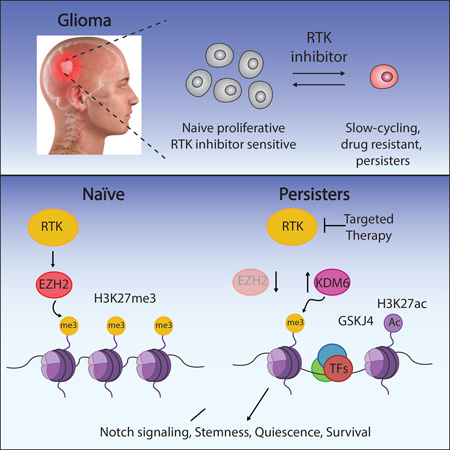
Summary
Glioblastoma, the most common and aggressive malignant brain tumor, is propagated by stem-like cancer cells refractory to existing therapies. Understanding the molecular mechanisms that control glioblastoma stem cell (GSC) proliferation and drug resistance may reveal opportunities for therapeutic interventions. Here we show GSCs can reversibly transition to a slow-cycling, persistent state in response to targeted kinase inhibitors. In this state, GSCs upregulate primitive developmental programs and are dependent upon Notch signaling. This transition is accompanied by widespread redistribution of repressive histone methylation. Accordingly, persister GSCs upregulate, and are dependent on, the histone demethylases KDM6A/B. Importantly, slow-cycling cells with high Notch activity and histone demethylase expression are present in primary glioblastomas before treatment, potentially contributing to relapse. Our findings illustrate how cancer cells may hijack aspects of native developmental programs for deranged proliferation, adaptation, and tolerance. They also suggest strategies for eliminating refractory tumor cells by targeting epigenetic and developmental pathways.
Graphical Abstract
Introduction
Glioblastoma (GBM), IDH wildtype, is the most frequent malignant primary brain tumor. Despite surgical resection, ionizing radiation, and chemotherapy, median survival remains less than fifteen months (Tanaka et al., 2012). Cancer genome-sequencing has catalogued a spectrum of genetic alterations in GBM (Brennan et al., 2013; McLendon et al., 2008) and identified potential ‘druggable’ targets. Receptor tyrosine kinases (RTKs) are the most commonly altered genes, with ~67% of adult GBMs harboring alterations of EGFR (57%), PDGFRA (13%), MET (2%), FGFR2/3 (3%) or other RTKs. RTK inhibitors have revolutionized treatment for certain cancer types with RTK alterations, but have failed to improve overall survival in GBM (Tanaka et al., 2012).
Therapeutic resistance and relapse in GBM relates to the extensive intratumoral genetic and phenotypic heterogeneity characteristic of these tumors (Eder and Kalman, 2014; Lathia et al., 2015). Evidence indicates that a subpopulation of stem-like cells, termed GBM stem cells (GSCs), underlie tumor propagation, drug resistance, and relapse (Bao et al., 2006; Lathia et al., 2015; Singh et al., 2004). The existence and functional importance of GSCs is supported by in vivo and in vitro evidence. First, a subpopulation of cells with stemness markers is present in human GBM, and is enriched upon treatment (Tamura et al., 2013). Second, primary tumor cells expressing stemness markers are highly tumorigenic when orthotopically xenotransplanted into mice (Singh et al., 2004). Third, stem-like cultures established from human tumors in serum-free conditions can propagate tumors, and have multipotent differentiation potential (Lee et al., 2006; Singh et al., 2004). GSCs are thus a critical model for the cancer stem cell field (Lathia et al., 2015).
Studies of gene regulatory circuits identified neurodevelopmental transcription factors (TFs) critical for GSC maintenance and tumorigenicity (Ikushima et al., 2009; Mehta et al., 2011; Rheinbay et al., 2013; Suvà et al., 2014). Cells co-expressing these TFs along with stemness markers are present in primary tumor specimens (Suvà et al., 2014). Furthermore, single-cell RNA-seq analysis of primary GBMs identified tumor cells with transcriptional circuits reminiscent of in vitro GSC models (Patel et al., 2014).
Despite their analogous neurodevelopmental states, in vivo stem-like cells differ markedly in their expression of cell cycle genes (Patel et al., 2014). In contrast to proliferative in vitro models, tumor cells have relatively low expression of cell cycle genes. This suggests that GSCs may adopt slow-cycling or quiescent states in vivo, consistent with the notion that cancer stem cells owe their refractory character to stable or transient quiescence (Sosa et al., 2014). Indeed, prior studies demonstrated that slow-cycling cells with stem-like properties promote tumor propagation and drug resistance in various cancer models (Chen et al., 2012; Roesch et al., 2010; Vanner et al., 2014).
We provide evidence that GSCs can adopt, and transition between, proliferative and slow-cycling states. We used the phenotype of reversible drug tolerance in RTK-dependent GSC lines to model the dynamic interconversion between these states. RTK inhibition led to rapid emergence of slow-cycling cells insensitive to RTK inhibitors and depleted for cell cycle gene expression programs. These persister cells (GSCPer) express increased levels of stemness markers and TFs, and develop dependence on Notch signaling. Transition to the persister state is accompanied by histone demethylase (KDM) upregulation and widespread redistribution of histone H3 lysine 27 trimethylation (H3K27me3). The H3K27 demethylases KDM6A/B are essential for the slow-cycling state, but largely dispensable for the proliferative state. Our study highlights key roles for chromatin reorganization and developmental plasticity in GBM biology, and suggests strategies for supplementing anti-proliferative therapies with modulators of epigenetic and developmental pathways.
Results
Sustained Inhibition of Kinase Signaling Enriches for Slow-Cycling GSCs
GSCs maintained in serum-free neurosphere culture conditions share features with neural stem cells (NSCs) and effectively initiate tumors in xenotransplantation assays (Lathia et al., 2015). To investigate proliferative programs in GSCs, we compared single-cell transcriptomes of primary GBM cells to in vitro GSCs (Patel et al., 2014). In primary tumors, only a small fraction of cells displays proliferative markers (2–20% Ki67+) (Louis et al., 2016) or express cell cycle signatures (Patel et al., 2014). When we compared developmental and cell cycle signatures, we found only a fraction of stem-like GBM tumor cells display proliferative signatures. In contrast, such signatures are evident in a large majority of in vitro GSCs (Figure 1A). While different GSC lines exhibit variable proliferation (Figure S1A) (Wakimoto et al., 2009), this potentially represents a critical distinction between in vitro and in vivo models.
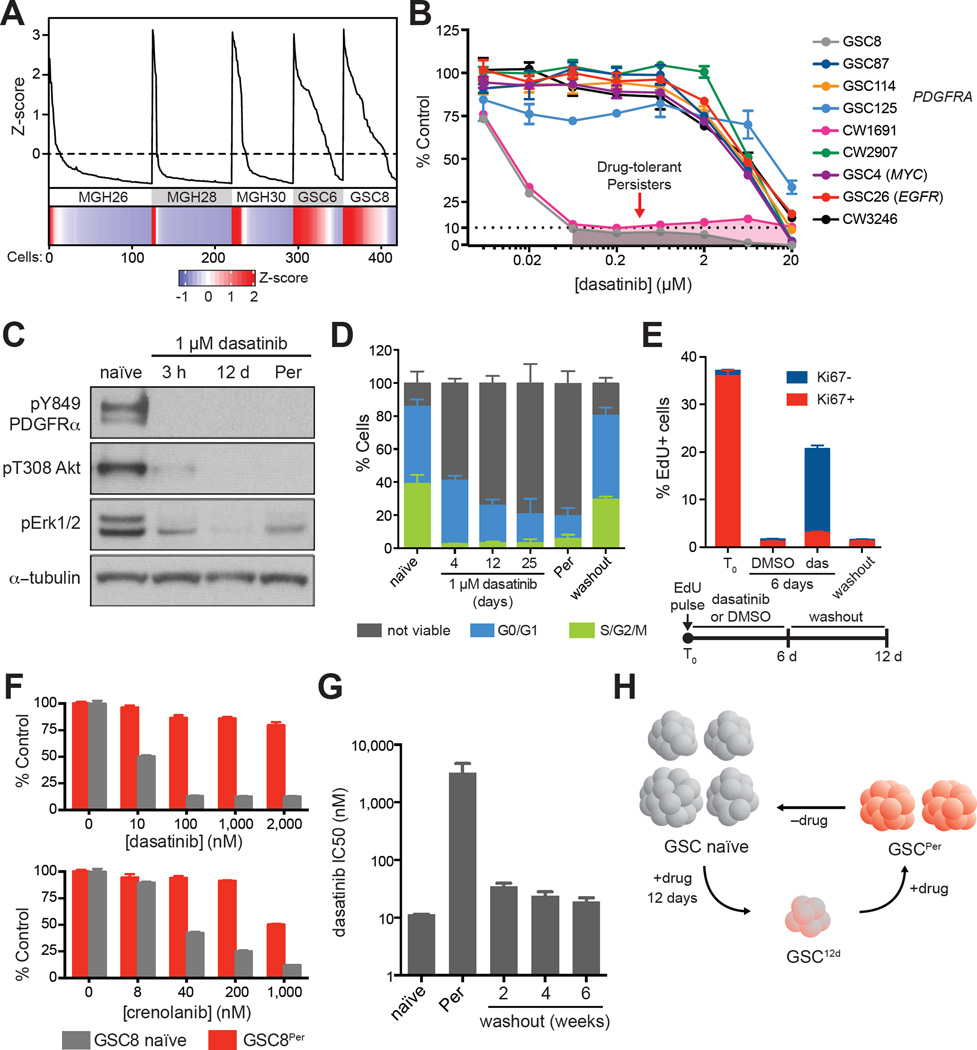
Figure 1. RTK Inhibition Prompts Emergence of Slow-Cycling Drug-Tolerant Persisters.
(A) Line graph shows cell cycle meta-signature z-scores (y-axis) for ordered individual cells (x-axis) for three primary tumors (MGH26, MGH28, MGH30) and two GSC lines (GSC6, GSC8). Lower panel: heatmap of cell cycle meta-signature z-scores. More cells in GSC lines display increased cell cycle expression in comparison to primary tumor specimens.
(B) Dose-response curves for treatment. Models treated for 4 days with the exception of CW1691 (6 days). PDGFRA amplified GSC8 and CW1691 display selective sensitivity (IC50 ~10 nM) in comparison to other lines tested. Error bars represent s.e.m. across three replicates. One of two biological replicates shown.
(C) Immunoblots show levels of phosphorylated PDGFRα, Akt, and Erk1/2 upon dasatinib treatment for 3 hours (3 h), 12 days (12 d), and >8 weeks (Per) in GSC8. Dasatinib treatment significantly reduced levels of phosphorylated proteins. One of two biological replicates shown.
(D) Stacked barplot shows the fraction of cells viable, in G0/G1, and in S/G2/M (y-axis), respectively, for GSC8 treated with dasatinib (1 µM) at various timepoints (x-axis). Washout refers to removal of dasatinib for >8 weeks. Error bars represent s.d. across at least three biological replicates.
(E) Stacked barplot summarizes flow cytometry data for Ki67 and EdU incorporation after EdU pulse (2 h) and subsequent treatments. Dasatinib treated cells maintained higher relative levels of EdU+ cells, which lose Ki67 positivity, compared to vehicle treated cells. Further 6-day washout of dasatinib depletes EdU+ cells. Error bars represent s.d. across three biological replicates.
(F) Barplots show the relative amount of cells (%, y-axis) after 4 day drug treatments at various doses (x-axis) in comparison to DMSO controls. GSC8Per tolerate higher concentrations of PDGFR inhibitors (dasatinib, crenolanib) in comparison to GSC8 naïve. Error bars represent s.e.m. across six replicates. One of two biological replicates is shown.
