Abstract
BRUCE is a highly conserved 528-kDa peripheral membrane protein of the trans-Golgi network. Owing to the presence of an N-terminal single baculovirus inhibitor repeat, BRUCE functions as an inhibitor of apoptosis protein and blocks apoptosis when overexpressed. In addition, due to the presence of a C-terminal ubiquitin-conjugating domain, BRUCE can covalently attach ubiquitin to substrates. Here we report the generation and characterization of BRUCE-deficient mice. Complete inactivation of the BRUCE gene resulted in perinatal lethality and growth retardation discernible after embryonic day 14. The growth defect is linked to impaired placental development and may be caused by insufficient oxygen and nutrient transfer across the placenta. Chorioallantoic placentation initiated normally, but the mutant placenta showed an impaired maturation of the labyrinth layer and a significant reduction of the spongiotrophoblast. No evidence for an elevated apoptosis rate was detectable in embryonic and extraembryonic tissues and in knockout fibroblasts. Thus, although BRUCE is broadly expressed in embryonic, extraembryonic, and adult mouse tissues, this bifunctional protein might play a unique role in normal trophoblast differentiation and embryonic survival.
Modification of proteins by covalent attachment of ubiquitin (ubiquitylation) occurs universally in eukaryotic cells (14). Ubiquitin is usually conjugated via its carboxy (C) terminus to lysine residues (ɛ-amino groups) of substrate proteins, thereby forming branched isopeptide-linked protein complexes. Substrates can be modified either by a single ubiquitin moiety (monoubiquitylation) or by a multiubiquitin chain, in which several ubiquitin moieties are linked together by isopeptide bonds (multiubiquitylation). Proteins modified by multiubiquitin chains are usually targeted to the 26 proteasome for degradation (14). In contrast, monoubiquitylation and modification by noncanonical (lysine 63-linked) multiubiquitin chains mediate nonproteolytic functions, including protein sorting, gene silencing, signal transduction, and DNA repair (19).
Ubiquitylation is ATP dependent and requires the sequential activities of usually three enzymes, termed ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (14). The E1 enzyme hydrolyses ATP and forms a thioester-linked complex between its active-site cysteine residue and the C-terminal glycine residue of ubiquitin. Ubiquitin is then transferred onto a cysteine residue of one of several E2 enzymes. These proteins are usually small proteins within the 14- to 35-kDa size range and are characterized by a so-called ubiquitin-conjugating (UBC) domain, which carries the active-site cysteine residue. Subsequently, an E3 enzyme binds both the E2 and the substrate and mediates the transfer of ubiquitin to a lysine residue of the substrate. The E3 enzymes also exist as a large protein family, and these enzymes in particular contribute to the substrate specificity of the system (14). Some substrates additionally require E4 enzymes for multiubiquitylation (16).
Previously, we cloned an unusual member of the UBC protein family from mouse by a homology-based strategy with sequences for yeast UBC domains as probes (12). This enzyme, called BRUCE (for baculovirus inhibitor of apoptosis repeat [BIR] repeat-containing ubiquitin-conjugating enzyme), is a giant 528-kDa protein which bears a UBC domain close to its C terminus. In addition, BRUCE carries a single BIR domain close to its amino (N)-terminal end. The BIR domain, a zinc-binding fold, is a hallmark of inhibitor of apoptosis proteins (IAPs), which bind and thereby inhibit caspases through this domain (9, 25, 27). IAPs belong to a larger group of so-called BIR domain-containing proteins, which also includes the mammalian protein survivin and the BIR1 proteins from the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe (27). In contrast to the IAPs, these proteins do not seem to block apoptosis but play a role in cytokinesis (27). BRUCE is highly conserved, and homologs have been identified in humans and Drosophila (1, 3, 28). The protein is absent in Caenorhabditis elegans, however. BRUCE is a peripheral membrane protein and localizes specifically to the trans-Golgi network (TGN) and vesicles (12). In neuronal cells, BRUCE is also found at vesicular structures in axons and dendrites (12).
BRUCE was recently shown to possess antiapoptotic activity (2, 3, 20). Similar to other IAPs, BRUCE binds and thereby inhibits caspases via its BIR domain (2). BRUCE inhibits most strongly the effector caspase 3 and binds to its processed, active form but not to the proform of the caspase. In HeLa and 293T cells, overexpression of BRUCE blocks apoptosis induced by various stimuli, including UV light and TRAIL (2). Moreover, antisense oligonucleotides against BRUCE sensitize cells against apoptosis induced by DNA-damaging agents (3), and downregulation by RNA interference or ubiquitin- or proteasome-dependent degradation of BRUCE promotes apoptosis (20). BRUCE also has activity to monoubiquitylate the proapoptotic, mitochondrion-derived protein Smac, which depends on the integrity of both the BIR and the UBC domains (2). Notably, this reaction is catalyzed by E1 and BRUCE alone and does not require additional E2 or E3 enzymes. Thus, BRUCE combines in a single large polypeptide E2 and E3 activities and functions as a chimeric E2/E3 ubiquitin ligase (2).
In this work we investigated the function of BRUCE and the consequences of its loss in vivo by generation and characterization of BRUCE knockout mice. We show that these mice exhibit perinatal lethality and growth deficiencies, which are linked to a defect in proper placental development. Surprisingly, no evidence for an elevated apoptosis rate was detectable in embryonic and extraembryonic tissues and knockout fibroblasts, which may be due to possible overlapping IAP activities. This suggests that the observed defects of BRUCE-deficient mice may not be exclusively linked to BRUCE's IAP activity but possibly may be linked to its ubiquitin ligase activity at endomembranes.
MATERIALS AND METHODS
Construction of the targeting vector.
Genomic 5′ sequence from the BRUCE locus of 129/SvJ mice was obtained by screening a λ phage library with cDNA fragments according to standard procedures. Three partially overlapping clones spanning exon 2 to 6 (λ5, λ7, and λ9) were subcloned into pBluescript SK(+) and analyzed by restriction mapping, Southern blotting, sequencing, and PCR. A 5-kb XbaI-EcoRI fragment from clone λ7 spanning parts of intron 3 and exon 4 was subcloned into pBluescript SK(+) and elongated by a 1-kb PCR fragment generated with primer jp71 (GCT GGA GAA GTA TTG CTT TGC) and mutagenic primer ko2XbaI*.la.asense (GGT CTA GAG GTA AGA CAA TCT GTC C) to introduce an XbaI site and a stop codon into exon 4. This construct was further elongated by a 2.2-kb PCR fragment generated with primers ko2XbaI*.la.sense (ACC TCT AGA CCA GTG CAC GTC CAG) and ko2SalItd.la.asense (GAC GTC GAC GAC AGC CTT ATG AGA CTA CCT GG) to introduce the XbaI site and the stop codon into exon 4 and a SalI site into intron 4. These three regions together represent the 5′ arm of homology. A neoR-tk cassette containing the neomycin resistance gene under control of the herpes simplex virus promoter, the 1.1-kb 3′ homology arm derived from exon 6 and intronic sequences by PCR with primers ko2XbaItd.sa.asense (CCG TCT AGA CTG TAC TGT AAA TAC TTC CAT GC) and ko2XbaItd.sa.sense (CCG TCT AGA TTA TGA TGT TAG TCC TAT GTC CTA C), and the negative selectable marker HSV-TK (herpes simplex virus thymidine kinase) was then placed into the SalI site of the pBluescript SK(+) construct. Thus, in the resulting targeting vector, exon 4 contains an introduced stop codon, whereas exon 5 is replaced with a neoR cassette.
