Abstract
RNA is involved in a variety of chromatin modification events, ranging from large-scale structural rearrangements to subtle local affects. Here, we extend the evidence for RNA–chromatin interactions to the centromere core. The data indicate that maize centromeric retrotransposons (CRMs) and satellite repeats (CentC) are not only transcribed, but that nearly half of the CRM and CentC RNA is tightly bound to centromeric histone H3 (CENH3), a key inner kinetochore protein. RNAs from another tandem repeat (180-bp knob sequence) or an abundant euchromatic retroelement (Opie) are undetectable within the same anti-CENH3 immune complexes. Both sense and antisense strands of CRM and CentC, but not small interfering RNAs homologous to either repeat, were found to coimmunoprecipitate with CENH3. The bulk of the immunoprecipitated RNA ranged in size from 40 to 200 nt. These data provide evidence for a pool of protected, single-stranded centromeric RNA within the centromere/kinetochore complex.
One of the most highly specialized yet poorly understood regions of the chromosome is the centromere (1). Tandem repeat arrays are the only identifying feature of most higher eukaryotic centromeres, and even these diverge at astonishing rates. In several well characterized centromeres, tandem repeats are completely absent (e.g., ref. 2). The rapid sequence evolution, combined with the fact that centromeres from one species can sometimes function in related species (3), suggests that epigenetic determinants are involved in establishing the centromeric state. Although the mechanisms that establish and maintain centromeres are not known, one of the first steps in the process must involve the deposition of the histone H3 variant, centromeric histone H3 (CENH3). CENH3 is generally viewed as the core of the centromere (1, 4), and is sufficient to recruit the major components of the inner kinetochore (5). How a cell targets CENH3 to the proper chromosomal location is a key question in centromere biology.
DNA replication occurs during S phase and is coincident with the deposition of the core histones. However, CENH3 is left out of this replication-coupled process and is deposited later in a replication-independent (RI) fashion (6, 7). CENH3 RI deposition requires specialized centromeric targeting as well as a poorly understood histone exchange reaction (8). An early event in histone replacement is likely to be transcription, which can disrupt nucleosomes (9) and, in principle, facilitate the incorporation of replacement histones (10). Indeed, an analysis of a human neocentromere (that had recently formed over a gene-containing region) demonstrated that 15 of the 51 genes present produced apparently normal transcripts (11). Similarly, rice centromere 8 contains 14 genes, four of which are actively expressed (12). Two other studies provide direct links between transcription and centromere activation: in humans, selection for transcription flanking an ectopic centromere was shown to promote CENH3 (CENP-A) recruitment (13), and in Schizosaccharomyces pombe, the GATA-like transcription factor Ams2 is required to initiate CENH3 (SpCENP-A) deposition and centromere formation (14).
There is also a considerable amount of evidence suggesting that RNA facilitates the targeting of chromatin-modifying complexes to specific regions of the genome (15). A major breakthrough was the demonstration by Volpe et al. (16) that RNA interference (RNAi) is required to establish a heterochromatic state in the pericentromeric domains that flank the (CENH3-binding) centromere core of S. pombe. Mutations in the RNAi pathway release pericentromeric repeats from transcriptional repression and perturb normal centromere function (16–19). Similarly, treatment of mouse cells with RNase causes the release of Heterochromatin Protein 1 and changes the spatial organization of histones in pericentromeric regions (20). These data and the discovery and characterization of the RNA-induced initiation of transcriptional silencing (RITS) effector complex (21) support a model whereby small interfering RNAs (siRNAs) directly target homologous DNA sequences for chromatin modification (16, 21). However, notably, RNAi mutations do not appear to affect the transcriptional status or protein composition of the functionally distinct CENH3-containing centromere core domains in either fission yeast or chicken cell lines (16, 19, 22).
Although kinetochores differ in morphology from species to species, recent data have established that an important group of kinetochore proteins are conserved from Saccharomyces cerevisiae to humans (23). The fact that budding yeast has many of the same kinetochore proteins found in more complex eukaryotes suggests that the large plant and animal centromeres represent multiple iterations of the simple Saccharomyces cerevisiae point centromere (24). A clue to higher order structure in humans came from chromatin immunoprecipitation (ChIP) experiments demonstrating that progressive micrococcal nuclease (MNase) digestion releases large complexes of constitutive centromere proteins before releasing the individual components (25). The molecules involved in the formation and stabilization of large-scale centromeric chromatin structure are not known. However, in several other chromatin protein complexes, RNA is an integral component. For instance, RNA is a known component of sex chromosome dosage compensation complexes in mammals and Drosophila (26, 27), of human pericentromeric heterochromatin (20), and of the yeast telomerase complex (28).
