Abstract
Leber congenital amaurosis (LCA4) has been linked to mutations in the photoreceptor-specific gene Aryl hydrocarbon interacting protein like 1 (Aipl1). To investigate the essential role of AIPL1 in retina, we generated a mouse model of LCA by inactivating the Aipl1 gene. In Aipl1–/– retinas, the outer nuclear layer develops normally, but rods and cones then quickly degenerate. Aipl1–/– mice have highly disorganized, short, fragmented photoreceptor outer segments and lack both rod and cone electroretinogram responses. Recent biochemical evidence indicates that AIPL1 can enhance protein farnesylation. Our study reveals that rod cGMP phosphodiesterase, a farnesylated protein, is absent and cGMP levels are elevated in AIPL1–/– retinas before the onset of degeneration. Our findings demonstrate that AIPL1 enhances the stability of phosphodiesterase and is essential for photoreceptor viability.
Leber congenital amaurosis (LCA) has the earliest onset and is the most severe form of inherited retinopathy in humans. LCA is genetically heterogeneous and is generally inherited in an autosomal recessive fashion. LCA is characterized by complete blindness and the near absence of electrical responses to light within 1 year of birth. LCA has been linked to mutations in the gene encoding Aipl1 (1–3). The vast majority of these mutations in Aipl1 are linked to LCA, but a few C-terminal mutations are linked to two other retinal diseases, cone–rod dystrophy and juvenile retinitis pigmentosa (3).
AIPL1 is expressed only in retina and pineal gland (1). In adult mouse retina, it is found within the outer plexiform layer and the inner segments of photoreceptors; in humans, AIPL1 is expressed in both developing rods and cones and mature rods (4, 5). Two possibilities for the essential role for AIPL1 in retina have been proposed (4). The first is that AIPL1 enhances an essential farnesylation reaction. Farnesylation is a specific type of prenylation, the addition of a farnesyl or geranylgeranyl residue to specific proteins. Several retinal proteins, cGMP phosphodiesterase (PDE), transducin, and rhodopsin (Rho) kinase (RK) are known to be farnesylated (6–9). Prenylation of retinal proteins is required for maintenance of retinal cytoarchitecture and photoreceptor structure. Inhibition of prenylation causes degeneration of photoreceptor outer segments (10). Prenylation also greatly enhances the stability of cGMP PDE, a protein essential for photoreceptor survival (11). Prenylation of PDE is also necessary for its membrane association (11). These studies suggest that AIPL1 is necessary for the maintenance of photoreceptors. The second possible essential role for AIPL1 is the control of photoreceptor proliferation and or differentiation. AIPL1 interacts with Nedd8-ultimate buster 1 (NUB1), a ubiquitously expressed protein thought to play an important role in regulating cell cycle progression (12). Based on the NUB1 interaction, early expression of AIPL1 in human retina, and the severity of AIPL1 mutations linked to LCA, it has been proposed that AIPL1 is essential for the initial development of photoreceptors (5).
To understand at the molecular level why a deficiency of AIPL1 causes LCA, we disrupted the Aipl1 gene in mice. In Aipl1 deficient mice, the initial development of photoreceptors proceeds normally. However, once outer segments begin to develop, photoreceptors undergo rapid and severe degeneration. In agreement with the characteristics of LCA, retinas from mice lacking AIPL1 produce no electrical responses to light. The biochemical basis for the rapid retinal degeneration appears to be destabilization of cGMP PDE.
Methods
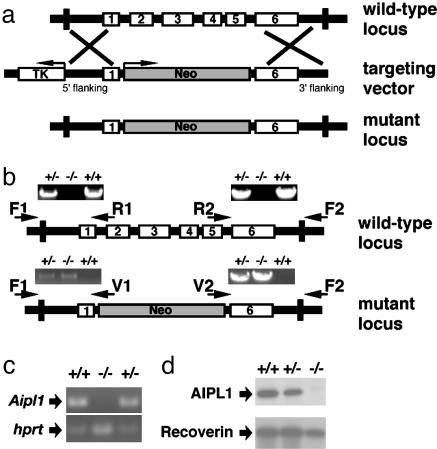
Generation of Aipl1-Targeted Mice. Genomic fragments containing the flanking regions of exon 1 (5.7 kb) and exon 6 (4.1 kb) of Aipl1 from genomic DNA of mouse strain 129Sv were amplified by PCR. To construct the targeting vector, we cloned the 5′ genomic flanking sequence (5.3 kb) as an HpaI/KpnI fragment and cloned the 3′ genomic flanking sequence (2.9 kb) as a PmeI/NotI fragment in pCG4619 targeting vector (gift of R. Palmiter, University of Washington). The targeting vector was linearized and electroporated into R1 embryonic stem (ES) cells, and neomycin-resistant colonies were selected. A correctly targeted clone, VR4, was identified by PCR analysis and confirmed by Southern blotting. ES cells heterozygous for the targeted mutation (VR4) were microinjected into C57BL/6J blastocysts to generate chimeras. The chimeras were mated with C57BL/6J females to obtain progeny that were heterozygous for the mutant allele. Heterozygous animals were inbred to generate homozygous animals that lack a functional Aipl1 gene. The targeted allele was maintained in 129/SvJ and C57BL/6 mixed backgrounds. Mice carrying the mutant allele were identified by PCR. Primers used to amplify wild-type allele from the N terminus of Aipl1 were F1 (5′-CTGGGGAGGTCAAAAGGTCATCAAAGT-3′) and R1 (5′-AAGAACCACGGGGTCCTATCT-3′) and from the C terminus were R2 (5′-CTGCTGAAGAAGGAGGAGTACTATG-3′) and F2 (5′-GGGCTAGAAGGCCTGTAGTCAT-3′). Primers used to amplify the knockout allele from the N terminus were F1 and V1 (5′-CTTTCCACACCCTAACTGACACAC-3′) and from the C terminus were V2 (5′-ATCGCCTTCTATCGCCTTCTTGAC-3′) and F2.
