cystatin-B | 0.492 | 0.328 |
| IPI:IPI00002280.1 | PCSK1N | ProSAAS | 0.313 | 0.328 |
| IPI:IPI00413674.1 | PHYHD1 | Isoform 1 of Phytanoyl-CoA dioxygenase domain-containing protein 1 | 0.511 | 0.302 |
| IPI:IPI00515081.4 | IGSF1 | Isoform 2 of Immunoglobulin superfamily member 1 | 0.366 | 0.296 |
| IPI:IPI00423460.3 | IGHA1 | Putative uncharacterized protein DKFZp686G21220 (Fragment) | 0.233 | 0.273 |
| IPI:IPI00218487.3 | GJA1 | Gap junction alpha-1 protein | 0.283 | 0.268 |
| IPI:IPI00022143.3 | ESYT1 | Isoform 1 of Extended synaptotagmin-1 | 0.384 | 0.265 |
| IPI:IPI00156689.3 | VAT1 | Synaptic vesicle membrane protein VAT-1 homolog | 0.360 | 0.238 |
| IPI:IPI00011200.5 | PHGDH | D-3-phosphoglycerate dehydrogenase | 0.261 | 0.217 |
| IPI:IPI00027442.4 | AARS | Alanyl-tRNA synthetase, cytoplasmic | 0.437 | 0.209 |
| IPI:IPI00010800.2 | NES | Nestin | 0.077 | 0.099 |
| IPI:IPI00001734.3 | PSAT1 | Phosphoserine aminotransferase | 0.067 | 0.086 |
| IPI:IPI00220301.5 | PRDX6 | Peroxiredoxin-6 | 0.078 | 0.082 |
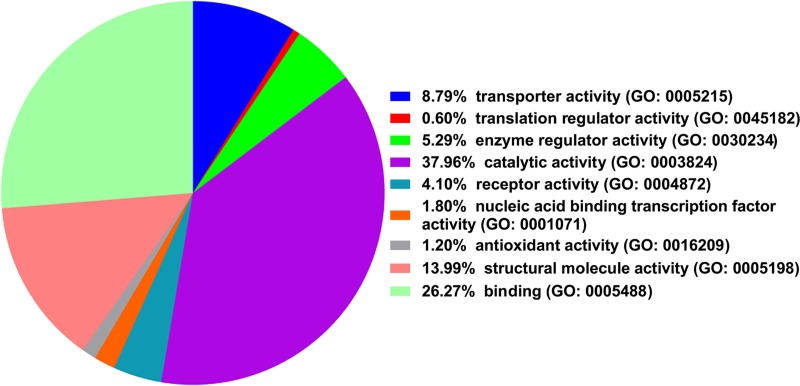
Using PANTHER classification system, the 153 proteins were divided into 9 functional categories including catalytic activity (38.0%), binding (26.3%), molecule-structuring activity (14.0%), transporter activity (8.8%), and enzyme regulation activity (5.3%) (Fig 1). 8 differentially expressed proteins, including FSCN1, CRMP1, NDRG1, DPYSL5, MAP4, FABP3, PRDX6 and PSAP were selected for further validation. The gene oncology terms of these 8 proteins were shown in S1 Table.
Fig 1. Molecule functional categories of 153 differentially exprssed proteins using the PANTHER Classification System.
qPCR in children having CCD with epilepsy
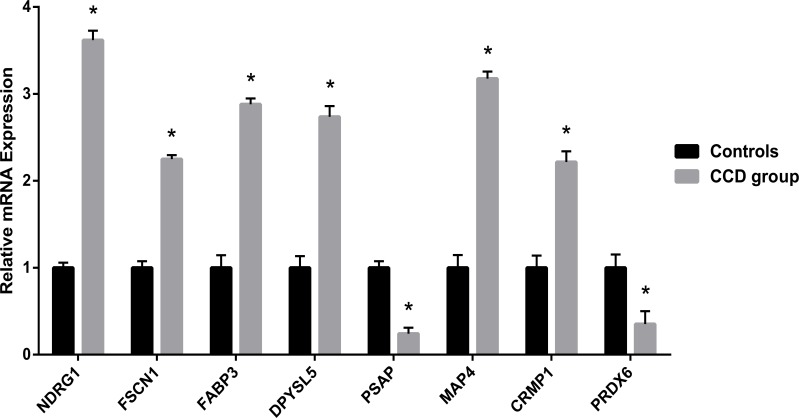
The levels of mRNA expression for fascin actin-bundling protein 1 (FSCN1), collapsin response mediator protein 1 (CRMP1), N-myc downstream regulated 1 (NDRG1), dihydropyrimidinase-related protein 5 (DPYSL5), peroxiredoxin 6 (PRDX6), prosaposin (PSAP), microtubule associated protein 4 (MAP4), and fatty acid binding protein 3 (FABP3) are presented in Fig 2. The expression of FSCN1, CRMP1, NDRG1, DPYSL5, MAP4, and FABP3 were found to be up-regulated in the CCD patients (Relative mRNA expression: CRMP1, 2.21±0.12; NDRG1, 3.61±0.11; DPYSL5, 2.73±0.12; MAP4, 3.17±0.08; FAPB3, 2.88±0.06. p<0.05 for each mRNA expression compared to controls), and the expression of PRDX6 and PSAP were down-regulated (Relative mRNA expression: PRDX6, 0.35±0.14; PSAP, 0.24±0.06. p<0.05 for both mRNA expression compared to controls) compared to controls.
