Abstract
A minimal amount of extranucleosomal DNA was required for nucleosome mobilization by ISW2 as shown by using a photochemical histone mapping approach to analyze nucleosome movement on a set of nucleosomes with varied lengths of extranucleosomal DNA. ISW2 was ineffective in repositioning or mobilizing nucleosomes with ≤20 bp of extranucleosomal DNA. In addition, ISW2 was able to slide nucleosomes to within only 10 to 13 bp of the edge of DNA fragments. The nucleosome mobilization was promoted by extranucleosomal single-stranded DNA with modest strand preference. Gaps (10 bp) just inside the nucleosome and in the extranucleosomal DNA showed that the transfer of torsional strain (twist) into the nucleosomal DNA region was not required for mobilizing nucleosomes. However, indications are that the extranucleosomal DNA immediately adjacent to the nucleosome has an important role in the initial stage of nucleosome movement by ISW2.
A high degree of DNA folding and compaction is required to fit genomic DNA into the eukaryotic cell. DNA is packed into the cell by means of association with architectural histone and nonhistone proteins, which constitute large DNA-protein complexes called chromatin. This association represents a significant obstacle to many important cellular processes requiring access to the DNA, such as transcription, replication, recombination, or DNA repair (34).
Eukaryotes have developed means for overcoming the repressive effect of chromatin structure. Two groups of enzymes that were shown to affect the accessibility of nucleosomal DNA and to alter the structure of chromatin are those which chemically modify histone proteins or use ATP hydrolysis to alter the chromatin structure through nucleosome mobilization. All ATP-dependent chromatin-remodeling complexes contain a subunit with a helicase-like ATPase domain, belonging to the SNF2-like family of proteins. Based on the sequence homology of this subunit and their functional properties, chromatin remodelers are subdivided into at least four major subfamilies: SNF2 (BRG1, BRM, Sth1) (2, 5, 22), ISWI (hSnf2L, dISWI, ISW1, and ISW2) (10, 23, 28-30), INO80 (26), and CHD (Mi-2) (27).
Common features of ATP-dependent chromatin remodelers are their abilities to reposition the histone octamer (7, 12, 18, 32) and to generate superhelical torsion (8). Members of the ISWI subfamily also have the ability to convert nucleosomal arrays from irregularly to regularly spaced (10, 29, 30). The Drosophila ATP-utilizing chromatin assembly factor (ACF) complex containing ISWI also facilitates the deposition of histones onto DNA in the presence of a histone chaperone (11). The repositioning of mononucleosomes by Drosophila ISWI varies depending on whether the ISWI is free or in a complex with other proteins. The free ISWI preferentially positions nucleosomes towards the ends of DNA, while ACF and chromatin accessibility complex (CHRAC) generate centrally positioned nucleosomes (4, 18). NURF (nucleosome remodeling factor) also does not slide nucleosomes to the ends of DNA but instead slides nucleosomes to their thermodynamically preferred positions (35).
Saccharomyces cerevisiae ISW2 was originally identified as a two-subunit complex (29) required for the repression of early meiotic genes (17). It possesses a helicase-like ATPase subunit that is highly homologous to that of Drosophila ISWI. Like Drosophila ACF and CHRAC, ISW2 preferentially positions mononucleosomes to the center of DNA and creates regularly spaced nucleosomal arrays (15, 29). ISW2 remodeling does not result in significant structural change of the path of DNA around the nucleosome, nor does it increase the accessibility of nucleosomal DNA (15).
ISW2 binds to ∼63 bp of extranucleosomal DNA, which in turn helps recruit and orient ISW2 on the nucleosome such that it interacts with the first 20 bp of nucleosomal DNA from the edge and ∼10 bp of DNA 1.5 helical turns from the dyad axis (14). ISW2 therefore approaches the nucleosome from one side perpendicular to the superhelical DNA axis and contacts two disparate sites on opposite sides of the nucleosome. For stable binding of ISW2, only 20 bp of extranucleosomal DNA was found to be critical, although longer DNA enhances the ability of ISW2 to bind and remodel nucleosomes under competitive conditions.
Here, we extend this data by determining the critical length and structural requirements of the extranucleosomal DNA for ISW2 to be able to mobilize nucleosomes by using site-specific histone-DNA photo-cross-linking to monitor the sliding of nucleosomes with single-bp resolution (15, 16). This method of mapping, along with gel shift assays, made it possible to track the movement of DNA on the surface of the nucleosome and to stably change the nucleosome translation. Portions of the extranucleosomal DNA at the entry site were also switched from double-stranded DNA (dsDNA) to single-stranded DNA (ssDNA) for determining if ISW2 required strand-specific translocation or transfer of torsional strain from these regions to mobilize nucleosomes.
MATERIALS AND METHODS
DNA probe synthesis.
End-labeled probes for histone-DNA cross-linking were generated by PCR amplification of pGEM-3Z-601 using AmpliTaq Gold polymerase (Applied Biosystems). Oligonucleotides, appropriate for a given length and placement of the nucleosome-positioning sequence (NPS), were obtained from Integrated DNA Technologies. Upstream or downstream oligonucleotides were labeled at the 5′ end by using T4 polynucleotide kinase (New England BioLabs) and [γ-32P]ATP (6,000 Ci/mmol) to obtain upper- or lower-strand-labeled DNA. DNA was purified with a QIAquick PCR purification kit (QIAGEN). The nine-nucleotide overhang probes were generated by digestion of PCR-generated DNA with TsrpI (3′ overhang) or NBstNB I (5′ overhang) and purified with a QIAquick PCR purification kit at 50°C to remove the smaller fragments. The probes were analyzed on a 6.5% polyacrylamide gel containing 8 M urea and were visualized by phosphorimaging.