(G) Barplot shows dasatinib IC50 values (y-axis) for GSC8 naïve, GSC8Per, and at different time points following dasatinib removal. Washout of dasatinib from GSC8Per leads to resensitization to dasatinib-mediated growth arrest. Error bars represent s.d. of three biological replicates from separately derived GSC8Per lines.
(H) Schematic illustrating formation of slow-cycling, drug-tolerant persisters.
See also Table S1, Figure S1, and Figure S2.
We therefore considered if proliferative in vitro GSCs could be induced to a non-proliferative state. We treated a panel of GSC lines with small molecule inhibitors of oncogenic signaling pathways (Figure 1B, S1A–S1B, and Table S1). GSC8 and CW1691, both of which harbor focal amplification of PDGFRA, are highly sensitive to dasatinib (IC50 ~ 10 nM, Figure 1B), a PDGFR/Src inhibitor. By contrast, four other models derived from PDGFRA-amplified tumors – GSC87, GSC114, GSC125, CW2907 – are largely insensitive to dasatinib.
In agreement with PDGFRα being the relevant target of dasatinib in the sensitive models, GSC8 and CW1691 are also sensitive to crenolanib, a PDGFR inhibitor that does not appreciably inhibit Src (Figure S1C). Furthermore, overexpression of a drug-resistant PDGFRA mutant (D842V) (Dewaele et al., 2008) partially rescues GSC proliferation in the presence of dasatinib or imatinib, another PDGFR inhibitor (Figure S1D–S1E), while shRNA knockdown of PDGFRA decreased GSC proliferation (Figure S1F). Dasatinib treatment reduced phosphorylation of PDGFRα (pY849), Akt (pT308) and Erk1/2 (pT202/pY204, pT185/pY187) (Figure 1C and S1G). While off-target effects of dasatinib cannot be ruled out, these findings support RTK inhibition as the predominant effector of drug treatment in these models. GSC8 is also sensitive to the MEK inhibitor PD0325901 (Figure S1B), suggesting that the effects of PDGFRa inhibition may operate via MAPK signaling.
Despite the effects of dasatinib, sustained exposure consistently yielded a persistent subpopulation that tolerates higher drug concentrations (Figure 1B). Specifically, treatment of naïve GSC8 (1 µM dasatinib, ~100 times IC50) leads to an acute drop in the fraction of cycling, viable cells followed by a slow recovery (Figure 1D and Figure S2A–S2D). EdU pulse labeling followed by 6-day dasatinib treatment confirmed that initially cycling cells (EdU+) retained label and transitioned to a Ki67− state (Figure 1E and Figure S2E). Subsequent washout of dasatinib resulted in rapid loss of EdU+ cells. These data suggest that while many cells acutely die upon exposure to dasatinib, a subset of proliferative cells undergoes cell cycle arrest, but remain viable and slowly expand despite the presence of drug. We refer to these drug persistent cells as GSC8Per. Notably, GSC8Per lines could also be derived with inhibitors of downstream kinases, including MEK and CDK4/6 (Figure S1B, S1H, and S2F). We also derived persisters from an EGFR-dependent line, GSC26, that tolerate higher concentrations of EGFR inhibitors (Figure S1I).
We isolated populations of GSC8Per cultured in dasatinib for at least 8 weeks. These cells are relatively insensitive to dasatinib and crenolanib (Figure 1F), and still lack PDGFRα pY849 (Figure 1C), suggesting no mutation of PDGFRα conferring resistance. We considered if other genetic mechanisms might underlie drug-tolerance in GSC8 persisters. It has been reported that drug-tolerance to EGFR inhibitors may be mediated by reduction of EGFR+ extrachromosomal DNA and EGFR expression levels (Nathanson et al., 2014). In GSC8Per, however, total levels of PDGFRα remained relatively constant or increased (Figure S1G and S1J), and low-coverage whole genome sequencing revealed no significant copy number changes at PDGFRA or other loci (Figure S1K). Most critically, the GSC8Per state is reversible. Withdrawal of dasatinib permits full recovery of growth (Figure 1D), as well as re-sensitization to acute drug-induced arrest even after >4 months of chronic dasatinib treatment (Figure 1G). The rapidity and reversibility of acquired resistance support an epigenetic rather than genetic tolerance mechanism, as demonstrated in several cancer cell lines (Easwaran et al., 2014; Knoechel et al., 2014; Koppikar et al., 2013; Sharma et al., 2010).
Persister GSCs Express Primitive Neurodevelopmental and Quiescence Signatures
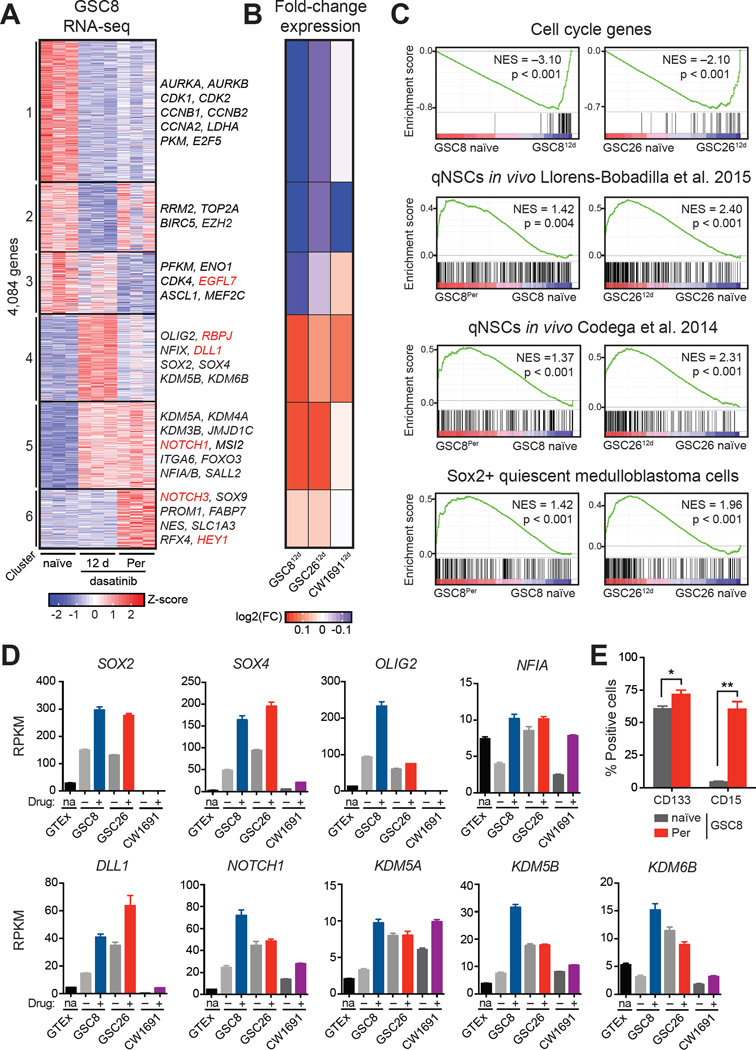
To investigate the regulatory circuits that sustain persister GSCs, we profiled gene expression in GSC8Per, in naïve GSCs, and in short-term treated GSCs (GSC812d) at maximal arrest (Figure 1H). Clustering of the 4,084 genes with highest variability across these states revealed gene sets with coherent expression changes (Figure 2A, Table S2). Genes depleted in GSC812d (clusters 1–3) and GSC8Per (clusters 1 and 3) were related to cell cycle and proliferation, consistent with reduced proliferation (Figure 2A, 2C, and S3A). By contrast, genes upregulated in GSC persisters (clusters 4–6) were enriched for signatures derived from quiescent NSCs (qNSCs) (Codega et al., 2014; Llorens-Bobadilla et al., 2015; Martynoga et al., 2013) and quiescent, stem-like medulloblastoma cells (Figure 2C and Figure S3B) (Vanner et al., 2014). Profiling of GSC26 and CW1691 treated with gefitinib or dasatinib, respectively, revealed similar changes in expression for the respective GSC8-derived gene clusters (Figure 2B), and enrichment of quiescent gene signatures (Figure 2C and Figure S3C).
Figure 2. Transcriptional Programs Related to Slow Proliferation and Stemness are Enriched in GSC8 Persisters.
(A) Heatmap shows expression profiles of the 4,084 most variably expressed genes across GSC8 naïve, GSC812d, and GSC8Per. K-means clustering was performed to distinguish sets of genes with coherent patterns of expression across the time course of dasatinib treatment. Data was generated from three biological replicates of separately derived GSC812d and GSC8Per cultures. Color represents z-scores of gene expression across rows.
(B) Heatmap shows average changes in gene expression of clusters 1–6 in GSC8, GSC26 and CW1691 after 12-day treatment with gefinitib and dasatinib, respectively, compared to corresponding untreated control. Similar trends of gene expression change are evident in GSC8, GSC26 and CW1691 persisters. Color represents log2(fold-change) gene expression, where the scale ranges from the minimum to maximum value observed in GSC2612d or CW169112d.
(C) Gene set enrichment analysis (GSEA) shows enrichment of a cell cycle meta-signature (upper panel), gene sets related to qNSCs (middle panels), and Sox2+ medulloblastoma cells (lower panel) across naïve and persister states for GSC8 and GSC26. The cell cycle gene signature is negatively enriched, while the quiescence gene signatures are positively enriched upon drug treatment.
(D) Barplots show the expression levels (reads per kilobase per million mapped reads (RPKM), y-axis) of SOX2, SOX4, OLIG2, NFIA, DLL1, NOTCH1, KDM5A, KDM5B and KDM6B in normal human brain (GTEx), GSC8 naïve, GSC812d, GSC26 naïve, GSC2612d, CW1691 naïve, and CW169112d. Most genes are upregulated in drug-treated cells. Error bars represent s.e.m.
(E) Barplot shows the fraction of cells (y-axis) CD133+ or CD15+ by flow cytometry. GSC8Per display significantly increased positivity for CD133 and CD15 in comparison to GSC8 naïve (Student’s t-test; *P < 0.05; **P < 0.01). Error bars represent s.e.m. across three biological replicates.
See also Table S2–S4 and Figure S3.
Gene sets upregulated in persisters contain stemness factors previously linked to GSCs (Gangemi et al., 2009; Ikushima et al., 2009; Mehta et al., 2011; Suvà et al., 2014), as well as regulators implicated in stem-like tumor cells in vivo (Patel et al., 2014) (Figure 2A, clusters 4–6, Figure 2D and S3D). GSC8Per also upregulate classical stemness markers, such as PROM1 (CD133) and SSEA-1 (CD15) (Singh et al., 2004; Son et al., 2009) (Figure 2A, 2E, and S3E). Many of these TFs and markers are already expressed in naïve GSCs but are upregulated on drug treatment (Figure 2D–2E and S3D). Notably, the persisters also upregulate multiple KDMs (Figure 2A, 2D, and S3F–S3I), including KDM3, KDM5, and KDM6 family members, some of which have been previously implicated in slow-cycling drug-resistant cell lines (Roesch et al., 2013; Sharma et al., 2010) and single-cell studies of primary GBM (Patel et al., 2014). Altogether, these expression changes are consistent with the notion that GSC persisters transition to a slow-cycling state and upregulate transcriptional programs shared with qNSCs.
