Abstract
Amplification of the human cytomegalovirus (HCMV) lytic origin (oriLyt) in human fibroblasts is dependent upon six core replication proteins and UL84, IE2, and UL36-38. Using a telomerase-immortalized human fibroblast cell line (T-HFs), oriLyt-dependent DNA replication no longer required the gene products of UL36-38. To determine the role of IE2 in DNA replication in human fibroblasts, we examined potential IE2-binding sites within HCMV oriLyt. We now show that a strong bidirectional promoter (oriLytPM) (nucleotides 91754 to 92030) is located in the previously identified core region of the origin and is required for efficient amplification of oriLyt. It was determined that a 14-bp novel DNA motif (oriLyt promoter activation element), which was initially identified as a binding element for the immediate-early protein IE2, was essential for oriLytPM activity. In Vero cells the oriLytPM was constitutively active and strongly repressed by IE2, but it was reactivated by UL84. In contrast, transfection of the oriLytPM into human fibroblasts resulted in a very low basal level of promoter activity that was dramatically up-regulated upon infection with HCMV. Cotransfection assays demonstrated that the transfection of UL84 along with IE2 transactivated the oriLytPM in human fibroblasts. Further activation was observed upon cotransfection of the set of plasmids expressing the entire replication complex. Efficient oriLyt amplification in the absence of IE2 in human fibroblasts was observed by replacing the oriLytPM with the simian virus 40 early promoter. Under these conditions, however, UL84 was still required for amplification of oriLyt. These results suggest that the mechanism of initiation of HCMV lytic replication in part involves transcriptional activation.
Human cytomegalovirus (HCMV) contains a single lytic origin for DNA replication, oriLyt, located near the center of the UL region and upstream of the open reading frame (ORF) encoding the single-stranded DNA-binding protein ppUL57 (1, 19, 40). The HCMV oriLyt is remarkable among viral replication origins for its apparent size and complexity. The entire oriLyt region of HCMV is located from nucleotides (nt) 90500 to 93930 and is composed of a core (nt 91751 to 93299) which contains two essential regions (1, 40, 66). These essential regions (I and II) contain a pyrimidine-rich sequence (Y-block), various reiterated sequences, several transcription factor-binding sites, direct and inverted repeat sequences, base composition biases and strand asymmetries, and RNA-DNA hybrid structures and is a site of active transcription (the small replication transcript [SRT]) (1, 22, 40, 49, 66). Despite the apparent exhaustive investigation of elements within oriLyt that contribute to DNA synthesis, few data exist with respect to the actual function of these elements in initiation of DNA replication.
Some information concerning the initiation of DNA synthesis can be inferred from the required virus-encoded transacting factors necessary to replicate oriLyt. In human fibroblasts, oriLyt-dependent DNA replication requires the core replication machinery and the gene products of IE2, UL36-38, and UL84 (50).
IE2 is the major transcription-activating protein of HCMV (41) and is also implicated in the autoregulation of the major immediate-early promoter (MIEP) (42). The IE2 protein interacts with itself (8, 14) and UL84 (57) as well as with specific cellular transcription factors (i.e., CREB, CBP, SP-1, and c-Jun) (34, 38, 52, 54, 61). Specific DNA-binding sites for IE2 have been found in the upstream regions of promoters activated by IE2 (2, 4, 53, 54) and at the cap site of the MIEP (26, 35, 39). Recently it was shown that potential IE2-binding sites are located within the HMCV oriLyt (21). Hence, IE2 may play a pivotal role in HCMV DNA synthesis as well as contributing a general transactivator function.
The proteins encoded by UL36-38 are necessary for oriLyt-dependent DNA replication in human fibroblasts. However, studies in Vero cells demonstrated that the UL36-38 locus can be replaced by HCMV UL69 in the cotransfection-replication assay; consequently, only UL84 is necessary for efficient oriLyt amplification in this cell type (50). The UL36-38 locus encodes proteins that were shown to have antiapoptotic properties and, therefore, their requirement in human fibroblasts may serve to establish a suitable cellular environment for transient DNA replication (17, 56).
UL84 is a nuclear protein and a component of DNA replication compartments in infected and transfected cells, and it facilitates the formation of these structures (50, 64). UL84 interferes with the transactivation activity of IE2 on the early promoter for UL112/113 (16). Despite its apparent critical role in DNA synthesis, very little is known about the function of UL84 in this context.
It appears that lytic origins fall into types. The first type, like herpes simplex virus types 1 and 2 along with human herpesviruses 6 (HHV6) and -7, interacts with a distinct origin-binding protein. This protein then imparts an enzymatic function that initiates DNA synthesis (5, 10, 11, 31-33, 43, 62). The second type, which includes Epstein-Barr virus (EBV) and HHV8, contains regions that are involved in transcriptional activation. Comparison of these lytic origins indicates a pattern of similar structures to those found within the HCMV oriLyt. Most lytic origins contain an AT-rich region and the presence of DNA elements that interact with virus-encoded transactivators. For example, the EBV oriLyt contains several Zta response elements (ZREs) and Rta response elements (RREs) (9, 18, 51). Some of these elements are present within an essential promoter-enhancer region of EBV oriLyt, indicating that transcription is involved in EBV oriLyt activation. For HHV8, an ORF50 response element (ORF50RE) located within the right half of the origin is responsive to K-Rta and subsequently activates a promoter element within oriLyt (3), again suggesting that transcription is involved in HHV8 oriLyt function.
In this report, we show that oriLyt-dependent DNA replication no longer required the gene products of UL36-38 when a telomerase-immortalized human fibroblast cell line was used. Hence, IE2 and UL84 were the only noncore proteins required for DNA synthesis in the transient-replication assay in human fibroblasts. In order to determine the role of these two proteins in DNA replication, we used UL84 and IE2 in transient-transfection assays, using regions within HCMV oriLyt as reporters. We show that a strong promoter with bidirectional activity is located within the core region of the oriLyt. The oriLyt promoter (oriLytPM) was constitutively active in Vero cells; however, in human fibroblasts this promoter was responsive to HCMV infection or to IE2 and UL84 in transient assays. Further activation was observed with the addition of plasmids expressing the core replication proteins. Targeted mutations within oriLytPM revealed that a 14-bp element (oriLyt promoter activation element [OPAE]) was responsible for promoter activity. The simian virus 40 (SV40) early promoter functionally substituted for the oriLytPM in transient-replication assays, strongly suggesting that oriLyt amplification is dependent upon transcriptional activation. Under these conditions, IE2 was no longer required for origin-dependent DNA replication in the cotransfection-replication assay in human fibroblasts, whereas UL84 was still necessary.
Taken together these results suggest that IE2, along with UL84, serves to activate the promoter in oriLyt and facilitate or trigger initiation of DNA synthesis via transcriptional activation. Also, it is apparent that UL84 has an additional, as-yet-undefined role in DNA replication, since it cannot be omitted from cotransfections even when IE2 is not required.
MATERIALS AND METHODS
Cells and virus.
Cos7, Vero, and human foreskin fibroblast (HFF) cells were maintained in Dulbecco's modification of Eagle's medium (catalog no. 50-003-PB; Cellgro) supplemented with 10% fetal bovine serum. HCMV strain AD169 cells (American Type Culture Collection) were maintained as frozen stocks. T-HFs were generated by infection with a retrovirus expressing hTERT (Geron Inc.) (29) followed by selection for G418-resistant colonies. Selected colonies were expanded and tested for telomerase activity using the trap assay (29, 58).
Plasmids.
The replication reporter pGEM-oriLyt, described previously (64), contains the PvuII-KpnI HCMV oriLyt fragment (nt 89796 to 94860) of AD169 (6) ligated into pGEM-7Zf(-) (catalog no. p2371; Promega). pZP13 containing full-length UL84 with its native promoter was constructed as previously described (44). pSI84 was generated by cleaving pZP13 with DraI and NotI, and the resultant 2-kb fragment containing the UL84 ORF was ligated into the mammalian expression vector pSI (catalog no. E1721; Promega). The IE2 expression construct pON2206, which contains the IE2 cDNA under the control of the SRα promoter, was a gift from E. Mocarski (Stanford University) (25).
The IE2-FLAG plasmid was constructed by PCR amplification of the IE2 ORF from pON2206 using PCR primer 5′-CGACTTAACAGATCTCGAGCTCAAGCTTCGAATTCATGGATTACAAGGATGACGACGATAAGGAGTCCTCTGCCAAGAGAAA-3′ and reverse primer 5′-CCCGGGCCCGCGGTACCGTCGACTGCAGAATTCTTACTGAGACTTGTTCCTCAGGTCCTGGATGG-3′. The reverse primer was constructed such that the IE2 was fused in frame with the FLAG epitope. The PCR product was then recombined into the phCMV Xi-cloning vector (catalog no. XC003120; Gene Therapy Systems, Inc.) as recommended by the manufacturer. pRL-CMV and pRL-SV40 were obtained from Promega (catalog nos. E2261 and E2231). The set of plasmid subclones encoding the HHV8 and HCMV replication proteins were made as described previously (3, 64).
Plasmids used in oriLyt promoter assays.