For generation of DNA fragments containing strand breaks, a solid phase DNA synthesis on immobilized templates using M-280 magnetic beads (Dynal Biotech) was adapted. The lower strand of the DNA fragment was biotinylated and immobilized on streptavidin-coated magnetic beads. DNAs were generated by PCR amplification of pGEM-3Z-601 by using AmpliTaq Gold polymerase (Applied Biosystems) with the oligonucleotide for synthesis of the lower strand being biotinylated at the 5′ end. DNA was bound to magnetic beads, and the upper strand was removed as described previously (25). Oligonucleotides starting just downstream of the strand break were annealed to the immobilized ssDNA and extended with the Klenow fragment of DNA polymerase I (New England BioLabs). DNA upstream of the strand break was synthesized through annealing and ligation of chemically synthesized oligonucleotides, and the full-length DNA fragment was released from the beads by cutting with an appropriate restriction endonuclease and purified by phenol-chloroform extraction and ethanol precipitation.
Nucleosome reconstitution.
The end-labeled probes were assembled with recombinant octamers (20) into nucleosomes by the rapid salt dilution method. Ten micrograms of recombinant octamer was mixed with 10 μg of sheared salmon sperm DNA (100 to 700 bp) and 1 pmol of end-labeled DNA in 2 M NaCl (total volume, 10 μl; 37°C), and the salt concentration was brought down to 300 mM by the stepwise addition of 6.8, 8.4, and 42 μl of 25 mM Tris-HCl (pH 7.5) at 10-min intervals.
ISW2 and ISW1a remodeling.
ISW2 was purified as described previously (29). Remodeling assays were conducted at 30°C for 30 min and contained 25 mM HEPES-KOH (pH 7.6), 5 mM MgCl2, 40 to 50 mM KCl, 0.1 mg of bovine serum albumin/ml, 6 mM Tris-HCl (pH 8.0), 5% glycerol, 30 mM NaCl, 300 μM ATP, 370 ng of nucleosomes, and 30 to 75 ng of ISW2 or 60 to 150 ng of ISW1a. For remodeling under ISW2-saturated conditions, nucleosomes were reconstituted with 5 pmol of a 156-, 166-, or 179-bp end-positioning DNA probe with an end-labeled upper strand and coupled with a p-azidophenacyl bromide (APB; Fluka) and purified on a 5 to 25% sucrose gradient. The amount of ISW2 needed to saturate nucleosomes was determined by gel shift assay on a 4% native polyacrylamide gel in 0.5× Tris-borate-EDTA (45 mM Tris-borate and 1 mM EDTA). Remodeling reactions were analyzed by gel shift assay on a 5% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 60/1) in 0.2× Tris-borate-EDTA at 10°C.
Histone-DNA cross-linking and mapping of H2B53 contacts on DNA.
H2B Ser53 was replaced with cysteine by site-directed mutagenesis and introduced into the recombinant octamer by refolding with H2A, H3, and H4 as described previously (20). The refolded octamer was reconstituted by the fast dilution method with 1 pmol of end-labeled DNA. The unique cysteine residue of H2B was coupled with APB. Two hundred micromolar APB was added to 10 μg of reconstituted nucleosomes and incubated for 3 h at room temperature in 1% dimethylformamide, 300 mM NaCl, 5% glycerol, and 10 mM Tris-HCl (pH 7.5). The excess APB was removed by dialysis against 100 mM NaCl, 5% glycerol, and 10 mM Tris-HCl (pH 7.5) or sucrose gradient centrifugation. Remodeling was conducted as described above, and cross-linking was performed by using an ultraviolet transilluminator (312 nm for 30 sec). Samples were denatured with 0.1% sodium dodecyl sulfate (SDS) and by heating for 20 min at 70°C. Histone-DNA conjugates were purified by phenol-chloroform (4/1) extraction. The organic phase was washed three times with 1% SDS and 1 M Tris-HCl (pH 8.0), precipitated with ethanol, and resuspended in 2% SDS, 20 mM NH4CH3COO, and 0.1 mM EDTA. The scission of DNA at the cross-linking site was triggered by incubation in 0.1 M NaOH for 45 min at 90°C. Cleaved samples were neutralized with 2 M HCl and ethanol precipitated. Samples were resuspended in formamide, resolved on a 6.5% polyacrylamide gel containing 8 M urea, and visualized by phosphorimaging. The amount of cleaved DNA present in each lane was normalized to the amount of total signal in the lane or to the amount of uncut DNA to facilitate the comparison of different lanes with similar samples before and after remodeling.
Exonuclease III mapping of nucleosome positions.
After nucleosome reconstitution and ISW2 remodeling as described above, the samples were supplemented with 0.03 or 0.1 U of exonuclease III and incubated for 10 min at 37°C. Reactions were stopped by the addition of formamide to a final concentration of 70%, resolved on a 6.5% polyacrylamide gel containing 8 M urea, and visualized by phosphorimaging.
RESULTS
Nucleosomes slid by ISW2 are not at their thermodynamically preferred positions.
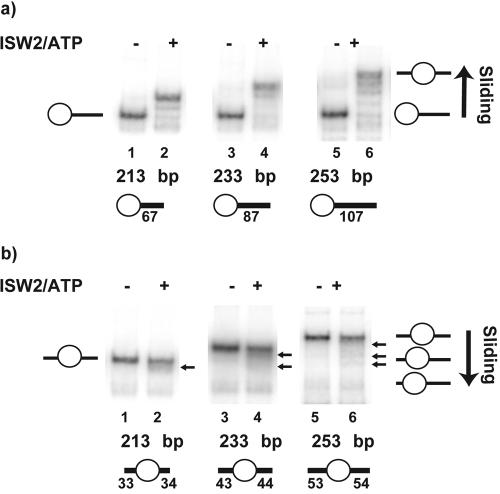
The NPS referred to as 601 and discovered by J. Widom and colleagues has ∼150 times higher affinity for nucleosomes than sea urchin 5S ribosomal DNA (19). The 601 sequence directs nucleosomes into a single predominant translational position on DNA fragments as long as 288 bp and has been used in the present study to assemble nucleosomes on DNA fragments of variable lengths with the NPS located at the end or in the center of the DNA (Fig. 1). ISW2 can quantitatively slide nucleosomes towards the center of DNA when they initially have been positioned at the end of DNA with 67, 87, or 107 bp of extranucleosomal DNA (Fig. 1a; compare lanes 2, 4, and 6 to lanes 1, 3, and 5). ISW2 has a preference for sliding nucleosomes from the end to the center of DNA fragments, as shown by nucleosomes not being slid as efficiently from a central position on DNA as from an end (Fig. 1).