The experiments described here were designed to test the hypothesis that the repeats of the maize centromere core are transcribed and that the resulting transcripts are bound in some fashion to centromeric chromatin (10, 29). In maize, CENH3 binds to 156-bp CentC repeat arrays and centromeric retrotransposable (CR) elements that are arranged in nearly continuous, intermingled arrays and clusters (3). Both CR elements and the centromeric satellite repeat CentC coimmunoprecipitate with maize CENH3, supporting the view that the retroelements cooperate with tandem repeat arrays to assemble a functional kinetochore (30). Here we used a variation of the sensitive native ChIP technique to show that CR and CentC RNA are tightly associated with the maize kinetochore. Subsequent analysis of the RNA revealed significant quantities of both strands of each repeat, ranging in size from 40 to 900 bp. The data show that, as within the S. pombe pericentromere (16), some level of transcription is a native feature of the centromere core and suggest a potential role for noncoding RNA in the specification of centromeric chromatin.
Methods
EST Analysis. Fourteen ESTs were characterized and listed below with respect to where they align on the consensus CR element from maize 2 (CRM2) element (GenBank accession no. AY129008). The EST CB278268 aligns to bases 800-1093 of CRM2, CB179846 aligns to bases 508-1062, CB278262 aligns to bases 797-1075, CB278268 aligns to bases 800-1093, AW076314 aligns to bases 646-1198, AW065493 aligns to bases 732-1204, AW076306 aligns to bases 657-1264, CB179288 aligns to bases 900-1376, CB278333 aligns to bases 760-1456, and BM660209 aligns to bases 7039–7391; each of these is homologous to the sense strand of CRM2 (31). The EST AW017992 aligns to bases 1458–847, and AW017999 aligns to bases 1458–890; these are homologous to the antisense strand of CRM2. BM335652 aligns to bases 6825–7300, and BM349031 aligns to bases 6825–7292; these sequences were derived from the sense strand and are terminated by poly(A) tracts within the 3′ LTR.
Immunoprecipitation and Slot-Blot Analysis. In our previously published ChIP protocol (30), we used MNase to digest the chromatin before immunoprecipitation. Although MNase is an excellent reagent for digesting maize chromatin, it is a known RNase (32). Preliminary studies established that, when MNase-prepared samples were blotted for RNA and probed with centromere repeats, the signal-to-noise (s/n) ratios were unacceptably low (<2 in three different experiments). Therefore, in all studies reported here, nuclei from the W23 inbred strain were isolated (30), and for each ChIP, ≈50 OD units (measured at 260 nm) were treated with RNase-free DNase I (Promega) for 10 min at 37°C and a concentration of 4 units/mg DNA. Fragments of ≈300–800 nt gave us the highest recovery (percentage of immunoprecipitation, %IP) of centromeric DNA.
To effectively separate DNA and RNA in our ChIP samples, we took advantage of the fact that nucleic acid hybridization occurs efficiently only on single-stranded molecules. When a sample is treated with formamide and heated slightly before blotting (68°C in 1× SSC/7% formamide/50% formaldehyde for 15 min), RNA is preserved, but the DNA remains double-stranded. Although both DNA and RNA will bind to a nylon membrane under these conditions (33, 34), only the RNA is freely available for hybridization (we used N+ Hybond membranes, Amersham Pharmacia). Conversely, when samples are treated with a high-pH denaturant (0.4 M NaOH) before blotting, DNA is well preserved and readily detected after hybridization, whereas RNA (which is unstable at high pH) is barely visible.
The %IP was defined as P/(P + S) from CENH3 Ab – P/(P+S) from the preimmune serum (S, supernatant; P, pellet). s/n ratios were calculated under the assumption that noise is the fraction of nuclear RNA immunoprecipitated by preimmune serum, and signal is the fraction immunoprecipitated by anti-CENH3 antibodies: s/n = [P/(P+S)] from anti-CENH3 treatment divided by the [P/(P+S)] from preimmmune control. For s/n calculations, preimmune %IP values were rounded up to 1.0 when the actual numbers were less. For the data shown in Fig. 2 A and D, immunoprecipitated samples were treated with RNaseA (0.4 μg/μl) at 37°C for 10 min after the sample had been treated with phenol/chloroform to remove chromatin proteins, antibodies, and other protein reagents.
Fig. 2.