Notch Signaling Underlies GSCPer Switch
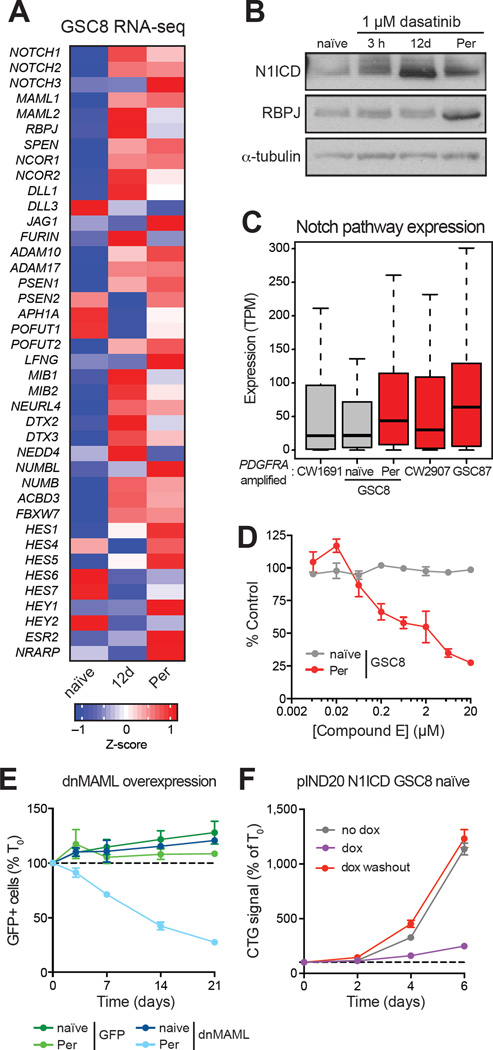
The strong upregulation of prominent Notch signaling genes prompted us to investigate this pathway (Figure 2A and 2D). Various notch pathway genes were upregulated in GSC8Per, including Notch ligands, receptors, and canonical downstream targets (Figure 3A and Figure S4A–S4B). Consistent with these changes, GSC8 persisters have high levels of Notch1 intracellular domain (N1ICD) (Figure 3B), the proteolytic cleavage product of activated Notch1 receptor that mediates gene induction. The dasatinib-insensitive PDGFRA-amplified models, GSC87 and CW2907, already express Notch pathway genes at higher levels, suggesting that they preexist in a Notch-high state (Figure 3C).
Figure 3. Notch Signaling is Activated in GSCPer.
(A) Heatmap shows average expression of genes implicated in Notch signaling (Kopan and Ilagan, 2009) for GSC8 naïve, GSC812d, and GSC8Per. Gene expression was averaged over replicates within each condition; color represents z-scores.
(B) Immunoblots show levels of N1ICD and RBPJ in GSC8 naïve and GSC8 treated with dasatinib for 3 h, 12 d, and >8 weeks (Per). One of two biological replicates is shown.
(C) Boxplots show Notch pathway gene expression (transcripts per million (TPM), y-axis) in different PDGFRA-amplified GBM lines. Notch signaling is increased in GSC8Per and is significantly higher in insensitive cell lines compared to sensitive cell lines (P < 10−7; Friedman test). Color indicates relative sensitivity (gray) or insensitivity (red) to dasatinib.
(D) Dose-response curves for 12-day Compound E treatment are shown. GSC8Per are preferentially sensitive in comparison to GSC8 naïve. Error bars represent s.e.m. across four replicates. One of three biological replicates is shown.
(E) Line graph shows fraction of GFP+ cells normalized to day zero (T0) (y-axis) over a time course following induced overexpression of dnMAML-GFP or GFP control. dnMAML overexpression selectively depleted GFP+ cells in GSC8Per. Error bars represent s.e.m. across three replicates. One of two biological replicates is shown.
(F) Line graphs show cell growth as relative Cell-titer Glo (CTG) values normalized to T0 (y-axis) over a time course (x-axis) following doxycycline (dox)-induced N1ICD overexpression. N1ICD overexpression in GSC8 naïve reduced proliferation, but is reversed upon dox washout. Error bars represent s.e.m. across three replicates. One of three biological replicates is shown.
See also Figure S4.
In considering whether Notch signaling is required for the slow-cycling state, we utilized a small molecule inhibitor of γ-secretase, a protease necessary for Notch activation (Kovall and Blacklow, 2010). Treatment with γ-secretase inhibitor (Compound E) reduced GSC8Per growth, but had little effect on naïve GSC8 (Figure 3D). Notch signaling can also be suppressed genetically using a dominant negative form of mastermind-like (MAML) proteins (dnMAML), critical co-factors for Notch-mediated gene activation (Kovall and Blacklow, 2010; Maillard et al., 2004). Overexpression of dnMAML markedly reduced GSC8Per proliferation, but had little effect on naïve GSC8 (Figure 3E). Conversely, overexpression of N1ICD was sufficient to induce a reversible growth reduction in GSC8 naïve (Figure 3F and S4D–S4F). These data suggest that Notch activation allows GSCs to transition to a slow-cycling, RTK-independent state.
Primary GBM Tumors Contain Notch-Positive Cells Depleted for Proliferation Markers
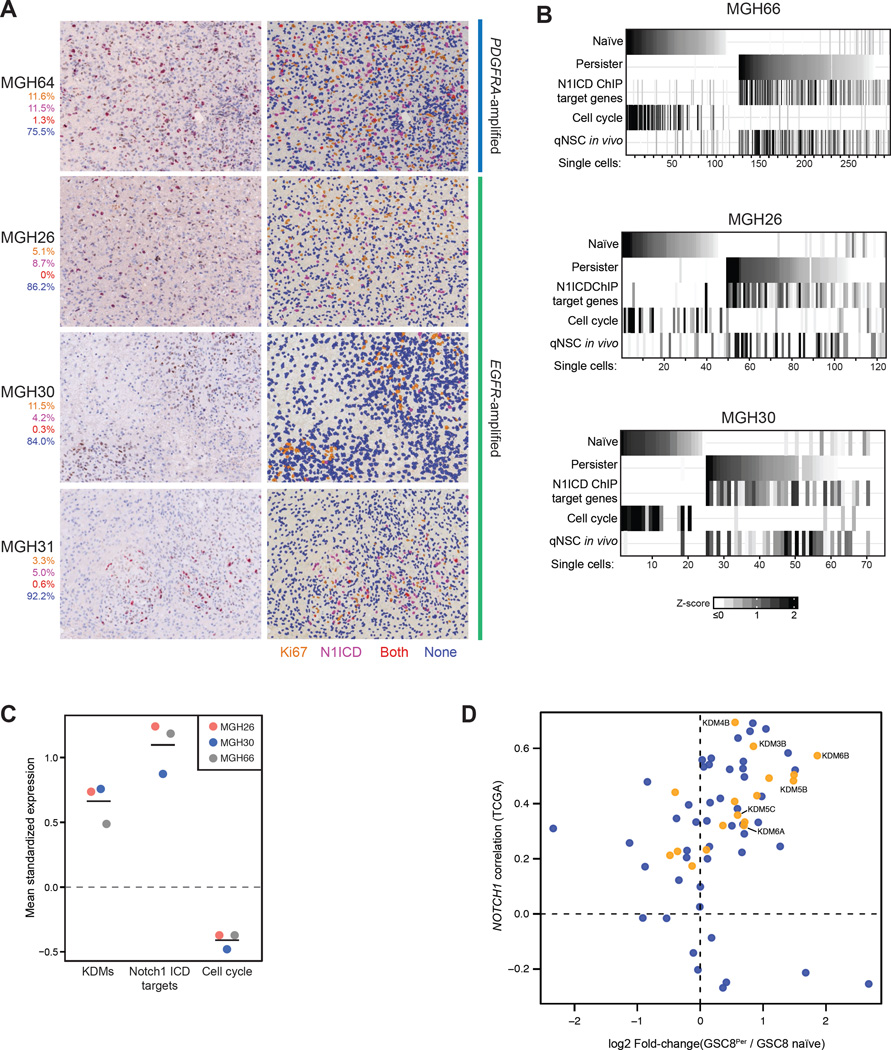
To address the in vivo relevance of our findings, we co-stained primary GBM specimens from PDGFRA- and EGFR-amplified tumors for the Notch marker N1ICD and the proliferation marker Ki67. A subset of N1ICD+ cells is evident in variable proportions (4.6 to 13%) in all specimens examined. Importantly, the majority of N1ICD+ cells (90–100%) were not Ki67+ (Figure 4A), suggesting that tumor cells with active Notch signaling are slow-cycling.
Figure 4. Slow-cycling, Notch-high Cells are Present in Primary GBM Tumors.
(A) Images show hematoxylin staining and immunohistochemistry for N1ICD and Ki67 in patient primary GBM tumors, demonstrating that a subpopulation of N1ICD+ cells are slow-cycling. Left hand panels: unenhanced images, right hand panels: enhanced pseudocolored images. Percentages represent the fraction of total cells that are Ki67+ only (orange), N1ICD+ only (magenta), double-positive (red), and double-negative (blue).
(B) Heatmap shows mean expression of gene sets (rows) derived from GSC8 naïve (clusters 1–3, Figure 2A) and GSC8Per (clusters 4–6, Figure 2A) as well as N1ICD GSC8Per target genes, a cell cycle signature, and a qNSC in vivo signature (rows) across single tumor cells (columns) in three EGFR-amplified tumors (MGH66, MGH30, MGH26). K-means clustering of columns identified groups of cells with similar expression profiles. Clusters were order based on expression of GSC8 naïve or GSC8Per signatures, respectively. Colors correspond to z-scores calculated across individual cells; in this representation negative z-scores are mapped to 0.
(C) Dotplot shows mean expression (y-axis) of KDM-, Notch-, and cell cycle signature gene sets within persister-like cells (top quintile) in MGH26, MGH30, and MGH66 (x-axis) (Figure 4B). Expression values were standardized (mean = 0; s.d. = 1) – observed deviations from zero reflect expression changes relative to all cells. Horizontal bars correspond to mean values for persister-like cells.
(D) For each annotated chromatin enzyme gene, scatterplot shows the correlation between its expression and NOTCH1 expression across all TCGA samples (y-axis) versus its expression change in GSC8Per relative to GSC8 naïve (x-axis). KDMs are highlighted in yellow.
See also Table S5.
We also integrated expression signatures from our in vitro GSC study with single-cell RNA-seq data for primary GBM tumors. We acquired RNA-seq profiles for 300 individual cells from one EGFR-amplified tumor (MGH66), and integrated published single-cell data from two additional EGFR-amplified tumors (Patel et al., 2014). We considered the following 5 gene signatures: (i) a naïve GSC signature comprising genes in clusters 1–3 (Figure 2A); (ii) a persister GSC signature comprising genes in clusters 4–6 (Figure 2A); (iii) a cell cycle signature; (iv) a Notch-target gene signature derived by mapping N1ICD binding sites (see below); and (v) an in vivo qNSC signature (Codega et al., 2014).
We scored each individual tumor cell for each of these 5 signatures. Signatures for naïve GSCs and cell cycle were commonly expressed in a small subset of tumor cells (Figure 4B), consistent with relatively low Ki67 positivity. Signatures for N1ICD-target genes and in vivo qNSCs scored in a distinct subset of persister-like tumor cells (Figure 4B–4C). Altogether, these data indicate that primary GBMs contain a subset of cells that shares features with persister GSCs, including Notch-positivity, stem-like gene expression, and depletion of proliferation markers.