FIG. 1.
ISW2 prefers to slide nucleosomes towards the center of DNA. Nucleosomes (125 ng) initially positioned at either the end (a) or the center (b) of DNA were mobilized with 30 ng of ISW2 and 3 mM ATP for 30 min at 30°C and then analyzed on a native 5% polyacrylamide gel.
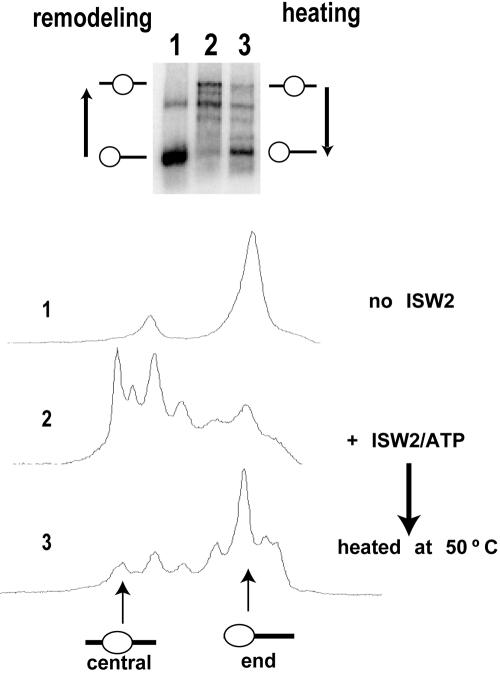
The positions to which nucleosomes were slid by ISW2 were not the thermodynamically preferred positions. After remodeling, ISW2 was stopped first by degradation of ATP with apyrase and then released from nucleosomes by competition with sonicated salmon sperm DNA (Fig. 2). Heating the nucleosomes at 50°C reversed the sliding of the nucleosome from the slid position to the starting and thermodynamically preferred position. ISW2 slides nucleosomes away from the thermodynamically preferred nucleosome position, unlike Drosophila NURF, which slides nucleosomes from the hsp70 promoter to a more thermodynamically preferred position (6).
FIG. 2.
ISW2 does not mobilize nucleosomes towards the thermodynamically preferred positions on DNA. Nucleosomes were positioned at the ends of DNA by using the nucleosome positioning sequence referred to as 601. Nucleosome samples were analyzed by gel shift on a native 5% polyacrylamide gel and quantitated as shown by phosphorimaging. Lane 1, samples with no ISW2; lane 2, samples after ISW2 remodeling; lane 3, reactions stopped after remodeling and then heated at 50°C for 30 min for equilibration to the thermodynamically preferred position(s). The samples contained 370 ng of nucleosomes and 60 ng of ISW2. After remodeling, ISW2 was inhibited by the addition of apyrase (0.1 U) and incubation for 20 min at 37°C, followed by the addition of 10 μg of DNA and 4 mM γ-S-ATP.
ISW2 requires greater than 20 bp of extranucleosomal DNA for efficient nucleosome repositioning.
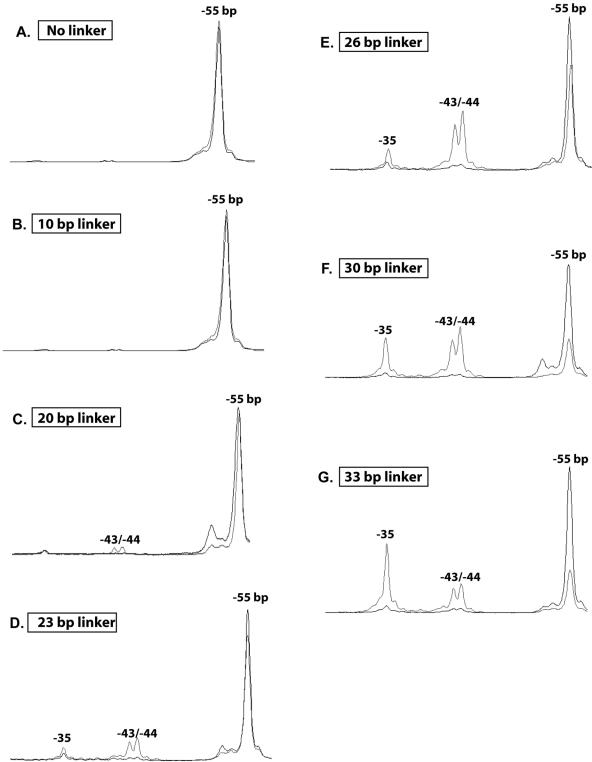
The minimal amount of extranucleosomal DNA required for nucleosome mobilization by ISW2 was determined by using nucleosomes assembled with DNA varying in length from 146 to 179 bp. All probes uniformly positioned the nucleosomes toward one end of the DNA and included 0 to 33 bp of extranucleosomal DNA. The extent of nucleosome sliding was determined by the photochemical mapping approach, so that even small changes in nucleosome position could be detected. The nucleosomes were constructed such that the cut site was located on the side of the dyad axis farthest in the DNA sequence from the linker DNA and was referred to as bp −55 with the dyad axis at bp 0. Nucleosomes slid 11, 12, or 20 bp towards the linker DNA when 33 bp of extranucleosomal DNA was present, as shown by the cut site moving from bp −55 to bp −35 and to bp −43 to −44, with as much as two-thirds of the nucleosomes being slid (Fig. 3G). No movement of the nucleosome was seen when no linker or 10 bp of linker DNA was present (Fig. 3A and B). Longer linker DNA initially promoted some movement of the nucleosome by ∼11 bp, as with 20 bp of linker DNA, but the majority (>95%) of the nucleosomes were still not changed (Fig. 3C). As the length of the linker DNA was increased to 23 and 26 bp, there was a gradual decrease at the original position of the nucleosome (∼70 to 80% remaining) that was associated with an increase in sliding by ∼11 bp at first and then by a total of 20 bp (Fig. 3D and E). A minimum of 30 bp of linker DNA was required for approximately two-thirds of the nucleosomes to slide from their original positions (Fig. 3F and G), whereas significant internal mobilization of the nucleosome was observed only with 26 bp of linker DNA (Fig. 3E).