Sequences within oriLyt were PCR amplified using pGEM-oriLyt as template and the following primers containing KpnI and XhoI restriction endonuclease sites at the 3′ and 5′ ends, respectively: P1, 5′-CCGCTCGAGCATTGGAAATCATGGTTGCCCAAATTTGGTAA-3′; P2, 5′-GGGGTACCCATTGGAAATCATGGTTGCCCAAATTTGGTAA-3′; P3, 5′-GGGGTACCCAGTCCGTTTTACGTATACCGGATGCTAGGCG-3′; P4, 5′-CCGCTCGAGGCGGTAGAATACAGCGATCCCTAGTGAAGCCAC-3′; P5, 5′-CCGTCTCGAGGCTTATGACGCGTATCCGGGAGTAGCGTCTACG-3′; P6, 5′-GGGGTACCGCGGTAGAATACAGCGATCCCTAGTGAAGCCAC-3′; P7, 5′-CCGCTCGAGCAGTCCGTTTTACGTATACCGGATGCTAGGCG-3′; P8, 5′-GGGGTACCGCTTATGACGCGTATCCGGGAGTAGCGTCTACG-3′. The oriLyt subclone pGL-ORI-A was amplified by using primers P1 and P6. pGL-ORI-B was generated using primers P1 and P3. pGL-ORI-BR was generated using primers P2 and P7. pGL-ORI-C was generated using primers P5 and P6. pGL-ORI-CR was generated using primers P4 and P8. pGL-ORI-AR was generated using primers P2 and P4. All the amplified fragments were subcloned into the luciferase reporter vector pGL2-Basic (catalog no. E1641; Promega).
As a control for IE2 transactivation, a luciferase reporter, pGL-112pr, was also constructed. The UL112/113 promoter was PCR amplified by using AD169 DNA as template and the following primer set: forward, 5′-CCGGTACCAGATCTCCGCGTCACCTTTCATCGAGT-3′; reverse, 5′-CCAAGCTTGGCCGTGGAGCGAGTGCCGCCGCAGCC-3′. The 750-bp PCR product was then cleaved with KpnI and HindIII and ligated into the luciferase reporter vector pGL2-Basic. The final construct, pGL-112pr, was used in the transfection and luciferase assays.
The pGEM-oriLytSVR and pGEM-oriLytSVL plasmids were generated by cleavage of pGEM-oriLyt with NotI and SgfI and removal of the intervening sequence, followed by ligation of the PCR-amplified SV40 early promoter. The SV40 early promoter region was amplified from the vector pSI (Promega) using PCR primers 5′-CCGCGGCCGCCTCGACAGATCTGCGCAGCACC-3′ and 5′-CCGCGATCGCGACTGTTGTGTCAGAAGAATCAAGCT-3′ (to generate pGEM-oriLyt SVR) or 5′-CCGCGATCGCCTCGACAGATCTGCGCAGCACC-3′ and 5′ CCGCGGCCGCGACTGTTGTGTCAGAAGAATCAAGCT-3′ (to generate pGEM-oriLytSVL). Primer pairs were designed such that a NotI site and a SgfI site were at the ends of the primers. Two sets of primers were used in order to ligate the promoter into pGEM-oriLyt in both directions. pGEM-oriLytSVR had the SV40 early promoter ligated into HCMV oriLyt such that the SV40 early promoter directed transcription in the right-to-left direction. pGEM-oriLytSVL had the SV40 early promoter ligated into HCMV oriLyt such that the SV40 early promoter directed transcription in the left-to-right direction.
Site-directed mutagenesis.
All mutations were generated using the QuikChange II site-directed mutagenesis kit (catalog no. 200523; Stratagene). Mutagenesis was performed according to the manufacturer's protocol.
pGL-ORI-Amut was generated using pGL-ORI-A plasmid as template and the primer set 5′-CCTTATCTACCAATCACGAGAAACGGATATA-3′ (forward) and 5′-GATTGTATATCCGTTTCTCGTGATTGGTAGATAAGG-3′ (reverse), where a single G nucleotide was inserted (underlined) into the wild-type sequence. pGEM-oriLyt+G was generated using the same primers as for pGL-ORI-Amut, except that the DNA template used for PCR was pGEM-oriLyt. All mutations were confirmed by DNA sequencing.
Chromatin immunoprecipitation (ChIP) assay.
A 225-cm flask of confluent T-HF cells (6 × 106 cells) was infected with AD169 at a multiplicity of infection of 1. At 48 h postinfection (hpi), cells were fixed with a 1% formaldehyde solution in 1× phosphate-buffered saline (PBS; pH 7.4) at room temperature for 10 min. The cells were washed twice with ice-cold PBS (pH 7.4) and harvested with 10 ml of PBS. The cells were then collected by centrifugation at 1,000 × g for 5 min. The pellet was resuspended in 750 μl of cell lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 2 mM MgCl, 0.2 mM EDTA, and 10 μl of protease inhibitor cocktail σ/ml) and sonicated five times at 10-s intervals. The lysis solution was diluted with PBS to a 5-ml volume and precleared with 2 μg of normal mouse immunoglobulin G-agarose (Santa Cruz Biotechnology) for 30 min at 4°C. The solution was again centrifuged to remove the normal mouse immunoglobulin G-agarose at 500 × g for 5 min. Two micrograms of IE2-specific antibody G13-12E2 (Vancouver Biotech, Ltd.) was added to 1 ml of the cell lysate and incubated for 1 h with rotation at 4°C. One milliliter of the lysate was also used for control immunoprecipitations that did not contain primary antibody. The solution was then incubated with 50 μl of protein A/G plus agarose beads (Santa Cruz Biotechnology) with rotation overnight at 4°C. The beads were pelleted and washed four times with ice-cold PBS. The beads were then resuspended in 100 μl of Tris-EDTA (TE) and treated with RNase A (50 μg/ml) for 30 min at 37°C. The complexes were treated with 5 μl of 10% sodium dodecyl sulfate and 500 μg of proteinase K/ml for 4 h at 37°C with occasional mixing. In order to disassociate the cross-linked protein-DNA complexes, the reaction mixtures were incubated at 65°C overnight. The supernatant was collected and extracted with phenol followed by chloroform and ethanol precipitation to isolate the immunoprecipitated DNA. The resulting pellet was resuspended in 20 μl of TE.
PCR was used to detect the immunoprecipitated DNA fragments. Two microliters of the TE solution was used as template for each PCR. To detect IE2 association with the cis repression signal (CRS) sequence found in the IE2 promoter, the two primers 5′-GCAGTACATCTACGTATTAGTCATCG-3′ and 5′-AGCTTAAGTTCGAGACTGTTGTGTC-3′ were used in the PCR.
To detect IE2 association with the OPAE within the HCMV oriLyt, primer pair A corresponding to nt 91928 to 91627 was used: 5′-CGTACACCGAGGACGGTGGAACCCTAACGGG-3′ and 5′-TCAACCCCGTGGCCGGCGAGGCGGTGAGCA-3′. In addition, as a negative control primer pair B was used, corresponding to nt 93128 to 92858 within oriLyt: 5′-ACGGCGCACATCTAGTGGAATTTTACCGACG-3′ and 5′-GATGGTGCTCCAGGGCGGTGGGACGGGCCCG-3′.
All reactions were tested for DNA contamination by running PCR without template DNA for the negative control. All reactions were also tested using AD169 as the template as a positive control.
Synthesis and purification of IE2-FLAG fusion protein.
Cos7 cells were plated into ten 100-mm tissue culture dishes 24 h prior to transfection. Transfection was performed using 10 μg of IE2-FLAG per plate with TransIt LT1 (catalog no. MIR2300; Mirus) according to the manufacturer's instructions. Total cell protein from all 10 dishes was harvested in 10 ml of lysis buffer (50 mM Tris HCl-150 mM NaCl-1 mM EDTA-1% Triton X-100; pH 7.4) at 48 h posttransfection (hpt). Cell lysates were sheared through an 18-gauge needle five times on ice, and cell debris was pellet at 16,000 × g for 10 min. FLAG fusion protein was purified by loading the supernatant onto a 1-ml EZview Red anti-FLAG M2 affinity gel column (product no. F2426; Sigma). The column was then washed with 5 ml of Tris-buffered saline (TBS; 50 mM Tris HCl-150 mM NaCl; pH 7.4) three times, and FLAG fusion protein was eluted with 100 μl of a 5-μg/μl solution of 3× FLAG peptide (product no. F4799; Sigma) in 5 ml of TBS. The fractions from the purification process were collected, resolved on a sodium dodecyl sulfate-10% polyacrylamide gel, and stained with Coomassie blue to monitor the protein purity and purification efficiency. Protein concentration of the purified IE2-FLAG was determined by Bradford assay.
Electrophoretic mobility shift DNA-binding assay (EMSA).
The DNA-binding probes representing the HCMV MIEP CRS were prepared by annealing the following complementary 30-mer CRS oligonucleotide pairs: 5′-GATCCCGGGAGCTCGTTTAGTGAACCGTCA-3′ and 5′-GATCTGACGGTTCACTAAACGAGCTCCCGG-3′. The OPAE site within oriLyt was represented by probes annealed using the oligonucleotide pair 5′-GATCCCTTATCTACCAATCACAGAAACGGATATA-3′ and 5′-GATCTATATCCGTTTCTGTGATTGGTAGATAAGG-3′. Labeling was accomplished by filling in the recessed ends with Escherichia coli DNA polymerase Klenow fragment and deoxynucleoside triphosphates in the presence of [α-32P]dCTP.