RNA Isolation and RT-PCR. Total RNA from a whole eye was isolated by TRIzol reagent (Invitrogen), following the manufacturer's guidelines. Oligo(dT)-primed reverse transcription reactions were performed with 1 μg of total RNA by using SuperScript II (Invitrogen) to obtain cDNA, which was used as a template in PCRs. Aipl1 was amplified with primers 5′-CTAGCCCAGCATGCTCCGGCA-3′ and 5′-CAGAAAGAGCCACAGCCTCTTGTC-3′, which yielded a product of 200 bp. PCR conditions for all of the reactions were 95°C for 1 min followed by 95°C for 45 sec, 55°C for 45 sec, and 72°C for 45 sec, for 30 cycles. Hprt, which was used as a control, was amplified by using primers 5′-CAAACTTTGCTTTCCCTGGT-3′ and 5′-CAAGGGCATATCCAACAACA-3′, and yielded a 250-bp product. For analyzing Pdeα transcripts, we used primers 5′-CTGGTTCTTTAACTGTCCAGTGCCA-3′ and 5′-ATGAGGAGATTACACCCATGCT-3′, which amplified a 300-bp product. Primers 5′-ACCAAATTGCTATAGGCAGAGTCC-3′ and 5′-AGCTGACGAGTATGAGGCCAAAGTCAAG-3′ amplified a 300-bp fragment of Pdeβ. Full-length Pdeγ was amplified by using 5′-CTGACAGAGTCCAGAAGCTAAGG-3′ and 5′-CTAGGGACTCAGGCTCAGGTTT-3′. All amplified products were confirmed by sequencing. For all experiments described in this study, we used littermates from heterozygous matings for heterozygous or wild-type controls.
Histology. Mouse eyes were enucleated under room illumination and fixed in Carnoy's fixative overnight at 4°C. Fixed whole eyes were embedded in paraffin and sectioned by using an ultramicrotome.
Immunohistochemistry. Mouse eyes were enucleated under room illumination and incubated in PBS with 4% paraformaldehyde at room temperature. Retinas were dissected after 10 min, and the incubation in 4% paraformaldehyde was continued at room temperature for 1 h. Fixed retinas were cryoprotected in 30% sucrose/PBS overnight at 4°C and frozen in optimal cutting temperature compound (OCT) on dry ice. Sections (10 μm) were cut and mounted on Superfrost Plus slides (Fisher Scientific) and stored at –20°C. For immunohistochemistry, slides were washed in PBS and incubated for 2 h in primary antibody. After extensive washing in PBS, slides were incubated in secondary antibody (Alexa anti-mouse 488 and Alexa anti-rabbit 568, 1:1,000; Molecular Probes) for 1.5 h. All incubations were done at room temperature in 5% goat serum/0.2% Triton X-100/PBS. 4′,6-Diamidino-2-phenylindole (DAPI) was used to visualize nuclei, and slides were cover-slipped with Fluoromount-G (Southern Biotechnology Associates).
Electron Microscopy. Enucleated mouse eyes were fixed with a mixture of 2.5% glutaraldehyde/2% paraformaldehyde in phosphate buffer (0.1 M, pH 7.4) for 10 min at room temperature. Eyecups were made by removing the cornea and the lens, and were fixed overnight at 4°C. The eyecups were postfixed in osmium tetroxide and embedded in Epon. Sections (0.1 μm) were cut from these blocks and used for electron microscopy.
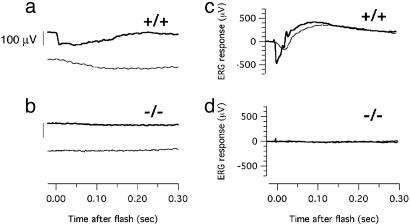
Electroretinography. A gold wire/contact lens electrode held on the cornea with 2–3% methylcellulose was used with a reference electrode in the mouth to record electroretinograms (ERGs). Mice were anesthetized with xylazine and ketamine and maintained on a heating block at 37°C. Light flashes from a photographic flash unit were delivered through a fiber optic bundle with a lens focused on the cornea. Stimuli were attenuated with neutral density filters. The output from the two intensities of white light flashes used in Fig. 3 were 300 nJ/cm2 and 300 μJ/cm2 measured at the cornea. ERG potentials were amplified ×1,000, filtered between 0.1 and 3,000 Hz, and digitized at 5,000 Hz. Data were collected in igor pro (Wavemetrics, Lake Oswego, OR) with an Instrutech ITC-16 analog to digital interface using a library of custom acquisition routines written by Fred Rieke (University of Washington).