Gene targeting in ES cells.
Fifty micrograms of linearized (NotI) plasmid DNA was electroporated into 107 R1 embryonic stem (ES) cells suspended in phosphate-buffered saline (PBS). The ES cells were then cultured on dishes treated with gelatin. Following double selection with G418 (300 μg/ml) (Sigma) and ganciclovir (2 μM) (Sigma), 64 ES cell clones were picked, expanded on mitomycin-treated embryonic fibroblasts, and analyzed by Southern blotting of genomic PvuII-digested DNA. An external probe comprising a 0.4-kb BamHI fragment of a PCR-amplified λ7 sequence (using primers jp101 [GAG CCT GTG ATT TAT AGT GTA C] and jp136 [CTT CAG CTG GAT AGT GAG TTC C]) was used to identify the 2.9-kb fragment of the wild-type (WT) allele and the 2.3-kb fragment of the mutated allele. In addition, a genomic PCR was performed to screen for homologous recombination events. One primer was chosen to be outside the targeting vector, and the other was chosen to be within the neoR cassette (outer.33, TCT TCT AAA TTA AGG CTT CAT AGT AGA TCT TGG; jp82, GCC TCT CCA CCC AAG CGG CCG GAG AAC CTG CGT GC). For higher sensitivity, a second round of PCR was performed to amplify a 1.5-kb genomic fragment (inner.22, GTT GTA CAC TAT AAA CAG G; jp83, GCA ATC CAT CTT GTT CAA TGG C).
Generation of BRUCE-deficient mice.
Two different ES cell clones with the targeted BRUCE allele were injected into 3.5-day-old C57BL/6 blastocysts and then reimplanted into 2.5-day pseudopregnant NMRI females. The spermatides of an infertile male chimera (germ line transmission was observed in the coat color of the F1 offspring) were used for in vitro fertilization of C57BL/6 oocytes, and fertilized oocytes were reimplanted into pseudopregnant B6D2F2 females. Male chimeras were then backcrossed to C57BL/6 females, and heterozygous male and female mice were interbred to produce homozygous mutant offspring. The genotypes of the mice were assessed by Southern blotting and by PCR analysis with genomic DNA from tail biopsies of 21-day-old pups with primers wt-0 (GTG TCT CCA CCT AAC CTA TGC) and wt-3.as (GGT GAT AAA ATC CAG CTT GAG C) to detect a 556-bp band for the WT allele and with additional primers neo4 (GGC TAT TCG GCT ATG ACT GGG C) and neo5 (GGG TAG CCA ACG CTA TGT CCT G) to amplify a 624-bp band for the mutated allele.
Weight study and histological examination.
After cervical dislocation of pregnant mice, embryos and placentas at different developmental stages (embryonic day 9.5 [E9.5] to E19.5) were dissected out of uterus and decidua and cleared of membranes and umbilical cords in PBS. Yolk sac was used for genotyping. After excess buffer was blotted with a paper towel, embryos and placentas were weighed. For histological examination, dissected tissue was fixed in Bouin's or 10% neutral buffered formalin overnight and then dehydrated and embedded in paraffin. Serial sections (4 to 8 μm thick) were stained with hematoxylin and eosin by using routine procedures.
Protein analysis and Western blotting.
Immunoblot analysis of BRUCE protein was performed either on E10 whole-embryo lysates obtained by solubilization of the embryo in sodium dodecyl sulfate (SDS) sample buffer or on extraembryonic tissue lysates obtained by homogenization of the tissue in PBS (500 μl per 100 mg of tissue, containing complete [Roche] protease inhibitor cocktail) followed by solubilization in SDS sample buffer. For fibroblast extracts, cells were harvested and counted, and equal numbers of cells were solubilized in SDS sample buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (in a 4 to 20% polyacrylamide gradient) and electroblotted onto polyvinylidene difluoride membrane (Immobilon P; Millipore). Blots were probed with a monoclonal antibody against the N terminus of BRUCE (Transduction Laboratories) and a rabbit polyclonal (affinity-purified) antibody recognizing the C terminus of BRUCE (12). Equal loading of the sample was assessed by probing blots with a goat antiserum against actin (I-19; Santa Cruz). Bands were visualized with peroxidase-conjugated secondary antibodies and the ECL detection kit (Amersham). Other antibodies used for immunoblotting were anti-XIAP (Transduction Laboratories), anti-cIAP1 (PharMingen), anti-cIAP2 (R&D Systems), antisurvivin (A-19; Santa Cruz), and anti-caspase 3 (K-19; Santa Cruz).
BrdU labeling, immunohistochemistry, and TUNEL assay.
Pregnant mice from heterozygous matings were injected intraperitoneally at E11.5, E13.5, E15.5, and E17.5 with 5-bromo-2-deoxyuridine (BrdU) (Sigma) at a dose of 50 μg per g of body weight. Pregnant mice were sacrificed 4 h after injection, and the uteri were removed. Embryos and corresponding placentas were shock frozen on dry ice and cut on a cryostat in 10- to 15-μm-thick sections. Detection of the incorporated BrdU was performed with a BrdU in situ detection kit (BD PharMingen) according to the manufacturer's instructions, followed by microscopic examination. For immunohistochemistry, the placenta was dissected on E9.5 to E17.5, fixed in 10% neutral buffered formalin, paraffin embedded, and serially sectioned (8 μm thick). Immunohistochemistry on dewaxed and rehydrated sections was performed with the Vectastain Universal Quick kit in combination with the M.O.M. Basic kit (Vector Laboratories) according to the manufacturer's protocol. Affinity-purified antilaminin antibody (1:25) (Sigma) was used as the primary antibody. Immune complexes were visualized with diaminobenzidine tetrahydrochloride precipitates, and the sections were subsequently counterstained with nuclear fast red. For analysis of programmed cell death, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed with the ApopTag in situ apoptosis detection kit (Oncor) according to the manufacturer's instructions. To determine apoptotic cell death, in addition to TUNEL staining, caspase 3 activity was detected by using an antibody that specifically binds to the cleaved and thereby activated form of caspase 3 (anti-active caspase 3 pAb; Promega). Immunohistochemistry was performed after permeabilization of the cells with 0.1% Triton X-100 by using the Vectastain Universal Quick kit (Vector Laboratories).
In situ hybridization.