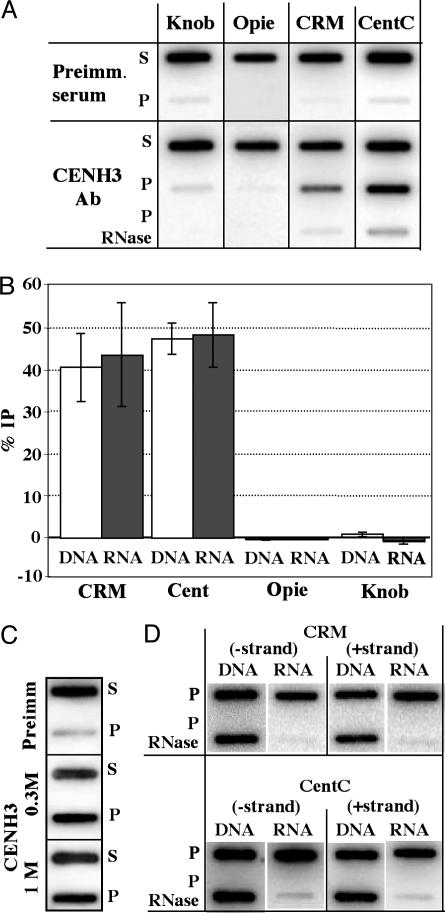
Centromere-encoded RNAs are coimmunoprecipitated with CENH3. (A) Chromatin samples were immunoprecipitated with anti-CENH3 antibodies, blotted for RNA, and probed with DNA probes for the sequences indicated. Supernatant (S) and pellet (P) fractions for the preimmune and anti-CENH3 treatments are shown. As shown in the bottom lane, RNaseA treatment removed the majority of RNA hybridization. These slot-blot images were used to acquire the numerical data in Table 1, experiment 1. (B) The RNA and DNA %IPs from five different experiments. The RNA %IP data (from Table 1) and associated DNA %IP values from each of five experiments are shown as mean ± SE. (C) High ionic strengths have a minimal impact on the immunoprecipitation of CentC RNA. The preimmmune (S and P) fractions are shown in the top two lanes, standard ChIP (using a 0.3 M NaCl wash) is shown in the middle two lanes, and standard ChIP after a 1 M NaCl wash is shown in the bottom two lanes. This experiment was not repeated in kind, but a second experiment with 0.7 M NaCl gave similar results. (D) Both strands of CRM and CentC are present after anti-CENH3-mediated ChIP. P fractions were blotted for the presence of DNA and RNA, either with or without prior RNaseA treatment. The sense and antisense strands are defined in Methods. This experiment was repeated three times with essentially identical results.
A plasmid containing a 1.7-kb fragment of the Opie GAG domain was obtained from Chris Della Vedova via Chris Lamb of the James Birchler laboratory (University of Missouri, Columbia). The clone is homologous to Opie B (GenBank accession no. AF466932, base pairs 41014–42769; ref. 35). All other DNA probes have been described (30). Strand-specific CRM and CentC probes were prepared by cloning the GAG.90 and CentC inserts (30) in both orientations into pBluescript (Stratagene), and transcribing the sequence with T7 polymerase (Riboprobe kit, Promega). The sense GAG.90 construct expresses the sense strand of the CRM element (GenBank accession no. AY129008), whereas the “sense” construct of CentC expresses the strand reported in GenBank under accession no. AF078923. Hybridization experiments involving single-stranded (M13-generated) DNA molecules (data not shown) and single-stranded RNA oligonucleotides (see Fig. 3A) demonstrated that the CRM riboprobes were essentially 100% strand-specific.
Fig. 3.
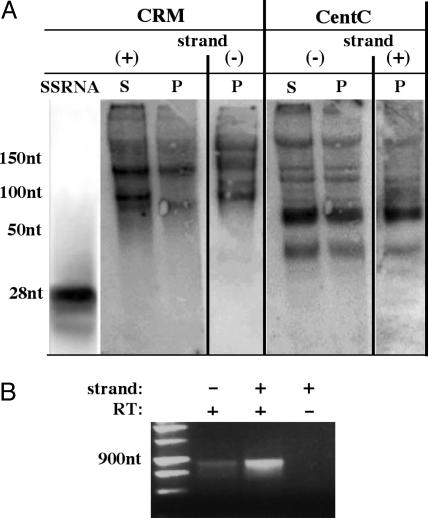
Centromeric siRNAs are not detected after CENH3-mediated ChIP. (A) The bulk of immunoprecipitated centromeric RNA ranges from 40 to 250 nt in length. The supernatant (S) and pellet (P) fractions are shown for both CRM and CentC. For the P fraction, hybridization with the opposite strand is shown. Hybridization to a 28-nt single-stranded synthetic RNA (SSRNA) homologous to the sense strand of the CRM GAG domain is shown at left. The 28-nt marker is underexposed relative to the other lanes. DNA markers (not shown, but indicated in nt) were used for higher molecular mass estimates. (B) Fragments (≈900 bp) of the CRM GAG domain were detected by strand-specific RT-PCR. This technique is not quantitative. The absence of product when no reverse transcriptase (RT–) was added indicates that the bands were derived from immunoprecipitated RNA, not DNA.
