Abstract
While mechanisms to detoxify plant produced, anti-herbivore compounds have been associated with plant host use by herbivores, less is known about the role of chemosensory perception in their life histories. This is especially true for generalists, including chelicerate herbivores that evolved herbivory independently from the more studied insect lineages. To shed light on chemosensory perception in a generalist herbivore, we characterized the chemosensory receptors (CRs) of the chelicerate two-spotted spider mite, Tetranychus urticae, an extreme generalist. Strikingly, T. urticae has more CRs than reported in any other arthropod to date. Including pseudogenes, 689 gustatory receptors were identified, as were 136 degenerin/Epithelial Na+ Channels (ENaCs) that have also been implicated as CRs in insects. The genomic distribution of T. urticae gustatory receptors indicates recurring bursts of lineage-specific proliferations, with the extent of receptor clusters reminiscent of those observed in the CR-rich genomes of vertebrates or C. elegans. Although pseudogenization of many gustatory receptors within clusters suggests relaxed selection, a subset of receptors is expressed. Consistent with functions as CRs, the genomic distribution and expression of ENaCs in lineage-specific T. urticae expansions mirrors that observed for gustatory receptors. The expansion of ENaCs in T. urticae to > 3-fold that reported in other animals was unexpected, raising the possibility that ENaCs in T. urticae have been co-opted to fulfill a major role performed by unrelated CRs in other animals. More broadly, our findings suggest an elaborate role for chemosensory perception in generalist herbivores that are of key ecological and agricultural importance.
Keywords: Tetranychus urticae, gustatory receptor, degenerin/epithelial Na+, channels, chemosensory receptor, herbivore
Introduction
Chemoreception is the process by which animals perceive their environment and tune their behavior according to the chemical stimuli they encounter. By recognizing chemical cues, they locate food sources, find mates, avoid predators and toxic substances, and modulate communication with conspecifics (Kaupp 2010; Cande et al. 2013). Chemosensory perception is comparatively well understood in vertebrates and insects, where several receptor families have been identified that recognize chemical stimuli (Nei et al. 2008; Touhara and Vosshall 2009; Kaupp 2010; Rytz et al. 2013; Freeman et al. 2014; Benton 2015; Jiang and Matsunami 2015). In the well-studied vertebrate olfactory system, compounds are perceived by seven transmembrane (TM) domain metabotropic G-protein coupled receptors (GPCRs), with ligand binding initiating an intracellular signaling cascade that leads to the opening of ion channels (Nei et al. 2008; Kaupp 2010). In contrast, whereas insect olfactory receptors (ORs) also have seven transmembrane domains, their membrane topology is inverted relative to vertebrate GPCRs, (Smart et al. 2008; Benton 2015), and insect ORs are thought to function directly as ionotropic, ligand (odorant) gated ion channels (Sato et al. 2008; Smart et al. 2008; Benton 2015).
In addition to insect ORs expressed in antennae, a second class of related receptors was subsequently identified in taste organs like labial palps (Clyne et al. 2000; Dunipace et al. 2001; Scott et al. 2001), and termed gustatory receptors (GRs) (Touhara and Vosshall 2009). While insect GRs are expressed in many nonantennal sensory organs that perceive contact chemical cues (e.g., sensilla on legs or taste bristles on wings; Dunipace et al. 2001), some GRs likely perceive volatile compounds as well (Jones et al. 2007; Nei et al. 2008). GRs have since been identified in arthropods outside the insects, including in the crustacean Daphnia pulex and the myriapod Strigamia maritima, although ORs are absent from both (Peñalva-Arana et al. 2009; Chipman et al. 2014). Among the major extant arthropod groups, phylogenetic studies suggest that Hexapoda (which includes insects) and Crustacea are a sister clade to Myriapoda (which includes centipedes and millipedes). Collectively, these groups compose the Mandibulata, to which the last major extant arthropod group, Chelicerata, is the sister taxon (see Edgecombe and Legg 2014). These observations suggest that the comparatively well-studied ORs are an insect-specific expansion of a GR lineage (Peñalva-Arana et al. 2009; Chipman et al. 2014; Benton 2015). Beyond arthropods, GR-related proteins have been found in diverse multicellular animals, and even plants, although the family is absent from chordates (Benton 2015; Robertson 2015). Following proposed nomenclature, we refer to these receptors in arthropods as GRs (with ORs as an insect-specific expansion), and those outside Arthropoda as GR-Like (GRL) (Benton 2015; Robertson 2015).
In addition to GRs, several other receptor classes have also been implicated as animal chemosensory receptors (CRs) (Cande et al. 2013; Freeman and Dahanukar 2015; Joseph and Carlson 2015). These include ionotropic receptors (IRs) that are present throughout the Protostomia (Croset et al. 2010), and that are related to the highly conserved ionotropic Glutamate Receptors (iGluRs) that perform synaptic roles (Benton et al. 2009). In insects, IRs have been implicated in both volatile odorant and taste perception (Joseph and Carlson 2015) and constitute the second well-studied receptor family mediating perception of chemical cues in insects. In addition, several other ion channel families have also been implicated as animal chemosensory receptors. These families include the small family of Transient Receptor Potential (TRP) cation channels (Fowler and Montell 2013), and also degenerin/Epithelial Na+ Channels (ENaCs) (Ben-Shahar 2011). The ENaC family is involved in various functions in metazoans, including osmoregulation, peptide signaling, and detection of different stimuli such as nociception, chemo- and mechanosensing (Kellenberger and Schild 2002; Ben-Shahar 2011). In Drosophila melanogaster, ENaCs encoded by the pickpocket (ppk) genes (Zelle et al. 2013) have functions in mechanosensing, but also in sensing chemicals including salts and water (Liu et al. 2003; Chen et al. 2010; Ben-Shahar 2011). The mechanisms of ENaC-mediated chemosensation are still enigmatic, and in some cases may be indirect (Ben-Shahar 2011). However, several studies have raised the possibility that PPK23 may function as a contact pheromone receptor, potentially in heteromultimeric complexes with other PPK members or other proteins (Lu et al. 2012; Thistle et al. 2012; Toda et al. 2012).
In addition to the use of divergent receptor types, a striking finding from genomic studies is that chemosensory receptor families have proliferated greatly in many animal genomes (Nei et al. 2008; Sanchez-Gracia et al. 2009; Rytz et al. 2013; Benton 2015; Montagné et al. 2015). The most extreme examples come from vertebrates, such as the elephant that has more than 4000 ORs (Niimura et al. 2014). In insects, the number of receptors is more moderate, but ORs and GRs are still among the largest gene families. For instance, the D. melanogaster genome harbors ∼60 ORs and GRs each (Gardiner et al. 2008). For ORs and GRs, the ratio of these receptor classes and the number of family members observed in D. melanogaster is representative of many insects with sequenced genomes (Nei et al. 2008; Benton 2015), although expansions to several-fold more receptors have occurred in some lineages, notably in the flour beetle (Tribolium castaneum) (Tribolium Genome Sequencing Consortium et al. 2008) and in many ants (Zhou et al. 2015). Because insects are the most species-rich group of animals, it has been suggested that ORs and GRs comprise the most member-rich group of metazoan proteins with distinct functions (Benton 2015). A similar mode of evolution has also been documented for insect IRs, which have expanded in D. melanogaster to 66 (Croset et al. 2010), and in the termite to about 150 (Terrapon et al. 2014). For ENaCs, expansions have been more moderate; nevertheless, whereas vertebrates typically have ∼10 ENaCs, the family has expanded in several insect taxa, reaching 31 members in D. melanogaster (Zelle et al. 2013). Given these dynamics, examining the tempo and mode of CR gene evolution, both across and within taxa as a function of variation in life history characteristics, has attracted much attention (Whiteman and Pierce 2008; Benton 2015). In both vertebrates and insects CR evolution is highly dynamic (Tribolium Genome Sequencing Consortium et al. 2008; Jiang and Matsunami 2015; Zhou et al. 2015). For instance, comparative studies of the genomes of 12 Drosophila species suggests that CR evolution is dominated by a fast birth/death process with some evidence of positive selection (Whiteman and Pierce 2008, and references therein).
Despite recent progress in insects and closely allied groups, our understanding of the evolution of olfaction and taste in other arthropods remains limited. In particular, little is known from the species-rich chelicerates—horseshoe crabs, scorpions, spiders, ticks and mites—the sister-taxon of mandibulates and the second most diverse arthropod group after hexapods (Dunlop 2010). As opposed to insects, chelicerates lack antennae and have more limited mobility, with potential repercussions to the evolution of chemosensing. The Acari—ticks and mites—are the most diverse clade within the chelicerates, with lifestyles ranging from parasitic to predatory to plant feeding (Grbić et al. 2011). Among chelicerates, the two-spotted spider mite (Tetranychus urticae) is an attractive species for understanding chemosensory processes. Its 90-Mb genome was sequenced using the Sanger method and is high quality (Grbić et al. 2011), an essential feature for the annotation of large gene families. Furthermore, T. urticae is an extreme generalist herbivore that has been documented to feed on over 1,000 plant species in 120 families (Migeon et al. 2010; Grbić et al. 2011), which contributes to its status as an important agricultural pest (Van Leeuwen et al. 2015). Although little is known about chemosensory processes in T. urticae, spider mites have neuron-rich setae harboring cuticular pores on the palpa and legs (Bostanian and Morrison 1973) that are reminiscent of functionally characterized chemosensory sensilla present on insect appendages (Sanchez-Gracia et al. 2009). As different plants produce many and diverse compounds to deter feeding (Howe and Jander 2008), T. urticae provides an opportunity to explore how exposure to a broad range of chemical stimuli has impacted CR evolution.