Hybridization of paraffin-embedded sections was performed essentially as described previously (6) 33P-labeled antisense and sense mRNA probes were transcribed in vitro by using T7, T3, or SP6 RNA polymerase (Roche) from plasmids containing cDNA sequences of mPl-1, Tpbp (4311), Flt-1, Mash 2, Gcm1, eHand, Flk-1, and BRUCE. For hybridization, sections were dewaxed and pretreated as described previously (6). Briefly, slides were pretreated with proteinase K (20 μg/ml) for 7 min, postfixed in 4% paraformaldehyde in PBS, and acetylated in acetic acid anhydride diluted 1:400 in 0.1 M triethanolamine (pH 8.0). After washing, slides were dehydrated in ethanol and air dried. Hybridizations were performed in defined hybridization buffer (InnoGenex) supplemented with ≥3 × 107 cpm of sense or antisense riboprobe per ml at 57.5°C overnight. Slides were washed by standard protocols, treated with RNase A (20 μg/ml) for 30 min at 37°C, washed again in SSC buffer (2×, 1×, and 0.5×; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and at least at 65°C in 0.1× SSC containing 0.1 mM dithiothreitol. Slides were finally rinsed, dehydrated, air dried, coated with autoradiographic emulsion (LM-1; Amersham), developed after 2 to 6 weeks, and counterstained with cresyl violet.
Cell culture of primary embryonic fibroblasts.
Primary mouse embryonic fibroblasts (MEFs) (at least four independent lines per genotype) were isolated from day 13.5 embryos. Part of the embryo was used for genotyping. The remaining embryonic tissue was minced by using a pair of scissors, syringe, and needle (19 gauge) and immersed in culture medium (Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 2 mM glutamine, 50 μg of penicillin per ml, and 50 μg of streptomycin per ml). Cells were then incubated at 37°C with 95% humidity and 7.5% CO2. A single-cell suspension of MEFs (excluding the embryonic cell clumps) was plated onto a new dish before reaching confluency, and these cells were regarded as passage 1 cells. In the growth curve experiments, passage 2 cells (four lines per genotype) were plated at 1 × 105, 3 × 105, and 9 × 105 cells per 6-cm-diameter culture dish and counted on successive days. In parallel the same MEF lines were immortalized as described previously (26). The number of cells in S phase in asynchronous cultures of fibroblasts was determined by using the BrdU in situ detection kit (BD PharMingen) according to the manufacturer's instructions, followed by microscopic examination of a randomly chosen area. For analysis of programmed cell death in asynchronous cultures of fibroblasts, TUNEL assays were performed with the ApopTag in situ apoptosis detection kit (Oncor) according to the manufacturer's instructions. Apoptosis was induced in primary MEFs by cultivating the cells for 0 to 36 h in growth medium containing different concentrations of various inducers of apoptosis (actinomycin D [Sigma], 0.01 to 2.5 μg/ml; anisomycin [Sigma], 0.05 to 2.5 μM; brefeldin A [Sigma], 0.1 to 20 μg/ml; calcimycin [Calbiochem], 0.05 to 5.0 μM; camptothecin [Calbiochem], 0.1 to 20 μg/ml, UV radiation, 10 to 500 J/m2; etoposide [Sigma], 0.5 to 100 μM; tunicamycin [Sigma], 0.1 to 10 μg/ml; H2O2 [Merck], 10 to 500 μM; tumor necrosis factor alpha [TNF-α] [R&D Systems], 1.0 ng/ml to 1.0 μg/ml [alone or in combination with 1.0 μg of cycloheximide]; cycloheximide [Sigma], 1.0 to 50 μg/ml; and hamster anti-mouse Fas [PharMingen], 1.0 to 100 ng/ml). Measurements of cell death were performed at different time points in a colorimetric assay with crystal violet (0.5% [wt/vol] in 40% methanol in PBS). Cell death was expressed as the optical density at 595 nm.
RESULTS
Targeted disruption of the mouse BRUCE gene.
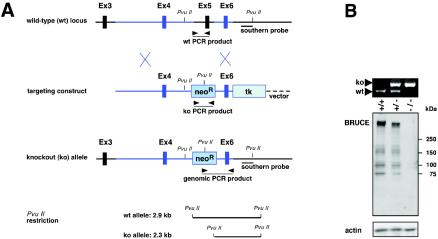
Murine BRUCE was originally identified by a homology-based strategy with DNA sequences for yeast ubiquitin-conjugating enzymes as a probe (12). The corresponding cDNA was cloned by cDNA walking with λ phage-based libraries. The genomic locus of BRUCE spans 175 kb on chromosome 17 and includes a 14,535-bp open reading frame (12). In order to design a knockout construct, we characterized the genomic organization of BRUCE's 5′ region. To this end, we cloned genomic sequences corresponding to the 5′ region from a λ library containing liver DNA of the mouse line 129/SvJ and characterized two nonconsecutive genomic regions in more detail. The first 20-kb region (R1) encompassed exon 1, whereas the second region of 30 kb (R2) contained exon 2 to exon 6 (Fig. 1A). We noticed that the first half of BRUCE's BIR domain is encoded by exon 5. Exon 6 corresponds to the metal-binding fold of the BIR domain; however it does not encode it completely. Region R1, and to a lesser extent also R2, contained multiple copies of B1 and B2 elements of the SINE family and were intensely rich in GC content. Based on this information, we chose a knockout strategy that took away a region encompassing exon 4 to exon 6, thereby eliminating essential portions of the BIR domain. In addition, a stop codon was introduced in the open reading frame of exon 4, which is expected to result in a complete null allele. The targeting construct (Fig. 1A) contained a neomycin resistance gene (neoR) flanked by sequences corresponding to segments containing exon 4 and exon 6, followed by a thymidine kinase cassette (TK).
FIG. 1.
Targeted disruption of the BRUCE gene by homologous recombination. (A) Schematic representation of the targeting strategy for BRUCE, showing the partial BRUCE locus with exons (Ex) 3 to 6, the targeting construct, and the targeted allele. Exon 5 is deleted and replaced with a neoR cassette. The probe used for Southern hybridization of neomycin-resistant ES cell clones and genomic DNA, which recognizes 2.9-kb WT and 2.3-kb mutant (ko) PvuII fragments, is indicated. Arrowheads represent primers used for screening ES cell DNA for homologous recombination events and for genotyping. (B) PCR analysis of yolk sac DNA from embryos derived from heterozygous matings (top panel). The positions of primer pairs for amplification of the 556-bp WT and the 624-bp mutant PCR fragments are shown in panel (A). Immunoblot analysis of BRUCE protein in E10 whole-embryo lysates from WT, heterozygous, and BRUCE null embryos by using a monoclonal antibody against the N terminus of the murine BRUCE protein (middle panel). Equal protein shown by an antiactin immunoblot (bottom panel).
Following transfection of R1 ES cells, we obtained correctly targeted ES clones at a frequency of 40.5%. Two clones were used to generate chimeras, but due to pronounced mortality among the littermates, only a single male chimera was available for crossing. Among the progeny, only one male mouse carried the knockout allele in the germ line. Because this animal was apparently infertile, progeny were obtained after in vitro fertilization of C57BL/6 oocytes and implantation into pseudopregnant B6D2F2 females. After crossing with C57BL/6 WT animals, the BRUCE alleles were transmitted in Mendelian ratio. BRUCE heterozygotes, identified by PCR (Fig. 1B) and Southern analysis using WT- and mutant-specific probes, were viable and fertile and exhibited no obvious phenotypic defects.
BRUCE deficiency results in retarded growth and perinatal lethality.
Phenotypically normal heterozygotes were intercrossed to produce BRUCE-deficient mice. We observed Mendelian inheritance up to the time of birth (Table 1). However, during or shortly after birth BRUCE null mice died, and no surviving knockout offspring could be obtained, demonstrating that the absence of BRUCE causes perinatal death. Tissue samples of knockout (BRUCE−/−) embryos verified that no detectable BRUCE protein was expressed (Fig. 1B), thus confirming that the targeting strategy led to a complete BRUCE knockout.