PCR. For the data shown in Fig. 1B, cDNA was prepared by using the Clontech SMART cDNA library construction kit. The library was verified as free of detectable DNA by amplifying a portion of the maize CenH3 known to contain an intron. PCR was carried out on the Clontech-prepared cDNA by using the following primers: GAG.65 (forward) 5′-AGGGAAATTCAGGACATCCTTGCTTA-3′ and (reverse) 5′-GATTCGGCAAAGATGCACCAGGAA-3′; GAG.90 (forward) 5′-CTGTTTGGTGGATAGAACATGGTAAGA-3′ and (reverse) 5′-GATTCGGCAAAGATGCACCAGGAA-3′; RT.42 (forward) 5′-ATGCAGCATTCTTTGCCTCCTGTTA-3 and (reverse) 5′-TCGTATGAAATTGGGAGATGAATGGAAA-3′; INT.56 (forward) 5′-TTGAATGTGATGCTAGTGGAATTGGA-3′ and (reverse) 5′-CCTCCATGCGCCTCCTGTAACAACAAAAG-3′; LTR.32 (forward) 5′-TTGGAATGTTCAAGCACAACATGGAA-3′ and (reverse) 5′-GCAAGTAGCGAGAGCTAAACTTGA-3′. Strand-specific RT-PCR (see Fig. 3B) was carried out on immunoprecipitated fractions that were digested twice with DNase (at 0.4 units/μl), by using primers for the CRM GAG.90.
Fig. 1.
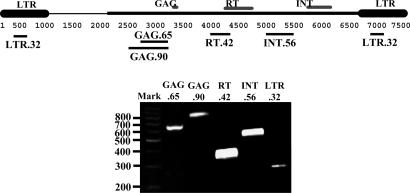
CRM is actively transcribed. (Upper) A map of CRM element (ref. 30, later called CRM2; ref. 31). The approximate locations of the GAG, reverse transcriptase (RT), and integrase (INT) domains are shown. (Lower) An agarose gel showing that several internal regions of CRM2 are readily amplified by RT-PCR. The locations of the amplified regions are indicated on the map.
RNA Detection by PAGE. For the data shown in Fig. 3A, samples were incubated for 10 min at 37°C with an excess of RNase-free DNase I (0.4 units/μl; >1,000 times the concentration used to digest chromatin for immunoprecipitation). It is difficult to estimate the quantity of RNA loaded, although each lane contains ≈20% of a ChIP experiment. Samples were added to RNA loading buffer containing formamide, heated for 5 min at 95°C, and electrophoresed on 15% denaturing polyacrylamide (7 M urea) gels. The marker lane contained 0.5 nmol of a 28-nt single-stranded RNA identical to the CRM GAG sense strand (5′-CCAAAUCUGCCCAGAAACCAGCAGGUA-3′). Gels were transferred to either N+ Hybond or Ambion Nylon 66 membranes and hybridized with strand-specific RNA probes. All data from ChIP samples were exposed to a PhosphorImager for 18–21 h. The exposure time for the 28-nt small RNA marker was 30 min.
Results
CRM Is an Expressed Retroelement. During our initial studies of CRM (30), we noticed that there were at least 18 CRM-homologous ESTs in GenBank (listed in Methods). The majority (14 of 18) of these ESTs are nonchimeric, suggesting that they were initiated from within the retroelement, and 12 of the nonchimeric transcripts are derived from the sense strand. Furthermore, at least two ESTs terminate in polyadenine tracts within the 3′ LTR, suggesting that a subset of the ESTs represent legitimate CRM-initiated cDNAs. Although we were not able to recover full-length cDNAs, at least five different regions of CRM were easily amplified by RT-PCR from poly(A)-selected mRNA (Fig. 1B).
Centromeric RNA Is Immunoprecipitated with CENH3 Ab. To test for the presence of RNA at maize centromeres, we developed and used a (RNase-free) DNase I-based chromatin preparation method. Chromatin was immunoprecipitated with preimmune and anti-CENH3 antisera, and RNA was immobilized on nylon membranes by using an RNA slot blotting protocol. Blots containing the supernatant (S) and pellet (P) fractions from both treatments were sequentially probed with the known centromeric sequences, CentC and CRM, and two negative controls: the 180-bp knob repeat (36), a tandemly arrayed sequence located exclusively on chromosome arms (37), and Opie B, a retrotransposon with a uniform euchromatic (generally noncentromeric) distribution (38, 39).
When RNA slot blots were probed with centromeric sequences, strong hybridization was observed in the pellet fractions. CRM and CentC signal intensities were far above background in all experiments (s/n ratios significantly exceeded 10; Table 1) and were nearly or completely RNaseA-sensitive (Fig. 2 A and D). In addition, the RNA was tightly bound within the CENH3 immune complex. Ionic strengths as high as 1 M, high enough to dissociate the majority of H2a–H2b dimers from canonical nucleosomes (40), had a surprisingly small effect on the recovery of RNA after ChIP (Fig. 2C).
