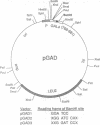
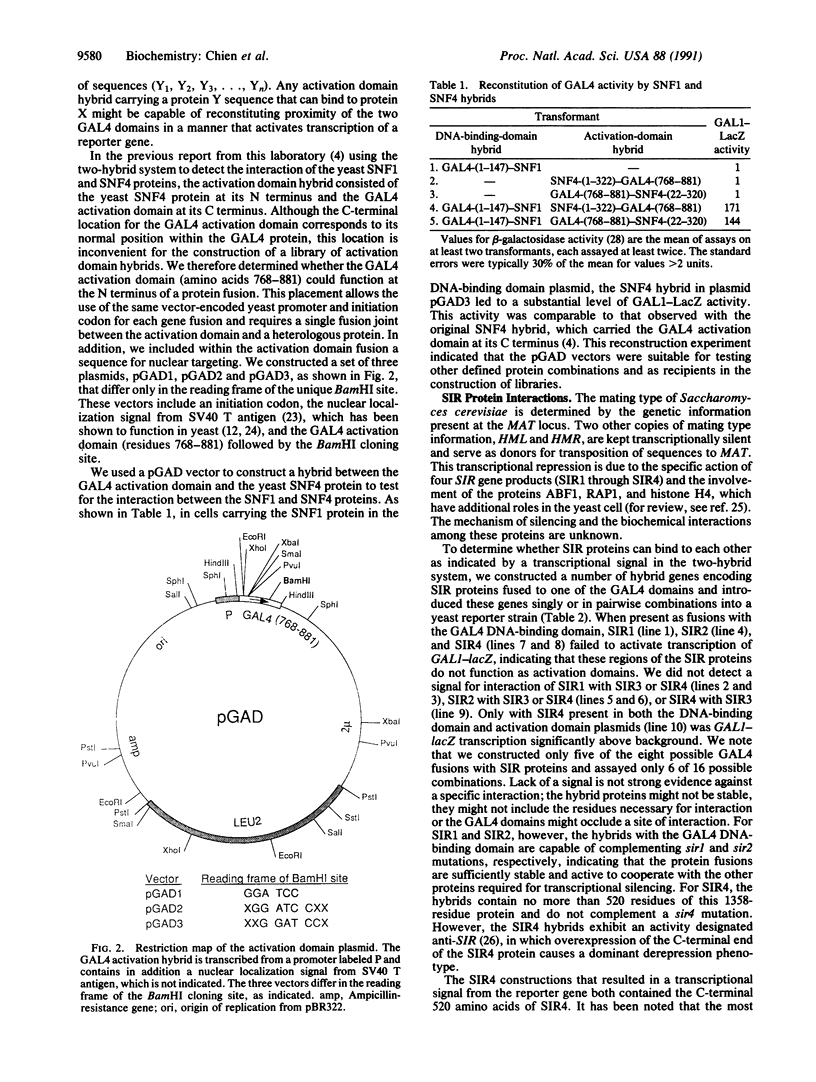
Abstract
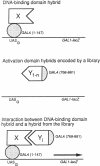
We describe a method that detects proteins capable of interacting with a known protein and that results in the immediate availability of the cloned genes for these interacting proteins. Plasmids are constructed to encode two hybrid proteins. One hybrid consists of the DNA-binding domain of the yeast transcriptional activator protein GAL4 fused to the known protein; the other hybrid consists of the GAL4 activation domain fused to protein sequences encoded by a library of yeast genomic DNA fragments. Interaction between the known protein and a protein encoded by one of the library plasmids leads to transcriptional activation of a reporter gene containing a binding site for GAL4. We used this method with the yeast SIR4 protein, which is involved in the transcriptional repression of yeast mating type information. (i) We used the two-hybrid system to demonstrate that SIR4 can form homodimers. (ii) A small domain consisting of the C terminus of SIR4 was shown to be sufficient to mediate this interaction. (iii) We screened a library to detect hybrid proteins that could interact with the SIR4 C-terminal domain and identified SIR4 from this library. This approach could be readily extended to mammalian proteins by the construction of appropriate cDNA libraries in the activation domain plasmid.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B., Sternglanz R. Gene expression. Chromatin contract to silence. Nature. 1990 Mar 15;344(6263):193–194. doi: 10.1038/344193a0. [DOI] [PubMed] [Google Scholar]
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Benton B. M., Eng W. K., Dunn J. J., Studier F. W., Sternglanz R., Fisher P. A. Signal-mediated import of bacteriophage T7 RNA polymerase into the Saccharomyces cerevisiae nucleus and specific transcription of target genes. Mol Cell Biol. 1990 Jan;10(1):353–360. doi: 10.1128/mcb.10.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Eng F. J., Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989 Nov;9(11):5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J. A., Ludlow J. W., Figge J., Shew J. Y., Huang C. M., Lee W. H., Marsilio E., Paucha E., Livingston D. M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988 Jul 15;54(2):275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Transcriptional silencing and lamins. Nature. 1989 Nov 2;342(6245):24–24. doi: 10.1038/342024a0. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kimmerly W. J., Rine J. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol Cell Biol. 1987 Dec;7(12):4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. P., Crawford L. V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979 Mar 15;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Marshall M., Mahoney D., Rose A., Hicks J. B., Broach J. R. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kayne P. S., Moerschell R. P., Tsunasawa S., Gribskov M., Colavito-Shepanski M., Grunstein M., Sherman F., Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989 Jul;8(7):2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelbock P., Dillon P. J., Perkins A., Rosen C. A. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science. 1990 Jun 29;248(4963):1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- Nelson M., Silver P. Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987 May;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Silver P. A., Keegan L. P., Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]