Abstract
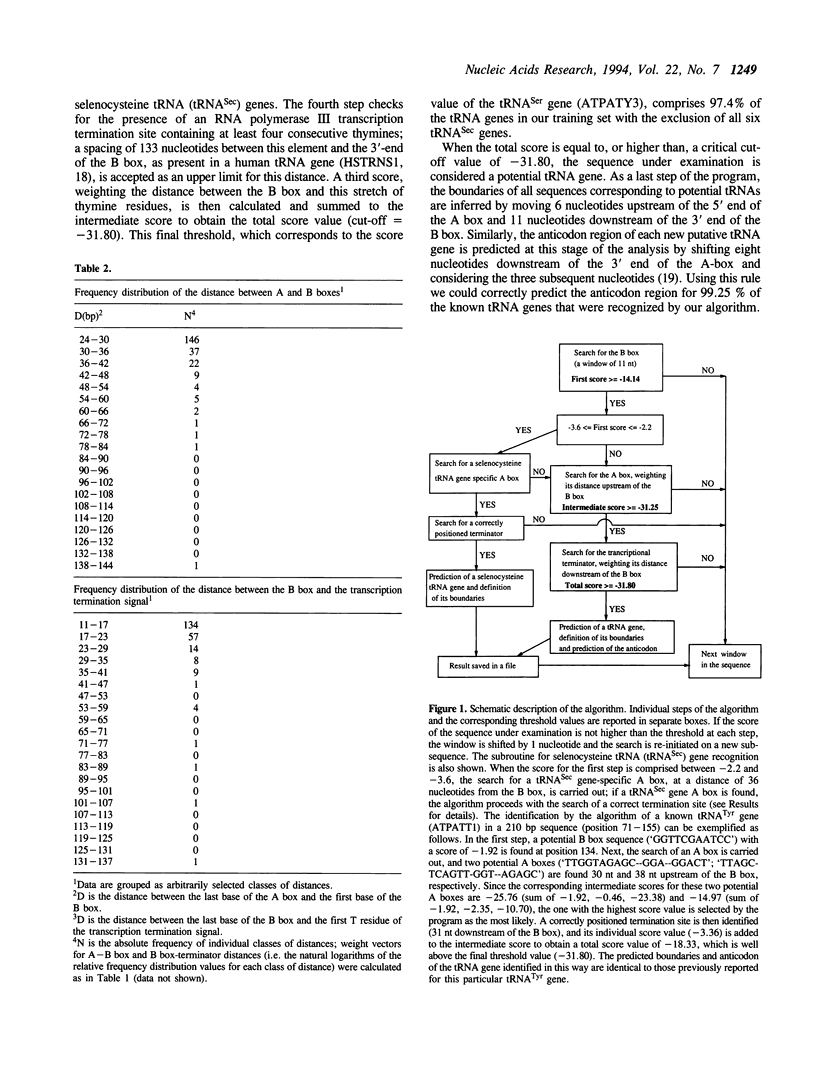
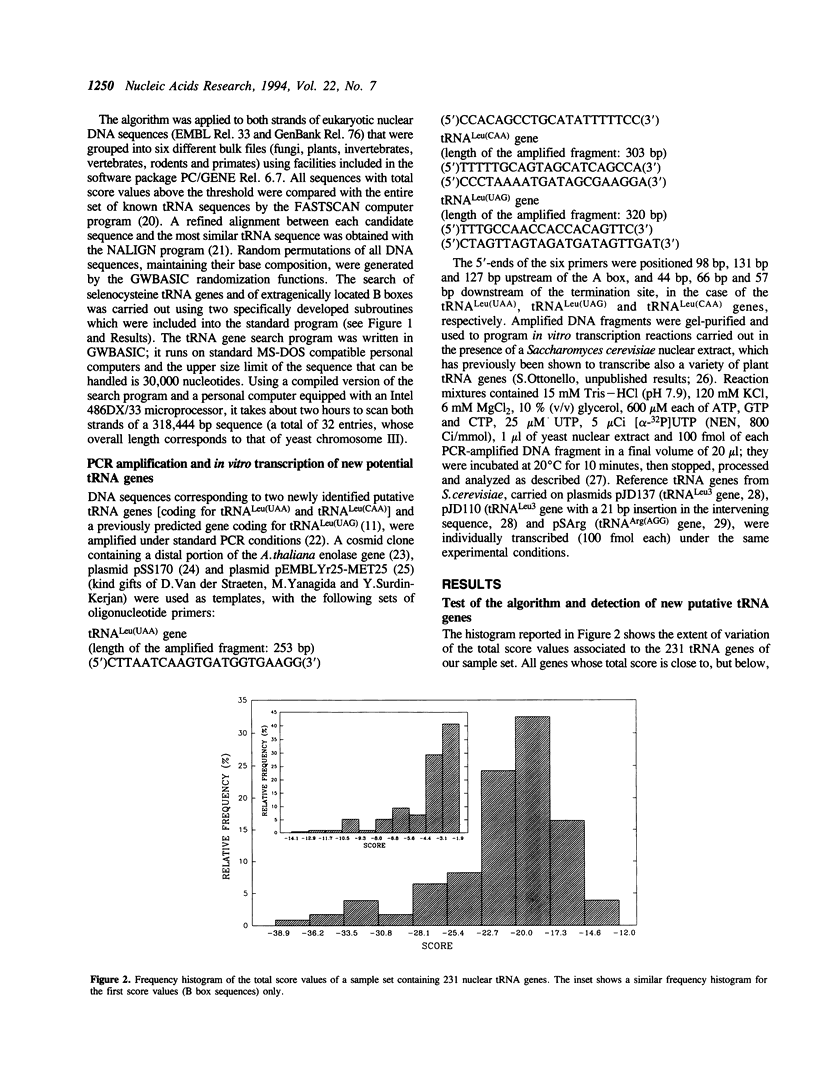
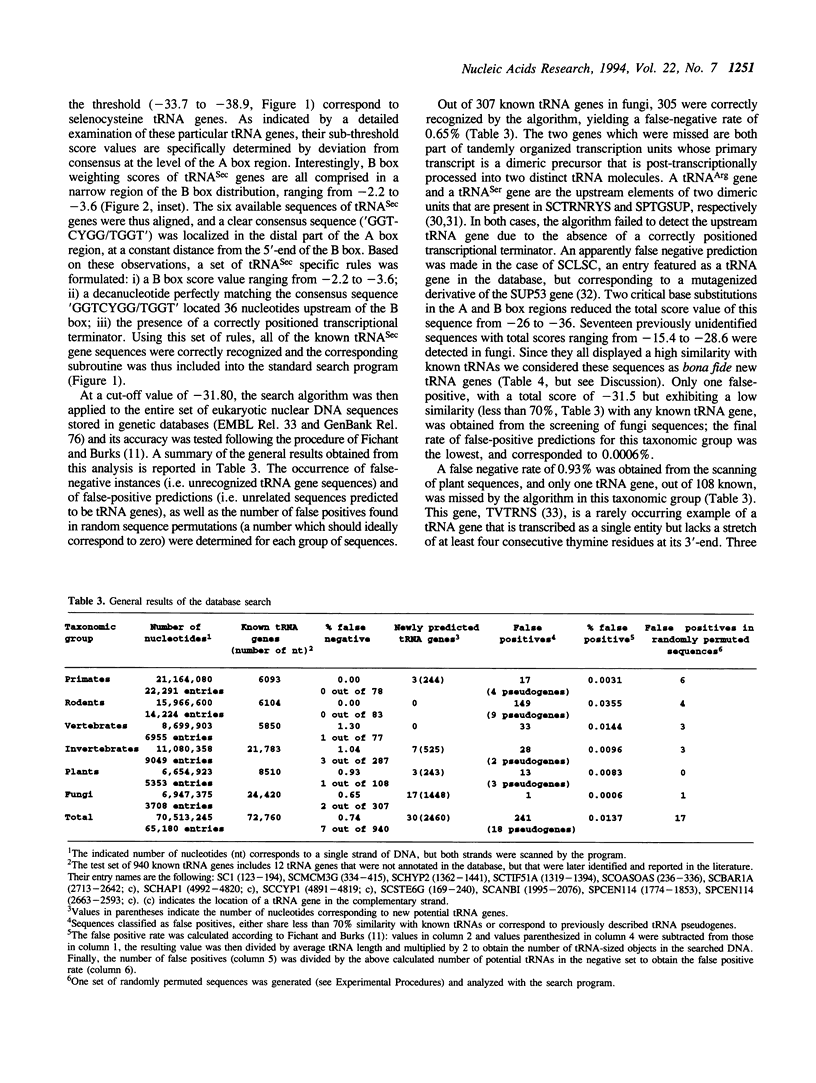
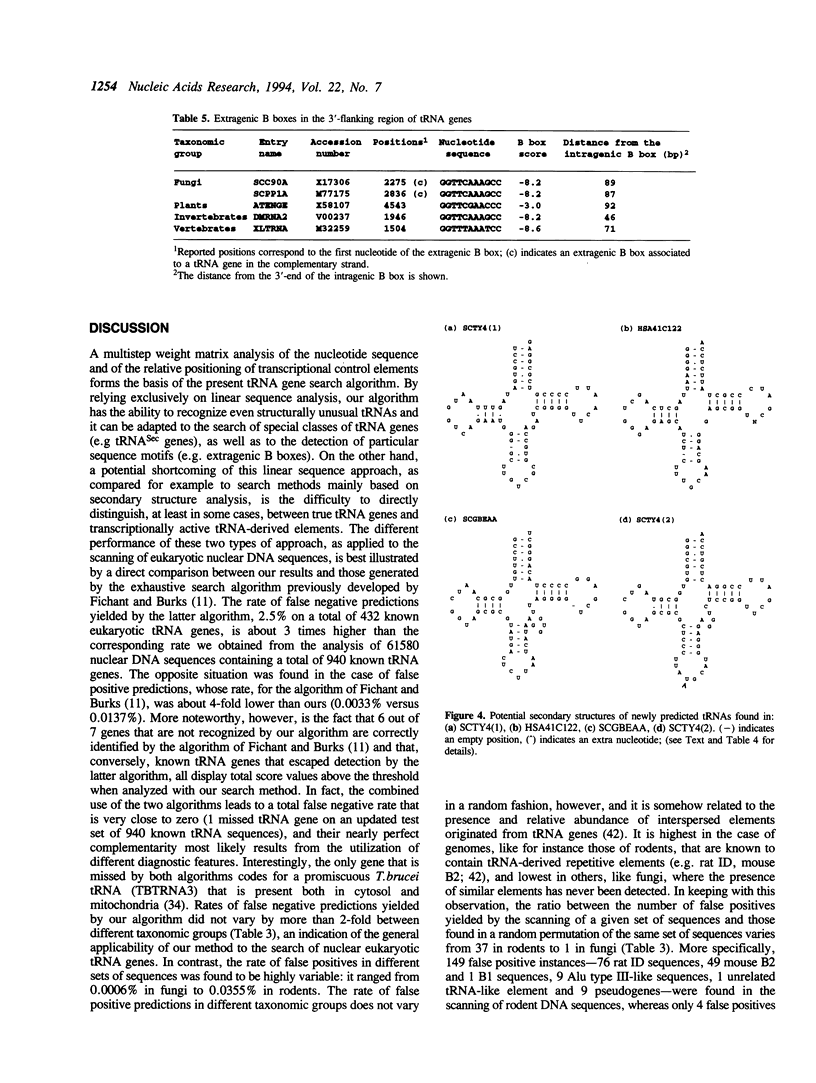
A linear method for the search of eukaryotic nuclear tRNA genes in DNA databases is described. Based on a modified version of the general weight matrix procedure, our algorithm relies on the recognition of two intragenic control regions known as A and B boxes, a transcription termination signal, and on the evaluation of the spacing between these elements. The scanning of the eukaryotic nuclear DNA database using this search algorithm correctly identified 933 of the 940 known tRNA genes (0.74% of false negatives). Thirty new potential tRNA genes were identified, and the transcriptional activity of two of them was directly verified by in vitro transcription. The total false positive rate of the algorithm was 0.014%. Structurally unusual tRNA genes, like those coding for selenocysteine tRNAs, could also be recognized using a set of rules concerning their specific properties, and one human gene coding for such tRNA was identified. Some of the newly identified tRNA genes were found in rather uncommon genomic positions: 2 in centromeric regions and 3 within introns. Furthermore, the presence of extragenically located B boxes in tRNA genes from various organisms could be detected through a specific subroutine of the standard search program.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D. A., Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990 Aug;4(8):1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- Burks C., Cinkosky M. J., Fischer W. M., Gilna P., Hayden J. E., Keen G. M., Kelly M., Kristofferson D., Lawrence J. GenBank. Nucleic Acids Res. 1992 May 11;20 (Suppl):2065–2069. doi: 10.1093/nar/20.suppl.