Fig. 3.
Electroretinography. Responses to moderate (300 nJ/cm2, lighter traces) and bright (300 μJ/cm2, darker traces) white light flashes were recorded from P12 Aipl1+/+ (a), P12 Aipl1–/– (b), P18 Aipl1+/+ (c), and P18 Aipl1–/– (d) retinas. (Scale bar in a and b represents 100 μV). No ERG responses could be elicited from any of the AIPL1–/– mice examined (n = 4 for each developmental stage).
Western Blotting. Whole eyes or retinas were homogenized in ROS buffer (20 mM Hepes, pH 7.4/60 mM KCl/2 mM MgCl2/30 mM NaCl/100 μM PMSF/1 mM DTT/5 mM 2-mercaptoethanol) and a mixture of protease inhibitors (Roche Diagnostics). Homogenized tissues were solubilized in 1× SDS lysis buffer (62.5 mM Tris·HCl, pH 6.8/2% SDS/10% glycerol/0.005% bromophenol blue/5% 2-mercaptoethanol) by heating the samples at 100°C for 5 min. Equal amounts (50 μg) of sample were separated by SDS/PAGE gel electrophoresis, transferred to poly(vinylidene fluoride) (PVDF) membranes (Immobilion-P, Millipore), and probed with antibodies (4). Total protein concentration was estimated by Bradford assay (Bio-Rad). The antibodies and the dilution used in this study were: AIPL1 polyclonal antibody at 1:5,000 dilution (4), Rho monoclonal antibody (4D2; gift of R. Molday, University of British Columbia, Vancouver) at 1:4,000 dilution, guanylyl cyclase (GC-E) and GC-F antibody (gift of D. L. Garbers, University of Texas Southwestern Medical Center, Dallas) at 1:5,000 dilution, transducin γ (Tγ) antibody (gift of N. Gautam, Washington University School of Medicine, St. Louis) at 1:1,000 dilution, RK monoclonal antibody (G8; Affinity BioReagents) at 1:5,000 dilution, recoverin antibody at 1:5,000 dilution, PDE antibody (MOE; gift of B. Fung, School of Medicine, Los Angeles) at 1:4,000 dilution, and Prkca antibody (Oxford Biomedical Research, Oxford, MI) at 1:5,000 dilution.
Measurement of PDE Activity and cGMP Levels. Retinas were dissected and homogenized in ROS buffer (three retinas in 100 μl of buffer). Homogenates (2 μg of total protein) were treated with 0.05 μg of trypsin for 4 min at room temperature, and then quenched with 2 μg of soybean trypsin inhibitor. PDE activity was measured in 80 mM Tris, pH 8/4 mM MgCl2/7.2 mM cGMP/10 nM CaCl2/0.2 μl of [3H]cGMP (specific activity, 15 Ci/mmol; 1 Ci = 37 GBq). The reaction mixture was incubated for 4 min at 30°C, and then stopped by heating for 2 min at 100°C. Two microliters of 5 mM GMP was added to each sample, and the samples were spun for 5 min at 16,000 × g in a microfuge. Five microliters of the reaction supernatant spotted on a TLC plate (PEI cellulose; EM Science), and GMP and cGMP were separated by using 0.2 M LiCl. Excised GMP spots were eluted in 1 ml of 2 M LiCl and counted in a Beckman scintillation counter. All reactions were performed in triplicate, and the experiment was repeated at least twice. We extracted cGMP from mouse retinas by boiling in 0.1 M HCl as described by Farber and Lolley (13). An aliquot of each extract before boiling was used to measure the protein concentration by Bradford assay (Bio-Rad) using BSA as the standard. The amount of cGMP in the extract was determined by a RIA kit (Amersham Pharmacia). Each reaction was performed in duplicate, and the experiment was repeated at least four times. To minimize the individual variations between mice, retinas were pooled from different mice for each time point assayed.
Results
Generation of AIPL1-Deficient Mice. To create a mouse model of LCA caused by mutations in AIPL1, we deleted most of the coding region (exons 2–5) of Aipl1 in the mouse genome by homologous recombination (Fig. 1a). A PCR-based method (Fig. 1b) and Southern blotting (data not shown) confirmed the deletion. RT-PCR analysis showed that Aipl1 mRNA is undetectable in retinas from Aipl1–/– mice (Fig. 1c). Immunoblots of retinal homogenates confirmed the absence of AIPL1 in the homozygous knockout and reduced AIPL1 protein levels in heterozygotes (Fig. 1d). The AIPL1 mutation is inherited with normal Mendelian distribution, indicating that lack of AIPL1 does not affect viability. Aipl1–/– and Aipl1+/– mice are healthy, fertile, and have no gross morphological abnormalities. Heterozygotes up to 8 months old do not exhibit any photoreceptor abnormalities detectable by ERG or histology (data not shown).
Fig. 1.