Fig 2. Relative mRNA expression levels of NDRG1, FSCN1, FABP3, DPYSL5, PSAP, MAP4, CRMP1, PRDX6.
*p<0.05 compared to controls.
Immunoblotting
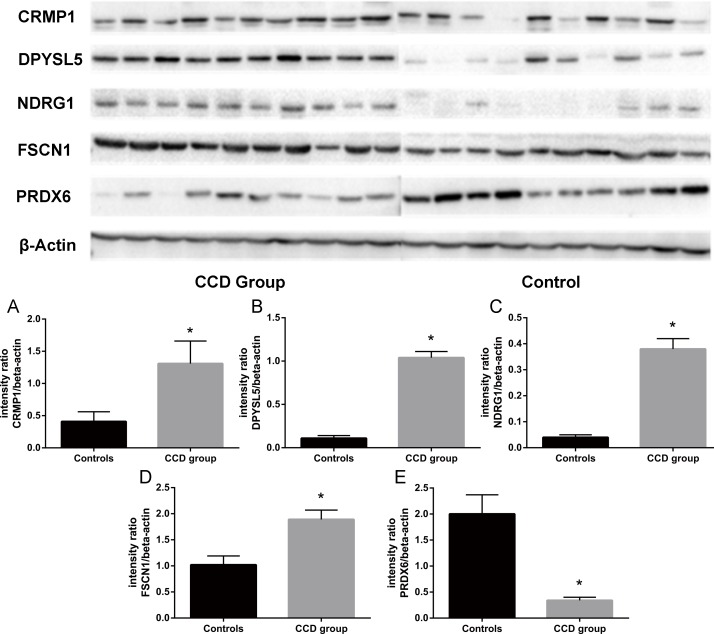
CRMP1, DPYSL5, FSCN1, NDRG1, PRDX6 were further validated with immunoblotting. In CCD patients, the protein levels of CRMP1 (CCD group: 1.31±0.35; Controls: 0.41±0.15. p<0.05), DPYSL5 (CCD group: 1.04±0.07; Controls: 0.11±0.03. p<0.05), FSCN1 (CCD group: 1.89±0.18; Controls: 1.02±0.17. p<0.05) and NDRG1 (CCD group: 0.38±0.04; Controls: 0.04±0.01. p<0.05) were increased, while the protein level of PRDX6 (CCD group: 0.34±0.06; Controls: 2.00±0.37. p<0.05) was decreased compared to controls (Fig 3).
Fig 3. Immunoblotting for CRMP1, DPYSL5, NDRG1, FSCN1 and PRDX6.
Quantification of protein levels showed increased expression of CRMP1 (A), DPYSL5 (B), NDRG1 (C) and FSCN1 (D) and decreased expression of PRDX6 (E) in brain tissue of childhood cortical dysplasia patients compared to controls. *p<0.05 compared to controls.