Notch Activity Targets Developmental Enhancers
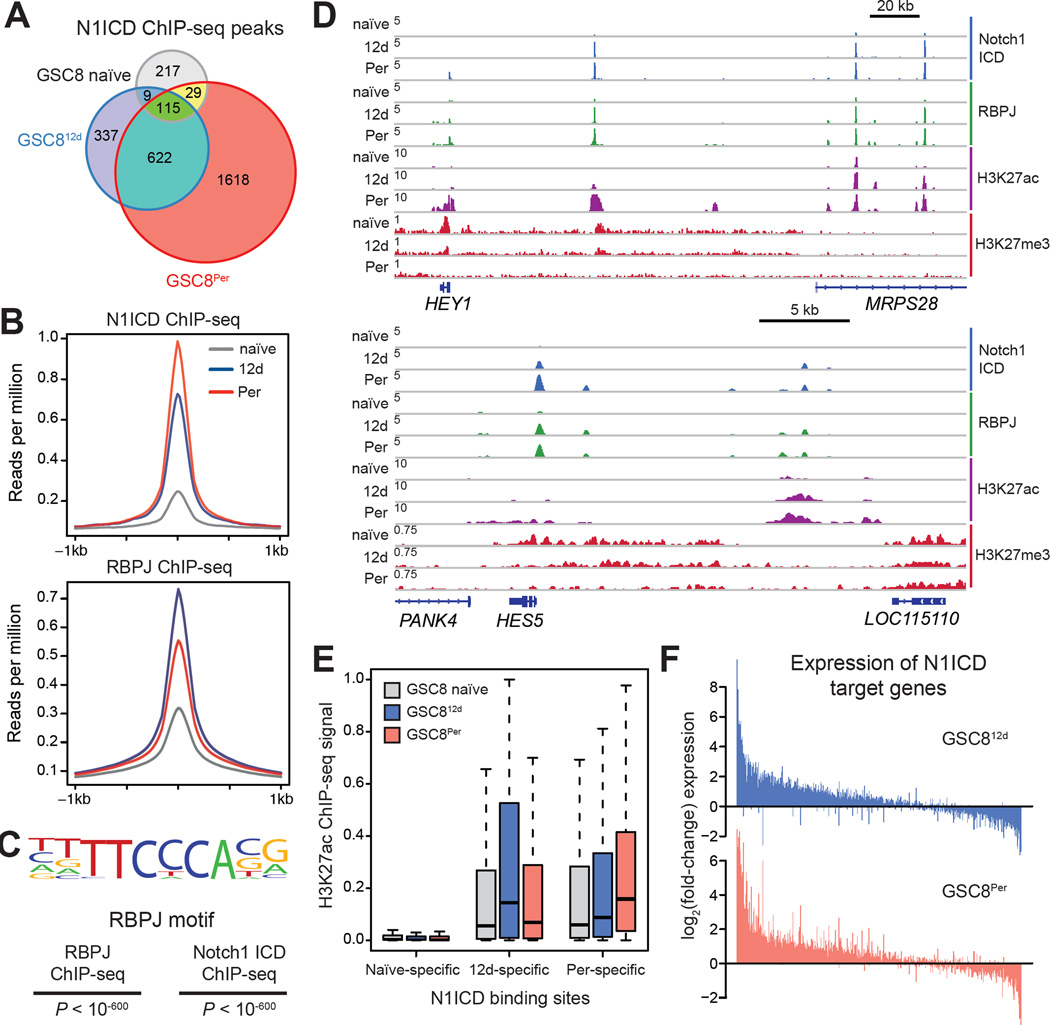
To investigate how Notch signaling sustains persister GSCs, we mapped N1ICD and its co-factor RBPJ by ChIP-seq across the three GSC8 states. Consistent with increased levels of N1ICD and RBPJ, we detected a ~6-fold increase in the number of N1ICD binding sites in GSC812d and GSC8Per, relative to GSC naïve (2,384 in GSC8Per vs. 370 in GSC naïve), as well as increased signal intensities over these sites (Figure 5A-5B). As expected, N1ICD binding sites largely overlapped RBPJ binding sites (1750/2384 in GSC8Per), and the underlying DNA sequences are enriched for RBPJ recognition motifs (Figure 5C). Consistent with prior studies in lymphoid cells (Wang et al., 2011), N1ICD binds primarily to distal sequences containing the active enhancer mark H3K27ac (Figure 5D–5E and S5A) (ENCODE Project Consortium et al., 2012).
Figure 5. N1ICD Binds to Neurodevelopmental Enhancers in GSC Persisters.
(A) Venn diagram shows the number and overlap of N1ICD ChIP-seq peaks across GSC8 naïve, GSC812d, and GSC8Per states. N1ICD localizes to fewer regions in GSC8 naïve (368 peaks) and shows increased binding upon drug treatment (1,083 and 2,384 in GSC812d and GSC8Per, respectively).
(B) ChIP-seq profile plots show ChIP-seq signal (y-axis, reads per million (rpm)) of N1ICD (upper panel) and RBPJ (lower panel) across peaks for each respective protein. The x-axis shows flanking regions of ±1 kb around the peak center.
(C) Consensus RBPJ motif logo detected in N1ICD and RBPJ ChIP-seq peaks in GSC8Per and the corresponding p-values are shown.
(D) ChIP-seq profiles show ChIP-seq signal (y-axis, rpm) of N1ICD, RBPJ, H3K27ac and H3K27me3 at the HEY1 (upper panel) and HES5 (bottom panel) locus.
(E) Boxplots show H3K27ac levels of GSC8 naïve, GSC812d and GSC8Per within 1.5 kb windows centered at NICD peaks identified within GSC8 naïve (left), GSC812d (middle), and GSC8Per (right). For each condition, H3K27ac levels were significantly higher within the corresponding condition-specific N1ICD peaks (GSC812d-specific NICD peaks: GSC8 naïve vs GSC812dP < 10−15; GSC8Per-specific NICD peaks: GSC8 naïve vs GSC8PerP < 10−16; Wilcoxon test).
(F) Barplots show log2(fold-change) in gene expression of N1ICD target genes specifically identified in GSC812d or GSC8Per for GSC812d (upper panel) and GSC8Per (lower panel). Genes were sorted decreasingly according to the added fold-change.
See also Figure S5.
To identify putative Notch target genes, we assigned N1ICD peaks detected in GSC8 naïve, GSC812d, or GSC8Per to the nearest gene promoter. We focused on 1,696 target genes identified in GSC812d and/or GSC8Per, but not in GSC8 naïve (Table S3). Consistent with a primary role in transcriptional activation, these candidate N1ICD targets were predominantly upregulated in GSC812d and GSC8Per, relative to GSC8 naïve (Figure 5E–5F), and enriched for Notch pathway and neurodevelopmental genes as well as various canonical Notch targets (Figure 5D and S5B–S5C). Illustrative examples are the HEY1 and HES5 loci (Figure 5D): HEY1 is associated with poor survival in GBM patients (Hulleman et al., 2009) and HES5 is implicated in qNSCs (Imayoshi et al., 2010). In addition to canonical targets, N1ICD bound H3K27ac-marked distal elements in the vicinity of many neurodevelopmental master regulators (e.g., SOX TFs, OLIG1/2) and several pro-survival genes (Figure S5D). These N1ICD and RBPJ binding patterns support a direct mechanistic role for Notch signaling in the activation of regulatory elements and neurodevelopmental genes in slow-cycling GSC persisters.
Dynamic Histone Acetylation and Demethylation Accompany GSCPer Switch
The dynamic acetylation of N1ICD-bound elements prompted us to investigate global chromatin alterations associated with the persister transition. We mapped H3K27ac genome-wide in naïve GSC8, GSC812d, and GSC8Per, and called enriched intervals, identifying an average of 12,938 sites in each GSC state (naïve: 12,412; GSC812d: 12,940; GSC8Per: 13,462). Approximately 63% of these sites are >5 kb away from any annotated transcriptional start site, thus representing candidate distal regulatory elements or enhancers. These enhancer-like elements are dynamic in activity (~25% common to all states, ~50% specific to one state).
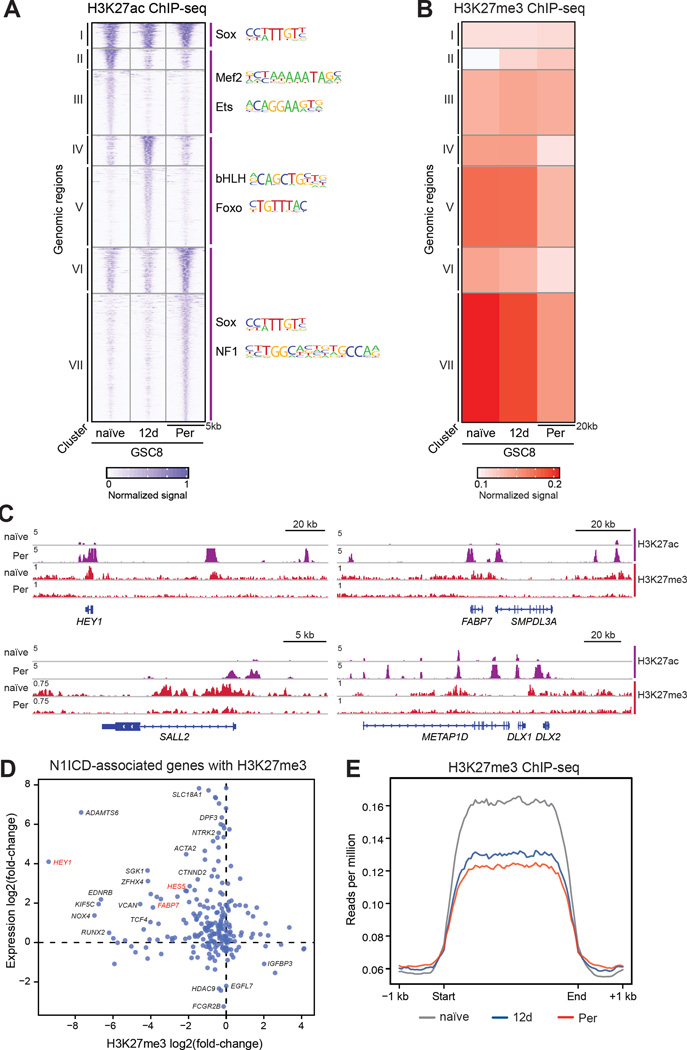
We used K-means clustering to distinguish sets of promoter- and enhancer-like elements with related patterns of activity across the three GSC8 states (Figure 6A). We scanned the DNA sequences underlying the corresponding clusters for over-represented TF motifs (Figure 6A). The resulting predictions of differential TF activity are largely concordant with the TF expression patterns derived by RNA-seq (Figure 2A). GSC8 naïve-specific elements (Figure 6A, cluster II-III) are enriched for MEF2 motifs, consistent with high expression of MEF2C and its dimerization partner ASCL1 in the naïve state (Figure 2A, cluster 3) (Black et al., 1996). ASCL1 has been previously implicated in proliferative stem cell populations (Rheinbay et al., 2013). GSC812d-specific elements (Figure 6A, cluster IV-V) are enriched for Foxo motifs, in agreement with upregulation of FOXO3 (Figure 2A: cluster 5, and S3D), a TF with established roles in qNSCs (Paik et al., 2009; Renault et al., 2009). GSC8Per-specific elements (Figure 6A, cluster VI-VII) are enriched for SOX and NFI motifs, consistent with increased expression of these TFs in GSC8Per. Thus, concordant differences in regulatory element activity and TF expression distinguish the respective GSC states, and implicate neurodevelopmental regulatory programs in persister GSCs.
Figure 6. H3K27me3 Redistribution Accompanies Re-activation of Neurodevelopmental Genes.