TABLE 1.
Genotypes of offspring from BRUCE+/− × BRUCE+/− matings after 10 generations of backcrossing with C57BL/6 wild-type animals
| Developmental stagea | No. of offspring with BRUCE genotype of:
|
||||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | NDb | Resorbed | |
| E9.5 | 2 | 4 | 1 | 1 | |
| E10.5 | 3 | 8 | 3 | 4 | 2 |
| E11.5 | 11 | 28 | 9 | 7 | |
| E12.5 | 25 | 48 | 26 | 3 | 5 |
| E13.5 | 29 | 70 | 27 | 3 | 20 |
| E14.5 | 28 | 49 | 23 | 8 | 18 |
| E15.5 | 8 | 16 | 9 | 2 | 3 |
| E16.5 | 2 | 11 | 5 | 4 | |
| E17.5 | 12 | 25 | 16 | ||
| E18.5 | 20 | 23 | 13 | 12 | |
| Prenatal totalc | 140 (21.7%) | 282 (43.7%) | 132 (20.4%) | 20 | 72 (11.1%) |
| P0 (1 h) | 12 | 27 | 7 | ||
| P0 (6 h) | 5 | 15 | 2 | ||
| P0 (12 h) | 7 | 11 | 1 | ||
| P0 (24 h) | 8 | 15 | 0 | ||
| P1 | 6 | 13 | 0 | ||
| Postnatal total | 38 | 81 | 10 | ||
E, day of embryonic development; P, postnatal day.
ND, not determined.
Expected ratio (+/+ to +/− to −/−), 1:2:1.
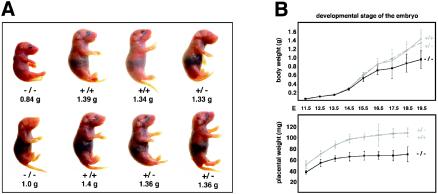
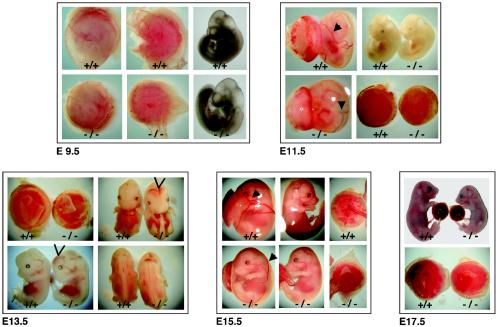
Newborn BRUCE−/− mice were significantly smaller than WT mice but otherwise appeared morphologically normal (Fig. 2A). To characterize the phenotype of mutant embryos, different stages of gestation were analyzed and the genotypes were determined by PCR analysis of genomic DNA obtained from the yolk sacs of each embryo (Fig. 1B). BRUCE mutants up to E12 were hardly distinguishable from their heterozygous and WT littermates. However, from about E13.5 onwards, BRUCE-deficient embryos showed increasing growth retardation, reflected by a slowed body weight gain (Fig. 2B). In addition, they appeared pale, and major blood vessels in mutant embryos and in the corresponding yolk sacs were sometimes not clearly visible (Fig. 3). This suggests that vascularization and/or blood supply was poor at this stage. Between E12 and E14, about 34% of the knockout embryos exhibited temporary bleeding in the brain (ventricle system) and neural tube, yet at later stages (E15.5 to birth), no remarkable morphological changes besides growth retardation were observed in 96% of the BRUCE mutants. However, 4% exhibited subcutaneous hemorrhages and generalized edema. Despite the growth retardation, we observed no consistent overt defects in vital organs and tissues that might result in embryonic lethality. Histological examination of major organs and tissues, such as heart, lung, liver, kidney, and neuronal tissue, of living BRUCE-deficient E18.5 embryos revealed no obvious abnormal architecture (data not shown).
FIG. 2.
Loss of BRUCE results in increasing growth retardation during embryonic development. (A) Growth-retarded knockout neonates within the offspring of a heterozygous mating (BRUCE+/− × BRUCE+/−). (B) Growth curves of WT (+/+), heterozygous (+/−), and mutant (−/−) BRUCE embryos and their placentas from mid- to late gestation. The average body and placental weights were determined over time; error bars indicate standard deviations. Males and females were pooled. The number of monitored pups was at least five. In most cases, 10 or more embryos per genotype were examined.
FIG. 3.
External appearance of WT (+/+) and mutant (−/−) BRUCE embryos and extraembryonic tissues at different embryonic and fetal stages. Knockout embryos are retarded in growth compared to WT littermates after E13.5. They appear paler, and major blood vessels are often not clearly visible. In 34% of the BRUCE null mutants, temporary bleedings in the ventricle system of the brain and in the neural tube could be observed between E12 and E14 (arrowheads). In the extraembryonic tissues, defects already appear at E9.5. Mutant placenta is paler and smaller than WT, and the major blood vessels in the yolk sac are less pronounced than in the WT (arrowheads).
BRUCE is required for normal placenta development.
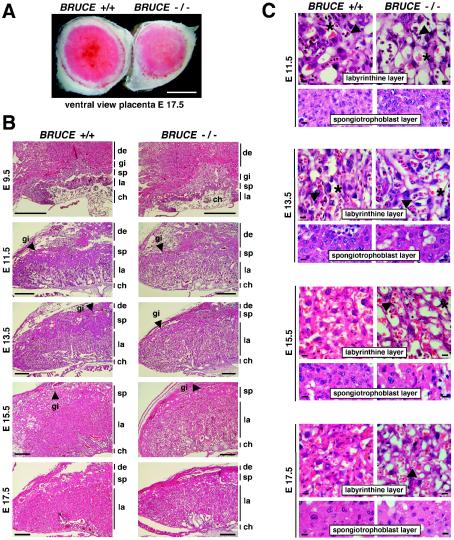
To determine the basis for the perinatal lethality of BRUCE mutants, we examined the morphology and histology of the extraembryonic tissue from E9.5 to E17.5 of progeny. The mouse placenta consists of the maternal decidual tissue and a component of embryonic origin. In the mature placenta the embryonic part is formed by three distinctive trophoblast cell structures that arise by E10 and remain until the end of gestation (5, 18). The inner labyrinthine layer, containing the network of fetal capillaries and maternal blood sinuses, develops into a highly folded and branched tissue, building up the maternal-fetal interface for nutrient and gas exchange. The mature labyrinth is established by E14.5, and after E17 it remains unchanged until birth (15, 18). A compact layer of trophoblast cells between the labyrinth and the outer giant cells, which derives largely from cells of the ectoplacental cone, forms the spongiotrophoblast zone (22). The trophoblast giant cells result from trophectoderm cells by endoreduplication and lie at the periphery of the placenta towards the maternal decidual tissue.