Immunohistochemistry
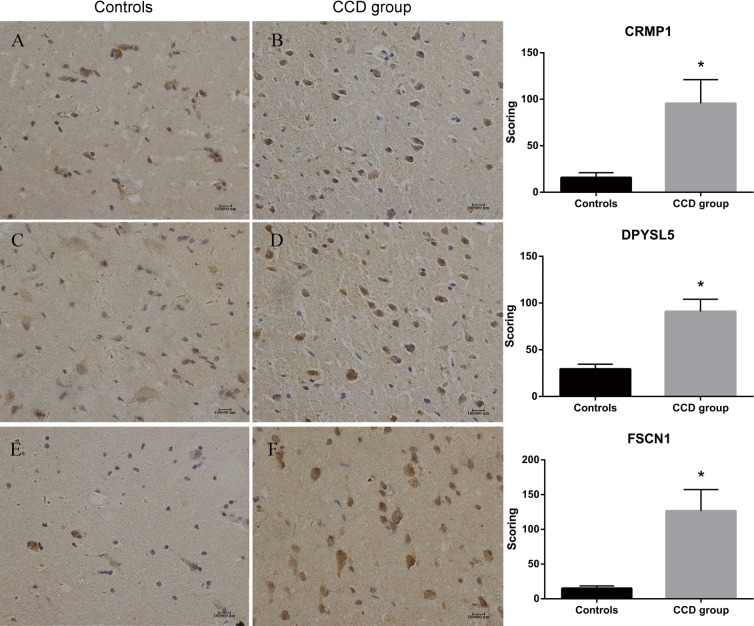
The expression of DPYSL5, CRMP1 and FSCN1 were further measured by IHC. There were increased immunoreactivity of CRMP1 (Scores: CCD group: 95.50±25.52; Controls: 15.76±5.28. p<0.05) (Fig 4), DPYSL5 (Scores: CCD group: 90.93±13.15; Controls: 29.33±5.21. p<0.05) (Fig 4), and FSCN1 (Scores: CCD group: 126.53±30.70; Controls: 15.06±3.45. p<0.05) (Fig 4) in the CDD group compared to controls.
Fig 4. Immunohistochemistry of CRMP1, DPYSL5 and FSCN1.
IHC score of CRMP1 (A & B), DPYSL5 (C & D) and FSCN1 (E & F) were significantly increased in brain tissues of childhood cortical dysplasia patients with epilepsy compared to controls. *p<0.05 compared to controls.
Discussion
In our study, 153 proteins were identified differentially expressed in brain tissues of CCD patients with epilepsy compared to controls using iTRAQ. According to the functional classification using PANTHER, the 153 differentially expressed proteins were divided into 9 categories, which were involved in activities of various biological process, including catalytic activity (38.0%), binding (26.3%), molecule-structuring activity (14.0%), transporter activity (8.8%), and enzyme regulation activity (5.3%).
Among the 153 proteins, the expression of FSCN1, CRMP1, NDRG1, DPYSL5, MAP4 and FABP3 was increased in the CCD patients compared to controls, while the expression of PRDX6 and PSAP was decreased in iTRAQ analysis. And these results were validated by real-time PCR, immunoblotting and immunohistochemistry.
FSCN1 is an actin-binding protein and can affect the formation and maintenance of cytoskeleton structure [22]. FSCN1 increases in neurogenesis and can help neurites maintain their normal shape, and it is considered as a candidate gene for developmental brain disorders [23, 24]. FSCN1-related pathways mainly participate in the migration of neurons, which was known as a key mechanism of cortical dysplasia [25]. So it is possible that, in our study, the increased level of FSCN1 indicates an abnormally enhanced neurogenesis, neurite outgrowth and neuronal migration, and thus, result in CCD and epilepsy. However, the effect of increased FSCN1 on neurons needs to be further researched.
Collapsin response mediator protein 1 (CRMP1) belongs to the collapsing response mediator protein family (CRMPs) which is involved in the Sema-3A signaling pathway [26–28], CRMP1 also regulates migration, neurite outgrouwth, and dendrite orientation of neurons, its loss can retard the radial migration and neurite outgrowth of neurons and lead to abnormal orientation of basal dendrites of neurons [29–31]. Similar to FSCN1, the increased level of CRMP may also possibly indicate an abnormally enhanced neuron migration and neurite outgrowth and abnormal orientation of dendrites, which may have roles in CCD. Interestingly, in temporal lobe epilepsy (TLE) patients and animal models, decreased CRMP1 expression was reported [32]. It is possible that CRMP1 plays different roles in CCD with epilepsy and TLE. The specific role of CRMP1 in CCD with epilepsy needs further evaluation.
NDRG1 is upregulated during cell differentiation, and its cellular distribution and molecular assembly changes with postnatal development, which is correlated with the maturation of brain [33]. NDRG1 exists in oligodendrocytes in cerebrum and decreases significantly at the end stage of myelin degradation [34, 35], and its mutation is found related to subcortical white matter abnormalities and severe demyelinating neuropathy [36]. Interestingly, in patients with cortical dysplasia, the change of oligodendrocytes and oligodendrocyte precursor cells is conflicting in previous reports [37, 38], and some patients with malformation of cortical development have reactive oligodendroglial hyperplasia [37]. These suggest complicated roles of oligodendrocytes and myelin sheath in cortical dysplasia. In our study, we found NDRG1 abnormally increased in CCD patients, which may suggest a possible mechanism of reactive oligodendroglial hyperplasia in CCD. However, whether NDRG1-mediated oligodendroglial change participate in the pathogenensis of CCD needs to be further illustrated.