For the binding reaction, IE2-FLAG-purified protein (500 ng) was preincubated in 20 μl of binding buffer (10 mM Tris-Cl, 0.5 mM dithiothreitol, 1 mM EDTA, 10% glycerol, 50 mM KCl, 8 mM MgCl2; pH 7.5) containing 40 μg of poly(dA-dT) · poly(dA-dT)/ml for 10 min at 20°C. 32P-labeled probe DNA (20,000 cpm) was then added for 20 min at 20°C. The mixtures were loaded onto 4% nondenaturing acrylamide gels and run at 100 V in 0.5× Tris-borate-EDTA buffer. After electrophoresis at 20°C, the gels were dried and autoradiographed. For cold competition assays, 20- or 50-fold molar excesses of nonradioactively labeled DNA fragments were added to the preincubation reaction mixtures. For antibody supershift assays, 1:100-diluted anti-IE1/2 antibody MAB810 (Chemicon) was added to the preincubation reaction mixtures.
Transient-cotransfection replication assay.
The HHV8 core replication proteins encoding helicase, primase, primase-associated factor, polymerase, polymerase accessory protein, and single-stranded DNA-binding protein (3) were cotransfected with and without UL84 and/or IE2 and/or UL36-38 expression constructs (44, 45) and pGEM-oriLyt, using the calcium phosphate coprecipitation assay (64). Total cellular DNA was harvested 5 days posttransfection, cleaved with EcoRI and DpnI, and resolved on a 0.8% agarose gel. The DNA was transferred to a Zeta-Probe nylon membrane and hybridized with 32P-labeled pGEM7zf(-). Blots were exposed to X-ray film for 5 to 15 h.
For replication assays involving pGEM-oriLytSVR(L), cells were cotransfected with pGEM-oriLyt along with pGEM-oriLytSVR(L) and infected with HCMV 24 hpt. Total cellular DNA was harvested and treated as described above. For the cotransfection-replication assay, cells were cotransfected with pGEM-oriLytSVR and pGEM-oriLyt along with the core replication proteins for HHV8 or HCMV, with and without UL84 and/or IE2 expression constructs. Total cellular DNA was harvested and treated as described above.
In the case of the transfection of pGEM-oriLyt+G, this plasmid was transfected into HFFs and cells were subsequently infected with HCMV. Total DNA was harvested 5 days postinfection and processed as described above.
Luciferase assay.
Vero or HFF cells were seeded into six-well tissue culture plates 24 h prior to transfection. Cotransfections were performed using TransIt LT1 (catalog no. MIR2300; Mirus) according to the manufacturer's instructions. Plasmids pON2206 (25) and pSI84 were cotransfected along with the various indicated luciferase reporter constructs. The total amount of DNA and the amount of each plasmid used in the cotransfection mixture was kept constant by using pGEM-7Zf(-) as control filler plasmid DNA. For cotransfections involving the core replication proteins, plasmids encoding HCMV core replication proteins were added at 2 μg each in addition to the reporter luciferase plasmid, with and without UL84 and IE2 expression plasmids. Total cell lysates were harvested 48 hpt or at various times postinfection as indicated in the case of HFFs, and luciferase assays were carried out with a luciferase assay system (catalog no. E4530; Promega) as recommended by the manufacturer.
RESULTS
The gene products of UL84 and IE2 are the only additional noncore proteins required for origin-dependent DNA replication in telomerase-immortalized HFF cells.
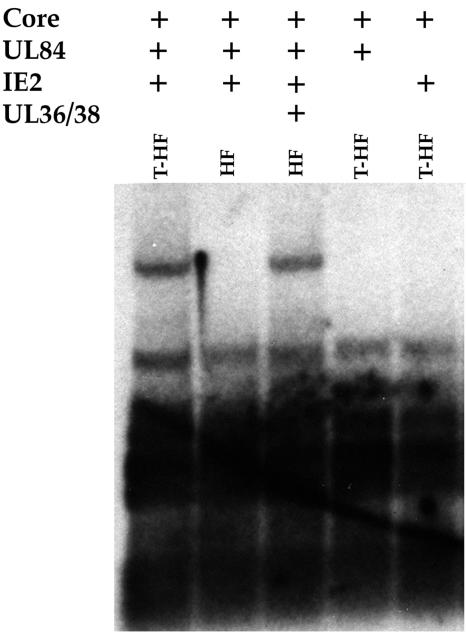
The original replication assay performed by Pari et al. demonstrated that the core replication proteins along with the gene products of UL112-113, UL36-38, IRS1, IE1/IE2, and UL84 were all required for efficient amplification of cloned oriLyt (44, 45). In a later study, when EBV core replication genes were used in the cotransfection assay, it was shown that, in Vero cells, UL84 was the only noncore protein required for origin-dependent DNA replication (50). In HFF cells, however, the gene products of IE2, UL36-38, and UL84 were all necessary for efficient amplification of oriLyt (50). Because recent evidence showed that the UL36-38 locus encodes proteins that have antiapoptotic properties (17, 56), we investigated the requirements for origin-dependent DNA replication in T-HFs. These cells have an unlimited life span in culture but retain an otherwise normal phenotype (data not shown). In order to address the issue of differences in protein requirements between telomerase-immortalized and wild-type HFF cells, we cotransfected both T-HFs and wild-type HFFs with plasmids expressing the HHV8 core replication proteins (3), UL84, IE2, and the UL36-38 proteins, and a plasmid containing the HCMV oriLyt, pGEM-oriLyt. HHV8 core replication proteins were used to enable us to focus on those HCMV proteins required for DNA replication and that perform noncore functions. HHV8 core proteins were fully capable of providing the essential enzymatic replication functions in the presence of UL84-, IE2- and, in the case of HFFs, UL36-38-encoded proteins (Fig. 1). Omission of either IE2 or UL84 expression constructs from transfection mixtures led to failure to amplify cloned oriLyt in T-HFs (Fig. 1). In wild-type HFFs, UL36-38 was absolutely required for oriLyt amplification (Fig. 1). However, UL36-38 was not necessary in T-HFs (Fig. 1). This indicated that in T-HFs, the only factors required for efficient oriLyt-dependent DNA replication were UL84 and IE2, implying that UL36-38 gene products do not directly participate in DNA replication and may function to increase the survival of primary cells when performing the cotransfection-replication assay.
FIG. 1.
Origin-dependent DNA replication in T-HFF cells. T-HFFs or normal HFFs were cotransfected with pGEM-oriLyt, the HHV8 core replication proteins, and various combinations of UL36-38, UL84, and IE2 expression plasmids. Cells were harvested 5 days posttransfection, and total cellular DNA was extracted and cleaved with EcoRI and DpnI. DNA was separated on an agarose gel and transferred to a nylon membrane. Replicated oriLyt was detected by hybridization to a radiolabeled pGEM plasmid DNA probe.
HCMV oriLyt contains a strong promoter with bidirectional activity that is constitutively active but repressed by IE2 in Vero cells.
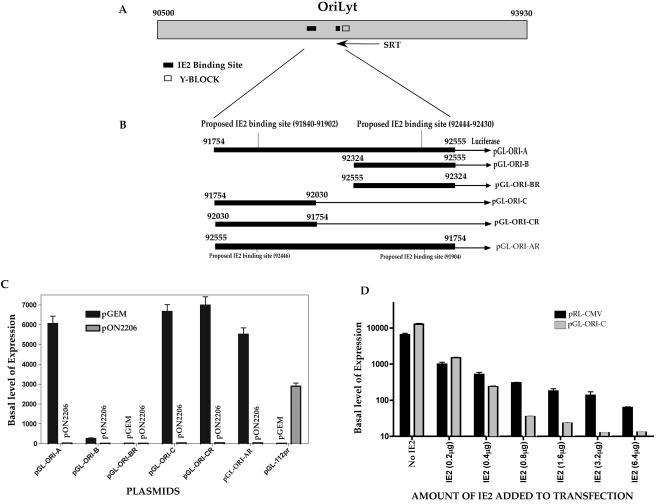
Since we determined that oriLyt-dependent DNA replication required only two additional gene products, namely IE2 and UL84, we next wanted to elucidate the function of IE2 and UL84 in lytic DNA replication. We chose to do this by examining the HCMV oriLyt region for any clues that may lead to a replication mechanism that involves a possible interaction of IE2 with oriLyt. Inspection of the oriLyt region revealed two potential IE2 binding sites. The first putative IE2 binding sequence, termed sequence-short, is located between nt 92430 and 92444 (5′-CGTTCTACGAAAACG-3′). This site is similar to the IE2 consensus-binding site CRS element found in the promoter region of IE2 (7, 36, 47). The second sequence, termed sequence-long, is located between nt 91840 and 91902. This sequence (5′-GCCACCTTATCTACCAATCACAGAAACGGATATACAATGACCCCTCCCTAGACTCCACCCCTT-3′) was identified by Huang et al. by using a modified approach coupling the methods of cyclic amplification and selection of targets to selectively amplify IE2-binding sites (21). Figure 2A is a schematic showing the positions of these two putative IE2-binding sites in relation to the Y-block and SRT within oriLyt. Since IE2 is the major transactivator protein in HCMV, we wanted to determine if the region of oriLyt containing the two putative IE2 binding elements could act as an IE2-responsive promoter. PCR fragments were generated such that these potential IE2-binding sites, along with some flanking sequence, were subcloned into the promoterless luciferase reporter vector pGL2-Basic (Promega) (Fig. 2B).
FIG. 2.