Recently, several CR families have been annotated in the draft genomes of Ixodes scapularis (the deer tick) and Metaseiulus occidentalis (a phytoseiid predatory mite) (Gulia-Nuss et al. 2016; Hoy et al. 2016). Within the Acari, these species are in the Parasitiformes, a sister order to the Acariformes to which T. urticae and diverse herbivorous mites belong. In I. scapularis and M. occidentalis, about 60 GRs were identified, with up to 65 IRs in M. occidentalis. (Gulia-Nuss et al. 2016; Hoy et al. 2016). These findings suggest an important role for chemosensory perception in chelicerates, with receptor numbers representative of many nonchelicerate arthropods (Nei et al. 2008; Peñalva-Arana et al. 2009; Sanchez-Gracia et al. 2009; Chipman et al. 2014; Benton 2015). In T. urticae, only TRP channels have been systematically annotated to date, and the TRPA channel associated with perception of noxious chemical stimuli is absent, raising the possibility that T. urticae does not perceive noxious compounds (Peng et al. 2015), or does so with other receptors.
To assess if the CR composition of I. scapularis and M. occidentalis is representative of all chelicerates, as well as to assess the mode of CR evolution in a generalist herbivore, we exhaustively mined and annotated T. urticae CRs. Strikingly, whereas few IRs are present, we identified 689 GRs (TuGRs), including pseudogenes, a number exceeding that reported in other arthropods to date. Moreover, we observed an unprecedented expansion of ENaCs, raising the possibility that this family has been co-opted to play a major role in chemosensation in T. urticae. Genomic organization, gene expression data, and polymorphism combine to suggest similar modes of dynamic gene family evolution for TuGRs and ENaCs, and shed light on the forces shaping chemosensation in generalist herbivores.
Materials and Methods
Tetranychus urticae Gustatory Receptor Gene Annotation
Crustacean (Daphnia) and insect gustatory or odorant receptors were used to perform tBLASTn searches against the draft T. urticae genome. A permissive E-value (at least as high as 1.0) was used initially in searches to identify putative receptor gene models, as numerous studies have shown that among arthropods primary sequences of GRs can be highly divergent (Peñalva-Arana et al. 2009; Chipman et al. 2014; Benton 2015; Robertson 2015; Gulia-Nuss et al. 2016; Hoy et al. 2016). Tetranychus urticae GR gene models—as assessed based on sequence homology and additional genic structural and membrane topological predictions (for a detailed discussion of criteria for arthropod GR annotation, see Robertson 2015)—located at tBLASTn hit genome regions were adjusted when necessary or new T. urticae GR gene models were created using GenomeView (Abeel et al. 2012). In assessing membrane topologies of receptors, both TMHMM (version 2.0c) (Krogh et al. 2001) and Phobius (version 1.01) (Käll et al. 2004) were used with default parameters. Subsequently, this process was repeated in an iterative manner using T. urticae GRs as queries for tBLASTn searches. Following the iterative tBLASTn searches and annotation, and to further ensure comprehensive identification of T. urticae gene models, we also performed HMMER searches (Eddy 2009) with a profile constructed from all the manually curated, intact T. urticae GRs, as well as the HMMER profile PF08395 (“7tm_7”) that is often detectable in arthropod GRs. These searches, which were carried out with the entire T. urticae proteome (the T. urticae annotation of October 29, 2015 was used throughout), identified no additional T. urticae GRs that had not been identified by tBLASTn searches (supplementary table S1, Supplementary Material online). Manually introduced GR gene models lack UTRs, and where GR models have UTRs from automated predictions (Grbić et al. 2011; Sterck et al. 2012), their reliability is hard to assess. We therefore limited all analyses of GR introns (number and position) to those within coding sequences (we applied this criterion as well to the analysis of CRs in other gene families).
Identification and Annotation of T. urticae iGluR/IRs
Using HMMER in combination with the Pfam domain PF00060 (Finn et al. 2010), T. urticae proteins were searched for iGluR/IRs, and hits with an E-value <1e-5 were initially considered as putative iGluRs or IRs, and used as queries for iterative tBLASTn searches as for GRs. Predicted T. urticae iGluR/IR gene models were adjusted as necessary or new T. urticae iGluR/IRs gene models were created using GenomeView. Conserved combinations of domains diagnostic for assessing iGluR and IR membership, including PF00060, PF10613, and PF01094 (Croset et al. 2010), were detected with InterProScan 5.19–58.0 (Jones et al. 2014). As we performed for GRs, and to ensure comprehensive discovery, we constructed a HMMER profile from the annotated T. urticae iGluR/IRs that was used to search the entire T. urticae proteome, identifying no additional family members (supplementary table S2, Supplementary Material online).
Identification and Annotation of T. urticae ENaCs
Drosophila melanogaster PPKs (Zelle et al. 2013) and a set of vertebrate and invertebrate ENaCs were used in BLASTp and tBLASTn searches against the T. urticae proteome and genome, respectively, to identify T. urticae ENaCs. E-values as high as at least 1.0 were applied initially, as like GRs, ENaCs are well documented to vary markedly in primary sequence among taxa (Zelle et al. 2013). Criteria for inclusion as ENaCs included the presence of the N- and C-terminal transmembrane domains, and diagnostic conserved cysteine and hinge residues in the extra-cellular loop region, as assessed against chicken ASIC1, an ENaC for which a crystal structure is available (Jasti et al. 2007). Predicted T. urticae ENaC gene models located at tBLASTn hit genome regions were adjusted when necessary or new T. urticae ENaC gene models were created using GenomeView. Additionally, and to ensure comprehensive discovery, we constructed a HMMER profile from all manually curated, intact T. urticae ENaCs. Further, many ENaCs can also be detected by the presence of the amiloride-sensitive sodium channel (ASC) domain, profile PF00858. HMMER searches with these profiles returned all the manually curated T. urticae ENaCs, and did not identify additional family members (supplementary table S3, Supplementary Material online).
Identification of ENaCs in I. scapularis and M. occidentalis
To assess if ENaCs in T. urticae are representative of other chelicerates in sequence and copy number, candidate ENaCs in the proteomes of the deer tick I. scapularis (assembly JCVI_ISG_i3_1.0, 20,486 proteins) (Gulia-Nuss et al. 2016) and the predatory mite M. occidentalis (“Gnomon set”, 18,338 proteins) (Hoy et al. 2016) were assessed with HMMER with domain PF00858 (ASC domain). An E-value threshold of 1.0 was used in HMMER searches. Many of the resulting hits were to sequences that are far too short to be full-length receptors, especially for I. scapularis, for which the large, repetitive genome is less well assembled than for T. urticae (Gulia-Nuss et al. 2016). To identify putative full-length ENaCs for phylogenetic analyses, a threshold length of 300 amino acids was applied, as was the presence of N- and C-terminal transmembrane domains that could be aligned to those of chicken ASIC1 (supplementary table S4, Supplementary Material online; alignments were performed as described for phylogenetic analyses).
Phylogenetic Analyses
GR, ENaC and iGluR/IR protein sequences were aligned using MAFFT v7.266 (Katoh and Standley 2013) with 1000 iterations with the options “E-INS-i” and “reorder”. Visualizations of alignments were performed with Jalview (Waterhouse et al. 2009). Two GR alignments were generated: one containing solely T. urticae GRs (447 intact T. urticae sequences) and one with a representative set of T. urticae GRs from clades A and B, all T. urticae GRs from clade C, 39 intact I. scapularis GRs (Gulia-Nuss et al. 2016), 57 intact M. occidentalis GRs (Hoy et al. 2016), and 58 D. melanogaster GRs (Robertson 2009). Intact T. urticae ENaCs (108 sequences) were aligned with those of I. scapularis (four sequences, supplementary table S4, Supplementary Material online), M. occidentalis (24 sequences, supplementary table S4, Supplementary Material online), D. melanogaster (Zelle et al. 2013) (31 sequences), C. elegans (Bazopoulou et al. 2007) (25 sequences; the “egas” subgroup consisting of four atypical ENaC sequences with an ASC domain, PF00858, and EGF repeats, InterPro domain IPR00742, were not included in the alignment), and chicken ASIC1 (Jasti et al. 2007). Intact T. urticae iGluR/IRs (18 sequences) were aligned with those of I. scapularis (Gulia-Nuss et al. 2016) (14 intact iGluR/IRs, and one incomplete IR, IscaIR25a), M. occidentalis (Hoy et al. 2016) (62 intact IR/iGluRs) and a representative set of D. melanogaster iGluR/IRs (Croset et al. 2010) (43 IR/iGluRs). The arthropod GR, ENaC and iGluR/IR alignments were trimmed using trimAl v1.4 (Capella-Gutierrez et al. 2009) as arthropod GR, ENaC and iGluR/IR protein sequences are known to be highly divergent across arthropods. The “gappyout” option was selected in trimAl v1.4 as it is one of the filtering methods with the least impact on tree accuracy (Tan et al. 2015). Model selection was done with ProtTest 3.4 (Darriba et al. 2011) and according to the Akaike information criterion JTT + G+F, JTT + I+G + F, WAG + I+G + F and LG + G+F were optimal for the phylogenetic reconstruction of T. urticae GR, arthropod GR, ENaC and iGluR/IR proteins, respectively. For each alignment a maximum likelihood analysis was performed using RAxML v8 HPC2-XSEDE (Stamatakis 2014) on the Cipres web portal (Miller et al. 2010) with 1,000 bootstrapping replicates. As the “estimate proportion of invariable sites (+I)” option is not recommended by the developer of RaxML v8 (page 59 of the RaxML v8.2.X manual) the JTT + G+F and WAG + G+F model was used instead for phylogenetic analysis of arthropod GR and ENaC protein sequences. The resulting GR and ENaC trees were midpoint rooted, whereas the iGluR/IR tree was rooted with NMDA receptors. Trees were visualized using MEGA 6.0 (Tamura et al. 2013) and edited with Adobe Illustrator software (Adobe Inc.).