Targeted disruption of Aipl1 in mouse. (a) Strategy for the targeted deletion of Aipl1. The coding exons (1–6) of Aipl1 are shown as open boxes. Exons 2–5 were replaced with the neomycin-resistance gene (Neo) cassette in the targeting vector. A copy of the thymidine kinase (TK) gene was placed at the 5′ end of the targeting vector for negative selection. (b) PCR analysis of mouse tail genomic DNA. The 6.1-kb wild-type N-terminal product (primers F1-R1) and the 5.1-kb wild-type C-terminal product (primers R2-F2) were detected only in +/– and +/+ mice. The –/– mice were identified by the presence of 5.2-kb mutant N-terminal product (primers F1-V1) and the 4.1-kb mutant C-terminal product (primers V2-F2). The +/– mice were identified by the presence of both mutant and wild-type PCR products. (c) RT-PCR of retinal RNA isolated from P12 littermates. (Upper) The expression levels of Aipl1.No Aipl1 expression is seen in Aipl1–/– mice. As a control, Hprt was amplified (30). (d) Immunoblot of protein extracts from mouse retina (P14). AIPL1 was detected by using a rabbit polyclonal antibody (4). Retinas from Aipl1+/– mice contained half the wild-type levels of AIPL1 protein, whereas no AIPL1 was detected in the retinas of Aipl1–/– mice. The levels of recoverin, a retinal protein, was checked as a control.
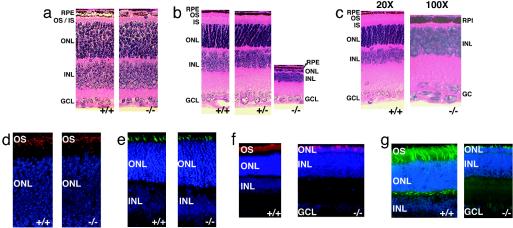
Retinal Structure and Rapid Degeneration of Photoreceptors. Deletion of Aipl1 does not cause any obvious morphological developmental changes in the retina. All retinal lamina develop appropriately, and there is no difference in the thickness of the outer nuclear layer before postnatal day 11 (P11, 10 rows of nuclei) (Fig. 2a and see Fig. 6a, which is published as supporting information on the PNAS web site). Immunofluorescence showed that the developing photoreceptor layer contains both rods and cones (Fig. 2 d and e). Phosphohistone immunostaining of retinas from P2 and -4 showed no difference in the number of mitotic cells between wild-type and Aipl1 knockout mice (data not shown). This finding suggests that AIPL1 does not play a role in cell proliferation.
Fig. 2.
Progressive degeneration of photoreceptors in the mice lacking AIPL1. Paraffin(5 μm) sections from whole eye were stained with hematoxylin and eosin. RPE, retinal pigment epithelium; OS, outer segments; IS, inner segments; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (a) At age P08, the thickness of the ONL was comparable between Aipl1–/– and wild-type littermates. (b) By P18, a single row of nuclei in the ONL are present in Aipl1–/– compared to 10–12 rows of nuclei in Aipl1+/– and wild-type littermates. (c) At P30, the degeneration of photoreceptors is complete with no ONL. The other retinal layers (INL and GCL) appear to be unaffected. For clarity, Aipl1–/– is shown at a ×100 magnification compared to wild-type littermate, which is shown at a ×20 magnification. (d) Immunostaining of P10 retinal sections with antibody against rod opsin (red) shows the normal formation of rods in mice lacking Aipl1. The distribution of rhodopsin appears to be normal in Aipl1–/– mice. (e) Staining of retinal sections from P10 with peanut agglutinin (PNA) conjugated to FITC (green) show the formation of cones in Aipl1–/– mice. (f) Immunostaining of P18 retinal sections with antibody against rod opsin (red) shows the rapid rod cell degeneration in mice lacking Aipl1. Only a single layer of opsin positive layer (red) is found in the retinal section from Aipl1–/– mice. (g) Staining of retinal sections from P18 with peanut agglutinin (PNA) conjugated to FITC (green) shows the rapid degeneration of cones in Aipl1–/– mice. Few cone cells (green) are found in the retinal section from Aipl1–/– mice. Increased PNA staining in cone pedicles of wild-type retinal section was observed in P18 in comparison to wild-type retinal section from P10 (g and e). In d–g, cell nuclei were stained with 4′,6-diamidino-2-phenylindole (blue).
Degeneration in Aipl1–/– retinas can first be detected by light microscopy at day 12, and it proceeds rapidly thereafter. By P14, the thickness of the photoreceptor layer is reduced by half (see Fig. 6b). At P18, only a single layer of photoreceptor nuclei remains (Fig. 2 b and f) and photoreceptor degeneration is complete by 4 weeks (Figs. 2c and 6c). Both rods and cones degenerate at a similar rate (Figs. 2 f and g and 6c). The degeneration is independent of light (data not shown) and specific to the photoreceptor layer (Fig. 2 b and c). As in other cases of photoreceptor degeneration (14, 15) glial fibrillary acidic protein is up-regulated in Aipl1–/– retinas (data not shown). Photoreceptor degeneration is more severe in central than peripheral retina. This finding indicates that degeneration proceeds from center to periphery (data not shown), consistent with previous models of retinal degeneration (16, 17). Staining of retinal sections with propidium iodide, an indicator of early stages of cell death (18) (see Fig. 7, which is published as supporting information on the PNAS web site), showed brightly stained apoptotic nuclei in the outer nuclear layer beginning at P9. This finding coincides with the initial elongation of photoreceptor outer segments. There are no signs of apoptosis before P9 (see Fig. 7).