A promoter region within oriLyt is constitutively active in Vero cells and is repressed by IE2. (A) Schematic of the oriLyt region showing the two IE2 putative binding sites and their relative location to the Y-block and SRT RNA. (B) Subcloning of HCMV oriLytPM into luciferase reporter constructs. The diagram shows the pGL-ORI-A, pGL-ORI-B, pGL-ORI-BR, pGL-ORI-C, pGL-ORI-CR, and pGL-ORI-AR constructs. pGL-ORI-A contains both sequence-short and sequence-long and their flanking sequences from nt 91754 to 92555. pGL-ORI-B contains only sequence-short and its flanking sequence from nt 92324 to 92555. pGL-ORI-BR contains the same sequence as pGL-ORI-B except it was ligated into the pGL2-Basic in the reverse orientation. pGL-ORI-C contains only sequence-long and its flanking sequence from nt 91754 to 92030. pGL-ORI-CR contains the same sequence as pGL-ORI-C, except it was ligated into the pGL2-Basic in the reverse orientation. pGL-ORI-AR contains the same fragment as pGL-ORI-A but was ligated into the pGL2-Basic in the reverse orientation. (C) Mapping of the oriLytPM region using luciferase reporter assay data. The graph shows the repression of oriLyt promoter activity by IE2 in Vero cells. pGL-ORI-A, pGL-ORI-B, pGL-ORI-BR, pGL-ORI-C, pGL-ORI-CR, and pGL-ORI-AR were each transfected into Vero cells in the presence or absence of an IE2 expression construct. As a control for IE2 transactivation, the pGL-112pr luciferase reporter construct containing the UL112/113 promoter region upstream of the luciferase gene was also used. Luciferase assays were carried out at 48 hpt. Data are shown as fold increase of the basal level of expression compared to that of the parent vector, pGL2-basic. Error bars are the standard deviations of six separate experiments. (D) Dose response of repression of the oriLyt promoter by IE2. Vero cells were transfected with either pGL-ORI-C or pRL-CMV and increasing amounts of the IE2 expression construct pON2206. Error bars are the standard deviations of three separate experiments.
Vero cells were transfected with pGL-ORI-A or pGL-ORI-AR, with or without the IE2 expression plasmid pON2206. These two luciferase reporter constructs contained both putative IE2-binding sites mapping to nt 91754 to 92555 but in different orientations (Fig. 2B). Cell lysates were harvested, and luciferase assays were carried out at 48 hpt. Both of these constructs displayed a high basal level of promoter activity in Vero cells (Fig. 2C, pGL-ORI-A or pGL-ORI-AR plus pGEM). The addition of IE2 to the transfection mixture resulted in a marked repression of the high basal luciferase activity for both constructs (Fig. 2C, compare pGL-ORI-A and pGL-ORI-AR plus IE2 to pGL-ORI-A and pGL-ORI-AR plus pGEM). Transfections containing control vector having the SRα promoter and pGL-ORI-A or pGL-ORI-AR had no effect on luciferase activity (data not shown). To further map this promoter region, constructs pGL-ORI-B (nt 92324 to 92555), which contained the downstream putative IE2-binding element (sequence-short), and pGL-ORI-C (nt 91754 to 92030), which contained the upstream putative IE2-binding element (sequence-long), were generated and used in the luciferase assay (Fig. 2B). As shown in Fig. 2C, only pGL-ORI-C displayed a high basal level of promoter activity in Vero cells (Fig. 2C, compare pGL-ORI-C plus pGEM to pGL-ORI-B plus pGEM), and this activity was again repressed by addition of IE2 to the transfection mixture (Fig. 2C, compare pGL-ORI-C plus IE2 to pGL-ORI-C plus pGEM). A strong luciferase signal was still observed when the DNA segment from nt 91754 to 92030 was reversed (Fig. 2, pGL-ORI-CR), whereas the reverse of the DNA segment from nt 92324 to 92555 still showed no activation (Fig. 2, pGL-ORI-BR). This result indicated that a strong constitutive promoter (oriLytPM) showing bidirectional activity was located within oriLyt between nt 91754 and 92030. This promoter, which contains a potential IE2-binding site, was repressed by the addition of IE2 to the transfection mixture when assayed in Vero cells.
As a control for IE2 activation of an early promoter, we cotransfected a luciferase construct pGL-112pr, which contained the UL112/113 promoter in the luciferase reporter vector pGL2-Basic, along with IE2 expression plasmid pON2206 into Vero cells. As expected, IE2 transactivated the UL112/113 promoter, which resulted in a marked increase in luciferase activity compared to that in the samples that lacked the IE2 expression construct pON2206 (Fig. 2C, pGL-112pr).
Since IE2 was shown to down-regulate its own expression by directly binding to the CRS element within the MIEP (7, 35, 36, 47, 48), we wanted to compare the observed IE2-mediated repression of the oriLyt promoter to the IE2-mediated repression of the HCMV MIEP in Vero cells. We again performed cotransfections using increasing amounts of pON2206 along with pGL-ORI-C or pRL-CMV. Basal luciferase activity from the vector pRL-CMV, which contains the MIEP along with the CRS element, was repressed in a dose-dependent manner in the presence of increasing amounts of IE2 expression plasmid (Fig. 2D). pGL-ORI-C also responded to increasing amounts of pON2206 similar to pRL-CMV, except that the degree of repression by IE2 was much greater as concentrations of pON2206 increased (Fig. 2D).
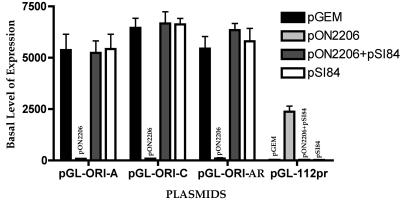
UL84 can release the IE2-mediated repression of the HCMV oriLytPM in Vero cells.
Since it was previously shown that UL84 interacted with IE2 in infected and transfected cells and this interaction was postulated to repress IE2 activation of the early promoter for UL112/113 (16, 57), we wanted to investigate the impact of UL84 on the IE2-mediated repression of the oriLytPM. Vero cells were transfected with various oriLyt-luciferase constructs, except that we now added a UL84 expression construct (pSI84) to the transfection mixture. IE2-mediated repression was inhibited when UL84 was added to the transfection mixture (Fig. 3, compare pGL-ORI-A, pGL-ORI-C, or pGL-ORI-AR plus pON2206 to pGL-ORI-A, pGL-ORI-C, or pGL-ORI-AR plus pON2206+pSI84). Cotransfection of the control parent vector, pSI, had no effect on IE2-mediated repression (data not shown). No change in promoter activity was observed when UL84 alone was transfected along with pGL-ORI-A, pGL-ORI-C, or pGL-ORI-AR (Fig. 3, compare pGL-ORI-A, pGL-ORI-C, or pGL-ORI-AR plus pGEM to pGL-ORI-A, pGL-ORI-C, or pGL-ORI-AR plus pSI84). This result indicated that the IE2-mediated repression of oriLytPM could be inhibited by the expression of UL84 in Vero cells. In contrast to the effect on the oriLytPM, IE2 activated the UL112/113 promoter (Fig. 3, pGL-112pr plus pON2206), and UL84 disrupted the IE2-mediated activation of this promoter (Fig. 3, pGL-112pr plus pON2206/pSI84), which was consistent with the results from previous studies (16).
FIG. 3.
UL84 relieves the IE2-mediated repression of oriLytPM in Vero cells. Vero cells were cotransfected with combinations of IE2 and UL84 expression plasmids along with the reporter constructs containing oriLytPM. The pGL-112pr luciferase reporter construct was also used as a control. Cells were harvested at 48 hpt, and luciferase activity was determined. Luciferase activity is shown as the fold increase in the basal level of expression over that of pGL2-Basic. Error bars are the standard deviations of six separate experiments.
Mutation of the novel DNA element within oriLytPM inhibits promoter activity in Vero cells.
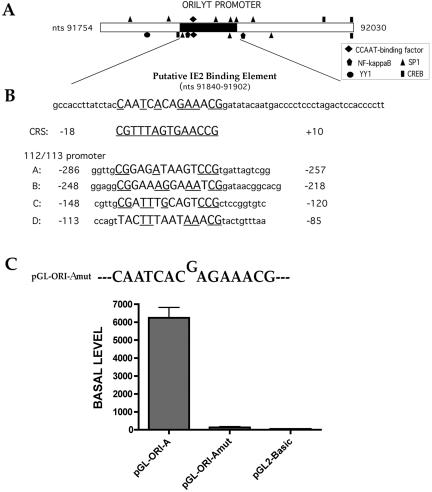
Investigation of the oriLytPM sequence using the on-line database Transfac (63) revealed that the oriLytPM contained various putative transcription factor-binding sites (Fig. 4A). These sites include a CCAAT factor-binding site, SP1, CREB, and an NF-κB binding site. Also, a transcriptional repressor, YY1, binding site was found within this region. Of course, the most salient feature with the oriLytPM was the presence of the novel putative IE2-binding element. The sequence of the putative oriLyt IE2-binding sites (nt 91840 to 91902) was compared to the previously described consensus IE2-binding CRS element(s) by using the Align on-line program (46). As shown in Fig. 4B, a CRS-like element having a 57% similarity to the consensus CRS sequence was present within the oriLytPM region. Inspection of other known IE2-binding sites (A, B, C, and D) (Fig. 4B) found within the UL112/113 promoter revealed 43, 57, 57, and 43% similarity to CRS, respectively (53).
FIG. 4.
A novel putative IE2-binding motif within oriLytPM is required for its promoter activation in Vero cells. (A) Schematic of the defined oriLytPM region with potential transcriptional factor-binding sites marked for the upper strand or the lower strand. The potential IE2-binding site (nt 91840 to 91902) is shown as a black bar on the sequence. (B) Sequence comparison of the potential IE2-binding site on oriLytPM with the CRS element and the IE2-binding sites located on the UL112/113 promoter. The previously published sequence of the CRS (35) is underlined. Capital letters represent the DNA domains that have similarity to the CRS element. Underlined sequences represent nucleotides in the CRS element that are conserved within the promoter sequence. (C) Graph of luciferase activity from the 1-bp insertion mutant construct transfected into Vero cells.