Detection and Analysis of CR Clusters
A sliding window approach (50-kb windows incremented in 10-kb steps) was used to identify clusters of each chemoreceptor gene family (or clade) throughout the genome. Genes were considered part of each sliding window cluster if any portion of them overlapped the 50-kb window. For this analysis, all members of each gene family (complete, incomplete and pseudogene) were included. Neighboring clusters sharing at least one gene were considered to be part of the same cluster and were subsequently merged into a single larger cluster (after Thomas 2005). The midpoints of the final clusters and the number of genes and pseudogenes contained within each cluster were used for plotting. To determine if genes in clusters are more closely related than expect by chance, for CR clusters by family or clade (two or more intact genes) the patristic distances as assessed from the respective phylogenies between all pairs of intact genes within a given cluster were averaged, and the mean of the resulting values was then assessed across all clusters genome wide. The resulting value was then compared with the distribution resulting from 10,000 permutations in which the metric was calculated with input datasets for which the identity of genes were randomly assigned to the genomic locations of CRs. These analyses were performed separately for TuGR-As, TuGR-Bs, and ENaCs. Patristic distances were assessed with DendroPy 4.1.0 (Sukumaran and Holder 2010).
Variation in Receptors among Strains
Genomic sequence reads generated previously with the Illumina method for the strains Montpellier (36-bp paired-end reads) (Grbić et al. 2011) and EtoxR (80-bp single end reads) (Van Leeuwen et al. 2012) were aligned to the reference London genome sequence using the read-mapper from the CLCBio assembly cell (command line executables, up to 10% mismatches allowed; www.qiagenbioinformatics.com/; last accessed August 23, 2016) to produce consensus sequences. Gene models of CRs from the London reference annotation were then assessed relative to the EtoxR and Montpellier alignment data. Both fixed and segregating differences within CRs could be assessed for each strain, neither of which is inbred (Van Leeuwen et al. 2012).
Detection of CR Gene Expression
About 1,200 adult fertilized T. urticae females from the London strain were placed on bean leaf arenas (±200 mites/leaf) and allowed to lay eggs for 4 h. Eggs were incubated at 27 °C, 65% RH and 16:8 h light:dark (LD) and resulting F1 virgin females were collected for RNA extraction at the age of 3 days. A similar procedure was followed to obtain males for RNA extraction, except that resulting F1 virgin females were transferred to new bean leaf arenas (±100 mites/leaf) and allowed to lay eggs for 16 h (with F1 virgin females transferred to a new bean leaf arena every 4 h). Eggs of F1 virgin females were incubated at 27 °C, 65% RH and 16:8 LD and resulting 1- to 2-day-old mature males (Krainacker and Carey 1989) were collected for RNA. Four RNA samples were obtained for each sex and RNA was extracted using the RNeasy mini kit (Qiagen, Belgium). Paired-end, strand-specific 100-bp long reads from four replicates each of male and female mites were aligned to the reference genome (Grbić et al. 2011) using STAR 2.4.1d (Dobin et al. 2013) in two-pass alignment mode with a maximum intron size of 20 kb. Read counts for individual, intact genes were quantified using HTSeq 0.6.1 (Anders et al. 2015). Only reads mapping uniquely to the CDS regions of genes were considered for downstream analyses. The effect of multi-mapped (repetitive) reads, albeit an initial concern for CR families with similar sequences among duplicates, was negligible for the majority of CRs (supplementary tables S1–S3, Supplementary Material online). Differential gene expression between male and female mites was assessed with DESeq2 1.12.3 (Love et al. 2014) applying a 5% false discovery rate.
Results
Expansions of Diverse Gustatory Receptor Lineages in the T. urticae Genome
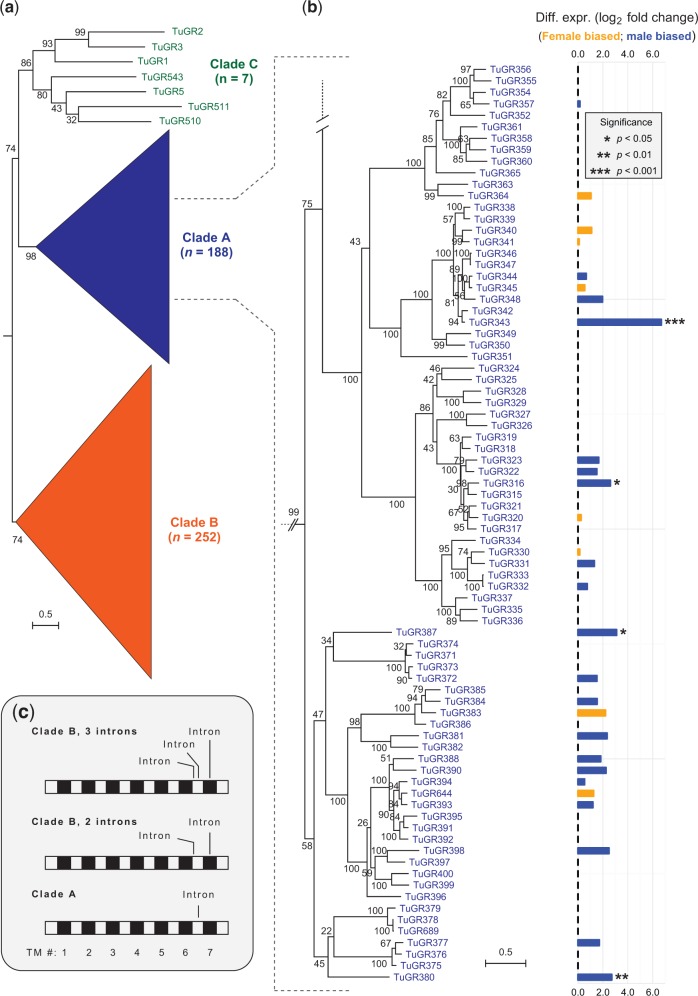
Mining of the T. urticae genome with arthropod GRs, followed by extensive manual checking, correction, implementation of new gene models, and subsequent iterative searching and reannotation, revealed a total of 689 TuGRs. Of these, 447 were intact, 220 were pseudogenes, and 22 were partial genes that extended into sequence gaps in the assembly (supplementary table S1, Supplementary Material online; unless otherwise noted, descriptions of gene families are limited to intact, and hence putatively functional, genes). A maximum-likelihood phylogenetic analysis revealed that the majority of TuGRs fell into two comparatively well-supported clades consisting of 188 and 252 intact genes (the TuGR-A and TuGR-B clades, respectively) while the remaining seven diverse TuGRs fell into a clade with only moderate support (hereafter TuGR-Cs) (fig. 1a and supplementary fig. S1, Supplementary Material online). Within each of the major A and B lineages, multiple well-supported clades with short branch lengths were apparent, indicative of recurrent episodes of proliferation (fig. 1b and supplementary fig. S1, Supplementary Material online). As revealed from a phylogenetic analysis including I. scapularis and M. occidentalis GRs, TuGR-As and TuGR-Bs reflect lineage-specific expansions in T. urticae (supplementary fig. S2, Supplementary Material online); the seven TuGR-Cs fell into a clade with low bootstrap support that included receptors from I. scapularis previously reported in “Ixodes expansion 2” (Gulia-Nuss et al. 2016) as well as a subset of M. occidentalis GRs. Despite the large repertoire of GRs, no T. urticae genes were identified with high similarity to insect ORs, including the highly conserved insect OR co-receptor (Orco). This finding further confirms a large body of evidence that arthropod ORs arose as a lineage-specific expansion in insects (Peñalva-Arana et al. 2009; Chipman et al. 2014; Gulia-Nuss et al. 2016; Hoy et al. 2016).