(A) Heatmap shows normalized H3K27ac ChIP-seq signal for GSC8 naïve, GSC812d, and GSC8Per across different genomic intervals (rows). K-means clustering of rows identified groups of H3K27ac regions that are shared (I), naïve-enriched (II-III), GSC812d-enriched (IV-V), and GSC8Per-enriched (VI-VII). Color corresponds to normalized ChIP signal.
(B) Heatmap shows average normalized ChIP-seq signal for H3K27me3 for groups of genomic intervals derived from clustering analysis in Figure 6A. H3K27me3 levels are depleted in cluster VI and VII. H3K27me3 signals were calculated within 20 kb windows centered around H3K27ac peaks, respectively. Color corresponds to normalized ChIP signal.
(C) ChIP-seq profiles show ChIP-seq signal (y-axis, rpm) for H3K27ac and H3K27me3 at genomic loci of HEY1, FABP7, DLX2, and SALL2.
(D) Scatterplot shows changes in expression (y-axis) and intragenic H3K27me3 levels (x-axis) of genes associated with an N1ICD peak in GSC8Per that contain at least one H3K27me3 peak in GSC naïve, GSC812d or GSC8Per. The y-axis represents log2(fold-change) in gene expression comparing GSC8Per to GSC8 naïve. The x-axis represents log2(fold-change) of intragenic H3K27me3 levels comparing GSC8Per to GSC8 naïve.
(E) ChIP-seq profile plots show H3K27me3 ChIP-seq signal (y-axis, rpm) across all H3K27me3 domains (>10 kb) in GSC8 naïve (grey), GSC812d (blue), and GSC8Per (red). The x-axis represents size scaled H3K27me3 domains, with ±1 kb flanking regions.
See also Figure S6.
Many neurodevelopmental and Notch pathway genes are established targets of Polycomb repressors and the associated histone mark H3K27me3, which restrain their activity in non-expressing cell types (Simon and Kingston, 2013). We noted that the H3K27 methyltransferase EZH2 is downregulated in GSC persisters, while the H3K27 demethylases KDM6A and KDM6B are amongst a large set of KDMs that are upregulated (Figure 2A and Figure S3G–S3H). We therefore mapped and compared the distribution of H3K27me3 across naïve GSC8, GSC812d, and GSC8Per. Many N1ICD-associated loci marked by H3K27me3 in naïve GSC8 undergo coincident H3K27 demethylation, upregulation, and H3K27 acetylation upon transition to the persister state (Figure 6D and Figure S5E). Examples include the HEY1 and HES5 loci, where increased binding of N1ICD and RBPJ is accompanied by gain of H3K27ac and loss of H3K27me3 (Figure 5D).
Broadly, we considered whether increased H3K27ac at enhancer-like elements in GSC8Per are associated with a reduction in H3K27me3. Indeed, many of these elements are initially marked by H3K27me3 but are strongly depleted for this repressive mark upon activation in GSC812d and/or GSC8Per cells (Figure 6A–6C cluster VII). Consistent with established roles for the respective modifications, genes near such regulated enhancer-like elements have higher expression in the persister cells (Wilcoxon test; P < 10−16).
Comparison of H3K27me3 profiles revealed an asymmetry between the GSC states. Whereas GSCPer-specific H3K27ac-marked elements tend to have H3K27me3 in naïve GSCs (Figure 6A–B, cluster VII), naïve GSC-specific H3K27ac elements have diminished H3K27me3 in GSCPer (Figure 6A–B, clusters II-III). This suggests that the transition from naïve to persister state is accompanied by a global loss of H3K27me3 from cis-regulatory elements. To gain more systematic insight, we called H3K27me3-enriched intervals genome-wide in each of the three GSC cell states. Remarkably, H3K27me3 levels across these intervals are markedly reduced in GSC812d and GSC8Per, relative to naïve (Figure 6E). A reduction in H3K27me3 levels also occurred following treatment of GSC8 naïve cells with MEK inhibitor PD0325901 (Figure S6A) and, to a lesser extent, following treatment of GSC26 cells with an EGFR inhibitor (Figure S6B). Altogether, these data suggest that the activation of neurodevelopmental loci and Notch pathway genes is accompanied by pervasive acetylation of cis-regulatory elements, and may be facilitated by a global redistribution of H3K27me3 repressive chromatin.
H3K27me3 Demethylases KDM6A/B are Essential for GSC Persisters
Multiple KDMs are coordinately upregulated in the persisters (Figure 2A), while Notch-high tumors and Notch-high tumor compartments also exhibit significantly higher KDM expression (Brennan et al., 2013; Patel et al., 2014; Verhaak et al., 2010) (Figure 4C–D). Moreover, widespread redistribution of H3K27me3 in cells exiting cycle is suggestive of active histone demethylation. Taken together, these data implicate the H3K27 demethylases KDM6A/B in the transition or maintenance of persister GSCs. We therefore separately knocked out KDM6A and KDM6B in GSC8 naïve and GSC8Per using CRISPR-Cas9 genome editing (Doench et al., 2014) and single-guide RNAs (sgRNAs) targeting their catalytic domains (Shi et al., 2015) (Figure S7A).
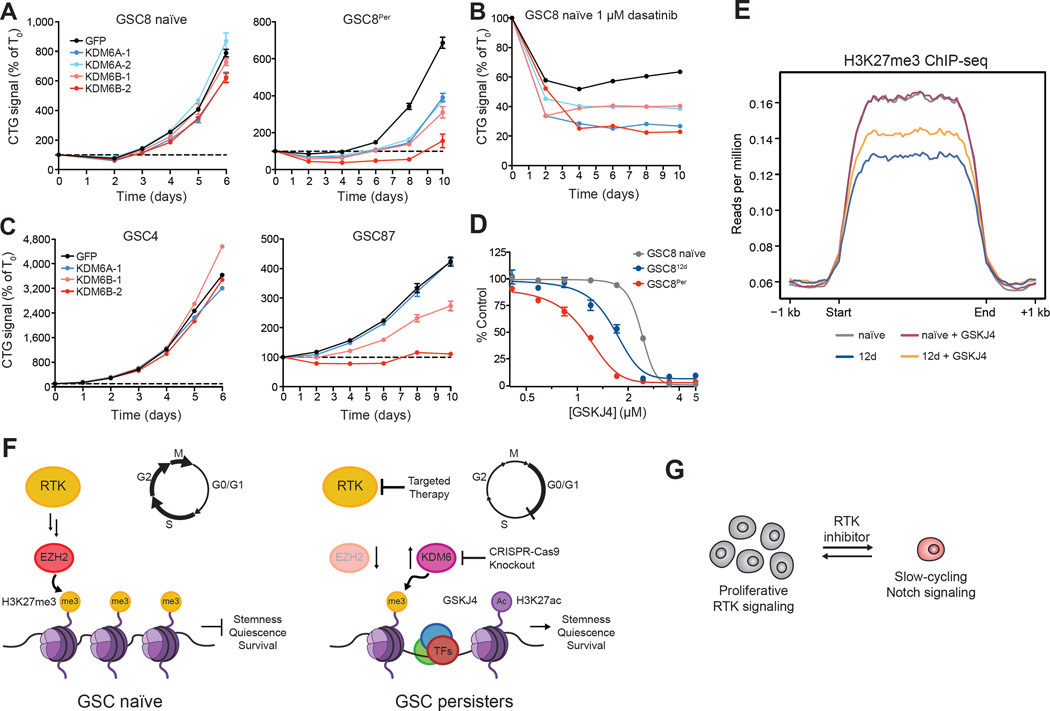
KDM6A and KDM6B knockout minimally affected GSC8 naïve cell growth (Figure 7A, left panel). By contrast, GSC8Per cells were highly sensitive to knockout of either demethylase (Figure 7A, right panel); growth reduction afforded by KDM6B knockout was most pronounced, consistent with a reported pro-oncogenic role for KDM6B (Hashizume et al., 2014; Ntziachristos et al., 2014). Moreover, GSC8 naïve cells lacking KDM6A or KDM6B were less effective at forming persisters upon dasatinib treatment (Figure 7B).
Figure 7. KDM6 is Essential in GSC Persisters.
(A–C) Line graphs show cell growth as relative CTG values normalized to T0 (y-axis) over a time course (x-axis) following CRISPR-Cas9 mediated knockout of respective genes. Knockout of KDM6A and KDM6B modestly affected the proliferation of GSC8 naïve (A, left panel) but significantly impaired proliferation of GSC8Per (A, right panel) and (B) emergence of persisters. Knockout of KDM6B preferentially affected the proliferation of GSC87 (C, right panel), which displays ‘persister-like’ characteristics. Error bars represent s.e.m. across at least three replicates. One of two biological replicates is shown.
(D) Dose-response curves for 4 day GSKJ4 treatment are shown. GSC812d and GSC8Per are preferentially sensitive to GSKJ4 in comparison to GSC8 naïve. Error bars represent s.e.m. across three replicates. One of three biological replicates is shown.
(E) ChIP-seq profile plots show H3K27me3 ChIP-seq signal (y-axis, rpm) across all H3K27me3 domains (>10 kb) in GSC8 naïve (grey), GSC812d (blue), GSC8 treated with GSKJ4 (8 d, 1.5 µM, purple), and GSC812d treated with GSKJ4 (8 d, 1.5 µM, orange) starting after 4 days of initial dasatinib treatment. The x-axis represents size scaled H3K27me3 domains, with ±1 kb flanking regions.
(F) Schematic illustrating a proposed model, whereby EZH2 loss and KDM6B upregulation may facilitate H3K27 remodeling and subsequent activation of stemness programs.
(G) Cartoon depicting the switch between RTK- and Notch-dependent states that may parallel antagonism between these pathways in normal neurodevelopment.
See also Table S4, Figure S6, and Figure S7.
To further explore this KDM6 dependency, we expanded our studies to other GSC models: GSC4 and GSC87. GSC4 is MYC-amplified, rapidly proliferating, and expresses a cell cycle gene signature akin to naïve GSC8 cells, while GSC87 is slow-cycling and expresses a persister-like gene signature at baseline (Figure S7B). GSC87 is highly CD133+, Notch-dependent, insensitive to dasatinib despite focal PDGFRA amplification, and displays similar H3K27ac profiles to GSC8Per (Figure 1B, S4C, and S7B–S7D). Knockout of either KDM6A or KDM6B had negligible effects on GSC4, but KDM6B knockout significantly impaired persister-like GSC87 (Figure 7C). These data support general roles for KDM6 demethylases in the transition and maintenance of slow-cycling, persister-like GSCs.
To evaluate the contributions of KDM6 enzymatic activity, we treated GSCs with the small molecule inhibitor ‘GSKJ4’ (Kruidenier et al., 2013) (Figure 7D–7E). GSKJ4 treatment (1.5 µM) had negligible effects on the preexisting H3K27me3 landscape of naïve GSC8 but rescued the H3K27me3 loss seen upon dasatinib treatment (Figure 7E and S6C). GSKJ4 treatment did not significantly increase H3K4me3 in these models, supporting its selectivity for KDM6A/B at the doses used (Heinemann et al., 2014) (Figure S6D–S6E). These results support the notion that KDM6 contributes to the global H3K27me3 alterations in dasatinib-treated GSCs.