Table 1. Association of RNA with CENH3.
| CRM
|
Opie
|
CentC
|
Knob
|
|||||
|---|---|---|---|---|---|---|---|---|
| Experiment | s/n | %IP | s/n | %IP | s/n | %IP | s/n | %IP |
| 1 | 26.3 | 26.0 | 1.0 | -0.3 | 47.1 | 46.3 | 1.2 | 0.1 |
| 2 | 12.9 | 12.6 | 1.0 | -0.9 | 22.4 | 21.6 | 1.0 | -0.2 |
| 3 | 41.5 | 46.2 | 1.0 | 0 | 53.7 | 52.8 | 1.3 | -0.6 |
| 4 | 42.1 | 46.9 | 1.0 | 0 | 53.7 | 52.8 | 1.0 | -4.0 |
| 5 | 86.4 | 86.4 | 0.8 | -1.0 | 22.4 | 69.1 | 1.0 | 0.2 |
Data are expressed as s/n ratio (s/n = [P/(P+S)] from anti-CENH3 treatment divided by [P/(P+S)] from preimmmune control) and %IP (%IP = [P/(P+S)] from anti-CENH3 treatment minus [P/(P+S)] from preimmune control). s/n ratio is an indication of the overall quantity of RNA, specificity of the antibody, the strength of interactions within the immune complex, and the overall efficiency of the procedure, and %IP is the percentage of nuclear RNA associated with CENH3 under the experimental conditions used. The s/n ratios were analyzed for statistical significance by using one-tailed t tests (P ≤ 0.025). For CRM and CentC, the s/n ratios were significantly >10, and for Opie and Knob, the s/n ratios were significantly <2.
The percentage of nuclear RNA associated with CENH3 was estimated by using background subtraction. An average of five independent experiments demonstrated that close to half of the CRM (44%) and CentC (48%) RNA in purified nuclei is associated with CENH3 (Table 1 and Fig. 2B). In one experiment, 86% of the CRM RNA and 69% of CentC RNA was recovered in the anti-CENH3 immune complex (Table 1). Conversely, the %IPs for knob and Opie RNA hovered around zero (Table 1 and Fig. 2 A and B), with s/n ratios significantly below 2 (Table 1).
Centromeric DNA was also measured and quantified after CENH3-mediated immunoprecipitation. As shown in Fig. 2B, the data indicate that the relative quantities of centromeric DNA and RNA mirror each other with remarkable accuracy.
Both Strands of CRM and CentC Are Associated with the CENH3 Immune Complex. In addition to DNA probes for the sequences shown in Fig. 2A, ChIP samples were hybridized with RNA probes specific for the forward and reverse strands of CRM (GAG) and CentC (Fig. 2D). Because the signal was almost entirely abolished by RNaseA treatment, these data confirm that single-stranded RNA homologous to both strands of CRM and CentC are coimmunoprecipitated with maize CENH3 antibodies. The reverse CRM transcripts may represent examples of transcription through nested CRM elements, because retroelements often insert into each other in reverse orientations (38). By using two forward CRM primers, we were able to recover several different PCR products from genomic DNA that, when sequenced, proved to represent reverse CRM–CRM insertions (data not shown). Similarly, CentC transcripts are likely derived from read-through transcription by such CRM elements (see Discussion).
The Majority of CENH3-Associated RNA Is >40 nt in Length. The simultaneous presence of forward and reverse transcripts is expected to activate the RNAi pathway and produce siRNAs (41). However, we were not able to detect centromeric RNA in the size range expected for siRNAs (22–30 nt) on polyacrylamide gels. Rather, the bulk of the CentC and CRM RNA was between 40 and 250 nt in length (Fig. 3A). The banding patterns in supernatant and pellet fractions were very similar, with both forward and reverse probes identifying several distinct bands (in four independent experiments). It is possible that the ≈40- and 60-nt bands homologous to CentC are dicer-like products, but to our knowledge, no bona fide siRNAs in this size range have been reported.
The apparent upper limit on the size of the RNA may be an artifact of the procedure, because much larger RNAs could be detected by using more sensitive methods. In two experiments, we were able to detect 900-nt RNAs from both strands of the CRM GAG domain by RT-PCR (Fig. 3B). Taken together, these data suggest that the centromere-associated RNAs are variable in size, but are rarely as small as would be expected if the RNAs were the product of RNAi.