Fig. 1.—
Phylogeny, expression, and structure of Tetranychus urticae gustatory receptors. (a) A midpoint rooted maximum-likelihood tree of 447 intact T. urticae gustatory receptors. Two lineage-specific expansions are apparent that are represented by 188 and 252 TuGRs, respectively (clade A and B TuGRs, grouped as triangles for display; the full ungrouped phylogeny is shown in supplementary fig. S1, Supplementary Material online). (b) A portion of the phylogeny corresponding to 80 TuGR-As with differential expression between males and females shown at the right (differential expression detected as presented in fig. 5). (c) Schematic of the canonical positions of phase-0 introns in TuGRs relative to the coding sequences (the location of the seven TM domains is as indicated, bottom). A subset of genes encoding TuGR-Bs have only two introns (middle), one less than for the other TuGR-Bs (top). As assessed from sequence alignments, the terminal intron positions corresponding to TM7 for all TuGR-Bs are identical (top and middle). The ancestral relationships for introns corresponding to the loop region between TMs 6 and 7 among the TuGR clades is less clear. The schematic is intended to show intron positions relative to TMs, and does not reflect relative sizes of the loop regions that vary among the receptors.
To provide additional support that TuGRs belong to the GR superfamily, and to further understand the sequence and structural relationships to GRs in other taxa, we examined four additional criteria that have been assessed for GR/GRL members (Robertson 2015): (1) membrane topology, (2) conserved intron positions, (3) the presence of the GR family “7tm_7 superfamily” conserved signature domain and homology to GRs from other species, and (4) sequence features of the TM7 domain. Gustatory receptors are unusual among animal proteins in having 7-TM topologies with intracellular N-termini. It is known that computational prediction of the seven TM domains in GRs is less robust than for 7-TM GPCRs, with several fewer or more TMs often predicted; nevertheless, where seven TMs are predicted, the N-termini are predicted to be intracellular in most cases (Benton 2015; Robertson 2015). As assessed with two TM prediction programs that gave similar findings—TMHMM and Phobius—TM number and topologies for TuGRs are consistent with those expected for GR family members (supplementary table S1, Supplementary Material online). For instance, with TMHMM nearly half of TuGRs were predicted to harbor seven TMs, and 88.8% were predicted to have between 6 and 8 TMs. Further, 93.6% of TuGRs with seven predicted TMs were also predicted to have intracellular N-termini.
In addition to membrane topology, GRs from other animals often have three phase-0 introns that map to the C-termini (Robertson 2015). The last of these introns corresponds to a conserved position in the final transmembrane helix, TM7, and is present in GRs in insects (Robertson et al. 2003), D. pulex (Peñalva-Arana et al. 2009), and even in some GRLs from taxonomically distant animals including C. elegans (Peñalva-Arana et al. 2009; Robertson 2015). This C-terminal intron organization is similar to the three phase-0 introns observed in 85.3% of clade B TuGRs (a sub-clade of 33 TuGR-Bs lack one of the introns in the last loop region; fig. 1c). In particular, the last phase-0 intron in TuGR-Bs corresponds to TM7, and therefore may reflect an ancestral intron widely distributed across the GR/GRL superfamily (Robertson 2015). In contrast, intron number and position are more variable in TuGR-Cs, and nearly all TuGR-As (99.4%) have a single phase-0 intron located in the loop between TM6 and TM7. Thus, the lineage-specific expansions of TuGR clades, as inferred from phylogenetic analyses (fig. 1a and supplementary fig. S1, Supplementary Material online), are also supported by diagnostic intron numbers and positions (fig. 1c). The median intron length for TuGR-As and TuGR-Bs is a mere 71 and 72 bp, respectively, a factor that contributes to their compact genic structure.
Despite conserved features of membrane topology and intron architecture, the TuGR clades are highly divergent from other arthropod GRs. For example, only 40 of 447 TuGRs (8.9%) had matches to the “7tm_7” domain with an E-value < 0.01 (37 TuGR-As, 1 TuGR-B and 2 TuGR-Cs as assessed with HMMER; supplementary table S1, Supplementary Material online). Likewise, as assessed by BLAST searches with full length TuGRs, only 15 of 447 receptors had hits to protein sequences in the nonredundant NCBI database with an E-value < 0.01. Nearly all the top hits, excluding hypothetical proteins, were to predicted GR proteins available from draft genome projects in other chelicerates (e.g., I. scapularis; the itch mite that causes scabies, Sarcoptes scabiei; or the Atlantic horseshoe crab, Limulus polyphemus). Secondary hits, or alternatively primary hits above an E-value of 0.01, harbored GRs from diverse insects as well (supplementary table S1, Supplementary Material online).
In addition to their divergence from other known GRs, the major TuGR lineages are also highly divergent from each other, with no residues universally conserved across all receptors (supplementary figs. S3 and S4, Supplementary Material online). Within the T. urticae A and B lineages, TM domains 5–7 tended to be most conserved (with TM7 having the highest conservation; supplementary figs. S3 and S4, Supplementary Material online). While not universal, many GRs from the major arthropod lineages, and even GRLs in nonarthropods, harbor the motif TYhhhhhQF (albeit often divergent) at the C-terminal end of TM7 (Robertson 2015). The motif is highly degenerate but still recognizable in some M. occidentalis GRs (Hoy et al. 2016), and is present in TuGR1, TuGR2, and TuGR3 (all TuGR-Cs). In contrast, the motif was not apparent (or too divergent to be easily recognized) in nearly all members of the expanded TuGR-A and TuGR-B clades. Within each of the TuGR A and B lineages, however, several residues are highly conserved or even nearly invariant in the C-terminal TMs (e.g., E and N residues in TuGR-Bs in the TM7 region; supplementary fig. S4, Supplementary Material online). Collectively, these analyses reveal that T. urticae harbors an exceptional repertoire of GRs that are highly divergent from those of other arthropods for which genome sequences are available.
Only a Few IRs
Ionotropic Receptors (IRs), which are related to ionotropic glutamate receptors (iGluRs) that play conserved roles in synaptic transmission, are implicated in chemosensing in diverse protostomes (Benton et al. 2009; Croset et al. 2010). Mining of the IR/iGluR family in the T. urticae genome uncovered 19 members, including one pseudogene, with conserved domain structures diagnostic of both iGluRs and IRs (supplementary table S2, Supplementary Material online) (Croset et al. 2010). As revealed by a phylogenetic analysis that included D. melanogaster, I. scapularis and M. occidentalis sequences (supplementary fig. S5, Supplementary Material online), the majority of T. urticae receptors fell in well-supported clades harboring iGluRs, with four, three, and seven TuiGluRs in clades with putative NMDA, AMPA, and kainate iGluRs, respectively. In contrast, a mere four T. urticae proteins are in the IR subfamily associated with chemosensation (supplementary fig. S5, Supplementary Material online), among the lowest reported to date in an arthropod genome. Three of these, TuIR1, TuIR3, and TuIR4, are present in a well-supported clade harboring D. melanogaster IR25a and its I. scapularis ortholog (IscaIR25a; Gulia-Nuss et al. 2016). The remaining IR (TuIR2) falls into a highly supported clade that includes D. melanogaster IR93a, as well as the previously reported I. scapularis IR93a ortholog (Gulia-Nuss et al. 2016).
A Lineage-Specific Expansion of T. urticae ENaCs Mirrors Those of TuGRs
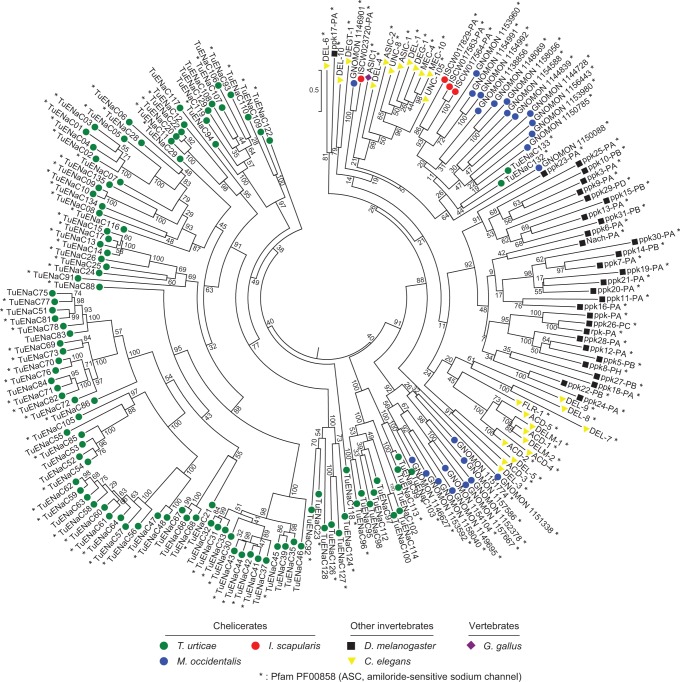
ENaCs have two TM domains with an intervening cysteine-rich extracellular loop, and are thought to function as multimeric complexes (Ben-Shahar 2011). With taxonomically diverse ENaC members including C. elegans proteins and D. melanogaster PPKs (Zelle et al. 2013), we identified homologs in the T. urticae genome (supplementary table S3, Supplementary Material online). In striking contrast to IRs, the T. urticae genome has 136 ENaCs of which 26 are pseudogenes and two are partial genes. A phylogenetic reconstruction with intact T. urticae ENaCs revealed two groups (fig. 2). Two T. urticae ENaCs—TuENaC132 and TuENaC133—fall into a clade with high bootstrap support that harbors C. elegans ENaCs, D. melanogaster PPKs, and chicken ASIC1, a vertebrate representative (Jasti et al. 2007). This suggests that TuENaC132 and TuENaC133 may have broadly conserved functional roles.