Consistent with our knockout data, GSKJ4 compromised proliferation of GSC812d and GSC8Per at doses tolerated by naïve GSC8 (Figure 7D). Likewise, GSC8 persisters derived from sustained MEK inhibition are also preferentially sensitive to GSKJ4 (Figure S7E). By contrast, GSC persisters displayed no preferential sensitivity to GSKJ5, an inactive isomer of GSKJ4 (Kruidenier et al., 2013), nor to ‘KDM5-C70’, a recently described KDM5-selective inhibitor (Johansson et al., 2016) (Figure S7E–S7F). Lastly, we found that GSC87, which exhibits persister-like features at baseline, also showed increased sensitivity to GSKJ4, relative to the PDGFRA-dependent line CW1691 (Figure S7G). Thus, preferential KDM6 dependency may be a general characteristic of persister-like GSCs. Altogether, these data suggest that reconfiguration of H3K27me3 landscapes by histone demethylases facilitate the activation of regulatory elements and neurodevelopmental gene expression programs required for the drug tolerant persister GSC state (Figure 7F).
Discussion
Reversible transitions between epigenetic states enable tumor cells to adapt to different pressures, including conventional and targeted therapy (Easwaran et al., 2014; Knoechel et al., 2014; Koppikar et al., 2013; Roesch et al., 2013; Sharma et al., 2010). Such transitions could involve de novo states accessible only to malignant cells, or may parallel aspects of normal developmental hierarchies. We show that patient-derived GSCs can evade RTK inhibition and other anti-proliferative therapies by regressing to a slow-cycling, Notch-dependent state. We also establish that the GSC persister transition is characterized by widespread remodeling of repressive chromatin and dependency on the H3K27 demethylases KDM6A/B.
An expanding body of literature supports a cancer stem cell model in GBM, wherein stemlike cells with differentiation potential mediate tumor propagation. GSCs are thought to resist current therapies and thus underlie inevitable relapse. Their refractory nature could relate in part to a subset of GSCs with a slow-cycling phenotype that is innately resistant to therapy (Chen et al., 2012; Lathia et al., 2015). Our analysis of clinical specimens by multi-spectral imaging and single-cell RNA-seq confirms that primary GBMs contain cells with persister-like features, including Notch-positivity, high KDM expression, and low cell cycle gene expression. The fact that these tumors were analyzed prior to therapy suggests that persister-like GSCs may preexist in GBMs. In agreement with this notion, one of our patient-derived models, GSC87, which is insensitive to PDGFR inhibitors despite harboring focal PDGFRA amplification, preexists in a persister-like state and is dependent on Notch signaling and KDM6. These findings provide a potential mechanistic explanation for innate drug resistance of GSCs, and emphasize the need to identify therapeutic strategies to eliminate this refractory tumor compartment.
Several cancer cell line models of reversible drug resistance, including lung cancer lines tolerant to EGFR inhibitor (Sharma et al., 2010) and melanoma lines resistant to BRAF inhibitors (Roesch et al., 2013), upregulate and become dependent on KDM5 enzymes, which remove an activating mark, H3K4 methylation. By contrast, while GSC persisters upregulate several KDMs, they show a particular dependency on KDM6 enzymes, which remove the repressive mark, H3K27me3. Consistently, the transition to the persister state is accompanied by widespread redistribution of this repressive modification, with striking loss occurring at developmental loci undergoing activation. We note that KDMs can also be induced by other stressors, including cell cycle arrest (Agger et al., 2009; Ene et al., 2012), DNA damage (Williams et al., 2014), hypoxia (Black et al., 2015; Xia et al., 2009), and inflammation (De Santa et al., 2007). The ensuing demethylation may allow activation of alternative cis-regulatory elements and pathways to support survival and adaptation (Easwaran et al., 2014). Chromatin structure is widely implicated in cell fate restriction, and its disruption by KDM-mediated demethylation appears central to the emergence of persister GSCs. Whether KDM6 enzymes play general roles in drug tolerance, or facilitate re-activation of primitive developmental programs in GSC persisters, awaits further study.
We also establish a critical and specific role for Notch signaling in the persister GSCs. Notch signaling has been shown to activate neurodevelopmental programs in NSCs (Aguirre et al., 2010; Imayoshi and Kageyama, 2011; Mizutani et al., 2007). Notch activity in GSCPer activates cis-regulatory elements that drive expression of master regulators of neurodevelopment and qNSCs. An important question remains whether the epigenetic switch between RTK- and Notch-dependent GSC states emulates the antagonism between these signaling programs in NSCs, wherein Notch signaling is associated with quiescence and RTK signaling with NSC activation. If so, proliferative heterogeneity within the tumor hierarchy may reflect aspects of a conserved developmental mechanism. A similar parallel was also suggested in low-grade glioma (Giachino et al., 2015), where Notch signaling may promote a less aggressive, slow-cycling population, suggestive of a tumor suppressor role. Yet in the context of drug treatment, this promotion of a slow-cycling state may have the untoward consequence of conferring resistance. The Notch-dependency of the refractory GSC state may thus portend opportunities for combination therapy, some of which are now being explored in pre-clinical and early clinical studies (Cenciarelli et al., 2014; Xu et al., 2016; Yahyanejad et al., 2016) .
In summary, we present a reversible epigenetic transition that enables GSCs to transition between proliferative and slow-cycling states, which may enable GBM tumors to propagate, adapt, and persist in the face of environmental and therapeutic pressures. Our work also reveals specific mechanisms by which developmental programs and epigenetic regulators facilitate this transition and sustain alternative states. A better understanding of these developmental programs and epigenetic mechanisms, and how they may be modulated, will be essential for developing effective therapeutic strategies that address malignant hierarchies and slow-cycling cancer cells in GBM and other tumors.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the lead contact Bradley Bernstein (Bernstein.bradley@mgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
GSC Derivation and Cell Culture
Patient-derived GBM culture lines were obtained from Massachusetts General Hospital (GSC4, GSC8, GSC26, GSC87, GSC114, GSC125) (Wakimoto et. al 2009) and Case Western Reserve University (CW1691, CW2907, CW3246). For CW1691, CW2907 and CW3246, GBM tissues were obtained from excess surgical materials from patients after review from a neuropathologist and used in accordance with an approved protocol by the Institutional Review Board at Cleveland Clinic 2559; GBM cells were derived immediately after dissociation of primary patient tumor (CW1961) or after transient xenograft passage (CW2907). GBM lines were maintained in Neurobasal medium (Life Technologies) supplemented with N2/B27, penicillin/streptomycin (Life Technologies), GlutaMAX (Life Technologies), recombinant human EGF (20 ng/mL, R & D systems), and recombinant human FGF2 (20 ng/mL, R & D systems). GSC114, GSC115, CW1691, CW2907, and CW3246 were additionally supplemented with recombinant human PDGF-AA and PDGF-BB (20 ng/mL each, Shenandoah Biotechnology). CW1691 was grown as adherent monolayers using laminin (5 µg/mL Engelbreth-Holm-Swarm laminin, Sigma).
GSC Persisters
For drug persister studies, fresh inhibitor and media were replenished every 4–6 days to GSC cultures. For downstream studies (e.g., ChIP, immunoblot, gene expression), viable cells were enriched using Lympholyte (Cedarlane) to viability levels comparable to untreated cultures.
Primary Patient Specimens
Single cell RNA-seq data for MGH26, MGH30, and MGH31 was taken from prior published studies (Patel et. al. 2014). Slides for immunohistochemistry for these tumors were cut from archived blocks kept in the MGH pathology tumor bank under Partners Institutional Review Board Protocol 2011P002334. For MGH64 and MGH66, samples were acquired fresh at time of surgical resection at the Massachusetts General Hospital. Patients were consented preoperatively according to the Institutional Review Board Protocol 1999P008145.
METHOD DETAILS
Cell Growth Assays
Freshly dissociated single cell suspensions were plated (96-well) in triplicate or quadruplicate at 1,000 to 10,000 cells/well for testing. For 4 day growth assays, CellTiter-Glo (Promega) was added to wells and end point luminescence was measured (BioTek Synergy HTX Platereader). For 12–15 day assays, 1 × compound in media was added at day 4–5 and all wells were replated with fresh media and compound at day 8–10 before viability was measured at day 12–15. For each inhibitor, n=3–5 replicates were used for each concentration, and each inhibitor was repeated in two to four independent experiments.
CRISPR-Cas9 and shRNA Experiments
CRISPR sgRNA sequences were designed according to Doench et. al. (Doench et al., 2014), selected to target the demethylase catalytic domain (Shi et al., 2015), and subcloned into lentiCRISPR v1. sgRNA targeting sequences are included in Table S4. Lentiviruses were produced using standard protocols. Briefly, CRISPR plasmids were cotransfected with GAG/POL and VSVG plasmids into 293T packaging cells using FuGENE HD (Promega) to produce virus. Viral supernatant was collected 72 h after transfection and concentrated using Lenti-X Concentrator (Clontech) following the manufacturer’s instructions. Approximately 1.25×106 adherently attached GSC cells (5 µg/mL Engelbreth-Holm-Swarm laminin, Sigma) were infected with concentrated virus for 24 h, and were selected 48 h after infection with puromycin (GSC4, GSC8: 1 µg/mL, GSC87: 2 µg/mL, Life Technologies) for 4 days. After 4–5 days of recovery, cells were dissociated and plated (96-well) in triplicate or quadruplicate at 2,500 to 5,000 cells/well. Growth was normalized to cells plated on day zero using an ATP standard curve measured at each time point. Each experiment was repeated in two to four independent experiments. Efficiency of genome editing was assessed by PCR amplification (PCR SuperMix, Life Technologies) of 50 ng of genomic DNA using primer sets surrounding the sgRNA-targeted region (~800 to 1,000 bp), followed by subjection of the resultant PCR products to SURVEYOR analysis according to the manufacturer’s protocol (Integrated DNA Technologies, 706020). PCR primers for SURVEYOR analysis are listed in Table S4. For shRNA knockdown experiments, the following lentiviral shRNAs were obtained from the RNAi Consortium in the pLKO.1 vector: PDGFRA (TRCN0000195132, TRCN0000196928, TRCN0000426196, TRCN0000419988) and GFP. Virus generation and growth testing for shRNA experiments followed essentially identical protocols outlined above except the growth assay was begun immediately after puromycin selection due to the immediacy of the phenotype. shRNA infection was performed in two biological replicates and the cell growth assay was performed with four replicates.
Overexpression Studies
The PDGFRA M260I ORF was PCR-amplified from p-DONR223-PDGFRA M260I using primers containing flanking Gateway attB sites and a stop codon, and then subcloned into p-DONR221. Site directed mutagenesis was performed using QuickChange Lightning Site-Directed Mutagenesis Kit according to the manufacturer’s instructions to generate wild-type PDGFRA (I260M) and PDGFRA D842V, which were subsequently subcloned into p-Inducer 20 (pIND20). Notch1 ICD ORF was PCR-amplified from Notch1 intracellular domain-pcw107 (Addgene, plasmid # 64621) using primers containing flanking Gateway attB sites and was subcloned into p-DONR221 and pIND20. dnMAML-GFP ORF was PCR-amplified from MigR1-dnMAML (Maillard et al., 2004) using primers containing flanking Gateway attB sites and was subcloned into p-DONR221 and pIND20. Virus production and infection of cells was carried out as described above, and GSCs were selected with G418 (500 µg/mL) for at least 1 week. pIND20 PDGFRA wt/D842V infected cells were induced with doxycycline (dox, 1 µg/mL) for at least 3 days before cell growth assays were performed. pIND20 dnMAML and pIND20 GFP infected GSC cells were induced with dox (1 µg/mL) for 4 days before mixed in equal proportion with uninfected GSC cells. Dox induction and mixing was performed independently in triplicate. The ratio of GFP+ to GFP− cells was measured by flow cytometry at the indicated timepoints. Experiment was performed in two biological replicates from the same pIND20 dnMAML and pIND20 GFP lines.