FIG. 3.
ISW2 requires >20 bp of extranucleosomal DNA for efficient nucleosome mobilization. Nucleosomes (370 ng) assembled at the end of DNA with 0 (A), 10 (B), 20 (C), 23 (D), 26 (E), 30 (F), or 33 (G) bp of extranucleosomal DNA were remodeled with 75 ng of ISW2 and 300 μM ATP for 30 min at 30°C. Nucleosome sliding was detected by site-directed histone-DNA cross-linking, analyzed on a 6.5% polyacrylamide gel containing 8 M urea, and visualized by phosphorimaging. Overlays of the nucleosomal cut sites before and after ISW2 remodeling are shown (black, nucleosome alone; gray, nucleosomes plus ISW2 plus 300 μM ATP).
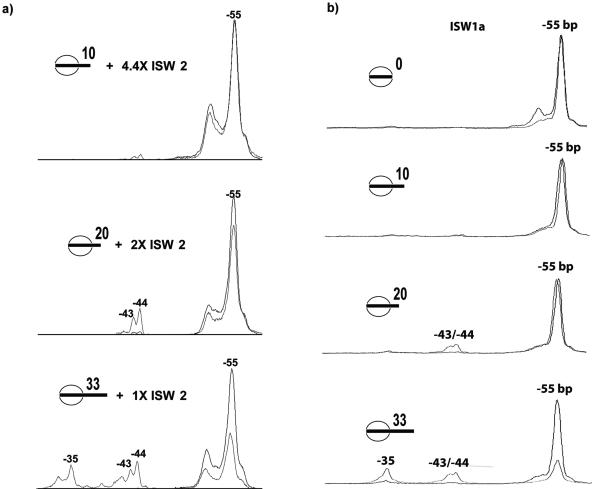
It was possible that the inability of ISW2 to change the translational position of nucleosomes with less than 26 to 30 bp of extranucleosomal DNA might have been due to inefficient binding of ISW2 (14, 33). Nucleosome sliding was done with saturating amounts of ISW2 to determine if the differences in nucleosome mobilization with shorter linker DNA were due to differences in ISW2 binding to nucleosomes. Nucleosomes with shorter or no linker DNA had 2 or 4.4 times more ISW2 added to compensate for differences in affinity (Fig. 4A). Although there was some increase of the 10-bp movement within nucleosomes containing 20 bp of extranucleosomal DNA when using more ISW2, over three-fourths of the nucleosomes still remained in the original position (Fig. 4A). A slight but detectable amount of a 10-bp movement was observed with 10 bp of extranucleosomal DNA, although no loss in the amount of nucleosomes at the original position was detected. The inability of ISW2 to slide nucleosomes with 10 and 20 bp of extranucleosomal DNA was thus shown not to be caused by an inability to bind nucleosomes.
FIG. 4.
The inability of ISW2 to slide nucleosomes with short extranucleosomal DNA is not due to an inability to bind. (a) Nucleosomes assembled with 156-, 166-, or 179-bp DNA fragments positioned towards one end were saturated by the addition of 2 μg, 900 ng, or 450 ng of ISW2, respectively, and remodeled for 10 min at 30°C. (b) ISW1a requires more than 20 bp of extranucleosomal DNA for efficient nucleosome mobilization. ISW1a was shown to slide nucleosomes with 0, 10, 20, and 33 bp of extranucleosomal DNA, as monitored by site-directed histone DNA cross-linking for ISW2 (see the legend for Fig. 3). Black lines, nucleosomes alone; gray lines, ISW1a ATP.
In order to determine if the properties of ISW2 are general for yeast ISWI complexes, the ability of ISW1a (29, 31) to slide nucleosomes was also examined. Extranucleosomal DNA of 20 bp or less was not sufficient to promote efficient displacement of nucleosomes from their original positions (Fig. 4B), the same result as with ISW2 (Fig. 3B). Similar to results with ISW2, ISW1a repositioned nucleosomes by 11, 12, and 20 bp increments when there was sufficient length of DNA (33 bp); although most of the nucleosomes remained at the original position at the intermediate length of 20 bp, some nucleosomes were moved by 11 to 12 bp. ISW1a was shown to have requirements similar to those of ISW2 for extranucleosomal DNA for nucleosome mobilization; the presence of extranucleosomal DNA is likely to be a general requirement for most or all of the ISWI-containing remodeling complexes.
ISW2 prefers not to slide nucleosomes to the very end of DNA fragment.
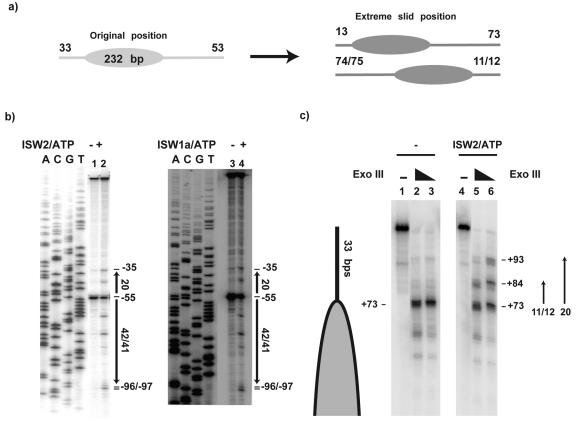
Although ISW2 preferentially slides nucleosomes towards the center of DNA fragments, a smaller percentage of nucleosomes are slid from the center towards the ends (Fig. 5a). Presumably because of the requirement for extranucleosomal DNA for nucleosome mobilization, nucleosomes were slid to within only 11 to 13 bp from DNA ends by ISW2 (Fig. 5b, lane 2) and ISW1a (lane 4). Earlier results had also shown that nucleosome repositioning on 183 bp DNA generated a single translational position with 15 and 16 bp of extranucleosomal DNA (15). All of these results demonstrate the mechanistic inability of ISW2 to slide nucleosomes to the very end of DNA.