Although the essential biochemical role of AIPL1 has not been established definitively, one function that has been suggested is protein transport (19). We evaluated this suggestion by examining the subcellular localization of several retinal proteins, including Rho, GC-E, RK, Tγ, PDE, and recoverin (Fig. 2d, supporting information, and data not shown). Each of these proteins localizes normally in Aipl1–/– photoreceptors before they degenerate. Therefore, it is unlikely that AIPL1 plays a general role in protein transport.
Electrophysiological Responses. We performed electroretinography to evaluate the ability of Aipl1–/– and Aipl1+/+ retinas to respond to light. At an early stage (P12) in retinal development, flash stimuli evoke small but reliable a-wave responses in Aipl1+/+ and Aipl1+/– retinas, indicating the onset of photoreceptor activity at the time of formation of outer segments (Fig. 3a). By P18, a- and b-waves in Aipl1+/+ retinas are strong (Fig. 3c). Responses from Aipl1+/– and Aipl1+/+ mice are indistinguishable at this age and at ages up to 90 days (data not shown). A paired flash analysis confirmed that cone responses are also normal in Aipl1+/– retinas (data not shown). In contrast, we were unable to evoke ERG responses from Aipl1–/– mice at any age with any illumination condition (Fig. 3 b and d). This finding indicates the absence of rod and cone activity in Aipl1–/– retinas and is consistent with the finding that children affected by the Aipl1 form of LCA have no recordable ERG within 1 year of birth.
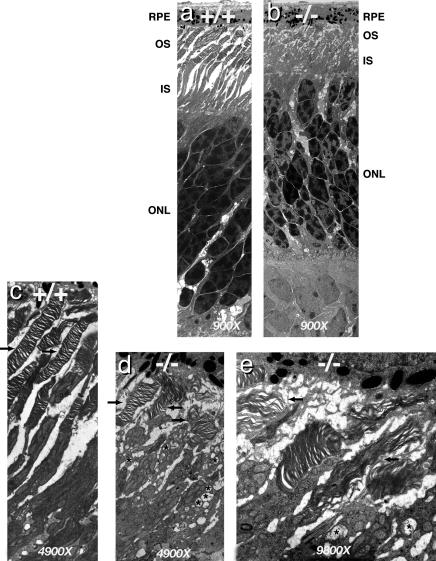
Disorganization of Photoreceptor Outer Segments. The absence of ERG responses prompted us to examine the ultrastructure of Aipl1–/– photoreceptor outer segments by transmission electron microscopy. At P8, when outer segments first appear, Aipl1+/+ and Aipl1–/– retinas are indistinguishable (data not shown). At P11, photoreceptor outer segments of Aipl1–/– mice are shorter than normal, highly disorganized, and fragmented (Fig. 4 a–d). Inner segments are enlarged and disorganized (Fig. 4 b and d). Vacuolar inclusions (indicated by asterisks), similar to those found in rd/rd mice, are present in the inner segments of photoreceptors in Aipl1–/– retinas (17) (Fig. 4 d and e).
Fig. 4.
Ultrastructure of photoreceptors of 11-day-old Aipl1-deficient mice and wild-type littermate. (a and b) Electron micrographs of wild-type and Aipl1–/– retina (magnification, ×900). Both outer and inner segments are considerably shorter in the Aipl1–/– retina. The nuclei in the outer nuclear layer of Aipl1–/– mice are not arranged as a regular array, but are missing some nuclei and show early signs of cell death. (c–e) Although the majority of photoreceptors in Aipl1–/– mice are short and highly disorganized (indicated by arrows in d and e), some photoreceptors with normal packing of disks and shorter outer segments are also found. In comparison, in wild-type retina most of the photoreceptors are organized and longer (indicated by arrows in c). Several vacuolar inclusions were observed only in the inner segments of Aipl1–/– mice (indicated by asterisks in d and e).
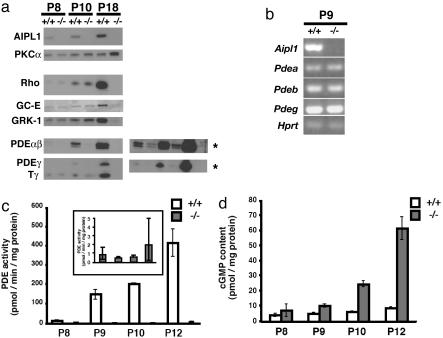
PDE Is Not Active in Mice Lacking AIPL1. We quantified the relative amounts of retinal proteins by immunoblot analysis. At P18, no photoreceptor-specific proteins are detectable because most of the photoreceptor layer has degenerated (Fig. 5a). At P8 and -10, well before significant degeneration has occurred, the levels of Rho, GC-E, RK, Tβγ, and recoverin are similar in Aipl1+/+ and Aipl1–/– retinas (Fig. 5a and data not shown). A striking exception is that the levels of all three subunits of PDE (α, β, and γ) were reduced by 90% in Aipl1–/– mice (n = 3). Reduced but detectable levels of all three PDE subunits are apparent at P8, the first day that they are normally expressed; however, the mRNA levels of all three PDE subunits are normal (Fig. 5b). This finding contrasts with the phenotype of the rd/rd mouse in which PDE-β protein is completely absent and mRNA is significantly reduced (20–23).