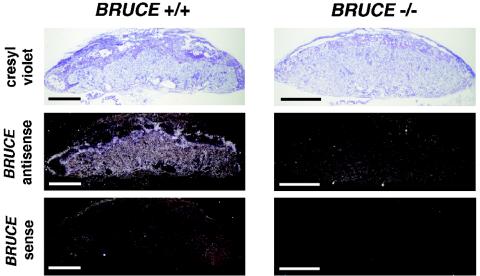
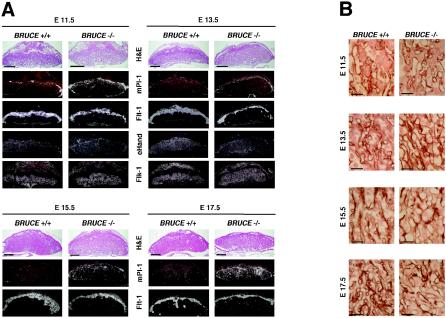
In situ hybridization analysis with a radiolabled antisense probe revealed a relatively high expression level of BRUCE in the different layers of the WT placenta, and also in the visceral yolk sac, throughout the different developmental ages (Fig. 4). Within the labyrinth and the chorionic plate, BRUCE mRNA is detectable in endothelial cells as well, but predominantly in labyrinthine and chorionic trophoblast cells. Increased levels of BRUCE mRNA were found in the spongiotrophoblast layer, whereas in trophoblast giant cells BRUCE expression was reduced. Notably, BRUCE expression is particularly abundant in those tissues that exhibit defects in the targeted mutants.
FIG. 4.
In situ hybridization analysis of BRUCE expression. Radiolabled antisense probe was reacted to E13.5 placenta of WT (BRUCE +/+) and mutant (BRUCE −/−) embryos to detect BRUCE mRNA levels in extraembryonic tissue. BRUCE is expressed predominantly in the diploid trophoblast of the placenta, including the labyrinth layer. Increased levels of BRUCE mRNA are found in the spongiotrophoblast layer, whereas in trophoblast giant cells BRUCE expression is reduced. The specificity of the BRUCE probe was demonstrated by parallel hybridization of WT tissue with a sense probe and by hybridization of knockout tissue with the antisense probe, which shows absence of BRUCE mRNA. Bars, 1 mm.
In comparison to placentas of WT and BRUCE heterozygous embryos, the homozygous mutant placenta appeared smaller and was frequently paler in color (Fig. 3 and 5A), suggesting a defect in vascularization and/or poor blood supply. The average weight of placenta of BRUCE-deficient embryos was 73% of that of WT embryos at E11.5, and this ratio further decreased to 65% until birth (Fig. 2B). The formation of the chorioallantoic connection appeared normal, and, except for a slight reduction of the spongiotrophoblast layer, no significant gross difference between WT and mutant placentas was detectable at early stages (Fig. 5B and C, E9.5 to E11.5). All structures, including the labyrinth as well as the trophoblast giant cells, appeared fairly normal, and the border of the spongiotrophoblast and the labyrinth layer was clearly noticeable. Also, infiltration of the labyrinth layer with embryonic blood vessels and maternal blood sinusoids occurred normally at this point.
FIG. 5.
Morphology and histology of WT and BRUCE mutant placentas. (A) Ventral view of a WT placenta (+/+) and a BRUCE mutant placenta (−/−) at E17.5, showing apparent decreased vascularization of the mutant placenta, as shown by its paler color. Bar, 0.5 cm. (B) Hematoxylin- and eosin-stained radial sections of WT and mutant placentas at different developmental ages. All characteristic layers are present in the mutant placenta (de, maternal decidual tissue; gi, trophoblast giant cells; sp, spongiotrophoblast layer; la, labyrinthine layer; ch, chorionic plate). Chorioallantoic fusion occurs normally, and decidua and trophoblast giant cell development appear to be normal. The spongiotrophoblast of the mutant placenta is significantly decreased in thickness, whereas the mutant labyrinth layer is not markedly reduced in size. Bars, 0.5 mm. (C) Higher magnification of the labyrinthine and spongiotrophoblast layers of WT and mutant placentas. Fetal blood vessels (arrowheads) and maternal blood sinuses (asterisks) are present in normal number and come into close contact at early developmental stages (e.g., E11.5). Later, the labyrinthine trophoblast of the mutant placenta appears less compact and fails to mature over time. The number of trophoblast cells within the labyrinth seems to be reduced. The spongiotrophoblast of the mutant placenta consists of a few cell layers only (in contrast to the WT), and the trophoblast cells are enlarged. Bars, 10 μm.
However, at later developmental stages (E13.5 to E17.5), the reduction in thickness of the spongiotrophoblast became more evident. This observation could also be confirmed by spongiotrophoblast marker analysis using in situ hybridization (Fig. 6A and http://www.biochem.mpg.de/jentsch/JentschDataLotz.html). The fetal fms-like tyrosine kinase Flt-1 or the spongiotrophoblast-specific marker Tpbp/4311 (17) can be used to monitor differentiation of ectoplacental cone cells into spongiotrophoblast cells from E9.5 onwards (13). In mutant placenta, the layer formed by Flt-1- and 4311-expressing cells was clearly reduced at all embryonic stages examined. By analyzing the expression profiles of the specific trophoblast giant cell marker Pl-1 (4) and proliferin (22), we observed that alongside the normal continuous giant cell layer, additional Pl-1- and proliferin-expressing cells can be detected in BRUCE-deficient placenta from E11.5 onwards. Initially, those cells are mainly spread over the spongiotrophoblast, but they also specifically appear within the labyrinth layer after E15.5. Although these cells seem to be slightly enlarged, they do not reach the typical giant cell shape.
FIG. 6.
Placental marker analysis of WT and BRUCE mutants at E11.5, E13.5, E15.5, and E17.5. (A) Specific antisense riboprobes were reacted with serial placental sections of WT (BRUCE +/+) and mutant (BRUCE −/−) embryos to discriminate the different placental layers. Placental lactogen (Pl-1) is highly expressed in trophoblast giant cells, and the fms-like tyrosine kinase Flt-1 was used to label spongiotrophoblast cells. The fetal liver kinase Flk-1 is specifically expressed in fetal endothelial cells and was therefore used as a marker for the labyrinth layer. Expression of the transcription factor eHand is detectable in diploid trophoblast cells of the labyrinth as well as in the spongiotrophoblast layer and at higher level in trophoblast giant cells. All layers are present in WT as well as in mutant placenta. Note the reduction of the spongiotrophoblast layer at all developmental stages examined and the increasing number of Pl-1-expressing cells in BRUCE-deficient placenta. Bars, 1 mm. (B) Development of the placental vascular network (from top to bottom). Immunohistochemical staining of basal lamina of fetal capillaries in the labyrinth layer by using an antibody to laminin shows the delay in maturation of the mutant placental vascular network. Thus, the fetal blood vessels in the labyrinth layer appear to be less branched and dilated, even at the end of gestation. Bars, 50 μm.
Additional markers were used to identify subsets of differentiated trophoblast and endothelial cells, such as Mash2 (10), Gcm1 (23), eHand (24), or Flk-1 (8). All cells were present in WT as well as in mutant placenta and were distributed in a similar manner throughout the placenta (Fig. 6A and http://www.biochem.mpg.de/jentsch/JentschDataLotz.html). In addition, no dysregulation in expression of the basic helix-loop-helix transcription factors (Mash2 and eHand), which are key regulators of spongiotrophoblast maintenance and giant cell differentiation (5), could be detected. However, correct organization of the labyrinth layer was apparently delayed in the mutant, resulting in a less dense structure, and the number of trophoblast cells within this layer seemed to be reduced. This phenotype became pronounced after E13.5, when a dilatation of fetal capillaries and maternal sinusoids could be detected, in contrast to WT tissue (Fig. 5C).