Fig. 2.—
Phylogeny of Tetranychus urticae ENaCs. A midpoint rooted maximum-likelihood tree of 108 intact T. urticae ENaCs along with those of other chelicerates (Ixodes scapularis and Metaseiulus occidentalis), the insect Drosophila melanogaster, and chicken ASIC1 is shown (species are as indicated at bottom). An asterisk denotes proteins for which the amiloride-sensitive sodium channel domain was detected as assessed with a HMMER search of all proteins included in the phylogeny (search criteria are as described in supplementary table S3, Supplementary Material online).
In contrast, the other 106 T. urticae ENaC members fall outside this clade (fig. 2). Unlike TuENaC132 and TuENaC133, which are intron rich with eight introns each, few of the remaining ENaCs harbored more than two introns, with 98 (92.5%) being intronless. Within this group, multiple highly supported clades of ENaCs with comparatively short branch lengths are apparent, a pattern suggesting recurrent, lineage-specific expansions mirroring that observed for T. urticae GRs (compare to fig. 1a and b and supplementary fig. S1, Supplementary Material online). The 106 ENaCs in the T. urticae specific clades are highly divergent from other animal ENaCs. For instance, while all D. melanogaster PPK proteins have the characteristic Pfam domain PF00858 (“ASC, amiloride-sensitive sodium channel”), the domain could be detected in only 75 of these T. urticae ENaCs (70.8% of the total; fig. 2 and supplementary table S3, Supplementary Material online). Nevertheless, T. urticae ENaCs with PF00858 are distributed within or subtending all T. urticae ENaCs clades, supporting membership in the family. Further, alignment of all T. urticae ENaCs with cASIC1, a vertebrate acid sensor for which a 3D structure has been established (Jasti et al. 2007), confirmed that domains and functionally important residues are conserved. These include the N- and C-terminal TM domains and invariant (or nearly invariant) cysteine residues in the extracellular loop. Additionally, key sequences in the “wrist” region at the interface of the predicted extracellular hand and the membrane spanning TMs are conserved (Jasti et al. 2007). These include residues in the PPPW sequence at positions 285–288 in cASIC1 for which the number of prolines can vary in other ENaCs, and for which the tryptophan can be a tyrosine (Jasti et al. 2007), a variation observed for most T. urticae ENaCs (supplementary fig. S6, Supplementary Material online).
To assess if the unprecedented expansion of divergent ENaCs in T. urticae is a general feature of chelicerates, we recovered putative full-length ENaCs from the genomes of I. scapularis and M. occidentalis (4 and 24 sequences were recovered, respectively; supplementary table S4, Supplementary Material online). None of these moderate number of I. scapularis and M. occidentalis sequences fell in clades harboring the 106 divergent T. urticae ENaCs, but rather are in the highly supported clade harboring TuENaC132, TuENaC133, and the ENaC sequences from other animals (fig. 2).
Genomic Organization of CRs in T. urticae
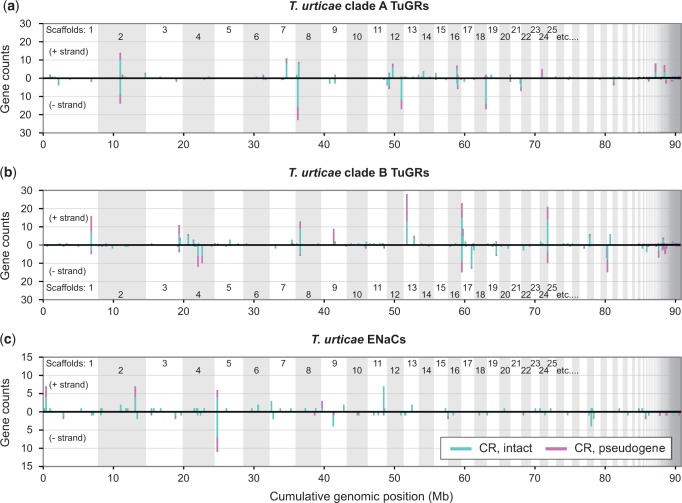
In both vertebrates and arthropods, the genomic organization of CRs has shed light on their origin and evolutionary dynamics (Nei et al. 2008). In T. urticae, the genes encoding both TuGR-As and TuGR-Bs are dispersed across the genome. Strikingly, as assessed with both intact GRs and pseudogenes, and allowing a single intervening non-CR gene to account for transpositions (or errors in the annotation), only 22.5% and 20.2% of TuGR-As and TuGR-Bs, respectively, are found as singletons. The remaining GRs are found in clusters distributed on many genome scaffolds (fig. 3a and b). Reflecting their independent expansions (fig. 1a and c), TuGR-A and TuGR-B clusters were observed at different genomic locations. Five clusters located on scaffolds 2, 8, 13, 17, and 24 harbored more than 20 GRs, with pseudogenes commonly located in the large clusters (fig. 3a and b). Interestingly, 62 TuGR genes are nested within the introns of 22 hosting genes, of which 17 are located within the large introns of tetur08g08289 (which encodes an unrelated protein). The expanded cluster of TuGRs within tetur08g08289 contributes to the size of this gene, which at 105 kb is the largest in the highly compact 90 Mb T. urticae genome.
Fig. 3.—
Genomic organization of Tetranychus urticae GRs and ENaCs. Genomic distribution of CRs by family or clade: (a) clade A TuGRs, (b) clade B TuGRs, and (c) ENaCs. In each case the distribution of CRs along the genome is shown with lengths of vertical line segments corresponding to counts in a gene cluster; gene counts for the forward (+) and reverse (−) strand orientations are as indicated. Clusters of CRs were calculated such that a given gene is represented only once, i.e., its count contributes to only one vertical line segment. Where clusters are observed, intact CRs are indicated by turquoise (line segments originating from the zero axis) with pseudogenes indicated in magenta (see legend). For plotting, partial genes were included in the pseudogene category. For display, the genome was concatenated from largest to smallest scaffolds (alternating scaffolds are indicated by shading).
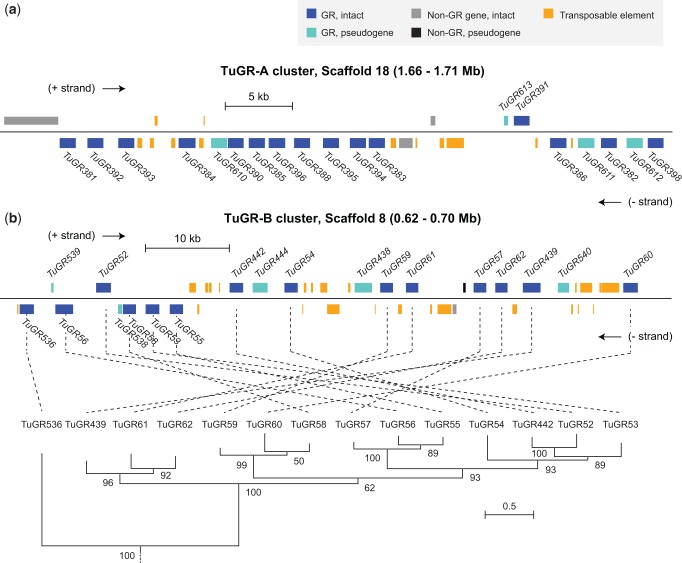
Within clusters, adjacent TuGR genes are typically found in head-to-tail orientations, e.g., as observed for a cluster on scaffold 18 that harbors 12 genes in such an arrangement beginning with TuGR381 and ending in TuGR383 (fig. 4a). However, many larger clusters also harbor genes in both orientations (figs. 3a and b and 4). An example of one such GR cluster is shown in figure 4b. While genes in this cluster are similar in sequence and are in the same well-supported, monophyletic clade—a general finding for TuGR clusters (supplementary fig. S7, Supplementary Material online)—the relationship between the genomic order of GR genes and the phylogenetic groupings suggests a complex history of intra-cluster rearrangements and potentially gene conversion among duplicate receptor genes. Strikingly, many GR clusters are also rich in transposable element (TE) sequences (fig. 4a and b).