Discussion
The centromere core is often viewed as a genetically inert domain of the chromosome (6, 42). However, recent results clearly indicate that genes within centromeres can be transcribed (11, 12) and suggest that transcription may contribute to centromere formation (13, 14). Here we add the observations that centromere repeats are actively transcribed, and that a significant fraction of the RNA is bound, directly or indirectly, to CENH3. The fact that CentC and CRM transcripts coimmunoprecipitate with native (not chemically cross linked) CENH3 complexes, at ionic strengths sufficient to partially disrupt nucleosomes, indicates that RNA is an integral component of centromeric chromatin (Table 1 and Figs. 2 and 3).
The Formation of Centromeric RNAs. To explain the origin of CentC transcripts, we refer to a convincing body of evidence indicating that retroelements can initiate aberrant read-through transcription of flanking DNA (43, 44). Although most retrotransposons are rare in centromeres (39, 45, 46), CRM elements are abundant and actively transcribed. CR elements can occupy as much 60% of the centromeric DNA in both maize and rice (3, 12), providing ample opportunity for transcripts to be initiated within the centromere. If the maize centromeric bacterial artificial chromosome 16H10 is used as a guide (31), roughly seven CRM/CentC junctions can be expected in a 90-kb CRM-rich region of centromeric DNA. We also found evidence of reverse CRM transcripts in the EST database, on blots (Figs. 2D and 3A), and by RT-PCR (Fig. 3B). These RNAs likely represent examples of the CRM promoter driving transcription through reverse-nested insertions. Such nested insertions, truncations, and other rearrangements known to be associated with retroelements have the potential to provide an array of forward and reverse templates for CRM and CentC. In principle, any promoter could initiate similar centromeric transcripts, whether they are from genes (11, 12) or other forms of transposable elements.
Size of the RNAs. Despite the presence of both strands of CentC and CRM, we did not detect canonical siRNA-sized molecules within the nucleus or in association with CENH3. It remains possible that low concentrations of centromeric siRNAs were present but not detected in our assays. We also note that, because our experiments were designed to study nuclear RNA, the possibility that centromeric siRNAs might exist within the cytoplasm was not addressed. However, in nuclei, the fact that larger RNAs from both strands of centromeric repeats were readily detected suggests that the RNAi machinery did not process these RNAs. A portion of the centromeric RNA may be kept in a single-stranded state within the kinetochore, or otherwise protected from the RNase-III like (dicer) enzymes that initiate RNAi.
RNAs ranging in size from 40 to 900 nt are recovered from immunoprecipitates (Fig. 3), but it is difficult to know how long the centromere-associated RNAs are in vivo. An analogy can be drawn to the Xist RNA that associates directly with chromatin during human X chromosome inactivation (27). Xist is 17 kb when transcribed, but only small fragments of the RNA (e.g., 242 bp) have been detected after immunoprecipitation with antibodies to macroH2A, a histone variant that is deposited on the inactive X chromosome subsequent to coating by Xist (47).
RNA in the Initiation and Maintenance of the Centromeric State. CENH3 is one of many replacement histones that are incorporated into chromatin after DNA replication (unlike core histones, which are assembled during DNA replication; refs. 6 and 7). Although the mechanisms of histone replacement are not well understood, it is likely that transcription, apparently a common feature of centromeres (refs. 11 and 12 and this report), facilitates the process. Transcription partially disassembles nucleosomes (9) and is correlated with the replacement of histone H3 with a variant known as histone H3.3 (48, 49). The fact that the S. pombe Ams2 transcription factor mediates CENH3 (SpCENP-A) localization (14) provides strong support for the idea that transcription is involved in establishing the centromeric state.
Our data also establish that centromeric RNA can remain bound to the centromere/kinetochore complex after transcription. Interestingly, early ultrastructural studies strongly support the idea that RNA is present at plant and animal kinetochores (50, 51). In its chromatin-bound capacity, centromeric RNA may have a targeting and/or stabilizing role. Telomerase RNA, for instance, not only targets the telomerase complex to the ends of chromosomes by base pairing, but also serves as a flexible scaffold for associated proteins (28). Zappulla and Cech (28), working with telomerase RNA, suggested that “the overall structure of telomerase is at least somewhat flexible” and that “telomerase RNA tethers [proteins] to the RNP rather than positioning them precisely within a highly structured complex” (28). In a broad sense, telomerase can be compared to RNA-induced initiation of transcriptional silencing (RITS), which is thought to use RNA to target (by base pairing) a three-protein ribonucleoprotein (RNP) complex on specific chromatin domains (16, 21). Similarly, we show that the relative quantity of centromeric and noncentromeric RNA mirrors the DNA within CENH3 immune complexes (Fig. 2B). Although our approach does not allow us to measure the stoichiometry or specificity of the RNA–DNA associations, the data are consistent with the idea that centromeric RNA interacts with cognate DNA sequences within CENH3-containing nucleosomes.