Increased expression of DPYSL5 can regulate dendritic development by mediating BDNF signaling in the central nervous system and modulate the function of CRMP2 by interacting with tubulin [39, 40], thus affect the cytoskeleton remodeling, which is important in CCD with epilepsy. It has been reproted that BDNF, a neurotrophin, plays an important role in dendritic arborzation and synaptic neurotransmission [41–43], and CRMP2, a signaling molecule of Semaphoring-3A and a repulsive guidance cue, can induce growth cone collapse and regulate neuronal polarity [28], axon elongation and multiple axon formation [44, 45]. These suggest that DPYSL5 may function in CCD with epilepsy via affecting BDNF and CRMP2.
MAP4 exists in brain and many other organs, one of its isoforms was found neural cell specific and it can inhibit the movement of the microtubules in a concentration-dependent manner and reduce microtubule-stabilizing activity [46–48]. MAP4 is also known associated with epilepsy [49]. Notably, microtubule-associated proteins were known important in regulating neuronal migration and brain development [50]. Defects of neuronal migration can lead to cortical malformation and consequently cause severe intellectual disability and refractory epilepsy [51]. Therefore, the increase of MAP4, as in our study, may inhibit the movement and activity of microtubles and thus impair neuronal migration which participate in CCD.
FABP3 is considered as a promising and sensitive marker for minor brian injury and Creutzfeldt-Jakob disease [52, 53]. FABP3 expression is very low in neonatal brains and gradually increases after birth until adulthood, its expression pattern is correlated with synaptogenesis, myelinogenesis, neurite formation and synapse maturation [54]. FABP3 regulate the incorporation of arachidonic acid into brain, and may also regulate gene expression via controlling the availability of fatty acid ligands required for PPAR and RXR activity [54]. In our study, a increased FABP3 level was found in CCD patients, possibly indicating early maturity of metabolism pattern in CCD patients, which may contribute to the formation of cortical dysplasia. Moreover, FABP3 deficiency in mice showed protective effect against experimental autoimmune encephalomyelitis [55], indicating a possible role of autoimmune inflammation in CCD.
PRDX6 is an antioxidant protein which mainly exists in glia and keeps increasing as growing, it may have important roles in alzheimer’s disease and parkinson’s disease [56, 57]. PRDX6 can clear reactive oxygen species, regulate gene expression in brain and protect against oxidative stress-induced neuronal death [58]. Whether the reduction of PRDX6 in CCD patients is causal or consequential factor of CCD remains to be further illustrated. It is possible that, reduction of PRDX6 is a result of enhanced oxidative stress, which has been reported in previous study [58]. However, it is also possible that reduction of PRDX6 may contribute to the pathogenesis of CCD, because oxidative stress has been associated with developmental brain disorders and epileptogenesis, although the specific role of oxidative stress in the pathogenesis of cortical dysplasia remains to be illustrated [59, 60].
PSAP is precursor of saposin and acts as a lysosomal protein and a potent secreted neurotrophic factor, its temporal pattern of expression in perinatal brain indicate its potential role in brain development [61]. Infants with PSAP deficiency presented multifocal myoclonus and cyanotic hypoxemia immediately after birth, grand-mal epilepsy in the following days, and cortical and white matter morphogenetic disorders [62, 63]. This deficiency is considered to cause such manifestations via impairing the lipid storage[62, 63]. Therefore, abnormally reduction of PSAP in CCD patients may indicate a possible role of PSAP in the pathogenesis of CCD. Moreover, in kainate-induced epilepsy models, PSAP reactively increases and protects against the neurotoxicity [64]. Thus, PSAP reduction in CCD may also participate in the neuronal damage in CCD.
In conclusion, we identified 153 differentially expressed proteins in CCD patients compared to controls. Among these proteins, FSCN1, CRMP1, NDRG1, DPYSL5, MAP4, FABP3, PRDX6 and PSAP were further validated. These proteins have not been related to CCD before. Mechanisms including neuronal migration, neurite growth, cytoskeleton remodeling, inflammation, oligodendroglia hyperplasia, metabolic pattern and lipid storage may be involved in CCD pathogenesis and/or pathophysiology via these proteins, providing potential targets and directions for future researches on cortical dysplasia. Our study also indicate a complicated pathogenetic background of CCD, as these differentially expressed proteins have various cellular distribution and function. Moreover, further study is needed to illustrate the specific effects of these differentially expressed proteins on CCD with epilepsy, considering the limited sample size due to the critical criteria of surgery in CCD patients, especially in children.
Supporting information
(DOC)
(XLSX)
(DOCX)
Acknowledgments
This study is supported by National Natural Science Foundation of China (grant number: 81571259, receiver: YC), and Chongqing Municipal Public Health Bureau, Chongqing People's Municipal Government (grant number: 2015ZDXM011, receiver: LC).