FIG. 5.
ISW2 and ISW1a are unable to reposition nucleosomes closer than 11 to 13 bp to the ends of DNA. (a) Nucleosomes were positioned on a 232-bp DNA such that there were 33 bp of extranucleosomal DNA on one side and 55 bp of extranucleosomal DNA on the other. After remodeling, the new translational positions closest to either end of DNA are 13 and 11/12 bp from the end. (b) Nucleosomes were mobilized with ISW2 (lane 2) and ISW1a (lane 4) and mapped by using site-directed histone-DNA cross-linking. (c) The last 13 bp of DNA on nucleosomes slid by ISW2 were not bound to the histone octamer, as shown by exonuclease III footprinting. Two discrete protections are evident when ISW2 (48 ng) and ATP (300 μM) were added to 179-bp end-positioned nucleosomes (300 ng) and incubated for 30 min at 30°C; they were not present with nucleosomes alone (compare lanes 5 and 6 to lanes 2 and 3).
It might be that ISW2 cannot slide nucleosomes over the last ∼10 bp of DNA because the DNA path in the remodeled nucleosome is such that this DNA region may already be associated with the nucleosome. Exonuclease footprinting of the edge of the nucleosome showed that as the nucleosome moved toward the DNA end, the terminal 13 bp of DNA remained accessible to exonuclease cleavage. Before remodeling, exonuclease digestion generates a single pause site 33 bp from the end of the DNA that corresponds well with the edge of the end-positioned nucleosome, as expected based on the previous site-directed mapping (Fig. 5C, lanes 2 and 3). ISW2 remodeling caused the appearance of two additional pause sites, located 11 and 20 bp from the original position (Fig. 5C, lanes 5 and 6). The ability of exonuclease to digest the DNA region in question verifies its expected accessibility and potential ability of ISW2 to bind to this region.
Strand-specific translocation of ISW2 along extranucleosomal DNA.
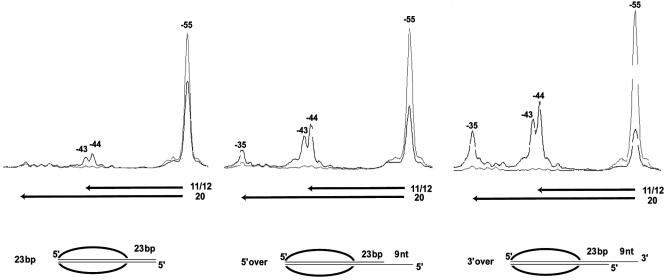
Single-stranded DNA was added in place of some of the extranucleosomal DNA to determine if only dsDNA would suffice for ISW2. A construct was made in which nine nucleotides of ssDNA was added onto 23 bp of extranucleosomal dsDNA to examine whether such extension would promote nucleosome mobilization by ISW2. A small amount of nucleosomes were slid 11 and 12 bp with 23 bp of extranucleosomal dsDNA, with 63% of the nucleosomes remaining in their original position, whereas the addition of nine nucleotides of ssDNA increased the efficiency and extent of nucleosome sliding (Fig. 6). The addition of nine nucleotides of ssDNA to the 5′ end enhanced movement away from the original position, with only 46% remaining and with a significant increase in the amount of nucleosomes moved 11, 12, and 20 bp. The addition of ssDNA to the 3′ end was even more stimulatory, with only 26% of the nucleosomes remaining in the original position and a larger number of nucleosomes moving 11, 12, and 20 bp away. The efficiency of nucleosome mobilization was comparable to that seen with 30 and 33 bp of extranucleosomal dsDNA (compare Fig. 6 with Fig. 3F and G). These results showed that the essential ISW2-extranucleosomal DNA interactions can be supported by ssDNA for nucleosome mobilization and demonstrate a modest strand preference for 3′ single-stranded extension. Recent results from others showed strand-specific translocation of ISWI by assaying for triplex displacement that was specifically blocked by placing 5- or 10-bp gaps in DNA in one strand and not the other (33). ISWI translocated along DNA away from the nucleosome in a 3′→5′ direction, as shown by gaps in the 3′→5′ strand preferentially interfering with triplex displacement by ISWI.
FIG. 6.
Single-stranded extranucleosomal DNA can promote ISW2 mobilization of nucleosomes with some strand specificity. Nucleosomes (370 ng) with 23 bp of double-stranded extranucleosomal DNA had nine nucleotides of ssDNA added onto either the 5′ or 3′ end of DNA to determine if ssDNA could substitute for dsDNA. Black lines, samples containing ISW2 (187 ng) and ATP (300 μM); gray lines, nucleosome alone; nt, nucleotides.
Generation of torsional strain in extranucleosomal DNA is not required for nucleosome mobilization.