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnol A. F., Margottin F., Huet J., Almouzni G., Prioleau M. N., Méchali M., Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993 Apr 1;362(6419):475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Rizos H., Henderson B. R., Baker R., Stewart T. S. Subfamilies of serine tRNA genes in the bovine genome. Mol Gen Genet. 1991 Dec;231(1):106–112. doi: 10.1007/BF00293828. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989 Jun 2;57(5):739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Cummins C. M., Donahue T. F., Culbertson M. R. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3565–3569. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Duimio L., Coda-Zabetta F., Sprague K. U., Ottonello S. A novel RNA polymerase III transcription factor fraction that is not required for template commitment. J Biol Chem. 1993 May 25;268(15):11199–11207. [PubMed] [Google Scholar]
- Fichant G. A., Burks C. Identifying potential tRNA genes in genomic DNA sequences. J Mol Biol. 1991 Aug 5;220(3):659–671. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Fuchs R., Stoehr P. J., Cameron G. N. The EMBL Data Library. Nucleic Acids Res. 1992 May 11;20 (Suppl):2071–2074. doi: 10.1093/nar/20.suppl.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J., Schumann G., Borschet G., Gösseringer R., Bach M., Bertling W. M., Marschalek R., Dingermann T. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5' and 3'-flanking regions. J Mol Biol. 1991 Dec 5;222(3):537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- Kerjan P., Cherest H., Surdin-Kerjan Y. Nucleotide sequence of the Saccharomyces cerevisiae MET25 gene. Nucleic Acids Res. 1986 Oct 24;14(20):7861–7871. doi: 10.1093/nar/14.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993 Jul;12(7):2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp J. L., Shu H. H., Martin N. C. Human tRNASer gene organization and a tRNASer gene sequence. Nucleic Acids Res. 1988 Jan 25;16(2):770–770. doi: 10.1093/nar/16.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. M., Clarke L., Carbon J. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1306–1310. doi: 10.1073/pnas.88.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lochmüller H., Stucka R., Feldmann H. A hot-spot for transposition of various Ty elements on chromosome V in Saccharomyces cerevisiae. Curr Genet. 1989 Oct;16(4):247–252. doi: 10.1007/BF00422110. [DOI] [PubMed] [Google Scholar]
- Léveillard T., Kassavetis G. A., Geiduschek E. P. Repression and redirection of Saccharomyces cerevisiae tRNA synthesis from upstream of the transcriptional start site. J Biol Chem. 1993 Feb 15;268(5):3594–3603. [PubMed] [Google Scholar]
- Marvel C. C. A program for the identification of tRNA-like structures in DNA sequence data. Nucleic Acids Res. 1986 Jan 10;14(1):431–435. doi: 10.1093/nar/14.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram J. C., Bell S. D., Nelson R. G., Barry J. D. tRNAs of Trypanosoma brucei. Unusual gene organization and mitochondrial importation. J Biol Chem. 1991 Sep 25;266(27):18313–18317. [PubMed] [Google Scholar]