Flow Cytometric Analysis
For cell cycle analysis, ~1×106 viable dissociated cells were plated and treated with 1 µM EdU for 2 h. Cells were then washed with PBS, incubated with Zombie Aqua viability dye (Biolegend) or Live/Dead fixable far red dead cell stain kit (Life Technologies) for 20 min, washed with PBS, and then fixed with ice-cold 70% ethanol. Staining for EdU was performed using the Click-iT EdU Flow Cytometry Assay kit (Life Technologies) according to the manufacturer’s protocol. DNA content was visualized using FxCycle Violet (Life Technologies). Ki67 (clone B56, BD Biosciences) was stained for 30 min at ambient temperature. For surface marker analysis, cells were washed with PBS and stained with antibodies against CD15 (VIMC6, Miltenyi Biotec), CD133/1 (AC133, Miltenyi Biotec), or PDGFRα (16A1, BioLegend) for 30 min at 4 °C . Gates were determined using an unst ained control, where ~2% of cells were considered positive. All experiments were performed with at least two biological replicates.
Immunohistochemistry and Spectral Imaging of Primary GBM Specimens
Immunohistochemical staining of formalin-fixed, paraffin-embedded sections of four primary GBM specimens was done on a Leica Bond III automated workstation. Dual staining for activated NOTCH1 (N1ICD) and Ki67 was performed following antigen retrieval using Bond Epitope Retrieval 2 solution for 40 min. Slides were first incubated with antibody against N1ICD (D3B8, Cell Signaling Technology, 4147) at a 1:50 dilution for 60 min at ambient temperature, followed by incubation with a rabbit-specific secondary antibody linked to horseradish peroxidase (Bond Polymer Refine Detection kit). N1ICD staining was developed with diaminobenzidine (Leica Detection kit). Slides were then incubated with a mouse monoclonal antibody specific for Ki67 (8D5) at a 1:8,000 dilution for 30 min. Binding of anti-Ki67 was detected using the mouse-specific Bond Polymer Refine Red Detection kit, which detects staining using Fast Red. Slides were then counterstained with hematoxylin, dehydrated, and coverslipped.
To generate spectral libraries, single-stained tissue sections were imaged using the Mantra multispectral imaging platform (PerkinElmer). The spectrally resolved individual profiles between 420–720 nm of 3,3′-diaminobenzidine (DAB; N1ICD), Fast Red (Ki67) and hematoxylin counterstain were used to deconvolute staining patterns in double-stained tissue sections. Five representative areas of each stained tissue section (n = 2 per patient sample) were imaged at 20´ magnification and deconvoluted using the Inform 2.1 software package (PerkinElmer).
Immunoblotting
Cells were lysed on ice using RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS) supplemented with fresh protease (HALT, Thermofisher; PMSF, 1 mM) and phosphatase inhibitors (Thermofisher). After 30 min, benzonase was added with MgCl2 (1 mM) to clarify the cell lysate. Immunobloting was performed according to standard procedures.
Tumor Dissociation
MGH66 patient sample was confirmed to be glioblastoma by frozen section diagnosis at the time of resection. Tissue was minced into <1mm pieces with a scalpel and enzymatic ally dissociated using the Brain Tumor Dissociation Kit (Miltenyi Biotec). Digested tissue was strained through a 100 micron cell strainer, washed in PBS and then layered carefully onto a 5mL density gradient (Lympholyte-H, Cedar Lane labs), which was centrifuged at 2,000 rpm for 10 min at room temperature. Dead cells, debris, and RBCs were pelleted and live cells collected from the interface and used for downstream applications including staining, flow cytometry, and single cell analysis. Viability was measured using trypan blue exclusion, which confirmed >90% cell viability.
Fluorescence-activated cell sorting of primary tumors
Tumor cells were suspended in 1% bovine serum albumin in Hanks buffered saline solution (BSA / HBSS) for blocking, and stained first with CD45-Vioblue direct antibody conjugate (Miltenyi Biotec) for 30 min at 4 °C . After washing in cold PBS, cells were resuspended in 1 mL of BSA/HBSS containing 1 µM calcein AM (Life Technologies) and 0.33 µM TO-PRO-3 iodide (Life Technologies) and stained for 30 minutes prior to cell sorting. Cells were run on a FACSAria Fusion Special Order System (Becton Dickinson) using 488nm (calcein AM, 530/30 filter), 640nm (TO-PRO-3, 670/14 filter), and 405nm (Vioblue, 450/50 filter) lasers. Singlet gating was performed using strict forward scatter height versus area criteria to eliminate doublets. Viable cells were identified by staining positive with calcein AM but negative for TO-PRO-3. Single cells were sorted into 96-well plates containing cold buffer TCL buffer (Qiagen) containing 1% β-mercaptoethanol, snap frozen on dry ice, and then stored at –80 °C prior to whole t ranscriptome amplification, library preparation, and sequencing.
Single-cell whole transcriptome amplification, library construction, and sequencing
Libraries from isolated single cells were generated based on the Smart-seq2 protocol (Picelli et al., 2014) with the following modifications. Agencourt RNAClean XP beads (Beckman Coulter) were used to purify single cell RNA prior to oligo-dT primed reverse transcription with Maxima reverse transcriptase and locked TSO oligonucleotide. Amplification of cDNA libraries was done with 20 cycle PCR amplification using KAPA HiFi HotStart ReadyMix (KAPA Biosystems) with subsequent Agencourt AMPure XP bead purification as described. Library preparation by tagmentation was done using the Nextera XT Library Prep kit (Illumina) with custom barcode adapters (sequences available upon request). Libraries from 384 cells from MGH66 with unique barcodes were combined and sequenced using a NextSeq 500 sequencer (Illumina).
Real-Time Quantitative RT-PCR and RNA-seq Library Preparation
Whole RNA was extracted from 1–3×106 cells using the QIAGEN RNeasy kit according to the manufacturer’s protocol. For qRT-PCR, total RNA was reverse-transcribed into cDNA (High Capacitiy cDNA Reverse Transcriptase Kit, Applied Biosystems) and qRT-PCR amplification was performed in triplicate using fast SYBR Green Master Mix (Life Technologies) with specific PCR primers for genes of interest and 18S as an endogenous control. qRT-PCR experiments were performed with two biological replicates. Relative quantification for each target was performed using the comparative Ct method (Applied Biosystems). Sequences for qRT-PCR primers used are listed in Table S4. For RNA-seq library preparation, Poly(A)+ RNA was enriched using magnetic oligo(dT)-beads (Life Technologies) and then ligated to RNA adaptors for sequencing. RNA-seq was performed with three biological replicates per GSC line and/or condition.
ChIP-seq
Briefly, formaldehyde-fixed cells were lysed and sheared (Branson S220) on wet ice. The sheared chromatin was cleared and incubated overnight at 4 °C with the following antibodies: H3K4me3 (Millipore, 07-473, Lot 2207275), H3K27me3 (Millipore, 07-449, lot 2382150), H3K27ac (Active Motif, 39133 lot 25812006), RBPJ (Abcam, ab25949, lot GR169397-1), and cleaved Notch1 (CST, 4147, lot #4). Antibody-chromatin complexes were immunoprecipitated with protein G magnetic Dynal beads (Life Technologies), washed, eluted, reverse crosslinked, and treated with RNAse A followed by proteinase K. ChIP DNA was purified using Ampure XP beads (Beckmann Coulter) and then used to prepare sequencing libraries for sequencing with the Next-Seq Illumina genome analyzer. Experiments were performed once.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the exact value of n, the definition of center, dispersion, and precision measures (mean ± SD or SEM) and statistical significance are reported in Figures and Figure Legends, as well as below. As a general rule, we considered data to be statistically significant when p < 0.05 by two-tailed Student’s T-Test, Wilcoxon rank-sum test or a Friedman test where appropriate.
Levels phosphorylated PDGFRα, Akt, and Erk1/2
For each assay n = 2 biological replicates involving separate drug treatments.
Cell growth assays (Cell-titer Glo)
N=3–4 replicates per cell line. N=2–4 independent experiments per compound, and 3–5 replicates per concentration (see figure legends for more detail). For overexpression studies n= 2. Relative CTG (Cell-tier Glo) values were normalized to T0 (y-axis) over a time course (x-axis) following treatment. Error was calculated as either s.d or s.e.m across all biological replicates. IC50 values: Calculated as standard, from at least three biological replicates.
CRISPR-Cas9 and shRNA experiments
Each CRISPR-Cas9 sgRNA experiment was repeated in two to four independent experiments involving separate infections; in each experiment, cells were plated in n = 3–5 replicates for the cell growth assay. shRNA infection was performed in two biological replicates and the cell growth assay was performed with n = 4 replicates. Growth was normalized to cells plated on day zero using an ATP standard curve measured at each time point.
Overexpression studies
Dox induction and mixing was performed independently in triplicate. The ratio of GFP+ to GFP− cells was measured by flow cytometry at the indicated timepoints, and error bars were calculated as s.e.m. across all three biological replicates (indicated in the figure legends). Experiment was performed in two biological replicates from the same pIND20 dnMAML and pIND20 GFP lines.
Flow Cytometric Analysis
PDGFRα staining was performed with two biological replicates, while staining for CD133 and CD15 were performed with three biological replicates. Gates were determined using an unstained control, where ~2% of cells were considered positive. Error bars were calculated as s.e.m. across three replicates and significance was determined using a two-tailed Student’s T-Test. Cell cycle analysis was performed with at least three biological replicates per time point, and gates were determined for each timepoint by the relative proportions of DAPI and EdU labelling.
Immunohistochemistry and Spectral Imaging of Primary GBM Specimens
Single slides from four tumors were stained with antibodies against Notch1 and Ki67 (see above methods) in two separate replicates. From each slide, five representative regions were quantified using spectral imaging, yielding >10,000 cells quantified per sample over two replicates.
Single-cell RNA-seq data analysis
Libraries from 384 cells from a single tumor (MGH66) were combined and sequenced. Paired-end, 38-base reads were mapped using Bowtie (Langmead et al., 2009) with parameters “-q --phred33-quals -n 1 -e 99999999 -l 25 -I 1 -X 2000 -a -m 15 -S -p 6” to the UCSC hg19 human transcriptome, which permits alignment of sequences with single base changes to account for point mutations. RSEM v1.2.3 (Li and Dewey, 2011) in paired end mode using parameters “--estimate-rspd --paired end -sam -p 6” was used to quantify expression in the form of normalized transcripts per million (TPM) for each gene in each single cell. Data for tumors MGH26, MGH28, and MGH30 were prepared as previously published in (Patel et al., 2014).
To remove low quality data for each tumor, we excluded all cells with less than 3,000 detectable genes and all genes with a mean TPM less than 10. The TPM values were subjected to base 2 logarithmic transformation after adding 1 to each value to avoid singularities. To account for global mean expression differences across cells, for each cell a size factor was calculated as described in (Anders and Huber, 2010) with the following modifications: i) computations were performed in log-space and adjusted accordingly; ii) the mean over all genes as opposed to the median was used ultimately. The cell-specific size factors were subtracted from the transformed TPM values. To assess the cell cycle state we used the cell cycle signature derived in our previous study (see (Patel et al., 2014) for details). To score single-cells for individual signatures (e.g. cell cycle or persister signatures) we computed mean TPM values for the corresponding signature gene sets and transformed resulting values into standard scores (z-scores). Gene signatures used are listed in Table S3.
Population RNA-seq analysis
Paired-end reads were aligned to UCSC transcriptome (hg19) using Bowtie (Langmead et al., 2009) (version 0.12.7) with the following parameters: --chunkmbs 512 -q --phred33-quals -n 0 -l 25 -I 1 -X 2000 -p 6 -a -m 15. Gene expression, quantified as transcript per million (TPM), was estimated using RSEM (Li and Dewey, 2011) with the following parameters: --fragment-length-max 1000 --estimate-rspd --paired-end.