Since sequence analysis reveled the presence of a novel putative IE2-binding element, we chose to mutate this element using site-directed mutagenesis. Based on a previous study showing that a 1-bp insertion within the CRS eliminated IE2 binding (8), we generated a construct that had a similar 1-bp insertion by a designing site-directed mutagenesis primer such that a single guanosine (G), between nt 91860 and 91861, was inserted into the novel IE2-binding element in pGL-ORI-A (Fig. 4C, pGL-ORI-Amut). This mutant construct was transfected into Vero cells, and luciferase activity was measured. Interestingly, the single nucleotide insertion completely abolished the constitutive activity of the oriLytPM in Vero cells (Fig. 4C, sample pGL-ORI-Amut). Promoter activity from pGL-ORI-Amut was unchanged upon transfection of IE2 and/or UL84 (data not shown). These results indicated that this putative IE2-binding motif was essential for the promoter activity in an IE2-independent manner and that IE2-mediated repression in Vero cells may be due to interaction with this element, such that activation is inhibited in some undefined mechanism. Consequently, we will refer to this element (CAATCACAGAAACG) as the OPAE.
The oriLyt promoter is activated in human fibroblasts by HCMV infection.
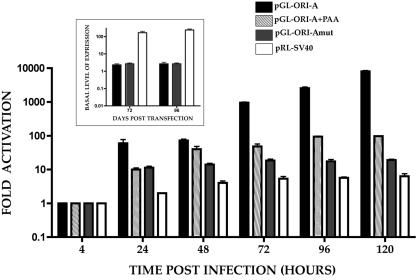
Since we determined that the oriLytPM was constitutively active in Vero cells, the next step was to examine the activity of this promoter in HCMV-permissive human fibroblast cells. HFFs were transfected with pGL-ORI-A in the presence or absence of the viral DNA synthesis inhibitor phosphonoacetic acid (PAA). Cells were subsequently infected with HCMV, and luciferase activity was assayed at various times postinfection. Transfection of pGL-ORI-A, in the absence of infecting virus, resulted in a low baseline level of luciferase activity (Fig. 5, inset). However, when transfected cells were infected with HCMV, promoter activity increased markedly as a function of time postinfection (Fig. 5). A significant increase in promoter activity was observed between 48 and 72 hpi, with the highest activity occurring at the last time point, 120 hpi (Fig. 5). Promoter activity from pGL-ORI-A was decreased over 100-fold at 120 hpi in the presence of PAA but was still 100-fold over baseline levels in the absence of virus (Fig. 5 and inset). This indicated that the oriLytPM responded to HCMV infection and showed the most activity at times associated with DNA replication. However, there was a 10-fold increase in promoter activity between 4 and 24 hpi in the presence of PAA (Fig. 5). This response increased to about 100-fold over baseline in the presence of PAA at 96 hpi. This suggested that the oriLyt promoter was responsive to viral factors present before and after the onset of DNA synthesis. This may indicate that an initial promoter response is due to immediate early and early proteins and not totally dependent upon DNA replication.
FIG. 5.
oriLytPM responds to HCMV infection in HFF cells. The graph shows luciferase activity from oriLytPM and control luciferase constructs transfected into HFF cells which were subsequently infected with HCMV. Luciferase activity was assayed at various times postinfection. Data are expressed as fold activation over basal level of activity (inset). (Inset) Luciferase constructs were transfected into HFFs, and basal levels were determined by comparing luciferase levels to pGL2-Basic or, in the case of pRL-CMV, to pRL-Null. Error bars are standard deviations from four separate experiments.
We also evaluated the activity of the mutated construct, pGL-ORI-Amut, in HFF cells. Cells were infected 24 hpt, and luciferase activity was measured at various times postinfection. The mutated construct, pGEM-ORI-Amut, responded poorly to infection, producing luciferase activity only about 10-fold higher than the basal activity observed in uninfected cells and over 100-fold less than the activity observed in infected cell samples (Fig. 5 and inset). This result was consistent with what was observed in Vero cells with respect to the depressed constitutive expression, indicating that the OPAE is essential for promoter function independent of cell type.
As a control for HCMV activation of nonspecific promoters we transfected pRL-SV40, which uses the SV40 early promoter to drive luciferase expression. This promoter did respond to HCMV infection over the 120-h infection period (Fig. 5 and inset). The response was almost 100-fold lower than that observed for the pGL-ORI-A plasmid at 120 hpi.
The presence of the core replication proteins is necessary for full activation of oriLytPM in human fibroblasts.
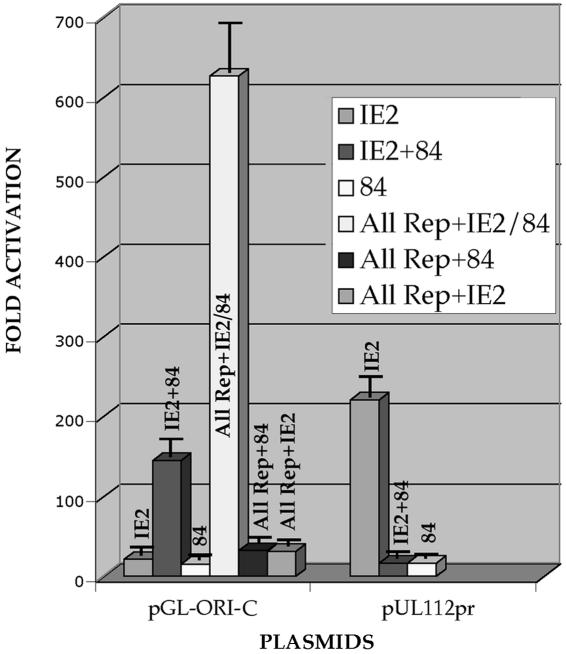
Since the oriLytPM was activated by HCMV infection in human fibroblasts, we performed cotransfections using plasmids encoding all of the necessary core replication factors for oriLyt-dependent DNA replication to determine which of these factors influenced oriLytPM activation in human fibroblasts. In addition, we wanted to determine if IE2 alone had any effect on the oriLytPM in human fibroblasts. Transfection of IE2 or UL84 expression plasmids alone had a minimal effect on oriLytPM activity (Fig. 6). However, when both UL84 and IE2 were added together to the cotransfection mixture, a 150-fold increase in promoter activity was observed (Fig. 6). This indicated that the combination of UL84 and IE2 had a stimulatory effect on promoter activity. Further, when plasmids encoding all the core replication proteins along with IE2 and UL84 were added to the mixture, activation of the oriLytPM increased considerably (Fig. 6). The omission of UL84 from the mixture decreased promoter activity to basal levels (Fig. 6, All rep + IE2). Likewise, when IE2 was omitted from the cotransfection mixture, a similar decrease in promoter activity was observed (Fig. 6, All rep + 84). These data revealed that in human fibroblasts the promoter region within oriLyt was responsive to UL84 and IE2 but required the entire replication complex for optimal activity, a result consistent with the infection data. This result was the opposite of what was observed in Vero cells, where the promoter was constitutively active and repressed by the addition of IE2 alone to the transfection mixture. However, the combination of UL84 and IE2 in Vero cells did produce a small increase in promoter activity over the high basal level.
FIG. 6.
UL84 and IE2 plus the core replication proteins are necessary for full activation of the oriLytPM in HFFs. HFF cells were cotransfected with expression plasmids expressing the core replication proteins and/or IE2 and/or UL84 along with the oriLytPM reporter plasmid pGL-ORI-C. The control reporter plasmid, p112/113pr, was also assayed for activity. The graph shows the fold activation over the pGL2-Basic vector. Error bars are the standard deviations of five separate experiments.
IE2 does not directly interact with OPAE.
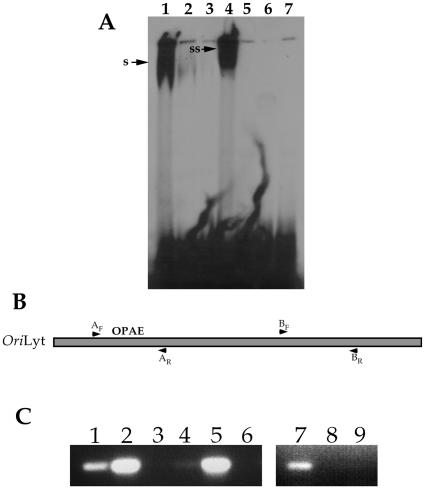
Once we determined that there was an IE2-responsive element within HCMV oriLyt, we next wanted to investigate if IE2 directly interacted with this element in vitro. IE2 purified from mammalian cells was incubated with either the CRS element or the OPAE, and an EMSA was performed. IE2 was able to interact with the CRS element in vitro in a specific manner, as demonstrated by cold competition experiments and the observed supershifted species with IE2-specific antibody (Fig. 7A, lanes 1 to 4). However, no specific IE2 interaction was detected using the OPAE oligonucleotides as substrate under the same conditions as with the CRS oligonucleotides (Fig. 7A, lane 5). This indicated that purified IE2 was unable to bind to the OPAE in vitro and suggested that additional factors may be required for interaction.
FIG. 7.