- Munz P., Amstutz H., Kohli J., Leupold U. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature. 1982 Nov 18;300(5889):225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Papanicolaou C., Gouy M., Ninio J. An energy model that predicts the correct folding of both the tRNA and the 5S RNA molecules. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):31–44. doi: 10.1093/nar/12.1part1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen A., Sarid S., Daniel V. Genes and pseudogenes in a reiterated rat tRNA gene cluster. Nucleic Acids Res. 1984 Jun 25;12(12):4893–4906. doi: 10.1093/nar/12.12.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sandmeyer S. B., Hansen L. J., Chalker D. L. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Shortridge R. D., Pirtle I. L., Pirtle R. M. IBM microcomputer programs that analyze DNA sequences for tRNA genes. Comput Appl Biosci. 1986 Apr;2(1):13–17. doi: 10.1093/bioinformatics/2.1.13. [DOI] [PubMed] [Google Scholar]
- Staden R. A computer program to search for tRNA genes. Nucleic Acids Res. 1980 Feb 25;8(4):817–825. [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Searching for patterns in protein and nucleic acid sequences. Methods Enzymol. 1990;183:193–211. doi: 10.1016/0076-6879(90)83014-z. [DOI] [PubMed] [Google Scholar]
- Stange N., Beier D., Beier H. Intron excision from tRNA precursors by plant splicing endonuclease requires unique features of the mature tRNA domain. Eur J Biochem. 1992 Nov 15;210(1):193–203. doi: 10.1111/j.1432-1033.1992.tb17408.x. [DOI] [PubMed] [Google Scholar]