Data processing was performed using R version ≥3.1.0 (R Core Team, 2014; http://www.Rproject.org/) and BioConductor (http://www.bioconductor.org). All heatmaps were generated using the R package ggplot2 (Wickham 2009). To identify variable genes across the time course of drug treatment, triplicate RNA-seq data sets were generated for each time point and processed as outlined above. The resulting TPM values were employed for differential gene expression analysis using the R package EBSeq (Leng et al., 2015). To identify variable genes the following pairwise comparisons were performed: GSC8 naïve versus GSC812d, GSC8 naïve versus GSC8Per, GSC812d versus GSC8Per. We considered all genes with a posterior probability of differential expression (PPDE) greater than 0.5 in at least one of the comparisons as variable genes. A table of all genes and significantly differentially expressed genes is provided in Table S2. GSEA version 2.1 (Subramanian et al., 2005) was carried out using signal-to-noise on log2 + 1 transformed TPM values as the metric; genes with mean TPM less than 10 under all given conditions were excluded from analysis. Gene signatures used for GSEA are listed in Table S3. For the comparison with GTEx expression data (GTEx Consortium, 2013), RPKM values were computed.
ChIP-seq data analysis
All experiments were performed once. Reads were aligned to hg19 using BWA (Li and Durbin, 2009) and identical ChIP-seq sequence reads were collapsed to avoid PCR duplicates. In order to avoid possible saturation biases, reads were downsampled to approximately similar numbers (~20 million reads). Peaks were called using HOMER v4.6 (Heinz et al., 2010) using matched inputs with the following parameters: H3K4me3, –histone –tagThreshold 30; H3K27ac, –histone –tagThreshold 50; H3K27me3, –histone –size 3000 –minDist 2500 –F 1.5 –L 0 –FDR 0.1; Notch1 ICD, –factor; and RBPJ, –factor. TF motif enrichment analysis was performed using HOMER v4.6 on 1 kb windows centered on previously called H3K27ac, Notch1 ICD, and RBPJ peaks (parameters: –size given –mask) and p-values presented were provided by the software. Quantification of the H3K27me3 ChIP-seq signals, using fore- and background normalization, was essentially performed as outlined in (Zhu et al., 2013) with few modifications. To exclude genomic regions with potential copy number aberrations, we quantified read counts derived from whole cell extracts ChIP-seq within 5 kb genomic windows. Read counts were performed using the R package GenomicRanges (Lawrence et al., 2013). To compute normalization constants, all genomic windows with read counts equal to 0 or greater than 3 standard deviations from the mean were excluded from the analysis. For a given H3K27me3 ChIP-seq data set, the background constant was estimated as the median of the read count distribution within 5 kb windows overlapping the 5% most highly expressed genes, as active genes are considered to be devoid of repressive chromatin modifications. The foreground signal was estimated as the median of the read count distribution within 5 kb windows centered around each called peak and subtracting the background signal. To quantify H3K27me3 signal within 5 kb regions of interest, the background signal was subtracted from the corresponding read count and the resulting value divided by the foreground signal. Values smaller or larger than 0 or 1 were mapped to 0 or 1, respectively. The fore- and background normalization of H3K27ac was performed analogously but with the following modifications. To account for the narrower signal, all operations were performed using 1 kb windows. For a given H3K27ac ChIP-seq data set, the background signal was estimated as the median of the read count distribution within all 1 kb windows across the genome but not overlapping with a called peak. To account for greater dynamic range, the foreground signal was estimated as the 0.95-quantile of the read count distribution within 1 kb windows centered around each called peak and subtracting the background constant. To estimate the H3K27ac signal within a 1 kb region of interest, the background constant was subtracted from the corresponding read count and the resulting value was divided by the foreground constant. Values smaller or larger than 0 or 1 were mapped to 0 or 1, respectively. In order to quantify ChIP-seq intensities within genomic regions of different sizes (e.g. genes), the obtained normalization constants were scaled accordingly. Association of peaks with neighboring genes was performed using the R package ChIPpeakAnno (Zhu et al., 2010).
ChIP-seq Binding Profiles
ChIP-seq profile plots were generated using ngs.plot (Shen et al., 2014). To assess H3K27me3 levels across broad domains, H3K27me3 metaprofiles were generated over the union of peaks, called in each of the corresponding GSC8 persister conditions, and having a minimum size of 10 kb.
Supplementary Material
Acknowledgments
We thank S. Muller-Knapp and the Structural Genomics Consortium (Oxford, UK), Russell Ryan, Nicolò Riggi, and Henry Pelish for helpful discussions. We thank David Sabatini and Kris Wood for Notch1 intracellular domain-pcw107, and William Hahn and David Root for p-DONR223-PDGFRA M260I. B.B.L. is supported by a Jane Coffin Childs Memorial Fund Postdoctoral Fellowship. C.S. is supported by a European Molecular Biology Organization (EMBO) fellowship (ALTF 654-2014) and a Swiss National Science Foundation fellowship. P.v.G is supported by an EMBO Fellowship (ALTF 1207-2014). A.P.P is supported by NIH/NINDS R25NS065743. W.A.F. is supported by an American Brain Tumor Association grant. B.E.B. is an American Cancer Society Research Professor. This research was supported by funds from the Howard Hughes Medical Institute, the National Human Genome Research Institute, the National Cancer Institute, and the National Brain Tumor Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw data files for the gene expression and ChIP-seq analysis is NCBI Gene Expression Omnibus: GSE74557.
Supplemental Information
Seven supplementary figures and five tables.
Author Contributions B.B.L., C.S., A.P.P., and B.E.B. designed the study and wrote the manuscript. B.B.L., A.P.P., L.K.D., S.M.G., T.E.M., A.S.V., C.H.H., C.D.C., S.J.R., S.J.S., and F.J.N. performed experimental work. C.S., B.B.L., and W.A.F. performed computational analyses. P.v.G., H.W., J.N.R., J.C.A, and M.L.S provided reagents and conceptual advice.
References
- Agger K, Cloos PAC, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–327. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Black BL, Ligon KL, Zhang Y, Olson EN. Cooperative transcriptional activation by the neurogenic basic helix-loop-helix protein MASH1 and members of the myocyte enhancer factor-2 (MEF2) family. Journal of Biological Chemistry. 1996;271:26659–26663. doi: 10.1074/jbc.271.43.26659. [DOI] [PubMed] [Google Scholar]
- Black JC, Atabakhsh E, Kim J, Biette KM, Van Rechem C, Ladd B, Burrowes PD, Donado C, Mattoo H, Kleinstiver BP, et al. Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 2015;29:1018–1031. doi: 10.1101/gad.259796.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenciarelli C, Marei HES, Zonfrillo M, Pierimarchi P, Paldino E, Casalbore P, Felsani A, Vescovi AL, Maira G, Mangiola A. PDGF receptor alpha inhibition induces apoptosis in glioblastoma cancer stem cells refractory to anti-Notch and anti-EGFR treatment. Mol. Cancer. 2014;13:247. doi: 10.1186/1476-4598-13-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, DeLeo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Dewaele B, Wasag B, Cools J, Sciot R, Prenen H, Vandenberghe P, Wozniak A, Schöffski P, Marynen P, Debiec-Rychter M. Activity of dasatinib, a dual SRC/ABL kinase inhibitor, and IPI-504, a heat shock protein 90 inhibitor, against gastrointestinal stromal tumor-associated PDGFRAD842V mutation. Clin. Cancer Res. 2008;14:5749–5758. doi: 10.1158/1078-0432.CCR-08-0533. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Tsai H-C, Baylin SB. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder K, Kalman B. Molecular heterogeneity of glioblastoma and its clinical relevance. Pathol. Oncol. Res. 2014;20:777–787. doi: 10.1007/s12253-014-9833-3. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, Lai C, Belova G, Cam M, Walling J, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS ONE. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi RMR, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 Silencing in Glioblastoma Tumor-Initiating Cells Causes Stop of Proliferation and Loss of Tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Giachino C, Boulay J-L, Ivanek R, Alvarado A, Tostado C, Lugert S, Tchorz J, Coban M, Mariani L, Bettler B, et al. A Tumor Suppressor Function for Notch Signaling in Forebrain Tumor Subtypes. Cancer Cell. 2015;28:730–742. doi: 10.1016/j.ccell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, Fang D, Huang X, Tom MW, Ngo V, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, Labelle M, Gerlach L-O, Birk P, Helin K. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514:E1–E2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulleman E, Quarto M, Vernell R, Masserdotti G, Colli E, Kros JM, Levi D, Gaetani P, Tunici P, Finocchiaro G, et al. A role for the transcription factor HEY1 in glioblastoma. J. Cell. Mol. Med. 2009;13:136–146. doi: 10.1111/j.1582-4934.2008.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential Roles of Notch Signaling in Maintenance of Neural Stem Cells in Developing and Adult Brains. Journal of Neuroscience. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R. The role of Notch signaling in adult neurogenesis. Mol. Neurobiol. 2011;44:7–12. doi: 10.1007/s12035-011-8186-0. [DOI] [PubMed] [Google Scholar]
- Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, Strain-Damerell C, Gileadi C, Philpott M, Burgess-Brown N, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014 doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, Liu F, Saunders LM, Mullally A, Abdel-Wahab O, et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2013;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- Kruidenier L, Chung C-W, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2013;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. C cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Leng N, Li Y, McIntosh BE, Nguyen BK, Duffin B, Tian S, Thomson JA, Dewey CN, Stewart R, Kendziorski C. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics. 2015;31:2614–2622. doi: 10.1093/bioinformatics/btv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Louis DN and International Agency for Research on Cancer. WHO classification of tumours of the central nervous system. 4th. Lyon: International Agency for Research on Cancer; 2007. [Google Scholar]
- Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urbán N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, et al. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, M Mastrogianakis G, Olson JJ, Mikkelsen T, Lehman N, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, Alberta JA, Kane MF, Theisen M, Ligon KL, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K-I, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J-H, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae S-S, Zheng H, Ying H, Mahoney J, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DAM, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinbay E, Suvà ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Körbel C, Laschke MW, Gimotty PA, Philipp SE, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J. Identification of human brain tumour initiating cells. Nature. 2004 doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam D-H, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al. Reconstructing and Reprogramming the Tumor-Propagating Potentialof Glioblastoma Stem-like Cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Aoyagi M, Ando N, Ogishima T, Wakimoto H, Yamamoto M, Ohno K. Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy. J. Neurosurg. 2013;119:1145–1155. doi: 10.3171/2013.7.JNS122417. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenuesfor glioblastoma: no longer a dead end? Nature Reviews Clinical Oncology. 2012;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, Kushida M, Head R, Morrissy S, Zhu X, et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26:33–47. doi: 10.1016/j.ccr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu T-C, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Research. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. 1st. New York: Springer; 2009. [Google Scholar]
- Williams K, Christensen J, Rappsilber J, Nielsen AL, Johansen JV, Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS ONE. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Shimizu F, Hovinga K, Beal K, Karimi S, Droms L, Peck KK, Gutin P, Iorgulescu JB, Kaley T, et al. Molecular and Clinical Effects of Notch Inhibition in Glioma Patients: A Phase 0/I Trial. Clin. Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyanejad S, King H, Iglesias VS, Granton PV, Barbeau LMO, van Hoof SJ, Groot AJ, Habets R, Prickaerts J, Chalmers AJ, et al. NOTCH blockade combined with radiation therapy and temozolomide prolongs survival of orthotopic glioblastoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.