IE2 interacts with OPAE in vivo. (A) IE2 failed to bind to OPAE in an EMSA. Purified IE2 was incubated with either CRS or OPAE oligonucleotides and analyzed by EMSA for protein binding. Lanes: 1, purified IE2 incubated with CRS oligonucleotide; 2, purified IE2 incubated with CRS oligonucleotide plus 20× cold CRS; 3, purified IE2 incubated with CRS oligonucleotide plus 50× cold CRS; 4, purified IE2 incubated with CRS oligonucleotide plus Mab810; 5, purified IE2 incubated with OPAE oligonucleotide; 6, purified IE2 incubated with OPAE oligonucleotide plus 50× cold OPAE; 7, purified IE2 incubated with OPAE oligonucleotide plus Mab810. s, the shifted oligonucleotide; ss, the supershifted species. (B) Schematic showing the relative location of the primer sets used in the ChIP assay within HCMV oriLyt. Nucleotide coordinates for primer sets are described in Materials and Methods. (C) IE2 interacts with a region of oriLyt containing OPAE in a ChIP assay. Infected cell extracts were immunoprecipitated with an antibody specific for IE2 (G13-12E2; Vancouver Biotech), and HCMV DNA fragments were amplified using specific PCR primers. Lanes: 1, amplification product from the ChIP assay using primers AF and AR; 2, positive PCR control for primers AF and AR; 3, amplification product from the ChIP assay using primers AF and AR, except the IE2-specific antibody was omitted from the reaction mixture; 4, amplification product from the ChIP assay using primers specific for the MIEP containing the CRS motif; 5, positive PCR control for CRS amplification primers; 6, amplification product from the ChIP assay using primers specific for the MIEP containing the CRS motif, except that the IE2-specific antibody was omitted from the reaction mixture; 7, positive PCR control for primers BF and BR; 8, amplification product from the ChIP assay using primers BF and BR; 9, amplification product from the ChIP assay using primers BF and BR, except that the IE2-specific antibody was omitted from the reaction mixture.
The negative result from the EMSA prompted us to determine if IE2 interacted with the OPAE in vivo. This would mean that IE2 would require additional cellular or viral factors to bind the OPAE. In order to determine if IE2 interacted with the oriLyt region containing the OPAE, we performed a ChIP assay using HCMV-infected HFF cell extract. PCR primers specific to regions within oriLyt that corresponded to the location of the OPAE or control primers specific for another region of the origin were used to assay for an IE2-specific interaction. The ChIP assay revealed that IE2 did interact with a region of oriLyt that contained the OPAE when the A primers were used (Fig. 7B and C, lane 1). No band was amplified from samples that omitted the IE2-specific antibody from the incubation mixture (Fig. 7C, lane 3). Also, when the B primers were used (Fig. 7B), which corresponded to a downstream region of oriLyt, no amplification product was detected (Fig. 7C, lane 8). This indicated that this region of oriLyt did not interact with IE2 and also served as a negative control for the ChIP assay. As a positive control, we used primers specific for the CRS element located within the upstream region of the MIEP. Using this primer set, an amplification product was detected, confirming that IE2 interacts with the CRS element in vivo (Fig. 7C, lane 4). These results suggested that IE2 interacts with the OPAE either as a complex with another viral or cellular factor or requires other factors to be present such that binding can occur.
An intact OPAE is essential for oriLyt amplification.
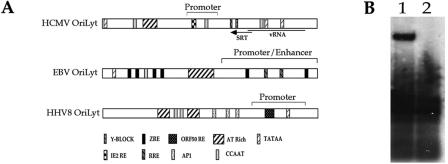
Based on the findings that a strong promoter region is present within HCMV oriLyt, we compared the HCMV origin with other herpesvirus lytic origins. Figure 8A is a comparison of the HCMV, HHV8, and EBV oriLyt regions. This diagram indicates that a significant part of HHV8 and EBV lytic origins contains promoter elements, and previous studies showed that promoter activity in these lytic origins was required for origin-dependent DNA replication (3, 18). Also shown is the specific viral protein-binding sites present within HHV8 and EBV oriLyt regions; these include ORF50RE for HHV8 and ZREs and RRE for EBV (Fig. 8A).
FIG. 8.
oriLyt promoter activity is required for DNA synthesis. (A) Comparison of the HCMV, EBV, and HHV8 oriLyt regions. The diagram is of the HCMV, EBV, and HHV8 oriLyt region(s) with potential transcriptional factor-binding sites and specific viral protein response element(s) marked. IE2 RE, IE2 response element. (B) A 1-bp insertion within the putative IE2-binding site inhibits oriLyt activity. HFF cells were transfected with pGEM-oriLyt+G or pGEM-oriLyt and infected with HCMV. Total cell DNA was harvested 5 days postinfection and cleaved with DpnI and EcoRI, and a Southern blot assay was performed to identify replication products. Lanes: 1, DNA from cells transfected with pGEM-oriLyt; 2, DNA from cells transfected with pGEM-oriLyt+G.
To determine if the oriLytPM was required for HCMV oriLyt-dependent DNA replication, we introduced the same lethal 1-bp mutation as in the oriLytPM luciferase construct (pGL-ORI-Amut) into the replication-competent oriLyt plasmid. This construct, pGEM-oriLyt+G, was transfected into HFF cells, which were subsequently infected with HCMV. Five days postinfection, total cellular DNA was harvested and replication products were examined as described in Materials and Methods. When the pGEM-oriLyt+G plasmid was used in the replication assay, no replication products were detected (Fig. 8B, lane 2), whereas the control transfection which contained the wild-type oriLyt (pGEM-oriLyt) yielded the expected replication product (Fig. 8B, lane 1). This indicated that the activity of this promoter was essential for efficient amplification of oriLyt.
The SV40 early promoter can functionally substitute for the HCMV oriLytPM.
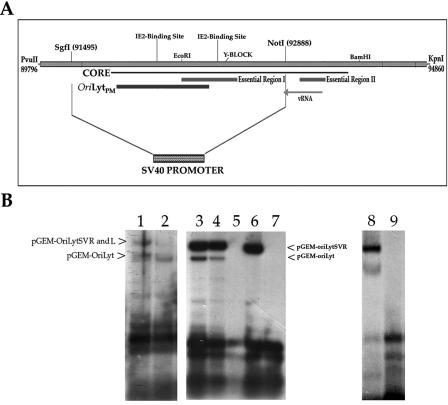
In the case of EBV, it was demonstrated that the promoter-enhancer region within oriLyt could be replaced by the HCMV immediate-early enhancer promoter (18). This indicated that, in at least EBV oriLyt, the presence of a nonnative strong constitutive promoter element was sufficient to replace the inducible native element. In HCMV, our study showed that oriLytPM mapped to a DNA sequence previously identified as the core and overlapped with one of the two essential regions (essential region I) (Fig. 9A) (66). Once we had determined that the promoter activity of the oriLytPM was essential for replication, we wanted to investigate the possibility that this region could be replaced with another strong constitutive promoter, such as in the case of EBV oriLyt.
FIG. 9.
A portion of the HCMV oriLyt can be replaced with the SV40 early promoter. (A) Schematic showing the oriLyt region where the SV40 early promoter was inserted. Also shown is the core region, essential regions I and II, and the location of the oriLytPM. (B) Replication of oriLyt plasmids containing the SV40 early promoter. Left: plasmids pGEM-oriLytSVR and pGEM-oriLytSVL were transfected into HFF cells along with pGEM-oriLyt. Cells were infected with HCMV and assayed 5 days later for oriLyt amplification. Lanes: 1, cotransfection of pGEM-oriLytSVR and pGEM-oriLyt; 2, cotransfection of pGEM-oriLytSVL and pGEM-oriLyt. Arrows show the positions of each replicated plasmid. Middle: cotransfection-replication assay with the core replication proteins in the presence and absence of UL84 or IE2. Lanes: 3, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HHV8 core replication proteins plus IE2 and UL84; 4, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HCMV core replication proteins plus IE2 and UL84; 5, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HHV8 core replication proteins plus IE2; 6, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HHV8 core replication proteins plus UL84; 7, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HHV8 core replication plus IE2 and UL84 and omission of the plasmid that encodes the putative helicase protein. Lanes in the right panel: 8, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HHV8 core replication proteins plus IE2 and UL84; 9, cotransfection of pGEM-oriLyt, pGEM-oriLytSVR, and the HHV8 core replication proteins plus IE2.
Since our initial oriLyt promoter studies involved a larger region of oriLyt in transient luciferase reporter assays, we replaced the region of oriLyt between the restriction sites SgiI (91495) and NotI (92888) with the SV40 early promoter (Fig. 9A). The SV40 early promoter was ligated into oriLyt such that two constructs were generated: one having the SV40 early promoter driving expression in the right-to-left direction (pGEM-oriLytSVR) and the other driving expression in the left-to-right direction (pGEM-oriLytSVL). These new constructs were transfected into HFF cells that were subsequently infected with HCMV. Five days postinfection cells were harvested, DNA was extracted, and a Southern blot assay was performed to examine replication products by cleaving with EcoRI and DpnI. Both the original pGEM-oriLyt and each of the new origin constructs were included in the transfection mixture, so that the replication efficiency of each plasmid could be compared directly to wild-type oriLyt. Since the new oriLyt constructs removed one EcoRI site, the amplified oriLyt-SV40 early promoter-containing plasmids migrated in the gel as a larger fragment than the wild-type pGEM-oriLyt plasmid. Plasmid construct pGEM-oriLytSVR replicated to approximately the same efficiency as the pGEM-oriLyt plasmid (Fig. 9B, lane 1), whereas the pGEM-oriLytSVL construct replicated at a decreased efficiency compared to the wild-type construct pGEM-oriLyt (Fig. 9B, lane 2). Amplification of the oriLyt constructs containing the SV40 early promoter in place of the promoter region within oriLyt indicated that the mechanism of DNA synthesis is, in part, due to the activation of transcription or assembly of the transcription machinery at a promoter element within oriLyt.
pGEM-oriLytSVR amplification in a cotransfection replication assay no longer requires IE2 in human fibroblasts.