We sincerely thank Professor Huaidong Hu and Professor Changlin Hu from The Second Affiliated Hospital of Chongqing Medical University for their advice and supports to this study. The authors declare that they have no conflict of interest.
The datasets generated and analysed during the current study are fully available in the supporting information (S1 Datasheet) attached to the manuscript, or via communication with the corresponding author.
Data Availability
The data underlying this study are available in the paper and its supporting information files. For any inquiries regarding the data, interested researchers may email the following address: lifen_chen@163.com.
Funding Statement
This study is supported by National Natural Science Foundation of China (grant number: 81571259, website of funder: http://www.nsfc.gov.cn/, receiver: YC), and Chongqing Municipal Public Health Bureau, Chongqing People's Municipal Government (grant number: 2015ZDXM011, website of funder: http://www.cqwsjsw.gov.cn/, receiver: LC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Crino PB, Chou K. Epilepsy and Cortical Dysplasias. Curr Treat Options Neurol. 2000; 2: 543–552. [DOI] [PubMed] [Google Scholar]
- 2.Sisodiya SM. Malformations of cortical development: burdens and insights from important causes of human epilepsy. Lancet Neurol. 2004; 3: 29–38. [DOI] [PubMed] [Google Scholar]
- 3.Lee SK, Kim DW. Focal cortical dysplasia and epilepsy surgery. J Epilepsy Res. 2013; 3:43–47. 10.14581/jer.13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaker T, Bernier A, Carmant L. Focal Cortical Dysplasia in Childhood Epilepsy. Semin Pediatr Neurol. 2016. May;23(2):108–19. 10.1016/j.spen.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 5.Mériaux C, Franck J, Park DB, Quanico J, Kim YH, Chung CK, et al. Human temporal lobe epilepsy analyses by tissue proteomics. Hippocampus. 2014. June;24(6):628–42. 10.1002/hipo.22246 [DOI] [PubMed] [Google Scholar]
- 6.Liu JY, Reeves C, Diehl B, Coppola A, Al-Hajri A, Hoskote C, et al. Early lipofuscin accumulation in Frontal Lobe Epilepsy. Ann Neurol. 2016. October 20. [DOI] [PubMed] [Google Scholar]
- 7.Xiao F, Chen D, Lu Y, Xiao Z, Guan LF, Yuan J, et al. Proteomic analysis of cerebrospinal fluid from patients with idiopathic temporal lobe epilepsy. Brain Res. 2009. February 19;1255:180–9. 10.1016/j.brainres.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Li H, Yang Y, Li S, Ren H, Zhang D, et al. Differential regulation of host genes including hepatic fatty acid synthase in HBV-transgenic mice. J Proteome Res. 2013; 12: 2967–2979. 10.1021/pr400247f [DOI] [PubMed] [Google Scholar]
- 9.Tong SW, Yang YX, Hu HD, An X, Ye F, Hu P, et al. Proteomic investigation of 5-fluorouracil resistance in a human hepatocellular carcinoma cell line. J Cell Biochem. 2012; 113: 1671–1680. 10.1002/jcb.24036 [DOI] [PubMed] [Google Scholar]
- 10.Blümcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, Palmini A, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011. January;52(1):158–74. 10.1111/j.1528-1167.2010.02777.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Li W, Hou Y, Niu Z, Zhong Y, Zhang Y, et al. Comparative membrane proteomic analysis between lung adenocarcinoma and normal tissue by iTRAQ labeling mass spectrometry. Am J Transl Res. 2014; 6: 267–280. [PMC free article] [PubMed] [Google Scholar]
- 12.See AL, Chong PK, Lu SY, Lim YP. CXCL3 is a potential target for breast cancer metastasis. Curr Cancer Drug Targets. 2014; 14: 294–309. [DOI] [PubMed] [Google Scholar]
- 13.Giorgianni F, Koirala D, Weber KT, Beranova-Giorgianni S. Proteome analysis of subsarcolemmal cardiomyocyte mitochondria: a comparison of different analytical platforms. Int J Mol Sci. 2014; 15: 9285–9301. 10.3390/ijms15069285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreda-Pineiro A, Garcia-Otero N, Bermejo-Barrera P. A review on preparative and semi-preparative offgel electrophoresis for multidimensional protein/peptide assessment. Anal Chim Acta. 2014; 836: 1–17. 10.1016/j.aca.2014.04.053 [DOI] [PubMed] [Google Scholar]