The development of the placental vascular network was further investigated immunohistochemically by using an antibody to laminin, which stains the basal lamina of fetal capillaries in the labyrinth layer. In mature WT placenta the fetal capillaries were elongated, of similar size, and evenly distributed throughout the labyrinth from the chorionic plate to the spongiotrophoblast layer. In contrast, the fetal blood vessels in mutant placenta showed less branching. They were highly variable in thickness and formed a less dense vasculature than in WT placenta (Fig. 6B). In addition, the elimination of the nuclei from red blood cells seemed to be delayed in BRUCE mutant placenta. In fact, in WT placenta, all red blood cells are nucleated before E11.5. However, between E12 and E15 these cells enucleate, and after E16 all red blood cells in the embryonic circulation are nonnucleated (15). In BRUCE mutants about 50% of cells in fetal blood vessels of E15.5 placenta were still nucleated, whereas in the WT almost all red blood cells had already lost their nuclei. Even at stage E17.5, single nucleated red blood cells were visible in the mutant labyrinth (Fig. 5C).
Apoptosis and proliferation.
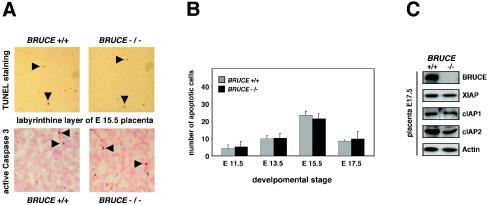
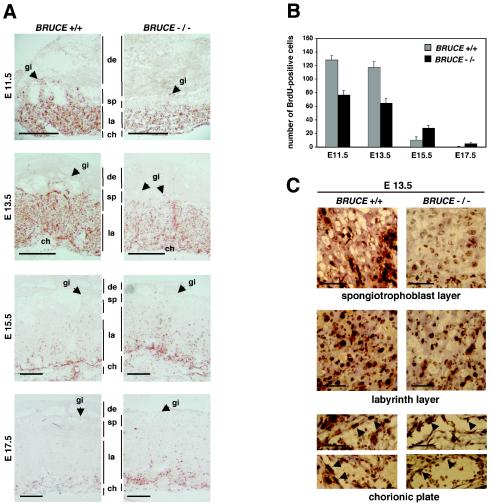
Recently, it was shown that BRUCE inhibits apoptosis, that it possesses IAP activity, and that overexpression of the protein blocks apoptosis (2, 3, 20). We therefore considered the possibility that the observed abnormalities of BRUCE-deficient mice are caused by an increase of apoptosis events. To address this question, the rate of apoptosis in WT and BRUCE mutant placenta, specifically within the spongiotrophoblast and the labyrinth layer, was analyzed. Yet, by using a TUNEL assay and immunohistochemistry to monitor activated caspase 3 levels, no significant differences between BRUCE-deficient and control placentas were found (Fig. 7A and B). Apoptosis could be observed at equally low rates in WT and mutant maternal decidual tissue, yolk sac, chorionic plate, spongiotrophoblasts, and labyrinth layer. Other IAP family members, which might overlap with BRUCE function, are expressed at equal levels in WT and knockout placenta (Fig. 7C). In contrast, we detected significant differences in cell proliferation between WT and BRUCE-deficient cells as monitored by BrdU incorporation. Analysis of E11.5 to E17.5 placenta indicated a changed BrdU index of BRUCE-deficient trophoblasts in comparison to WT littermate controls (Fig. 8A and B). At early stages, the number of proliferating cells was significantly reduced within the mutant labyrinth and almost no proliferating cells were visible within the spongiotrophoblast layer, whereas in the corresponding WT layers high proliferation rates could be observed. The proliferating activity in the labyrinth of knockout placenta was reduced to 60% at E11.5 and to 55% at developmental stage E13.5. At later stages (E15.5 and E17.5), when cells in the mature WT placenta already ceased proliferating, BrdU-positive cells were still detectable in the mutant labyrinth (Fig. 8B). This effect seems to be predominantly restricted to trophoblast cells, since the numbers of proliferating endothelial cells within the chorionic plates of WT and knockout placenta were similar (Fig. 8C). However, intensive trophoblast proliferation and branching are a prerequisite for vascularization of the labyrinth (22), and thus the number of proliferating endothelial and trophoblast cells within the labyrinth layer seems to be reduced to the same extent. From these findings we conclude that the abnormalities observed in BRUCE mutant placenta are perhaps caused by an altered proliferation and/or differentiation pattern of trophoblast cells and are not due to enhanced apoptosis in this tissue.
FIG. 7.
Apoptosis in extraembryonic tissues. (A) Similar rates of apoptotic cell death in the placentas of WT and BRUCE null mutants, as shown by immunohistochemical detection of caspase 3 activity and TUNEL staining (positive cells are marked by arrowheads). Staining methods were performed on sections of at least three different placentas per indicated genotype. The stained labyrinth layer of E15.5 WT and mutant placentas is shown as representative area. (B) For quantification, TUNEL-positive cells were counted in five randomly chosen labyrinth areas (200-fold magnification) of at least three different placentas of WT and knockout genotypes at the indicated developmental stages. Each bar represents the average ± standard deviation of apoptotic cells per area. (C) Levels of IAP proteins in WT (+/+) and BRUCE-deficient (−/−) placental tissue of E17.5 embryos. Expression levels of proteins were determined by Western blotting of placental lysates (excluding maternal tissue). Equal protein loading was shown by antiactin staining.
FIG. 8.
Cell proliferation in placenta. (A) Proliferating cells were visualized by immunohistochemical detection of BrdU incorporation (labeling of placental layers as for Fig. 5). The proliferation pattern is changed in placenta of BRUCE null mutants. There are almost no proliferating cells detectable in the strongly reduced spongiotrophoblast layer. Proliferation is also impaired in the labyrinthine layer. There are clearly fewer proliferating cells at early stages (e.g., E11.5 and E 13.5), whereas at later stages (e.g., E15.5 and E17.5), when cells in the mature WT placenta stop proliferating, cells still proliferate in the labyrinth layer of the mutant placenta. BrdU incorporation was monitored in three different WT and three mutant placentas. A representative sample of each phenotype is shown. Bars, 1 mm. (B) Higher magnification (×400) of the spongiotrophoblasts, the labyrinthine layers, and the chorionic plates of WT and mutant placentas. Note that the proliferating activity of endothelial cells of fetal blood vessels within the chorionic plate of mutant placenta does not seem to be altered. Bars, 50 μm. (C) For quantification, BrdU-positive cells were counted in five randomly chosen labyrinth areas (400-fold magnification) of at least three different placentas of WT and knockout genotype at the indicated developmental stages. Each bar represents the average ± standard deviation of cells in S phase per area.
Analysis of embryonic fibroblasts cultured in vitro.