Fig. 5.
Analysis of protein expression levels in Aipl1–/– and wild-type control retina. (a) Immunoblots of retinal homogenates from P8, P10, and P18 wild type and mice lacking AIPL1. Equal amounts of retinal proteins (50 μg) were loaded in each lane. As expected, AIPL1 was not expressed in Aipl1–/– retina. Protein kinase C-α (PKCα), which is expressed in glial, rod bipolar, and some amacrine cells, was unaltered in P8 or P10 Aipl1–/– retina. A slight up-regulation in the expression of PKCα was observed in P18 Aipl1–/– retina, after the outer nuclear layer was completely degenerated. The expression of Rho, GC-E, and rhodopsin kinase (GRK1) showed no alteration in protein levels at P8 and P10. Once the photoreceptor layer degenerated completely by P18, none of the photoreceptor proteins, such as Rho, GC-E, GRK1, and Tγ, were present in P18 Aipl1–/– retina. In the Aipl1–/– mice at P8, PDE subunits (α, β, and γ) were reduced by 90% before the degeneration started. When immunoblots were exposed to film for longer times, all three PDE subunits could be detected on the Western blot from P8 and P10 Aipl1–/– retina (as indicated by asterisks). This shows that the full-length PDE subunits are made in Aipl1–/– retina. (b) RT-PCR analysis of PDE subunits at P9 shows that mRNA levels of all three subunits are not altered. The level of hprt was also examined as a control. (c) Trypsin-activated PDE activated from light-adapted retinas of control wild-type and Aipl1–/– mice at different stages of retinal development (24). (Inset) The low levels of PDE activity observed in Aipl1–/– mice. The results are the mean ± SEM of two to four samples, where each measurement was done in triplicate. (e) cGMP levels in the light-adapted retinas of control wild-type and Aipl1–/– mice during retinal development. The results are the mean ± SEM of four to six samples, where each measurement was done in duplicate.
To confirm the reduced levels of PDE, we assayed PDE activity after limited trypsin proteolysis, a treatment that specifically destroys the inhibitor but not the catalytic subunits of PDE (24). We found no significant PDE activity in Aipl1–/– retinas compared to Aipl1+/+ retinas at P8-P12 (Fig. 5c and Inset). This finding suggests that the low amount of PDE (10% of Aipl1+/+) detected immunologically in Aipl1–/– retinas at this age is not enzymatically active. We also measured the cGMP content of retinas. Consistent with the absence of cGMP hydrolytic activity in Aipl1 –/– retinas, cGMP accumulates to higher levels (7- to 9-fold higher at P12) in Aipl1–/– retinas than in Aipl1+/+ retinas (Fig. 5d). The elevation of cGMP levels precedes the retinal degeneration.
Rapid retinal degeneration and elevated levels of retinal cGMP are characteristic phenotypes of an extensively studied mouse mutant, rd/rd (17). However, there are distinct differences between the phenotypes of Aipl1–/– and rd/rd. Inactivation of the PDE-β subunit gene in rd/rd reduces the level of PDE-β mRNA and causes complete loss of the PDE-β subunit (20–23). In contrast, PDE mRNA levels are normal, and all three PDE subunits are destabilized in Aipl1–/– retinas before the retina degenerates. Full-length PDE-β subunit can be detected at P8–P10 of Aipl1–/– retinas (Fig. 5a). Full-length PDE-β is undetectable at this age in rd/rd retinas because PDE-β is truncated by a stop codon in the rd gene. Aipl1–/– mice also differ from rd/rd mice in that no detectable ERG response can be recorded from Aipl1–/– mice at early stages of retinal development (P12), whereas rd/rd mice produce reduced but significant responses at that age (17). Retinal degeneration in Aipl1–/– mice has remained completely linked to the inactivated Aipl1 locus on chromosome 11 through three generations of outcrosses (data not shown). The PDE catalytic subunits are on different linkage groups. Only the PDE-γ gene is in linkage group 11. To eliminate the possibility that a spurious mutation in PDE-γ subunit could contribute to retinal degeneration, we sequenced the entire gene and found no mutations. The PDE-β gene linked to rd is on chromosome 5. To make certain that the rd mutations were absent from our strains, we sequenced the entire PDE-β gene from our Aipl1–/– mice. Neither of the two PDE-β mutations linked to rd, Y347Stop or the proviral insertion in intron 1 (25–27), was present in the Aipl1–/– mice that we characterized (see Fig. 8, which is published as supporting information on the PNAS web site).