- 15.Schleicher TR, VerBerkmoes NC, Shah M, Nyholm SV. Colonization state influences the hemocyte proteome in a beneficial squid-Vibrio symbiosis. Mol Cell Proteomics. 2014; 13: 2673–2686. 10.1074/mcp.M113.037259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim S, Choong LY, Kuan CP, Yunhao C, Lim YP. Regulation of macrophage inhibitory factor (MIF) by epidermal growth factor receptor (EGFR) in the MCF10AT model of breast cancer progression. J Proteome Res. 2009; 8: 4062–4076. 10.1021/pr900430n [DOI] [PubMed] [Google Scholar]
- 17.Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol Cell Proteomics. 2008; 7: 853–863. 10.1074/mcp.M700251-MCP200 [DOI] [PubMed] [Google Scholar]
- 18.Gan CS, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J Proteome Res. 2007; 6: 821–827. 10.1021/pr060474i [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Choong LY, Lin Q, Philp R, Wong CH, Ang BK, et al. Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol Cell Proteomics. 2007; 6: 2072–2087. 10.1074/mcp.M700395-MCP200 [DOI] [PubMed] [Google Scholar]
- 20.Chong PK, Lee H, Zhou J, Liu SC, Loh MC, So JB, et al. Reduced plasma APOA1 level is associated with gastric tumor growth in MKN45 mouse xenograft model. J Proteomics. 2010; 73: 1632–1640. 10.1016/j.jprot.2010.04.005 [DOI] [PubMed] [Google Scholar]
- 21.Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK, et al. Novel breast cancer metastasis-associated proteins. J Proteome Res. 2009; 8: 583–594.31. 10.1021/pr8007368 [DOI] [PubMed] [Google Scholar]
- 22.Cohan CS, Welnhofer EA, Zhao L, Matsumura F, Yamashiro S. Role of the actin bundling protein fascin in growth cone morphogenesis: localization in filopodia and lamellipodia. Cell Motil Cytoskeleton. 2001; 48: 109–120. [DOI] [PubMed] [Google Scholar]
- 23.Kraft R, Escobar MM, Narro ML, Kurtis JL, Efrat A, Barnard K, et al. Phenotypes of Drosophila brain neurons in primary culture reveal a role for fascin in neurite shape and trajectory. J Neurosci. 2006. August 23;26(34):8734–47. 10.1523/JNEUROSCI.2106-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Megiorni F, Indovina P, Mora B, Mazzilli MC. Minor expression of fascin-1 gene (FSCN1) in NTera2 cells depleted of CREB-binding protein. Neurosci Lett. 2005. June 10–17;381(1–2):169–74. 10.1016/j.neulet.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 25.Bast T, Ramantani G, Seitz A, Rating D. Focal cortical dysplasia: prevalence, clinical presentation and epilepsy in children and adults. Acta Neurol Scand. 2006. February;113(2):72–81. 10.1111/j.1600-0404.2005.00555.x [DOI] [PubMed] [Google Scholar]
- 26.Veyrac A, Reibel S, Sacquet J, Mutin M, Camdessanche JP, Kolattukudy P, et al. CRMP5 regulates generation and survival of newborn neurons in olfactory and hippocampal neurogenic areas of the adult mouse brain. PLoS One. 2011; 6: e23721 10.1371/journal.pone.0023721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells. 2005; 10: 165–179. 10.1111/j.1365-2443.2005.00827.x [DOI] [PubMed] [Google Scholar]
- 28.Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995; 376: 509–514. 10.1038/376509a0 [DOI] [PubMed] [Google Scholar]
- 29.Yamashita N, Uchida Y, Ohshima T, Hirai S, Nakamura F, Taniguchi M, et al. Collapsin response mediator protein 1 mediates reelin signaling in cortical neuronal migration. J Neurosci. 2006;26(51):13357–133 62. 10.1523/JNEUROSCI.4276-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higurashi M, Iketani M, Takei K, Yamashita N, Aoki R, Kawahara N, et al. Localized role of CRMP1 and CRMP2 in neurite outgrowth and growth cone steering. Dev Neurobiol. 2012. December;72(12):1528–40. 10.1002/dneu.22017 [DOI] [PubMed] [Google Scholar]
- 31.Takaya R, Nagai J, Piao W, Niisato E, Nakabayashi T, Yamazaki Y, et al. CRMP1 and CRMP4 are required for proper orientation of dendrites of cerebral pyramidal neurons in the developing mouse brain. Genes Cells. 2016. September;21(9):994–1005. [DOI] [PubMed] [Google Scholar]
- 32.Luo J, Zeng K, Zhang C, Fang M, Zhang X, Zhu Q, et al. Down-regulation of CRMP-1 in patients with epilepsy and a rat model. Neurochem Res. 2012. July;37(7):1381–91. 10.1007/s11064-012-0712-6 [DOI] [PubMed] [Google Scholar]