There are also strong parallels between our data, telomerase, and the RNAs that regulate dosage compensation in humans and Drosophila. In both species, long RNAs interact with regulatory proteins to form a complex known in Drosophila as the compensasome (27). RNA-containing compensasomes spread along entire chromosomes to either shut down (humans) or double (Drosophila) gene expression (27). One working model is that compensasome RNA provides low-affinity contacts that facilitate higher-order interactions among chromatin proteins (26, 27). Amrein (26) and Wutz (27) again point to the fact that RNA is more flexible than protein, and can tolerate rapid sequence divergence while still maintaining function. Unlike telomerase, however, dosage compensation complexes function exclusively in cis, spreading to linked sites but never from one chromosome to another.
The available data suggest that centromere transcription may contribute to both the deposition (initiation) and stabilization (maintenance) of kinetochore chromatin structure. Transcription could initiate the process by opening chromatin to allow the replacement of histone H3 with CENH3, a key early event in centromere specification (8, 52). Secondly, centromere-associated RNA could provide a flexible scaffold that brings together and stabilizes the proteins of the inner kinetochore. Centromeric targeting may be conferred by base pairing (consistent with the roles of siRNA or telomerase RNA; refs. 16 and 21) or by a cis-acting/spreading mechanism (as exemplified by the compensasome; refs. 26 and 27). Importantly, an RNA scaffold could also help to explain how centromeric repeats evolve so rapidly (1) but maintain their function. If RNA has an important functional role in maintaining the centromeric state, then DNA sequence may be less important than the secondary structure of the folded RNA derived from it.
Acknowledgments
We thank Roger Deal for helping to establish the conditions used for DNase I ChIP and Amy Bouck for useful statistics discussions. This work was supported by National Science Foundation Grant 9975827 (to R.K.D.).
Author contributions: R.K.D. designed research; C.N.T. and C.X.Z. performed research; R.K.D. and C.N.T. analyzed data; and R.K.D. and C.N.T. wrote the paper.
Abbreviations: CENH3, centromeric histone H3; RNAi, RNA interference; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; MNase, micrococcal nuclease; CR, centromeric retrotransposable; CRM, maize CR; s/n, signal to noise; %IP, percentage of immunoprecipitation.
References
- 1.Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- 2.Amor, D. J., Kalitsis, P., Sumer, H. & Choo, K. H. A. (2004) Trends Cell Biol. 14, 359–368. [DOI] [PubMed] [Google Scholar]
- 3.Jin, W., Melo, J. R., Nagaki, K., Talbert, P. B., Henikoff, S., Dawe, R. K. & Jiang, J. (2004) Plant Cell 16, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo, K. H. A. (2000) Trends Cell Biol. 10, 182–188. [DOI] [PubMed] [Google Scholar]
- 5.Hooser, A. V., Ouspensik, I., Gregson, H., Starr, D., Yen, T., Goldberg, M., Yokomori, K., Earnshaw, W., Sullivan, K. & Brinkley, B. (2001) J. Cell Sci. 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad, K. & Henikoff, S. (2002) Proc. Natl. Acad. Sci. USA 99, 6477–6484. [Google Scholar]
- 7.Shelby, R. D., Monier, K. & Sullivan, K. F. (2000) J. Cell Biol. 151, 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan, K. F. (2001) Curr. Opin. Genet. Dev. 11, 182–188. [DOI] [PubMed] [Google Scholar]
- 9.Boeger, H., Griesenbeck, J., Strattan, J. S. & Kornberg, R. D. (2003) Mol. Cell 11, 1587–1598. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, J., Birchler, J. A., Parrott, W. A. & Dawe, R. K. (2003) Trends Plant Sci. 8, 570–575. [DOI] [PubMed] [Google Scholar]
- 11.Saffery, R., Sumer, H., Hassan, S., Wong, L. H., Craig, J. M., Todokoro, K., Anderson, M., Stafford, A. & Choo, K. H. (2003) Mol. Cell 12, 509–516. [DOI] [PubMed] [Google Scholar]
- 12.Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P. B., Kim, M., Jones, K. M., Henikoff, S., Buell, C. R. & Jiang, J. (2004) Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- 13.Nakano, M., Okamoto, Y., Ohzeki, J. & Masumoto, H. (2003) J. Cell Sci. 116, 4021–4034. [DOI] [PubMed] [Google Scholar]
- 14.Chen, E. S., Saitoh, S., Yanagida, M. & Takahashi, K. (2003) Mol. Cell 11, 175–187. [DOI] [PubMed] [Google Scholar]
- 15.Grewal, S. I. & Moazed, D. (2003) Science 301, 798–802. [DOI] [PubMed] [Google Scholar]