Discussion
Our analysis of Aipl1–/– mice shows that normal expression of AIPL1 is required for photoreceptor survival. Although rods and cones form in the absence of AIPL1, they do not respond to light. Degeneration of both rods and cones is complete within 3 weeks of birth. The PDE holoenzyme is destabilized by the AIPL1 deficiency. Degeneration appears to be caused by this destabilization. Support for this idea comes from the observation that direct inactivation of the PDE-β subunit gene in rd/rd mice also causes photoreceptor degeneration. Like the PDE mutation in rd/rd mice, the Aipl1–/– mutation also reduces cGMP hydrolytic activity and elevates cGMP, causing rapid degeneration of photoreceptor cells (17).
What is the link between AIPL1 and PDE stability? Our previous study revealed that AIPL1 can enhance protein farnesylation (4). At least three proteins in photoreceptors are known to be farnesylated, PDE-α, Tγ, and RK (6–9). Of these, only PDE-α requires farnesylation for its stability (9, 11). A previous study showed that mutations that block farnesylation cause degradation of PDE-α protein in cultured cells without loss of PDE-α mRNA (11). Loss of protein without loss of mRNA is a phenotype specific to defects in farnesylation of PDE-α (11). In our study of Aipl1–/– retinas, we found that mRNA levels for all three PDE subunits are normal. In contrast, mutations that block geranylgeranylation of the PDE-β subunit reduce the stability of both protein and mRNA (11). Altogether, these observations indicate that AIPL1 directly influences the farnesylation of PDE-α. In the absence of AIPL1, PDE-α is not efficiently farnesylated and is unstable and the entire trimeric PDE complex degrades. It is also possible that AIPL1, through its interaction with the farnesylated PDE-α subunit, may act as a chaperone to promote the folding and assembly of PDE subunit complex. In this case, absence of AIPL1 results in defective subunit assembly, and the whole complex is destabilized. It is possible that AIPL1 plays a role in promoting the proper intracellular localization of PDE in outer segments. However, it is unlikely that AIPL1 is involved in the transport of PDE because the residual PDE found in AIPL1 knockout retinas localizes to photoreceptor outer segments (Fig. 9, which is published as supporting information on the PNAS web site). Further studies are needed to distinguish the mechanism by which AIPL1 affects the stability of the PDE heterotrimer. Irrespective of the mechanism by which AIPL1 affects the stability of active PDE, reduction in PDE results in the consequent elevation of cGMP, causing rapid apoptosis of photoreceptor cells.
Our analysis of the phenotype of the Aipl1–/– mouse has revealed a molecular pathway that can contribute to photoreceptor degeneration in LCA linked to mutations in AIPL1. We have shown that the Aipl1–/– mouse is a suitable animal model with which to further enhance the understanding of LCA. In humans, mutations in AIPL1 cause LCA, a disease that affects both rods and cones. LCA has a more severe effect on the retina and an earlier onset than retinitis pigmentosa linked to mutations in PDE subunits (28, 29). This finding suggests that there may be significant functions for AIPL1 in addition to stabilizing PDE. Further studies will be needed to identify those additional activities.
Supplementary Material
Acknowledgments
We thank G. Froelick, S. Kikkawa, D. Possin, M. Roberts, L. Siverts, K. Shimizu, M. Taylor, and C. Ware for technical support; J. Saari, C. Tucker, and Hurley laboratory members for critical reading of the manuscript and for constructive suggestions; and J. Chen (University of Utah, Salt Lake City), B. Fung, N. Gautam, D. L. Garbers, R. Molday, R. Palmiter, M. Simon (California Institute of Technology, Pasadena), and G. Travis (University of California, Los Angeles) for reagents used in this study. This work was supported by National Institutes of Health Grant EY013572 (to J.B.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LCA, Leber congenital amaurosis; PDE, phosphodiesterase; Rho, rhodopsin; RK, Rho kinase; Pn, postnatal day n;Tγ, transducin γ; GC-E, guanylyl cyclase; ERG, electroretinogram.