- 33.Wakisaka Y, Furuta A, Masuda K, Morikawa W, Kuwano M, Iwaki T. Cellular distribution of NDRG1 protein in the rat kidney and brain during normal postnatal development. J Histochem Cytochem. 2003. November;51(11):1515–25. 10.1177/002215540305101111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuda T, Kokame K, Miyata T. Differential expression patterns of NDRG family proteins in the central nervous system. J Histochem Cytochem. 2008. February;56(2):175–82. 10.1369/jhc.7A7323.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruïne AP, Baldwin HS, et al. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J. 2010. November;24(11):4153–66. 10.1096/fj.09-151464 [DOI] [PubMed] [Google Scholar]
- 36.Echaniz-Laguna A, Degos B, Bonnet C, Latour P, Hamadouche T, Lévy N, et al. NDRG1-linked Charcot-Marie-Tooth disease (CMT4D) with central nervous system involvement. Neuromuscul Disord. 2007. February;17(2):163–8. 10.1016/j.nmd.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 37.Scholl T, Mühlebner A, Ricken G, Gruber V, Fabing A, Samueli S, et al. Impaired oligodendroglial turnover is associated with myelin pathology in Focal Cortical Dysplasia and Tuberous Sclerosis Complex. Brain Pathol. 2016. October 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepherd C, Liu J, Goc J, Martinian L, Jacques TS, Sisodiya SM, et al. A quantitative study of white matter hypomyelination and oligodendroglial maturation in focal cortical dysplasia type II. Epilepsia. 2013. May;54(5):898–908. 10.1111/epi.12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashita N, Mosinger B, Roy A, Miyazaki M, Ugajin K, Nakamura F, et al. CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells. J Neurosci. 2011; 31: 1773–1779. 10.1523/JNEUROSCI.5337-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brot S, Rogemond V, Perrot V, Chounlamountri N, Auger C, Honnorat J, et al. CRMP5 interacts with tubulin to inhibit neurite outgrowth, thereby modulating the function of CRMP2. J Neurosci. 2010; 30: 10639–10654. 10.1523/JNEUROSCI.0059-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao WQ, Zheng JL, Karihaloo M. Neurotrophin-4/5 (NT-4/5) and brain-derived neurotrophic factor (BDNF) act at later stages of cerebellar granule cell differentiation. J Neurosci. 1995; 15: 2656–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz PM, Borghesani PR, Levy RL, Pomeroy SL, Segal RA. Abnormal cerebellar development and foliation in BDNF-/- mice reveals a role for neurotrophins in CNS patterning. Neuron. 1997; 19: 269–281. [DOI] [PubMed] [Google Scholar]
- 43.Carter AR, Chen C, Schwartz PM, Segal RA. Brain-derived neurotrophic factor modulates cerebellar plasticity and synaptic ultrastructure. J Neurosci. 2002; 22: 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005; 120: 137–149. 10.1016/j.cell.2004.11.012 [DOI] [PubMed] [Google Scholar]
- 45.Inagaki N, Chihara K, Arimura N, Ménager C, Kawano Y, Matsuo N, et al. CRMP-2 induces axons in cultured hippocampal neurons. Nat Neurosci. 2001; 4: 781–782. 10.1038/90476 [DOI] [PubMed] [Google Scholar]
- 46.Matsushima K, Aosaki M, Tokuraku K, Hasan MR, Nakagawa H, Kotani S. Identification of a neural cell specific variant of microtubule-associated protein 4. Cell Struct Funct. 2005. February;29(5–6):111–24. [DOI] [PubMed] [Google Scholar]
- 47.Tokuraku K, Noguchi TQ, Nishie M, Matsushima K, Kotani S. An isoform of microtubule-associated protein 4 inhibits kinesin-driven microtubule gliding. J Biochem. 2007. April;141(4):585–91. 10.1093/jb/mvm063 [DOI] [PubMed] [Google Scholar]
- 48.Hasan MR, Jin M, Matsushima K, Miyamoto S, Kotani S, Nakagawa H. Differences in the regulation of microtubule stability by the pro-rich region variants of microtubule-associated protein 4. FEBS Lett. 2006. June 12;580(14):3505–10. 10.1016/j.febslet.2006.05.028 [DOI] [PubMed] [Google Scholar]
- 49.Wu Q, Liu J, Fang A, Li R, Bai Y, Kriegstein AR, et al. The dynamics of neuronal migration. Adv Exp Med Biol. 2014;800:25–36. 10.1007/978-94-007-7687-6_2 [DOI] [PubMed] [Google Scholar]