Fig. 4.—
Structures of GR clusters in the Tetranychus urticae genome. Representative organization of GR clusters on (a) scaffold 18 (TuGR-As) and (b) scaffold 8 (TuGR-Bs). Genomic features are as indicated in the legend (top right). Within each cluster, genes above the black line are transcribed in the forward direction (+ strand), whereas genes below are in the reverse orientation (- strand). Introns are omitted for display, but all intact GRs indicated have the canonical intron structures for their respective GR clade, excepting TuGR439 that has an additional intron. For the TuGR-B cluster (b), the phylogenetic relationships among GRs is as shown at the bottom (all of the TuGR-B genes correspond to a monophyletic clade as taken from the phylogeny of all TuGRs, see supplementary fig. S1, Supplementary Material online).
We also examined genomic distributions for T. urticae ENaCs which, while less abundant than TuGRs, have as many members as the combined number of ORs and GRs in many insects (Nei et al. 2008). Albeit less extreme, the patterns observed for T. urticae ENaCs mirror those of TuGRs (fig. 3c and supplementary figs. S7 and S8, Supplementary Material online). While genes encoding ENaCs are distributed throughout the T. urticae genome, and 52.2% are found as singletons (a 2.4-fold increase over that observed for TuGRs), 65 genes, including those encoding ENaCs in intronless T. urticae specific clades, are found in clusters. The largest of these includes 17 genes on scaffold 5. As observed for GRs, ENaC pseudogenes were commonly observed in clusters, some of which harbor substantial insertions of TE sequences (fig. 3 and supplementary fig. S8, Supplementary Material online).
The remaining T. urticae CR genes that encode the small TuGR-C clade and the IR family were found throughout the genome as singletons (excepting TuGR5, TuGR510 and TuGR543, which are nearby each other on scaffold 11).
Intra-Specific Variation in T. urticae CRs
High birth and death rates characterize many large gene families in animals and plants (Michelmore and Meyers 1998; Thomas 2006; Clark et al. 2007). Several observations suggest a similar scenario for T. urticae GRs and ENaCs. For instance, for both T. urticae GRs and ENaCs, many pseudogene reconstructions are full length or nearly so (fig. 4 and supplementary fig. S8 and table S1, Supplementary Material online). In fact, ∼20% of GRs annotated as pseudogenes harbor only one or a few inactivating mutations (e.g., a frameshift or stop codon; supplementary table S1, Supplementary Material online). This pattern is indicative of very recent inactivation, and therefore suggests that inactivating changes may segregate in T. urticae populations, potentially contributing to phenotypic variation.
To test the former, we examined low-coverage resequencing data for two additional T. urticae strains, Montpellier (maintained in Europe) (Grbić et al. 2011) and EtoxR (from Japan) (Van Leeuwen et al. 2012); the reference strain, London, originated from Canada (Grbić et al. 2011). A limitation of the Montpellier and EtoxR strain data is that the strains were sequenced with single end or short 36 bp paired end reads, and were not inbred (Grbić et al. 2011; Van Leeuwen et al. 2012), a combination that makes larger structural variant prediction unreliable. Restricting our analysis to SNP and small indel changes, and visual inspection of aligned sequences to the London reference, we found that 21 TuGRs that were pseudogenes in the London strain appear to be intact in the Montpellier strain, the EtoxR strain, or in both (supplementary table S5, Supplementary Material online). In a few cases, allelic variation for inactivating sequence changes was observed within these strains. Conversely, 10 TuGR genes that are intact in the London strain appear to be pseudogenes, or to segregate for inactivating mutations, in the Montpellier and EtoxR strains. ENaCs also show allelic variation. For example, while TuENaC13 is an intact gene in the London reference sequence, and TuENaC18 is a pseudogene, both segregate for putative functional and nonfunctional copies in the Montpellier strain.
Expression of Chemosensory Receptors
Comparatively little is known about the regulation of CR expression in arthropods. To understand the expression dynamics of T. urticae GRs and ENaCs, including sex-specific expression differences, we generated deep, strand-specific RNA-seq data with 4-fold biological replication from stage-matched adult males and females. The RNA was collected from whole bodies, as dissection of putative mite chemosensory structures (Bostanian and Morrison 1973), which has been performed for insects (e.g., Matthews et al. 2016), was deemed not feasible (adult female mites are only ∼500 microns in length, and males are substantially smaller).
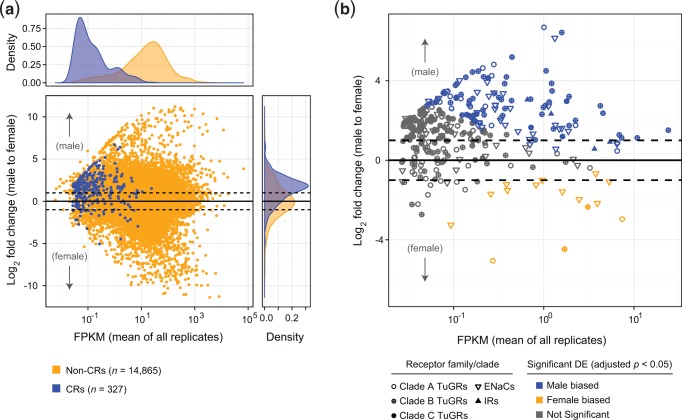
In insects, most CRs are expressed in a small number of neurons in sensory structures like the antenna, labellum, or legs (Benton 2015; Joseph and Carlson 2015). A prediction is therefore that CR expression would be very low given that whole mites were sampled for RNA-seq (i.e., chemosensory neurons account for a minute fraction of all cells). We therefore initially assessed expression by combining data across all sexes and replicates. As opposed to most non-CR coding genes, many TuGR-As and TuGR-Bs either lacked expression support entirely, or were supported by a tiny number of aligned RNA-seq reads (fig. 5 and supplementary tables S1 and S6, Supplementary Material online). The low number of RNA-seq reads for many TuGR genes raises the question of whether the expression is specific (e.g., a small number of reads could reflect genomic contamination). However, even for genes supported by 5 or fewer read pairs, spliced reads were identified in 30.8% of cases, and the canonical intron structures associated with both TuGR-As and TuGR-Bs (fig. 1c) were overwhelmingly supported. Strikingly, the pattern observed for TuGRs was mirrored for most ENaCs, where a substantial fraction of genes were either expressed at a low level, or not expressed (fig. 5 and supplementary table S3, Supplementary Material online).
Fig. 5.—
Expression levels and sex specificity of expression of CRs. (a) Expression levels of intact (nonpseudogenized) GRs, ENaCs, and IRs as assessed against other coding genes and as a function of sex. Most CRs are lowly expressed (density plot at top; see also supplementary tables S1–S3 and S6, Supplementary Material online) and with higher levels in male samples (right). (b) Expression of CR genes shown in panel a broken down by CR family or clade with significant sex-specific expression as indicated (a false discovery rate of 5% was used). For genes in both plots, only those passing the criteria for estimation of differential expression by DESeq2 are shown (supplementary table S6, Supplementary Material online).
Low (or no) expression precluded tests of differential expression between sexes for 41.5% of CRs (supplementary table S6, Supplementary Material online). Where differential expression could be assessed, 19 of 79 TuGR-As (24.1%), 61 of 164 TuGR-Bs (37.2%), 4 of 7 TuGR-Cs, 43 of 73 ENaCs (58.9%), and all four IRs were significantly differentially expressed (fig. 5 and supplementary table S6, Supplementary Material online). Relative to non-CR genes, differential expression across all CR families was moderately male biased (fig. 5). However, for most CR genes we believe it is unlikely that this reflects biologically relevant sex-specific expression differences in the neurons of chemosensory structures. Briefly, males are markedly smaller than females, for which much of the body mass is associated with the female reproductive system, so neuronal contributions to whole-body RNA are almost certainly proportionally less in female mites. Supporting this conjecture, homologs of more general, well-characterized C. elegans neuronal markers also tended to be higher in males with log2 fold changes of ∼1.0–3.0 (supplementary table S7, Supplementary Material online), a fold-change range consistent with that observed for a large fraction of CRs (fig. 5). Nevertheless, a moderate number of CRs—including both GRs and ENaCs—are candidates to mediate sex-specific behaviors as they exhibited highly sex-specific expression (fig. 5b and supplementary tables S6, Supplementary Material online).
Within genomic clusters, expression levels of both GRs and ENaCs vary greatly. In particular, although a moderate number of CRs within clusters are robustly expressed, and/or exhibit sex biases in expression, many are either not expressed at all or expressed at very low levels (fig. 1b and supplementary tables S1 and S3, Supplementary Material online). Finally, understanding the spatial and temporal regulation of CR expression has attracted much interest (Benton 2015). In this context, we observed substantive antisense expression at some CR loci (supplementary tables S1–S3, Supplementary Material online), potentially suggesting the presence of noncoding antisense RNAs that have been implicated in the control of gene expression in some other organisms (Ietswaart et al. 2012).