References
- 1.Sohocki, M. M., Bowne, S. J., Sullivan, L. S., Blackshaw, S., Cepko, C. L., Payne, A. M., Bhattacharya, S. S., Khaliq, S., Qasim Mehdi, S., Birch, D. G., et al. (2000) Nat. Genet. 24, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohocki, M. M., Daiger, S. P., Bowne, S. J., Rodriquez, J. A., Northrup, H., Heckenlively, J. R., Birch, D. G., Mintz-Hittner, H., Ruiz, R. S., Lewis, R. A., et al. (2001) Hum. Mutat. 17, 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohocki, M. M., Perrault, I., Leroy, B. P., Payne, A. M., Dharmaraj, S., Bhattacharya, S. S., Kaplan, J., Maumenee, I. H., Koenekoop, R., Meire, F. M., et al. (2000) Mol. Genet. Metab. 70, 142–150. [DOI] [PubMed] [Google Scholar]
- 4.Ramamurthy, V., Roberts, M., van den Akker, F., Niemi, G., Reh, T. A. & Hurley, J. B. (2003) Proc. Natl. Acad. Sci. USA 100, 12630–12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Spuy, J., Kim, J. H., Yu, Y. S., Szel, A., Luthert, P. J., Clark, B. J. & Cheetham, M. E. (2003) Invest. Ophthalmol. Vis. Sci. 44, 5396–5403. [DOI] [PubMed] [Google Scholar]
- 6.Anant, J. S., Ong, O. C., Xie, H. Y., Clarke, S., O'Brien, P. J. & Fung, B. K. (1992) J. Biol. Chem. 267, 687–690. [PubMed] [Google Scholar]
- 7.Lai, R. K., Perez-Sala, D., Canada, F. J. & Rando, R. R. (1990) Proc. Natl. Acad. Sci. USA 87, 7673–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukada, Y., Takao, T., Ohguro, H., Yoshizawa, T., Akino, T. & Shimonishi, Y. (1990) Nature 346, 658–660. [DOI] [PubMed] [Google Scholar]
- 9.Inglese, J., Glickman, J. F., Lorenz, W., Caron, M. G. & Lefkowitz, R. J. (1992) J. Biol. Chem. 267, 1422–1425. [PubMed] [Google Scholar]
- 10.Pittler, S. J., Fliesler, S. J., Fisher, P. L., Keller, P. K. & Rapp, L. M. (1995) J. Cell Biol. 130, 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin, N. & Baehr, W. (1994) J. Biol. Chem. 269, 3265–3271. [PubMed] [Google Scholar]
- 12.Akey, D. T., Zhu, X., Dyer, M., Li, A., Sorensen, A., Blackshaw, S., Fukuda-Kamitani, T., Daiger, S. P., Craft, C. M., Kamitani, T. & Sohocki, M. M. (2002) Hum. Mol. Genet. 11, 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farber, D. B. & Lolley, R. N. (1982) Methods Enzymol. 81, 551–556. [DOI] [PubMed] [Google Scholar]
- 14.Ekstrom, P., Sanyal, S., Narfstrom, K., Chader, G. J. & van Veen, T. (1988) Invest. Ophthalmol. Vis. Sci. 29, 1363–1371. [PubMed] [Google Scholar]
- 15.Sheedlo, H. J., Jaynes, D., Bolan, A. L. & Turner, J. E. (1995) Brain Res. Dev. Brain Res. 85, 171–180. [DOI] [PubMed] [Google Scholar]
- 16.Carter-Dawson, L. D., LaVail, M. M. & Sidman, R. L. (1978) Invest. Ophthalmol. Vis. Sci. 17, 489–498. [PubMed] [Google Scholar]
- 17.Farber, D. B., Flannery, J. G. & Bowes-Rickman, C. (1994) Prog. Retinal Eye Res. 13, 31–65. [Google Scholar]
- 18.Rich, K. A., Zhan, Y. & Blanks, J. C. (1997) J. Neurobiol. 32, 593–612. [DOI] [PubMed] [Google Scholar]
- 19.van der Spuy, J., Chapple, J. P., Clark, B. J., Luthert, P. J., Sethi, C. S. & Cheetham, M. E. (2002) Hum. Mol. Genet. 11, 823–831. [DOI] [PubMed] [Google Scholar]
- 20.Bowes, C., Danciger, M., Kozak, C. A. & Farber, D. B. (1989) Proc. Natl. Acad. Sci. USA 86, 9722–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelan, J. K. & Bok, D. (2000) Exp. Eye Res. 71, 119–128. [DOI] [PubMed] [Google Scholar]
- 22.Lee, R. H., Navon, S. E., Brown, B. M., Fung, B. K. & Lolley, R. N. (1988) Invest. Ophthalmol. Vis. Sci. 29, 1021–1027. [PubMed] [Google Scholar]
- 23.Tsang, S. H., Gouras, P., Yamashita, C. K., Kjeldbye, H., Fisher, J., Farber, D. B. & Goff, S. P. (1996) Science 272, 1026–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurley, J. B. & Stryer, L. (1982) J. Biol. Chem. 257, 11094–11099. [PubMed] [Google Scholar]
- 25.Pittler, S. J. & Baehr, W. (1991) Proc. Natl. Acad. Sci. USA 88, 8322–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowes, C., Li, T., Frankel, W. N., Danciger, M., Coffin, J. M., Applebury, M. L. & Farber, D. B. (1993) Proc. Natl. Acad. Sci. USA 90, 2955–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimenez, E. & Montoliu, L. (2001) Lab. Anim. 35, 153–156. [DOI] [PubMed] [Google Scholar]
- 28.McLaughlin, M. E., Sandberg, M. A., Berson, E. L. & Dryja, T. P. (1993) Nat. Genet. 4, 130–134. [DOI] [PubMed] [Google Scholar]
- 29.Huang, S. H., Pittler, S. J., Huang, X., Oliveira, L., Berson, E. L. & Dryja, T. P. (1995) Nat. Genet. 11, 468–471. [DOI] [PubMed] [Google Scholar]
- 30.Mears, A. J., Kondo, M., Swain, P. K., Takada, Y., Bush, R. A., Saunders, T. L., Sieving, P. A. & Swaroop, A. (2001) Nat. Genet. 29, 447–452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.