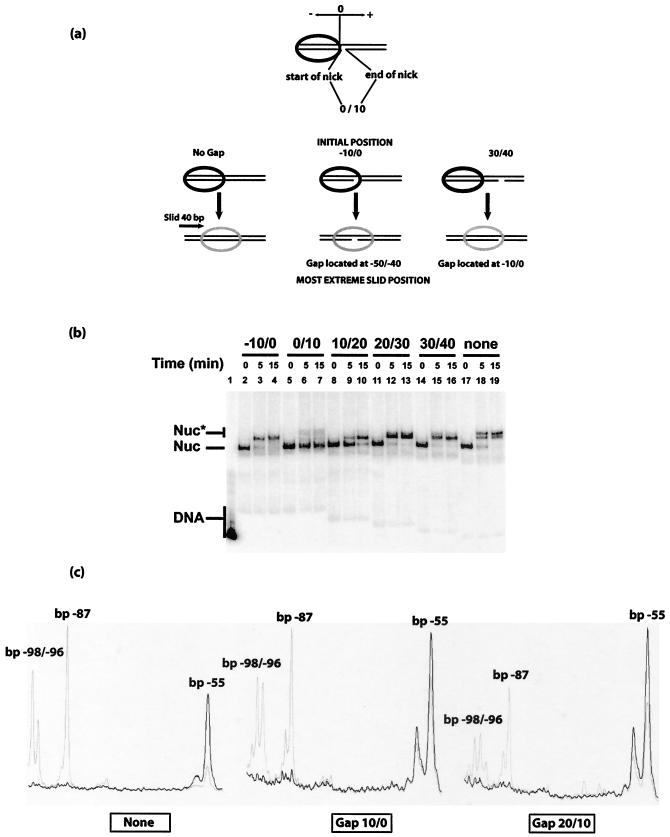
Strand-specific translocation of the remodeling complex away from the nucleosome was proposed in order to create torsional strain at the entry site of the nucleosome, which might cause nucleosome movement either by twist propagation through the DNA or by displacing DNA from the entry site to create a bulge on the surface of the nucleosome that can move around the surface in a wave-like manner (24). The need for torsional strain in the extranucleosomal DNA for nucleosomes to move was tested in a manner similar to that of Whitehouse et al. (33), by placement of 10-bp gaps in the 3′→5′ strand at locations throughout the extranucleosomal DNA and just within the nucleosomal bound region (Fig. 7a). The gaps were placed in 10-bp increments starting from just within the nucleosome to 30 bp from the edge of the nucleosome. These nucleosome constructs are referred to as −10/0 to 30/40, with the numbers referring to the numbers of base pairs from the edge of the nucleosome; the first and second numbers are the start and end positions of the gap (Fig. 7a). A negative number indicates that the gap is within the nucleosomal bound region.
FIG. 7.
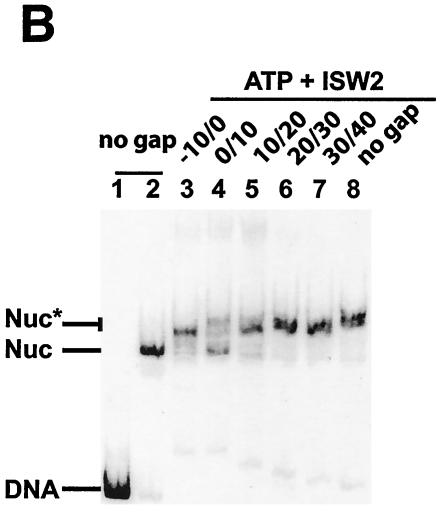
Gaps in extranucleosomal DNA immediately adjacent affect nucleosome sliding by ISW2. (a) Gaps of 10 bp were placed in the 3′ to 5′ strand in a region spanning from just within the nucleosomal bound region to 30 bp from the entry site. The position of the gap is indicated by the first and second numbers, indicating the start and end of the gap with regard to the edge of the nucleosome. The edge of the nucleosomeis the zero reference point; any position inside the nucleosome is negative, and any position in the extranucleosomal DNA is positive. After remodeling, nucleosomes were slid as much as 40 bp from their original positions, with the gap moving to a position inside the nucleosome as shown. (b) Mobilization of these nucleosomes was monitored by gel shift assay on a native 5% polyacrylamide gel after 5 or 15 min incubation at 30°C. The locations of free DNA and nucleosomes that were (Nuc*) and were not (Nuc) moved to the center are indicated on the left. The initial locations of the gap are indicated above the lanes, as described for panel a. (c) Movement of nucleosomes after remodeling by ISW2 with no gap or gaps at bp 10/0 and 20/10 were monitored by site-directed histone-DNA cross-linking. Black lines, original cut sites before ISW2 remodeling; gray lines, original cut sites after ISW2 remodeling.
The sliding of nucleosomes positioned towards the far end of a 204-bp DNA strand was monitored by gel shift assays (Fig. 7b). Most of the nucleosomes without gaps were slid from the end to a central position on DNA within 5 min (compare lanes 17 and 18). Gaps either just inside the nucleosome or 20 bp or more from the edge did not interfere with the translocation of nucleosomes (Fig. 7b, lanes 2 to 4 and 11 to 16). These nucleosomes moved 30 and 40 bp from their original positions as determined by site-directed mapping (Fig. 7c and results not shown), with the gap being slid inside the nucleosome two to three helical turns away from the dyad axis to just within the nucleosome (Fig. 7a). Gaps in the extranucleosomal DNA closest to the nucleosome (0 to 20 bp) did affect nucleosome mobilization either by slowing down (Fig. 7b, compare lanes 9 and 10) or by blocking their movement (Fig. 7b, lanes 5 to 7). The DNA gap immediately adjacent to the edge of the nucleosome showed the most severe effects on nucleosome sliding.