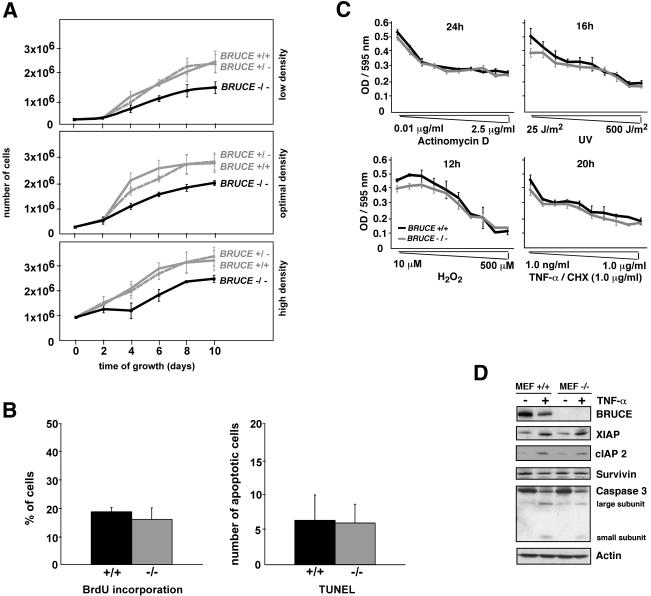
We further investigated possible defects in proliferation and apoptosis in primary cell culture. We established primary MEFs prepared from E13.5 WT, heterozygous, and BRUCE knockout fetuses. The morphology and size of the mutant MEFs were indistinguishable from those of WT and heterozygous cells at the beginning of the culture period, but the number of cells obtained from knockout fibroblast cultures was significantly lower. Comparing growth behavior of fibroblasts at different suboptimal high and low and at optimal growth densities, we observed that knockout cells grow slower than WT and heterozygous cells and, in addition, that mutant MEFs reached only ∼70% of the normal culture density of WT cells (Fig. 9A).
FIG. 9.
Characterization of BRUCE-deficient fibroblast cultures. (A) Growth curves of WT (BRUCE +/+), heterozygous (BRUCE +/−), and BRUCE-deficient (BRUCE −/−) primary fibroblasts. Cells were inoculated at suboptimal high (9 × 105 cells per 6-cm-diameter dish) and low (1 × 105 cells per 6-cm-diameter dish) densities and at optimal growth density (3 × 105 cells per 6-cm-diameter dish) and counted after the indicated period of growth. The average number of isolated cells (± standard deviation) from four individual littermates of each genotype, determined in duplicate, is shown. (B) Proliferation (left) and apoptosis (right) in asynchronous cultures of primary WT (+/+) and knockout (−/−) fibroblasts. The ordinate shows the percentage of cells that had incorporated BrdU (left) or the number of TUNEL-positive cells (right) in a randomly chosen area. Each point represents the average ± standard deviation from at least two cell lines from individual littermates of the indicated genotype, each determined in quadruplicate. (C) Effect of different apoptosis-inducing stimuli on fibroblasts from WT (MEF +/+) and mutant (MEF −/−) littermates. Cells were treated for the indicated time with the different apoptotic stimuli (actinomycin D, UV light, H2O2 [oxidative stress], and TNF-α in combination with 1.0 μg of cycloheximide [CHX] per ml). Cell death is shown as a function of concentration of the inducers of apoptosis and was assessed by measuring crystal violet staining of surviving cells (optical density [OD] at 595 nm). Primary fibroblasts isolated from three individual littermates of the indicated genotypes were analyzed in the experiment, and data represent the means ± standard deviations. (D) Levels of IAP proteins and caspase 3 in WT (+/+) and BRUCE-deficient (−/−) fibroblasts. Either cells were untreated or apoptosis was induced by treatment with 100 ng of human TNF-α per ml for 16 h. Expression levels of proteins were determined by Western blotting of whole-cell lysates. Equal protein loading was shown by antiactin staining.
To investigate the consequence of BRUCE deficiency for apoptosis and proliferation of fibroblasts, the number of cells in S phase (determined by BrdU labeling) and the number of apoptotic cells (determined by TUNEL staining) in continuously growing cultures of WT and knockout fibroblasts were determined (Fig. 9B). The numbers of S-phase cells in WT and mutant cultures were indistinguishable. Fluorescence-activated cell sorting analysis of asynchronous cells also revealed similar distributions of G1, S, and G2 cell cycle phases in knockout and WT cultures (not shown). The numbers of apoptotic cells in WT and mutant cultures were similar as well (Fig. 9B). In addition, primary BRUCE-deficient fibroblasts were not more susceptible than WT cells to apoptosis induced by different apoptotic stimuli, such as actinomycin D, oxidative stress, or TNF-α (Fig. 9C) or Fas, brefeldin A, or camptothecin (not shown). Next, we asked whether other IAP family members are able to compensate for the lack of BRUCE, as, for example, reported for the knockout of XIAP (11). To address this question, we investigated the levels of XIAP, c-IAP2, and survivin in WT or BRUCE-deficient MEFs that were either untreated or treated with TNF-α. No increase of IAP levels could be detected in mutant cells compared to the WT control (Fig. 9D). Moreover, caspase 3 levels also were similar in WT and knockout fibroblasts after induction of apoptosis.
DISCUSSION
In this study we showed that BRUCE-deficient mice die perinatally and that surviving neonates are strongly retarded in growth. We provide evidence that these phenotypes are most likely linked to defects in placental development. The placenta is crucial during mammalian development, as it functions as a maternal-fetal interface for gas, nutrient, and waste exchange. In BRUCE-deficient placenta, the chorioallantoic fusion occurs normally and the typical placental trophoblast structures are well distinguishable. However, the spongiotrophoblast layer is markedly reduced, and the labyrinth layer fails to mature properly. This leads to a reduced vascular density, which seems to reduce the surface and thereby the efficiency of nutrient and oxygen exchange. The observed temporal hemorrhages in brain and neural tube of several knockout pups, which are specifically pronounced between stages E12.5 and E14.5, might be secondary effects caused by the defects in extraembryonic tissues. Notably, the temporal abnormalities are paralleled by BRUCE expression. BRUCE mRNA levels are ubiquitous and high at early stages but vanish to a minimum around E15 and specifically rise again at late embryonic developmental stages (12).
The basis for the observed placenta defect of BRUCE−/− mice is currently unclear. BRUCE combines in a single protein two distinct activities (2, 12). It functions as an antiapoptotic IAP due to the presence of an N-terminal BIR domain, and it possesses ubiquitylation activity, which depends on the C-terminal UBC domain. Due to the design of our targeting vector, we generated a complete BRUCE null mutant, and therefore both activities of BRUCE are absent in the mutant. Downregulation of BRUCE in HeLa cells by RNAi treatment promotes apoptosis (20), and loss-of-function mutants of dBruce, the Drosophila homolog, also are prone to apoptosis in certain settings (1, 28). However, we observed no evidence for a higher apoptosis rate in BRUCE-deficient embryonic and extraembryonic tissues. Our data rather suggest that the observed placenta deficiencies are perhaps linked to differential proliferation or differentiation defects of diploid trophoblast cells, which may affect the normal branching of the embryonic labyrinthine vascular system.