- 50.Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruïne AP, Baldwin HS, et al. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J. 2010. November;24(11):4153–66. 10.1096/fj.09-151464 [DOI] [PubMed] [Google Scholar]
- 51.Liu JS. Molecular genetics of neuronal migration disorders. Curr Neurol Neurosci Rep. 2011. April;11(2):171–8. 10.1007/s11910-010-0176-5 [DOI] [PubMed] [Google Scholar]
- 52.Pelsers MM, Hanhoff T, Van der Voort D, Arts B, Peters M, Ponds R, et al. Brain- and heart-type fatty acid-binding proteins in the brain: tissue distribution and clinical utility. Clin Chem. 2004. September;50(9):1568–75. 10.1373/clinchem.2003.030361 [DOI] [PubMed] [Google Scholar]
- 53.Steinacker P, Mollenhauer B, Bibl M, Cepek L, Esselmann H, Brechlin P, et al. Heart fatty acid binding protein as a potential diagnostic marker for neurodegenerative diseases. Neurosci Lett. 2004. November 3;370(1):36–9. 10.1016/j.neulet.2004.07.061 [DOI] [PubMed] [Google Scholar]
- 54.Liu RZ, Mita R, Beaulieu M, Gao Z, Godbout R. Fatty acid binding proteins in brain development and disease. Int J Dev Biol. 2010;54(8–9):1229–39. 10.1387/ijdb.092976rl [DOI] [PubMed] [Google Scholar]
- 55.Reynolds JM, Liu Q, Brittingham KC, Liu Y, Gruenthal M, Gorgun CZ, et al. Deficiency of fatty acid-binding proteins in mice confers protection from development of experimental autoimmune encephalomyelitis. J Immunol. 2007. July 1;179(1):313–21. [DOI] [PubMed] [Google Scholar]
- 56.Shim SY, Kim HS, Kim EK, Choi JH. Expression of peroxiredoxin 1, 2, and 6 in the rat brain during perinatal development and in response to dexamethasone. Free Radic Res. 2012. March;46(3):231–9. 10.3109/10715762.2011.649749 [DOI] [PubMed] [Google Scholar]
- 57.Goemaere J, Knoops B. Peroxiredoxin distribution in the mouse brain with emphasis on neuronal populations affected in neurodegenerative disorders. J Comp Neurol. 2012. February 1;520(2):258–80. 10.1002/cne.22689 [DOI] [PubMed] [Google Scholar]
- 58.Singh SP, Chhunchha B, Fatma N, Kubo E, Singh SP, Singh DP. Delivery of a protein transduction domain-mediated Prdx6 protein ameliorates oxidative stress-induced injury in human and mouse neuronal cells. Am J Physiol Cell Physiol. 2016. January 1;310(1):C1–16. 10.1152/ajpcell.00229.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi M. Oxidative stress in developmental brain disorders. Neuropathology. 2009. February;29(1):1–8. 10.1111/j.1440-1789.2008.00888.x [DOI] [PubMed] [Google Scholar]
- 60.Aguiar CC, Almeida AB, Ara0jo PV, de Abreu RN, Chaves EM, do Vale OC, et al. Oxidative stress and epilepsy: literature review. Oxid Med Cell Longev. 2012;2012:795259 10.1155/2012/795259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xue B, Chen J, Gao H, Saito S, Kobayashi N, Shimokawa T, et al. Chronological changes in prosaposin in the developing rat brain. Neurosci Res. 2011. September;71(1):22–34. 10.1016/j.neures.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 62.Elleder M, Jerábková M, Befekadu A, HrebBefekadu A, Ledvinová J, et al. Prosaposin deficiency—a rarely diagnosed, rapidly progressing, neonatal neurovisceral lipid storage disease. Report of a further patient. Neuropediatrics. 2005. June;36(3):171–80. 10.1055/s-2005-865608 [DOI] [PubMed] [Google Scholar]
- 63.Motta M, Tatti M, Furlan F, Celato A, Di Fruscio G, Polo G, et al. Clinical, biochemical and molecular characterization of prosaposin deficiency. Clin Genet. 2016. September;90(3):220–9. 10.1111/cge.12753 [DOI] [PubMed] [Google Scholar]
- 64.Nabeka H, Uematsu K, Takechi H, Shimokawa T, Yamamiya K, Li C, et al. Prosaposin overexpression following kainic acid-induced neurotoxicity. PLoS One. 2014. December 2;9(12):e110534 10.1371/journal.pone.0110534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLSX)
(DOCX)
Data Availability Statement
The data underlying this study are available in the paper and its supporting information files. For any inquiries regarding the data, interested researchers may email the following address: lifen_chen@163.com.