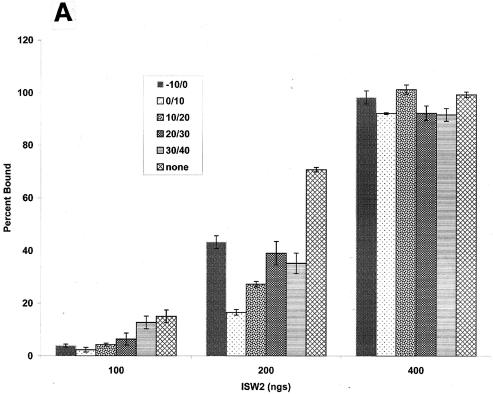
High-resolution mapping of the position of the 0/10- and 10/20-bp gapped nucleosomes showed that there was indeed a significant amount of the original nucleosome position present after remodeling when the gap was immediately adjacent to the edge of the nucleosome (Fig. 7c). Whereas 75% of the nucleosomes were slid from their original position when there was no gap, only 26 and 45% of nucleosomes were slid when the gap was at bp 0/10 and 10/20, respectively. The overall change in nucleosome position mapped in this manner was consistent with that observed by gel shift assay. However, the lack of movement from the original nucleosome position did not appear to be due to an inability to mobilize nucleosomes, as a significant amount of nucleosomes were shifted by 30 and 40 bp. These data suggest that ISW2, with the placement of these gaps, was unable to stably push the nucleosome in one direction and instead shuttles nucleosomes rapidly back and forth between the original and slid positions. Potentially, the effects of DNA gaps on nucleosome mobilization may be caused by effects on ISW2 binding to nucleosomes. ISW2 was found to bind generally less well to gapped nucleosomes than to normal nucleosomes and, in particular, the 0/10 gap in the extranucleosomal DNA interfered the most with ISW2 binding (by factors of as much as approximately four to six) (Fig. 8A). The reduced affinity of ISW2 for the gapped nucleosomes was, however, readily compensated for at higher ISW2 concentrations. Since the previous nucleosome mobilization assays were done at low, substoichiometric amounts of ISW2, the reduced sliding could have been due to reduced binding of ISW2. The nucleosome mobilization experiments were repeated with saturating amounts of ISW2 to determine if the inability to slide persisted even when reduced binding of ISW2 was compensated for. After remodeling, ISW2 was stripped off the nucleosomes by using excess competitor DNA and nucleosome movement was determined by gel shift assay. As before, when the gap was located at bp 0/10, nucleosome mobilization was severely affected (Fig. 8B; compare lane 4 to lanes 2, 3, and 5 to 8). The gap at bp 10/20 did not show much of an effect, likely because the previously observed lower rate of sliding was compensated for due to the saturating amounts of ISW2 and/or the time of incubation. These results show that defects in nucleosome mobilization with nucleosomes containing gaps at bp 0/10 are not due to an overall reduced binding of ISW2 to nucleosomes.
FIG. 8.
The effect of the gap at bp 0/10 on the mobilization of nucleosomes by ISW2 is not due to reduced binding of ISW2. (A) Quantification of gel shift assays of ISW2 binding to gapped nucleosomes are shown at three different amounts of ISW2 (100, 200, and 400 ng). Binding assays were performed as described in Materials and Methods. (B) Sliding assays of nucleosomes with or without 10-bp gaps were done as described for Fig. 7b, except that saturating amounts of ISW2 (400 ng) were added and, after remodeling excess, competitor DNA was added to compete ISW2 away from the labeled nucleosomes.
DISCUSSION
The ISWI group of chromatin remodelers provides an ideal model system for studying ATP-dependent chromatin remodeling. ISWI complexes are relatively abundant in the cell and are significantly smaller than many of the other ATP-dependent chromatin-remodeling complexes, such as SWI/SNF, RSC, NURD, and INO80. A general underlying property of ATP-dependent chromatin-remodeling complexes is their ability to translocate or slide the histone octamer along DNA, thereby changing its translational position (3). It is not clear how these complexes are able to slide the nucleosome bound to 147 bp of DNA to new positions on DNA with no dissociation of the histone octamer from DNA. Studies from several laboratories have indicated that extranucleosomal DNA immediately adjacent to the nucleosome is important for nucleosome sliding by ISWI-type remodeling complexes (1, 33). Recently, it was shown that extranucleosomal DNA enhances the overall recruitment of ISW2 to the nucleosome and promotes the site selective binding of ISW2 to the core nucleosome particle (14). Here, we demonstrate that aside from its ability to facilitate binding of ISW2, there is a minimal requirement of >20 bp of extranucleosomal DNA for efficient nucleosome mobilization. Using a high resolution site-directed mapping approach, we showed that ISW2 did not mobilize nucleosomes well with 10 or 20 bp of extranucleosomal DNA even when sufficient ISW2 was added to completely bind nucleosomes. Intermediate levels of nucleosome movement were observed with 23 and 26 bp of extranucleosomal DNA, with the most optimal being 30 and 33 bp. Another piece of evidence for ISW2 requiring extranucleosomal DNA for nucleosome mobilization was that it can slide nucleosomes to within only ∼11 to 13 bp of the ends of DNA.
The requirement for a minimal amount of extranucleosomal DNA suggests that it has a functional role in the underlying process of nucleosome sliding by ISW2. Nucleosome mobilization by ISWI, RSC, SWI/SNF, and Rad54 appears to occur by the complex translocating along extranucleosomal DNA in a strand-specific manner (13, 24, 33). Such translocation could create torsional strain in the DNA at the entry site and cause rotational twisting of DNA through the nucleosome and/or planar bulging of DNA from the surface of the nucleosome (24, 33). These two changes would cause the DNA to move around the nucleosome either in a screw-type motion, 1 bp at a time, or in a wave-like motion propagating around the surface of the nucleosome.
In order to see if ISW2 has strand-specific translocation activity, we first showed that ssDNA was able to substitute for dsDNA in the extranucleosomal region. These data suggest that ISW2 has a modest 5′→3′ strand preference for translocating along DNA in the extranucleosomal DNA region. In another report, the recombinant Drosophila ISWI protein was found to function in a highly 3′→5′ strand-specific manner by using 5- and 10-bp DNA gaps (33). One reason for these differences could be the difference of the free catalytic subunit versus the complete complex with the catalytic subunit activity being regulated by physical interactions with accessory subunits. These differences are evident in the different directional preferences of free ISWI in sliding nucleosomes to the ends of DNA and of CHRAC and ACF in sliding nucleosomes to the center of DNA. ISW2 appears to function similarly to CHRAC in that they have identical sliding and spacing activities and have in common two orthologous histone fold subunits (unpublished data from our lab and references 9 and 21). The differences in strand specificity could also be due to inherent differences in the two assays. The translocation of ISWI in a strand-specific manner was observed with nucleosomes in which the translocation on DNA was uncoupled from the sliding of the nucleosome by using nucleosomes terminally positioned at DNA ends that were not able to be slid by ISWI. The evidence for translocation of ISWI on DNA was its ability to displace a triplex-forming oligonucleotide (TFO) from DNA. It might be that the ability to displace the TFO is not connected with the nucleosome sliding activity of ISWI, such that the two assays may be measuring different activities of ISWI and that the translocation activity observed for ISWI is not a requirement for nucleosome sliding.