- Stråby K. B. A yeast tRNA(Arg) gene can act as promoter for a 5' flank deficient, non-transcribable tRNA(SUP)6 gene to produce biologically active suppressor tRNA. Nucleic Acids Res. 1988 Apr 11;16(7):2841–2857. doi: 10.1093/nar/16.7.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B., Kubli E. tRNA(Tyr) genes of Drosophila melanogaster: expression of single-copy genes studied by S1 mapping. Mol Cell Biol. 1988 Aug;8(8):3322–3331. doi: 10.1128/mcb.8.8.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska-Kulińska Z., Jarmołowski A., Augustyniak J. A nuclear tRNA(UGASer) gene from the wheat Triticum vulgare var. Aria. Gene. 1989 Apr 15;77(1):163–167. doi: 10.1016/0378-1119(89)90370-3. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Niwa O., Yanagida M. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J Mol Biol. 1991 Mar 5;218(1):13–17. doi: 10.1016/0022-2836(91)90867-6. [DOI] [PubMed] [Google Scholar]
- Van der Straeten D., Rodrigues-Pousada R. A., Goodman H. M., Van Montagu M. Plant enolase: gene structure, expression, and evolution. Plant Cell. 1991 Jul;3(7):719–735. doi: 10.1105/tpc.3.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y., Matsumoto S., Kojima S., Ohshima K., Okada N., Machida Y. Molecular characterization of a short interspersed repetitive element from tobacco that exhibits sequence homology to specific tRNAs. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S., Rivier D. H., Sprague K. U. Sequences far downstream from the classical tRNA promoter elements bind RNA polymerase III transcription factors. Mol Cell Biol. 1991 Mar;11(3):1382–1392. doi: 10.1128/mcb.11.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]