BRUCE−/− animals display a slight delay in erythrocyte maturation, characterized by increased numbers of nucleated red blood cells, predominantly in the peripheral circulation. In addition, knockout embryos are visually paler than the littermate controls. But, as shown for Rb or SOCS3-deficient mice (7, 21) erythropoeisis defects might be secondary to placental defects. Oxygen and/or nutrient deprivation, as well as a possibly altered placental secretion of erythropoietic differentiation factors, could increase the number of immature red blood cells.
Growth defects could also be observed in cultures of BRUCE-deficient primary embryonic fibroblasts. However, BRUCE−/− MEFs also are not detectably prone to apoptotic stimuli compared to MEFs from WT mice. A possible explanation for the lack of discernible apoptosis phenotypes in BRUCE-deficient cells is that other IAPs may compensate for a BRUCE deficiency in certain cells. Indeed, previous work has shown that IAPs have overlapping functions. For example, mice deficient in XIAP, the strongest IAP known, do not exhibit higher apoptosis rates, due to an apparent overlap with cIAP1 and cIAP2 (11). In case of BRUCE deficiency, we observed no upregulation of other IAPs such as XIAP or cIAP2 in extraembryonic tissue as well as in isolated fibroblasts. However, this might not be surprising given the fact that BRUCE is about 10-fold less potent than XIAP in blocking apoptosis (2). Alternatively, as shown for Drosophila dBruce (1), murine BRUCE might act as a local inhibitor of specific activated caspases that also function in nonapoptotic pathways.
Another possibility is that the observed defects are caused by the absence of BRUCE's ubiquitylation activity. Currently the only known substrate for BRUCE-dependent ubiquitylation is Smac. However, analogous to other ubiquitin ligases, BRUCE may have additional substrates. Notably, Smac is only monoubiquitylated by BRUCE. Since monoubiquitylation does not usually promote proteasomal degradation, BRUCE's activity could play a nonproteolytical role. BRUCE localizes specifically to the TGN and vesicular structures. Therefore, it will be interesting to see whether BRUCE's IAP and ubiquitylation activity is specifically important for TGN function, for example, TGN maintenance or protein sorting.
Acknowledgments
G.P. constructed the BRUCE knockout mouse, and K.L. characterized the phenotype.
We especially thank K.-A. Nave and M. Klugmann for their generous help in introducing us to mouse knockout techniques. We thank F. Zimmermann for excellent transgenic and in vitro fertilization service; J. Putzke, B. Kunkel, N. Borchers, and M. Moser for help with histology techniques; C. Hitz (Flt-1 and Flk-1), D. I. Linzer (Pl-1), J. Rossant (4311), A. Joyner (Mash2), and M. Wegner (Gcm1) for kindly providing cDNA hybridization probes; and T. Bartke, C. Pohl, K.-A. Nave, W. Wurst, T. Floss, and C. Hitz for discussions.
The work was supported by grants from the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie (to S.J.).
REFERENCES
- 1.Arama, E., J. Agapite, and H. Steller. 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4:687-697. [DOI] [PubMed] [Google Scholar]
- 2.Bartke, T., Pohl, C., G. Pyrowolakis, and S. Jentsch. 2004. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell 14:801-811. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., M. Naito, S. Hori, T. Mashima, T. Yamori, and T. Tsuruo. 1999. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem. Biophys. Res. Commun. 264:847-854. [DOI] [PubMed] [Google Scholar]
- 4.Colosi, P., F. Talamantes, and D. I. Linzer. 1987. Molecular cloning and expression of mouse placental lactogen I complementary deoxyribonucleic acid. Mol. Endocrinol. 1:767-776. [DOI] [PubMed] [Google Scholar]
- 5.Cross, J. C. 2000. Genetic insights into trophoblast differentiation and placental morphogenesis. Semin. Cell Dev. Biol. 11:105-113. [DOI] [PubMed] [Google Scholar]
- 6.Dagerlind, A., K. Friberg, A. J. Bean, and T. Hokfelt. 1992. Sensitive mRNA detection using unfixed tissue: combined radioactive and non-radioactive in situ hybridization histochemistry. Histochemistry 98:39-49. [DOI] [PubMed] [Google Scholar]
- 7.de Bruin, A., L. Wu, H. I. Saavedra, P. Wilson, Y. Yang, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2003. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc. Natl. Acad. Sci. USA 100:6546-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumont, D. J., G. H. Fong, M. C. Puri, G. Gradwohl, K. Alitalo, and M. L. Breitman. 1995. Vascularization of the mouse embryo: a study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev. Dyn. 203:80-92. [DOI] [PubMed] [Google Scholar]
- 9.Goyal, L. 2001. Cell death inhibition: keeping caspases in check. Cell 104:805-808. [DOI] [PubMed] [Google Scholar]
- 10.Guillemot, F., A. Nagy, A. Auerbach, J. Rossant, and A. L. Joyner. 1994. Essential role of Mash-2 in extraembryonic development. Nature 371:333-336. [DOI] [PubMed] [Google Scholar]
- 11.Harlin, H., S. B. Reffey, C. S. Duckett, T. Lindsten, and C. B. Thompson. 2001. Characterization of XIAP-deficient mice. Mol. Cell. Biol. 21:3604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauser, H. P., M. Bardroff, G. Pyrowolakis, and S. Jentsch. 1998. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J. Cell Biol. 141:1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirashima, M., Y. Lu, L. Byers, and J. Rossant. 2003. Trophoblast expression of fms-like tyrosine kinase 1 is not required for the establishment of the maternal-fetal interface in the mouse placenta. Proc. Natl. Acad. Sci. USA 100:15637-15642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann, M. H. 1992. The atlas of mouse development. Academic Press Ltd., London, United Kingdom.
- 16.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635-644. [DOI] [PubMed] [Google Scholar]
- 17.Lescisin, K. R., S. Varmuza, and J. Rossant. 1988. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 2:1639-1646. [DOI] [PubMed] [Google Scholar]
- 18.Ma, S., J. Charron, and R. L. Erikson. 2003. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol. Cell. Biol. 23:6936-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickart, C. M. 2000. Ubiquitin in chains. Trends Biochem. Sci. 25:544-548. [DOI] [PubMed] [Google Scholar]
- 20.Qiu, X. B., S. L. Markant, J. Yuan, and A. L. Goldberg. 2004. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 23:800-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts, A. W., L. Robb, S. Rakar, L. Hartley, L. Cluse, N. A. Nicola, D. Metcalf, D. J. Hilton, and W. S. Alexander. 2001. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proc. Natl. Acad. Sci. USA 98:9324-9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber, J., E. Riethmacher-Sonnenberg, D. Riethmacher, E. E. Tuerk, J. Enderich, M. R. Bosl, and M. Wegner. 2000. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol. 20:2466-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott, I. C., L. Anson-Cartwright, P. Riley, D. Reda, and J. C. Cross. 2000. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell. Biol. 20:530-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9:459-470. [DOI] [PubMed] [Google Scholar]
- 26.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhagen, A. M., E. J. Coulson, and D. L. Vaux. 2001. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2:REVIEWS3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernooy, S. Y., V. Chow, J. Su, K. Verbrugghe, J. Yang, S. Cole, M. R. Olson, and B. A. Hay. 2002. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr. Biol. 12:1164-1168. [DOI] [PubMed] [Google Scholar]