Once we had determined that oriLyt constructs containing the SV40 early promoter were able to replicate, we next wanted to investigate if IE2 was still required for lytic replication in the cotransfection replication assay in human fibroblasts. T-HF cells were transfected with both pGEM-oriLyt and pGEM-oriLytSVR along with all required core replication proteins and combinations of UL84 and IE2 expression plasmids. Both pGEM-oriLyt as well as pGEM-oriLytSVR replicated in the presence of either HHV8 core replication proteins or HCMV core replication proteins UL84 and IE2 (Fig. 9B, lanes 3 and 4, respectively). However, when IE2 was omitted from the transfection mixture, a replication product was still detected for the amplification of pGEM-oriLytSVR (Fig. 9B, lane 6, top band). As expected, no replicated product was observed for wild-type oriLyt (Fig. 9B, lane 6). The inclusion of UL84 in the transfection mixture was still necessary for amplification of both pGEM-oriLyt as well as pGEM-oriLytSVR, since a cotransfection omitting the UL84 expression plasmid failed to produce any detectable replication products (Fig. 9B, lane 5). No replication products were detected when one of the core replication proteins, helicase, was omitted from the transfection mixture (Fig. 9B, lane 7). This result indicated that the requirement of IE2 in oriLyt-dependent DNA replication was to activate the promoter within oriLyt. Because lane 5 contained a smaller amount of DNA sample, we wanted to confirm that UL84 was still required for oriLyt amplification when the SV40 promoter was substituted for the oriLyt promoter. Consequently, we again performed the cotransfection assay omitting UL84. Control samples containing all the required replication proteins plus UL84 and IE2 replicated both pGEM-oriLyt as well as pGEM-oriLytSVR (Fig. 9, lane 8). However, when UL84 was omitted, neither replication origin was amplified (Fig. 9, lane 9).
DISCUSSION
We have identified a strong bidirectional promoter within the HCMV oriLyt. Activation of this promoter depends on an intact novel element, the OPAE. Several lines of evidence suggest that transcriptional activation is essential for oriLyt function: (i) a 1-bp insertion inhibits both promoter activity and oriLyt amplification, (ii) the SV40 early promoter can functionally substitute for the native oriLytPM, and (iii) the transactivator protein IE2 is required for activation of the oriLytPM and amplification of the oriLyt in human fibroblasts. We have shown that it is highly likely that the requirement for IE2 in the cotransfection replication assay in human fibroblasts is to activate the oriLytPM in conjunction with the formation of the replication complex in a UL84-dependent manner.
The oriLytPM appeared to show bidirectional promoter activity and was constitutively active in Vero cells in transient-transfection assays. Promoters with bidirectional activity have been described in mammalian cells (20, 23, 24, 27, 55, 59, 65). In most cases, these bidirectional promoters control gene expression for two divergent proteins. Essential elements controlling the activity of these mammalian bidirectional promoters include CCAAT boxes and SP1 (GC box) transcription factor-binding sites. These sites are also present within the oriLytPM region (Fig. 4A). The bidirectional nature of the oriLytPM may be necessary to initiate lytic DNA replication in the native oriLyt, although substitution studies using the SV40 early promoter suggest that transcriptional activation in one direction is sufficient for efficient replication of oriLyt.
Surprisingly, in Vero cells cotransfection of the oriLytPM with an IE2 expression construct resulted in a marked decrease in luciferase activity, and the addition of UL84 to the transfection mixture relieved the observed IE2-mediated repression of oriLytPM, whereas in human fibroblasts the addition of UL84 and IE2 was necessary for activation of the oriLytPM, and the addition of the core replication proteins produced a further, more robust increase in activity. It is interesting to note the difference in activation of the HCMV oriLytPM in different cell types. Vero cells appear to encode factors that can constitutively transactivate the oriLytPM. Therefore, IE2 in the cotransfection-replication assay in these cells is not required, and in fact it suppresses promoter activation. In human fibroblasts, it is apparent that IE2, along with UL84, transactivates the promoter region in oriLyt, and this might be the primary function for IE2 in lytic replication. Therefore, IE2 plays a key role in facilitating the activation of the promoter element in oriLyt, and a substantial portion of the lytic origin is involved in transcriptional activation. The difference in promoter activity between cell types may be the reason why previous studies have shown that IE2 is not required for amplification of oriLyt in Vero cells (50). Our data are consistent with the results of Gebert et al., who reported that the overexpression of UL84 resulted in a decrease in IE2-mediated activation of the UL112-113 early promoter and a decrease in down-regulation of the MIEP (16). In the case of the activity of the oriLytPM in Vero cells, UL84 acted in a similar manner to that observed in U373 cells; however, in human fibroblasts UL84 augmented the transactivation activity of IE2.
Although the OPAE within oriLytPM shows some homology with the IE2 consensus-binding sequence of the CRS, our results suggest that this element may interact with other transcription factors (known or unknown) in the context of oriLyt in Vero cells. Mutagenesis of this putative IE2-binding element resulted in a dramatic decrease in promoter activity in Vero cells, even in the absence of IE2. In human fibroblasts, the OPAE was also required for promoter activation, since cells transfected with the mutated construct did not show an increase luciferase activity upon infection. The 1-bp insertion mutation, which inhibited promoter activity (in both Vero cells and human fibroblasts), also abolished the ability of oriLyt to replicate in human fibroblasts. Therefore, it is likely that in human fibroblasts, the OPAE may be recognized by an IE2-UL84 complex and may mediate binding of these proteins in either a direct or indirect manner or facilitate the binding of other transcription factors. This initial protein interaction may further recruit other proteins involved in DNA synthesis, as evidenced by the observation that the presence of the entire replication machinery enhanced the activity of this promoter and further stimulated transcriptional activity. This implied that assembly of the replication complex may facilitate transcription or recruit transcription factors and/or the transcription machinery. This appears to be the case for Zta, where a superactivation of the oriLyt promoter for EBV was observed when plasmids expressing the helicase-primase proteins were added to transfection mixtures in the transient-reporter assays (15). Therefore, it is apparent that initial transactivation by activator proteins can be enhanced by the assembly of enzymes involved in the DNA synthesis. Although we have not identified any specific mRNA start site(s) for the oriLytPM, an evaluation of the oriLyt region using Promoter 2.0 software (30) has identified two highly likely mRNA cap sites at nt 91729 (upstream of the oriLytPM) and 92190 (downstream of the oriLytPM). Future studies will focus on the evaluation of this region for the presence of mRNA transcripts.
It is apparent from the ChIP assay that IE2 requires other factors to interact with oriLyt. We could find no direct binding of IE2 to the OPAE by using EMSA under the same conditions that we observed binding of IE2 with the CRS element. This result is consistent with our findings in the transient assay, where oriLyt promoter activation was dependent upon the presence of IE2 and UL84 with further stimulation occurring in the presence of the entire replication complex. An indirect interaction of IE2 with OPAE may indicate that both IE2 and UL84 may bind to oriLyt as part of a multiprotein complex, possibly in conjunction with a cellular factor.
A detailed analysis of transcription has been performed within the oriLyt region (22). Further downstream from the oriLytPM, a small transcript was identified. This transcript, known as the SRT, is in the opposite strand polarity from the oriLytPM. Other transcripts were also identified within the promoter region (22). Recent evidence has indicated that another promoter, outside of the core region, activates oriLyt (28). This promoter is upstream of the oriLyt promoter described in this report and comprises the cis regulatory region for the UL57 gene. The initiator region for the UL57 promoter was able to activate the minimal core oriLyt, indicating that transcriptional activation plays a role in oriLyt function. In the study presented here, we have also shown that oriLyt amplification involves a mechanism that is dependent upon transcriptional activation. The replacement of the native oriLytPM with the SV40 early promoter resulted in a robust replication signal, indicating that this region of the HCMV oriLyt is primarily involved in facilitating transcription. Although the oriLytPM appears to act in a direction-independent manner, insertion of the SV40 early promoter such that expression is directed in the left-to-right direction resulted in a weaker replication signal in the infection assay. This apparent weaker replication signal was also observed in the cotransfection replication with the same construct (data not shown). This observation suggests that promoter activity in the right-to-leftward direction may be the predominant contributor to initiation of DNA synthesis. Another interesting observation was that the degree of replication of the pGEM-oriLytSVR construct was much higher than that of pGEM-oriLyt in the cotransfection assay. This was not the case in the transfection-infection assay, where replication signals for both wild-type oriLyt as well as the SV40 early promoter-containing oriLyt produced comparable but decreased signals. This may have been the result of some promoter interference in the infection assay, where the presence of virus may have reduced the activity of the SV40 early promoter.