Discussion
Almost 700 GRs, including pseudogenes, are present in the T. urticae genome assembly. This number exceeds the count (ORs and GRs together) observed in the most GR family member-rich insect genomes, like that of T. castaneum (586 receptors) (Tribolium Genome Sequencing Consortium et al. 2008) and ant species (i.e., 470 receptors in Camponotus floridanus; Zhou et al. 2015). Therefore, the comparatively modest number of GRs annotated in currently available genomes from Crustacea (D. pulex; Peñalva-Arana et al. 2009), Myriapoda (S. maritima; Chipman et al. 2014), and Chelicerata (e.g., I. scapularis and M. occidentalis; Gulia-Nuss et al. 2016; Hoy et al. 2016) are not representative of all noninsect arthropods. Mirroring patterns of lineage-specific expansions of GRs observed among taxa (Robertson et al. 2003; Peñalva-Arana et al. 2009; Chipman et al. 2014; Zhou et al. 2015; Gulia-Nuss et al. 2016; Hoy et al. 2016), the lineage-specific expansions of the TuGR-As and TuGR-Bs within T. urticae are also striking. These two lineages are nearly as diverse from each other in sequence and intron structure as they are to other arthropod GRs, as well as to GRLs of more basal animals (Benton 2015; Robertson 2015). In particular, the TYhhhhhQF motif in TM7 that is present in many arthropod GRs—including M. occidentalis GRs (albeit divergent) (Hoy et al. 2016)—is recognizable in only a handful of TuGRs.
In contrast to other arthropod species, including the chelicerates I. scapularis and M. occidentalis (Gulia-Nuss et al. 2016; Hoy et al. 2016), T. urticae has few IRs, three of which are related to D. melanogaster IR25a, with the fourth falling in a clade with IR93a. Only the IR25a lineage is widely present in protostome species (Croset et al. 2010), with IR93a (or IR93a-like proteins) broadly distributed among the major arthropod subphyla (Croset et al. 2010; Chipman et al. 2014; Gulia-Nuss et al. 2016; Hoy et al. 2016). These patterns are consistent with a limited contribution of IRs to the perception of chemical cues in T. urticae. In contrast to the paucity of IRs, an unanticipated finding is that ENaCs have expanded greatly in T. urticae. In particular, a divergent lineage of 106 ENaCs is reminiscent of, albeit far more extreme than, the expansion observed for PPKs in D. melanogaster, for which diversification to facilitate signaling by unknown ligands has been postulated (Zelle et al. 2013). By analogy, T. urticae ENaCs may contribute prominently to the species’ chemosensory ability. Although speculative, our findings raise the possibility that the prominent role in chemoreception played by IRs in other arthropods has been co-opted in T. urticae by ENaCs (in concert with, potentially, the expanded set of GRs).
The genome organization of T. urticae CRs provides additional insights into their evolution. The widespread genomic distribution of GRs suggests an ancient proliferation in the genome, whereas the large clusters are consistent with recent bouts of proliferation. This latter pattern contrasts with that observed in insects like D. melanogaster, for which ORs and GRs are dispersed in the genome in isolation or as small clusters, suggestive of more ancient proliferative episodes (Robertson et al. 2003). In this sense, genomic patterns for T. urticae GRs more closely resemble those observed for vertebrate ORs or C. elegans CRs for which a high percentage of receptors are in large clusters, many are pseudogenes, and gene conversion may play a role in generating diversity (Robertson 2000, 2001; Glusman et al. 2001; Zhang and Firestein 2002; Niimura et al. 2014; Jiang and Matsunami 2015). For T. urticae GRs, these dynamics may have been facilitated by their compact structure, i.e., few introns of small size, which may have facilitated pairing and nonallelic homologous recombination between duplicates to produce copy number variation (Hastings et al. 2009). Further, in many cases TEs are inserted into T. urticae GR clusters. As TEs can facilitate genomic rearrangements (Cordaux and Batzer 2009), they may have played a role in the observed structural complexity of some large TuGR clusters (e.g., intra-cluster rearrangements). Although the T. urticae ENaC family is smaller than the GR family, the patterns of genomic organization are strikingly similar to GRs, indicative of related underlying evolutionary dynamics.
More generally, TE insertions are often deleterious for host genes via coding sequence disruption or effects on expression (Cordaux and Batzer 2009). This likely reflects relaxed selection on individual T. urticae GRs, consistent with their high rate of pseudogenization, and relaxation of purifying selection has been well established for CRs in both vertebrates and invertebrates (Nei et al. 2008). In agreement with this, many T. urticae CRs appear to be either not expressed or expressed at a low level. Some genes in CR clusters do have robust expression, however, consistent with functional roles. This provides a potential explanation for why CR clusters—even those rich in pseudogenes—are not lost en masse by large deletions. It is noteworthy that a small number of CRs in different families are expressed in a highly sex-specific manner. These may mediate sex-specific behaviors, i.e., CRs with male-specific expression may perceive pheromones produced by T. urticae females (Oku et al. 2015). In contrast, receptors expressed higher in females may detect overcrowding or deteriorating plant hosts, conditions that elicit female-specific dispersal behaviors (Smitley and Kennedy 1985). In addition, we note that a small number of TuGRs are expressed highly in both sexes. Whether these serve as co-receptors in multimeric complexes, a documented role for Orco in insects (Carraher et al. 2015), warrants additional investigation. Further, some GRs may perform roles other than perception of environmental cues, as observed for a small number of GRLs (Benton 2015; Saina et al. 2015), and a subset of T. urticae ENaCs likely also have other roles (e.g., in mechanosensing; Kellenberger and Schild 2002; Ben-Shahar 2011).
Nevertheless, adjusting for morphological differences, most T. urticae CRs show little evidence for sex-specific expression. This contrasts with that observed for ORs in some insects with sex-divergent behaviors or ecology (Andersson et al. 2014). However, males and females in T. urticae populations share similar abiotic and biotic environments, consistent with CR expression in both sexes if most CRs perceive shared environmental cues. It is currently unknown if the proliferations of GRs and ENaCs observed in T. urticae are a general feature of herbivorous acariform mites. However, while a phylogenetic signal is possible, the proliferation of T. urticae CRs and shared expression between sexes may reflect the action of selection associated with the species’ extraordinarily broad plant host range. It is noteworthy that studies with Drosophila genomes have suggested that CR losses, coupled with positive selection on a subset of receptors, correlates with the evolution of specialization (McBride 2007; McBride et al. 2007; Whiteman and Pierce 2008), although demographic factors may confound this conclusion (Gardiner et al. 2008). Further, changes in a small number of ORs are associated with a dramatic host shift from microbe feeding to specialist herbivory in flies in the Scaptomyza genus (Goldman-Huertas et al. 2015).
As compared with generalist drosophilid flies, the host range for T. urticae is far more extreme (Migeon et al. 2010). In T. urticae, gene families associated with plant secondary compound detoxification have undergone major expansions (Grbić et al. 2011; Dermauw et al. 2013; Van Leeuwen and Dermauw 2016). Additionally, the rate of the evolution of pesticide resistance in T. urticae is exceptional, a correlate of robust detoxification pathways (Dermauw et al. 2013). Given these observations, and the limited mobility of mites, a plausible a priori hypothesis is that perception of diverse chemical cues might be comparatively unimportant in T. urticae. However, while detoxification pathways are undoubtedly critical, our data nonetheless suggest a major role for chemosensory perception. One area of future investigation is to unravel the roles that CRs play, either by behavioral (indirect) mechanisms or possibly more direct chemical cue coupled signaling, to affect the large-scale changes in T. urticae detoxification gene networks that have been documented to occur upon plant host shifts (Grbić et al. 2011; Dermauw et al. 2013).
Conclusions
Our characterization of T. urticae CRs adds to a growing body of evidence that lineage-specific expansions of CRs—both within and between receptor families—feature prominently in fulfilling chemosensory roles among diverse animal taxa. However, establishing the relevance of given expansions to life history traits remains a challenge. In this context the Tetranychus genus affords many opportunities. In addition to the extreme generalist T. urticae, extant sister species are present across the entire host range spectrum. For example, T. evansi feeds on many species within one plant family (Solanaceae), while T. lintearius and T. ezoensis are extreme specialists that feed predominantly on a single plant species (Migeon et al. 2010). Therefore, genomic studies with sister species should shed light on the impact of plant host range on CR dynamics. Further, T. urticae populations have been documented to vary in host plant performance (Fellous et al. 2014, and references therein), and our work shows that T. urticae CRs can vary (intact or disrupted) among populations. This provides an exciting opportunity to understand the microevolutionary forces underlying both the diversification as well as the maintenance of CRs in a genetically tractable herbivore.
Supplementary Material
Supplementary figures S1–S8 and tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Erik Jorgensen for suggesting neuronal marker genes, and Vojislava Grbić for helpful comments on the article. This work was funded by the National Science Foundation (DEB-1457346 to R.M.C.); the Fund for Scientific Research Flanders (FWO) (G009312N to T.V.L., G053815N to T.V.L. and W.D., and B/08974 to P.C.T.N.); the Genome Canada and the Ontario Genomics Institute (OGI-046 to M.G.); the Ontario Research Fund–Global Leadership in Genomics and Life Sciences (GL2-01-035 to M.G.); and the National Sciences and Engineering Research Council of Canada (STPGP322206-05 to M.G.). R.G. was funded by National Institutes of Health Genetics Training Grant T32 GM07464. W.D. is a postdoctoral fellow of the Fund for Scientific Research Flanders. The statements herein represent the views of the authors, and not necessarily those of the funding agencies.