- 16.Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I. & Martienssen, R. A. (2002) Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 17.Volpe, T., Schramke, V., Hamilton, G. L., White, S. A., Teng, G., Martienssen, R. A. & Allshire, R. C. (2003) Chromosome Res. 11, 137–146. [DOI] [PubMed] [Google Scholar]
- 18.Hall, I. M., Noma, K. & Grewal, S. I. (2003) Proc. Natl. Acad. Sci. USA 100, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukagawa, T., Nogami, M., Yoshikawa, M., Ikeno, M., Okazaki, T., Takami, Y., Nakayama, T. & Oshimura, M. (2004) Nat. Cell Biol. 6, 784–791. [DOI] [PubMed] [Google Scholar]
- 20.Maison, C., Bailly, D., Peters, A. H., Quivy, J. P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T. & Almouzni, G. (2002) Nat. Genet. 30, 329–334. [DOI] [PubMed] [Google Scholar]
- 21.Verdel, A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S. I. & Moazed, D. (2004) Science 303, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partridge, J. F., Borgstrom, B. & Allshire, R. C. (2000) Genes Dev. 14, 783–791. [PMC free article] [PubMed] [Google Scholar]
- 23.Westermann, S., Cheeseman, I. M., Anderson, S., Yates, J. R., III, Drubin, D. G. & Barnes, G. (2003) J. Cell Biol. 163, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blower, M., Sullivan, B. & Karpen, G. (2002) Dev. Cell 2, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ando, S., Yang, H., Nozaki, N., Okazaki, T. & Yoda, K. (2002) Mol. Cell. Biol. 22, 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amrein, H. (2000) Genome Biol. 1, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wutz, A. (2003) BioEssays 25, 434–442. [DOI] [PubMed] [Google Scholar]
- 28.Zappulla, D. C. & Cech, T. R. (2004) Proc. Natl. Acad. Sci. USA 101, 10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawe, R. K. (2003) Plant Cell 15, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong, C. X., Marshall, J. B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J. A., Jiang, J. & Dawe, R. K. (2002) Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagaki, K., Song, J., Stupar, R., Parokonny, A. S., Yuan, Q., Ouyang, S., Liu, J., Dawe, R. K., Buell, C. R. & Jiang, J. (2003) Genetics 163, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telford, D. & Stewart, B. (1989) Int. J. Biochem. 21, 127–137. [DOI] [PubMed] [Google Scholar]
- 33.Khandjian, E. W. & Meric, C. (1986) Anal. Biochem. 159, 227–232. [DOI] [PubMed] [Google Scholar]
- 34.Shihara, H. & Shikita, M. (1990) Anal. Biochem. 184, 207–212. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P. & Bennetzen, J. L. (2002) Plant Cell 14, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peacock, W. J., Dennis, E. S., Rhoades, M. M. & Pryor, A. J. (1981) Proc. Natl. Acad. Sci. USA 78, 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawe, R. K. & Hiatt, E. N. (2004) Chromosome Res. 12, 655–669. [DOI] [PubMed] [Google Scholar]
- 38.SanMiguel, P., Tikhonov, A., Jin, Y. K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P. S., Edwards, K. J., Lee, M., Avramova, Z. & Bennetzen, J. L. (1996) Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- 39.Mroczek, R. J. & Dawe, R. K. (2003) Genetics 165, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burton, D. R., Butler, M. J., Hyde, J. E., Phillips, D., Skidmore, C. J. & Walker, I. O. (1978) Nucleic Acids Res. 5, 3643–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 42.Pidoux, A. L., Richardson, W. & Allshire, R. C. (2003) J. Cell Biol. 161, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitelaw, E. & Martin, D. I. (2001) Nat. Genet. 27, 361–365. [DOI] [PubMed] [Google Scholar]
- 44.Kashkush, K., Feldman, M. & Levy, A. A. (2003) Nat. Genet. 33, 102–106. [DOI] [PubMed] [Google Scholar]
- 45.Sun, X., Wahlstrom, J. & Karpen, G. (1997) Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schueler, M. G., Higgins, A. W., Rudd, M. K., Gustashaw, K. & Willard, H. F. (2001) Science 294, 109–114. [DOI] [PubMed] [Google Scholar]
- 47.Gilbert, S. L., Pehrson, J. R. & Sharp, P. A. (2000) J. Biol. Chem. 275, 36491–36494. [DOI] [PubMed] [Google Scholar]
- 48.McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. (2004) Proc. Natl. Acad. Sci. USA 101, 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. (2004) Cell 116, 51–61. [DOI] [PubMed] [Google Scholar]
- 50.Braselton, J. P. (1975) Cytobiologie 12, 148–151. [Google Scholar]
- 51.Rieder, C. L. (1979) J. Cell Biol. 80, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choo, K. H. A. (2001) Dev. Cell 1, 165–177. [DOI] [PubMed] [Google Scholar]