The presence of promoters within lytic origins is not unique to HCMV: promoter sequences and associated activity have been found in other herpesvirus oriLyt sequences (Fig. 8A). In HHV8, the viral transactivator ORF50 was shown to bind to and transactivate a promoter region within the HHV8 oriLyt (3). ORF50 also activates various early gene promoters via direct binding, a function similar to that of HCMV IE2 (37, 60). Interestingly, the ORF50 protein interacts with the proposed origin-binding protein, K-bZIP (RAP). K-bZIP also serves to regulate the activity of ORF50 with respect to the up-regulation of early gene promoters, similar to the function of UL84 and the interaction with IE2. In EBV, the oriLyt region is composed of two elements. The first is a core element that contains ZREs, which are the substrates for the transactivator protein Zta (18, 51). These regions are absolutely required for oriLyt replication in transient-transfection assays (12, 13, 51). The second element is an enhancer-promoter region containing the DNA-binding sites for two virus-encoded transactivators, Rta and Zta (9). This element can be replaced with the enhancer-promoter loci from the HCMV MIEP (18).
Earlier studies showed that in human fibroblasts, UL84, IE2, and UL36-38 were all required for efficient oriLyt amplification, whereas in Vero cells UL84 was the only protein, in addition to the core proteins, required for origin-dependent DNA replication (50). Using an HFF cell line immortalized by the stable expression of the catalytic subunit of telomerase (hTERT) and the HHV8 core replication proteins, we demonstrated that UL36-38 was no longer necessary for oriLyt replication. Although the role of UL36-38 in replication is unknown, it was shown that proteins encoded by this locus inhibit apoptosis (17, 56). It is possible that the function of the UL36-38 proteins is to promote cell survival and therefore facilitate a more efficient cellular environment for replication, a process that is not strictly required in transformed cells such as Vero cells or in primary cells that express telomerase. We also showed that when the SV40 early promoter was substituted for the oriLytPM, IE2 was no longer required for oriLyt-dependent replication. This indicates that the main function of IE2 in oriLyt-dependent DNA replication is to activate the promoter in oriLyt. One other important result revealed here is that UL84 was still required for efficient amplification of the SV40 early promoter-containing oriLyt, implying that UL84 may have a dual role in replication. Although it appears that the IE2-UL84 interaction may function to initiate DNA synthesis by activating transcription within oriLyt, UL84 itself may bind to oriLyt and supply an additional function, perhaps enzymatic, in lytic DNA replication.
Acknowledgments
This work was supported by NIH Public Health Service grant AI45096.
REFERENCES
- 1.Anders, D. G., M. A. Kacica, G. Pari, and S. M. Punturieri. 1992. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J. Virol. 66:3373-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., T. Albrecht, and E. A. Thompson. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075-22082. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner, R. C., J. J. Crute, M. S. Dodson, and I. R. Lehman. 1991. The herpes simplex virus 1 origin binding protein: a DNA helicase. J. Biol. Chem. 266:2669-2674. [PubMed] [Google Scholar]
- 6.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol 154:125-169. [DOI] [PubMed] [Google Scholar]
- 7.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus IE2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiou, C. J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dewhurst, S., D. M. Krenitsky, and C. Dykes. 1994. Human herpesvirus 6B origin: sequence diversity, requirement for two binding sites for origin-binding protein, and enhanced replication from origin multimers. J. Virol. 68:6799-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias, P., C. M. Gustafsson, and O. Hammarsten. 1990. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oris. J. Biol. Chem. 265:17167-17173. [PubMed] [Google Scholar]
- 12.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furnari, B. A., E. Poma, T. F. Kowalik, S. M. Huong, and E. S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, Z., A. Krithivas, J. E. Finan, O. J. Semmes, S. Zhou, Y. Wang, and S. D. Hayward. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 19.Hamzeh, F. M., P. S. Lietman, W. Gibson, and G. S. Hayward. 1990. Identification of the lytic origin of DNA replication in human cytomegalovirus by a novel approach utilizing ganciclovir-induced chain termination. J. Virol. 64:6184-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, J. J., P. Bross, M. Westergaard, M. N. Nielsen, H. Eiberg, A. D. Borglum, J. Mogensen, K. Kristiansen, L. Bolund, and N. Gregersen. 2003. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum. Genet. 112:71-77. [DOI] [PubMed] [Google Scholar]
- 21.Huang, C. H., and J. Y. Chen. 2002. Identification of additional IE2-p86-responsive cis-repressive sequences within the human cytomegalovirus major immediate early gene promoter. J. Biomed. Sci. 9:460-470. [DOI] [PubMed] [Google Scholar]
- 22.Huang, L., Y. Zhu, and D. G. Anders. 1996. The variable 3′ ends of a human cytomegalovirus oriLyt transcript (SRT) overlap an essential, conserved replicator element. J. Virol. 70:5272-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda, S., H. Ayabe, K. Mori, Y. Seki, and S. Seki. 2002. Identification of the functional elements in the bidirectional promoter of the mouse O-sialoglycoprotein endopeptidase and APEX nuclease genes. Biochem. Biophys. Res. Commun. 296:785-791. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, S., A. Mochizuki, A. H. Sarker, and S. Seki. 2000. Identification of functional elements in the bidirectional promoter of the mouse Nthl1 and Tsc2 genes. Biochem. Biophys. Res. Commun. 273:1063-1068. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins, D. E., C. L. Martens, and E. S. Mocarski. 1994. Human cytomegalovirus late protein encoded by ie2: a trans-activator as well as a repressor of gene expression. J. Gen. Virol. 75:2337-2348. [DOI] [PubMed] [Google Scholar]
- 26.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai, Y., K. Asai, Y. Miura, Y. Inoue, M. Yamamoto, A. Moriyama, N. Yamamoto, and T. Kato. 2003. Structure and promoter activity of the human glia maturation factor-gamma gene: a TATA-less, GC-rich and bidirectional promoter. Biochim. Biophys. Acta 1625:246-252. [DOI] [PubMed] [Google Scholar]
- 28.Kiehl, A., L. Huang, D. Franchi, and D. G. Anders. 2003. Multiple 5′ ends of human cytomegalovirus UL57 transcripts identify a complex, cycloheximide-resistant promoter region that activates oriLyt. Virology 314:410-422. [DOI] [PubMed] [Google Scholar]
- 29.Kirchoff, V., S. Wong, S. St. Jeor, and G. S. Pari. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147:321-333. [DOI] [PubMed] [Google Scholar]
- 30.Knudsen, S. 1999. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics 15:356-361. [DOI] [PubMed] [Google Scholar]
- 31.Koff, A., J. F. Schwedes, and P. Tegtmeyer. 1991. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J. Virol. 65:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krug, L. T., N. Inoue, and P. E. Pellett. 2001. Differences in DNA binding specificity among roseolovirus origin binding proteins. Virology 288:145-153. [DOI] [PubMed] [Google Scholar]
- 33.Krug, L. T., N. Inoue, and P. E. Pellett. 2001. Sequence requirements for interaction of human herpesvirus 7 origin binding protein with the origin of lytic replication. J. Virol. 75:3925-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. USA 90:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masse, M. J., S. Karlin, G. A. Schachtel, and E. S. Mocarski. 1992. Human cytomegalovirus origin of DNA replication (oriLyt) resides within a highly complex repetitive region. Proc. Natl. Acad. Sci. USA 89:5246-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mocarski, E. J., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 42.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1 (491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1988. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc. Natl. Acad. Sci. USA 85:5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 47.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pizzorno, M. C., P. O'Hare, L. Sha, R. L. LaFemina, and G. S. Hayward. 1988. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prichard, M. N., S. Jairath, M. E. Penfold, S. St. Jeor, M. C. Bohlman, and G. S. Pari. 1998. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J. Virol. 72:6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schepers, A., D. Pich, and W. Hammerschmidt. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367-376. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seki, Y., S. Ikeda, H. Kiyohara, H. Ayabe, T. Seki, and H. Matsui. 2002. Sequencing analysis of a putative human O-sialoglycoprotein endopeptidase gene (OSGEP) and analysis of a bidirectional promoter between the OSGEP and APEX genes. Gene 285:101-108. [DOI] [PubMed] [Google Scholar]
- 56.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spector, D. J., and M. J. Tevethia. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 68:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szatmari, I., S. Tokes, C. B. Dunn, T. J. Bardos, and J. Aradi. 2000. Modified telomeric repeat amplification protocol: a quantitative radioactive assay for telomerase without using electrophoresis. Anal. Biochem. 282:80-88. [DOI] [PubMed] [Google Scholar]
- 59.Trinklein, N. D., S. F. Aldred, S. J. Hartman, D. I. Schroeder, R. P. Otillar, and R. M. Myers. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wara-aswapati, N., Z. Yang, W. R. Waterman, Y. Koyama, S. Tetradis, B. K. Choy, A. C. Webb, and P. E. Auron. 1999. Cytomegalovirus IE2 protein stimulates interleukin 1β gene transcription via tethering to Spi-1/PU. 1. Mol. Cell. Biol. 19:6803-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weir, H. M., and N. D. Stow. 1990. Two binding sites for the herpes simplex virus type 1 UL9 protein are required for efficient activity of the oriS replication origin. J. Gen. Virol. 71:1379-1385. [DOI] [PubMed] [Google Scholar]
- 63.Wingender, E., X. Chen, R. Hehl, H. Karas, I. Liebich, V. Matys, T. Meinhardt, M. Pruss, I. Reuter, and F. Schacherer. 2000. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, Y., K. S. Colletti, and G. S. Pari. 2002. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76:8931-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, L. F., J. H. Ding, B. Z. Yang, G. C. He, and C. Roe. 2003. Characterization of the bidirectional promoter region between the human genes encoding VLCAD and PSD-95. Genomics 82:660-668. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, Y., L. Huang, and D. G. Anders. 1998. Human cytomegalovirus oriLyt sequence requirements. J. Virol. 72:4989-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]