These experiments also demonstrated that torsional strain created within the extranucleosomal DNA or twist diffusion initiated at the extranucleosomal DNA and its propagation into the nucleosome are not likely to be required for nucleosome sliding, as the extra flexibility conferred by the gaps would interfere due to DNA being able to freely rotate. Since gaps just inside the nucleosome at bp −10/0 did not affect nucleosome movement by ISW2, the transfer of torsional strain into the nucleosome should not be critical for nucleosome mobilization. Also, gaps placed at bp 20/30 and 30/40 did not affect nucleosome mobilization even though the gaps pass by the edge of the nucleosome and into the nucleosomal bound region during the sliding process. These results strongly suggest that gaps do not affect nucleosome sliding, yet having the initial gap at the edge of the nucleosome does affect nucleosome sliding. It seems that having the gap near the edge of the nucleosome only affects nucleosome mobilization when it is there at the initiation of sliding or translocation. Therefore, ISW2-DNA interactions within the first 10 or 20 bp from the edge of the nucleosome appear to have a role in the initiation of nucleosome sliding, possibly in determining the direction of movement. The initiation role of this region is apparently distinct from the ability to mobilize nucleosomes, as the DNA was still moved on the surface of the nucleosome as observed by site-directed mapping, but not in a directed manner away from the original position.
The strand-specific activity observed by Whitehouse et al. with Drosophila ISWI also had the gap immediately adjacent to the nucleosome, with the triplex farther away from the edge of the nucleosome. The observed strand-specific interference of TFO displacement by ISWI may have been caused by disrupting the same initiation step as in that observed here and not by an inability of ISWI to move the DNA on the surface of the nucleosomes.
Although we have shown that torsional strain and twist diffusion are not required in the extranucleosomal region, it is possible that the generation and propagation of torsional strain at a different location in the nucleosome may be required. Since in our experiments the gap is moved fairly far into the nucleosome, it seems that if there is such a requirement, it is likely to be at a position farther inside the nucleosome. It is likely that such an activity does exist, since these chromatin remodelers have been shown to create torsional strain, but where on the nucleosome this twisting action is required is yet to be determined.
Acknowledgments
This work was supported by Public Health Science grant GM48413 from the National Institute of General Medical Sciences and by grant RPG-99-199-01-GMC from the American Cancer Society.
We thank Stefan Kassabov for helpful discussions and comments, Vamsi Gangaraju for ISW1a, and other members of the Bartholomew lab.
REFERENCES
- 1.Brehm, A., G. Langst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 3.Clapier, C. R., K. P. Nightingale, and P. B. Becker. 2002. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 30:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corona, D. F., G. Langst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 5.Dingwall, A. K., S. J. Beek, C. M. McCallum, J. W. Tamkun, G. V. Kalpana, S. P. Goff, and M. P. Scott. 1995. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamiche, A., J. G. Kang, C. Dennis, H. Xiao, and C. Wu. 2001. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. USA 98:14316-14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamiche, A., R. Sandaltzopoulos, D. A. Gdula, and C. Wu. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833-842. [DOI] [PubMed] [Google Scholar]
- 8.Havas, K., I. Whitehouse, and T. Owen-Hughes. 2001. ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci. 58:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iida, T., and H. Araki. 2004. Noncompetitive counteractions of DNA polymerase ɛ and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 11.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaskelioff, M., I. M. Gavin, C. L. Peterson, and C. Logie. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaskelioff, M., S. Van Komen, J. E. Krebs, P. Sung, and C. L. Peterson. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212-9218. [DOI] [PubMed] [Google Scholar]
- 14.Kagalwala, M. N., B. J. Glaus, W. Dang, M. Zofall, and B. Bartholomew. 2004. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 23:2092-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassabov, S. R., N. M. Henry, M. Zofall, T. Tsukiyama, and B. Bartholomew. 2002. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 22:7524-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassabov, S. R., B. Zhang, J. Persinger, and B. Bartholomew. 2003. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell 11:391-403. [DOI] [PubMed] [Google Scholar]
- 17.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 19.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119:1-16. [DOI] [PubMed] [Google Scholar]
- 21.McConnell, A. D., M. E. Gelbart, and T. Tsukiyama. 2004. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 24:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson, C. L., A. Dingwall, and M. P. Scott. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91:2905-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poot, R. A., G. Dellaire, B. B. Hulsmann, M. A. Grimaldi, D. F. Corona, P. B. Becker, W. A. Bickmore, and P. D. Varga-Weisz. 2000. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19:3377-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha, A., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta, S. M., J. Persinger, B. Bartholomew, and C. L. Peterson. 1999. Use of DNA photoaffinity labeling to study nucleosome remodeling by SWI/SNF. Methods 19:434-446. [DOI] [PubMed] [Google Scholar]
- 26.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 27.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 29.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 31.Vary, J. C., Jr., V. K. Gangaraju, J. Qin, C. C. Landel, C. Kooperberg, B. Bartholomew, and T. Tsukiyama. 2003. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 23:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 33.Whitehouse, I., C. Stockdale, A. Flaus, M. D. Szczelkun, and T. Owen-Hughes. 2003. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 23:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolffe, A. 1998. Chromatin: structure and function, 3rd ed. Academic Press, San Diego, Calif.
- 35.Xiao, H., R. Sandaltzopoulos, H. M. Wang, A. Hamiche, R. Ranallo, K. M. Lee, D. Fu, and C. Wu. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8:531-543. [DOI] [PubMed] [Google Scholar]