Literature Cited
- Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y. 2012. GenomeView: a next-generation genome browser. Nucleic Acids Res. 40:e12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MN, et al. 2014. Sex-and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae). BMC Genomics 15:501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazopoulou D, Voglis G, Tavernarakis N. 2007. The role of DEG/ENaC ion channels in sensory mechanotransduction In: Wang DH, editor. Molecular sensors for cardiovascular homeostasis. New York, US: Springer; p. 3–31. [Google Scholar]
- Ben-Shahar Y. 2011. Sensory functions for degenerin/epithelial sodium channels (DEG/ENaC). Adv Genet. 76:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R. 2015. Multigene family evolution: perspectives from insect chemoreceptors. Trends Ecol Evol. 30:590–600. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanian NJ, Morrison FO. 1973. Morphology and ultrastructure of sense organs in the twospotted spider mite (Acarina: Tetranychidae). Ann Entomol Soc Am. 66:379–383. [Google Scholar]
- Cande J, Prud’homme B, Gompel N. 2013. Smells like evolution: the role of chemoreceptor evolution in behavioral change. Curr Opin Neurobiol. 23:152–158. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraher C, et al. 2015. Towards an understanding of the structural basis for insect olfaction by odorant receptors. Insect Biochem Mol Biol. 66:31–41. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wang Q, Wang Z. 2010. The amiloride-sensitive epithelial Na+ channel PPK28 is essential for Drosophila gustatory water reception. J Neurosci Off J Soc Neurosci. 30:6247–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman AD, et al. 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, et al. 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317:338–342. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. 2000. Candidate taste receptors in Drosophila. Science 287:1830–1834. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, et al. 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermauw W, et al. 2013. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc Natl Acad Sci. 110:E113–E122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Meister S, McNealy C, Amrein H. 2001. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol. 11:822–835. [DOI] [PubMed] [Google Scholar]
- Dunlop JA. 2010. Geological history and phylogeny of Chelicerata. Arthropod Struct Dev. 39:124–142. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform Int Conf Genome Inform. 23:205–211. [PubMed] [Google Scholar]
- Edgecombe GD, Legg DA. 2014. Origins and early evolution of arthropods. Palaeontology 57:457–468. [Google Scholar]
- Fellous S, et al. 2014. Combining experimental evolution and field population assays to study the evolution of host range breadth. J Evol Biol. 27:911–919. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler MA, Montell C. 2013. Drosophila TRP channels and animal behavior. Life Sci. 92:394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman EG, Dahanukar A. 2015. Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol. 34:140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman EG, Wisotsky Z, Dahanukar A. 2014. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc Natl Acad Sci. 111:1598–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner A, Barker D, Butlin RK, Jordan WC, Ritchie MG. 2008. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol Ecol. 17:1648–1657. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. 2001. The complete human olfactory subgenome. Genome Res. 11:685–702. [DOI] [PubMed] [Google Scholar]
- Goldman-Huertas B, et al. 2015. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc Natl Acad Sci. 112:3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M, et al. 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009. Mechanisms of change in gene copy number. Nat Rev Genet. 10:551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. 2008. Plant immunity to insect herbivores. Annu Rev Plant Biol. 59:41–66. [DOI] [PubMed] [Google Scholar]
- Hoy MA, et al. 2016. Genome sequencing of the phytoseiid predatory mite Metaseiulus occidentalis reveals completely atomised Hox genes and super-dynamic intron evolution. Genome Biol Evol. 8:1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C. 2012. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28:445–453. [DOI] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. 2007. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449:316–323. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Matsunami H. 2015. Mammalian odorant receptors: functional evolution and variation. Curr Opin Neurobiol. 34:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445:86–90. [DOI] [PubMed] [Google Scholar]
- Joseph RM, Carlson JR. 2015. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31:683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 338:1027–1036. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB. 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 11:188–200. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, Schild L. 2002. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 82:735–767. [DOI] [PubMed] [Google Scholar]
- Krainacker DA, Carey JR. 1989. Reproductive limits and heterogeneity of male twospotted spider mites. Entomol Exp Appl. 50:209–214. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Liu L, et al. 2003. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron 39:133–146. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, LaMora A, Sun Y, Welsh MJ, Ben-Shahar Y. 2012. ppk23-dependent chemosensory functions contribute to courtship behavior in Drosophila melanogaster. PLoS Genet. 8:e1002587.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, McBride CS, DeGennaro M, Despo O, Vosshall LB. 2016. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics 17:32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS. 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci U S A. 104:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR, O’Meara BC. 2007. Five drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177:1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8:1113–1130. [DOI] [PubMed] [Google Scholar]
- Migeon A, Nouguier E, Dorkeld F. 2010. Spider mites web: a comprehensive database for the Tetranychidae Trends in Acarology. In: Sabelis M, Bruin J, editors. Proceedings of the 12th International Congress. Dordrecht, The Netherlands: Elsevier. 557–560. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees In: Gateway Computing Environments Workshop (GCE), 2010. IEEE; Red Hook, NY: Curran Associates inc. p. 1–8. [Google Scholar]
- Montagné N, de Fouchier A, Newcomb RD, Jacquin-Joly E. 2015. Advances in the identification and characterization of olfactory receptors in insects In: Glatz R, editor. Progress in molecular biology and translational science. Vol. 130 London, UK: Elsevier; p. 55–80. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 9:951–963. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku K, Weldegergis BT, Poelman EH, De Jong PW, Dicke M. 2015. Altered volatile profile associated with precopulatory mate guarding attracts spider mite males. J Chem Ecol. 41:187–193. [DOI] [PubMed] [Google Scholar]
- Peñalva-Arana DC, Lynch M, Robertson HM. 2009. The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol. 9:79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Shi X, Kadowaki T. 2015. Evolution of TRP channels inferred by their classification in diverse animal species. Mol Phylogenet Evol. 84:145–157. [DOI] [PubMed] [Google Scholar]
- Robertson HM. 2000. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 10:192–203. [DOI] [PubMed] [Google Scholar]
- Robertson HM. 2001. Updating the str and srj (stl) families of chemoreceptors in Caenorhabditis nematodes reveals frequent gene movement within and between chromosomes. Chem Senses 26:151–159. [DOI] [PubMed] [Google Scholar]
- Robertson HM. 2009. The insect chemoreceptor superfamily in Drosophila pseudoobscura: molecular evolution of ecologically relevant genes over 25 million years. J Insect Sci Online 9:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. 2015. The insect chemoreceptor superfamily is ancient in animals. Chem Senses 40:609–614. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci. 100:14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz R, Croset V, Benton R. 2013. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 43:888–897. [DOI] [PubMed] [Google Scholar]
- Saina M, et al. 2015. A cnidarian homologue of an insect gustatory receptor functions in developmental body patterning. Nat Commun. 6:6243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gracia A, Vieira FG, Rozas J. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103:208–216. [DOI] [PubMed] [Google Scholar]
- Sato K, et al. 2008. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452:1002–1006. [DOI] [PubMed] [Google Scholar]
- Scott K, et al. 2001. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104:661–673. [DOI] [PubMed] [Google Scholar]
- Smart R, et al. 2008. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 38:770–780. [DOI] [PubMed] [Google Scholar]
- Smitley DR, Kennedy GG. 1985. Photo-oriented aerial-dispersal behavior of Tetranychus urticae (Acari: Tetranychidae) enhances escape from the leaf surface. Ann Entomol Soc Am. 78:609–614. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck L, Billiau K, Abeel T, Rouzé P, Van de Peer Y. 2012. ORCAE: online resource for community annotation of eukaryotes. Nat Methods 9:1041–1041.. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, et al. 2015. Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst Biol. 64:778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrapon N, et al. 2014. Molecular traces of alternative social organization in a termite genome. Nat Commun. 5:3636.. [DOI] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. 2012. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149:1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH. 2005. Analysis of homologous gene clusters in Caenorhabditis elegans reveals striking regional cluster domains. Genetics 172:127–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH. 2006. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 16:1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda H, Zhao X, Dickson BJ. 2012. The Drosophila female aphrodisiac pheromone activates ppk23+ sensory neurons to elicit male courtship behavior. Cell Rep. 1:599–607. [DOI] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. 2009. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 71:307–332. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium, et al. 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, Dermauw W. 2016. The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu Rev Entomol. 61:475–498. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, et al. 2012. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc Natl Acad Sci. 109:4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen T, Tirry L, Yamamoto A, Nauen R, Dermauw W. 2015. The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pestic Biochem Physiol. 121:12–21. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Pierce NE. 2008. Delicious poison: genetics of Drosophila host plant preference. Trends Ecol Evol. 23:473–478. [DOI] [PubMed] [Google Scholar]
- Zelle KM, Lu B, Pyfrom SC, Ben-Shahar Y. 2013. The genetic architecture of degenerin/epithelial sodium channels in Drosophila. G3 GenesGenomesGenetics 3:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. 2002. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 5:124–133. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. 2015. Chemoreceptor evolution in hymenoptera and its implications for the evolution of eusociality. Genome Biol Evol. 7:2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.