Abstract
Dnmt3a and Dnmt3b are responsible for the establishment of DNA methylation patterns during development. These proteins contain, in addition to a C-terminal catalytic domain, a unique N-terminal regulatory region that harbors conserved domains, including a PWWP domain. The PWWP domain, characterized by the presence of a highly conserved proline-tryptophan-tryptophan-proline motif, is a module of 100 to 150 amino acids found in many chromatin-associated proteins. However, the function of the PWWP domain remains largely unknown. In this study, we provide evidence that the PWWP domains of Dnmt3a and Dnmt3b are involved in functional specialization of these enzymes. We show that both endogenous and green fluorescent protein-tagged Dnmt3a and Dnmt3b are particularly concentrated in pericentric heterochromatin. Mutagenesis analysis indicates that their PWWP domains are required for their association with pericentric heterochromatin. Disruption of the PWWP domain abolishes the ability of Dnmt3a and Dnmt3b to methylate the major satellite repeats at pericentric heterochromatin. Furthermore, we demonstrate that the Dnmt3a PWWP domain has little DNA-binding ability, in contrast to the Dnmt3b PWWP domain, which binds DNA nonspecifically. Collectively, our results suggest that the PWWP domains of Dnmt3a and Dnmt3b are essential for targeting these enzymes to pericentric heterochromatin, probably via a mechanism other than protein-DNA interactions.
DNA methylation is catalyzed by DNA methyltransferases, which transfer a methyl group (-CH3) from S-adenosyl-l-methionine to the C-5 position of cytosine residues. Three active DNA methyltransferases, Dnmt1, Dnmt3a, and Dnmt3b, have been identified in humans and mice (5). Dnmt1 is ubiquitously expressed in proliferating cells, localizes to replication foci, and interacts with the proliferating cell nuclear antigen (PCNA) (8, 22). Although Dnmt1 can methylate both unmethylated and hemimethylated DNA in vitro, it has a 10- to 50-fold preference for hemimethylated substrates (30, 41). Inactivation of Dnmt1 in embryonic stem (ES) cells and mice leads to global demethylation of genomic DNA but has little effect on de novo methylation of newly integrated retrovirus DNA (21, 24). In contrast to Dnmt1, Dnmt3a and Dnmt3b are highly expressed in ES cells, early embryos, and developing germ cells but are downregulated in somatic tissues of postnatal animals (29). The Dnmt3 enzymes have an equal preference for hemi- and unmethylated DNA substrates in vitro (1, 29). Inactivation of both Dnmt3a and Dnmt3b by gene targeting blocks de novo methylation in ES cells and early embryos as well as de novo methylation of imprinted genes in the germ cells (16, 28). These findings led to the widely accepted view that Dnmt3a and Dnmt3b function primarily as de novo methyltransferases that establish DNA methylation patterns during embryogenesis and gametogenesis, whereas Dnmt1 functions primarily as a maintenance methyltransferase that copies the parental-strand methylation pattern onto the daughter strand after each round of DNA replication.
The distinct biochemical properties and biological functions exhibited by the de novo and maintenance methyltransferases are partly due to the structural differences of these enzymes. Both the Dnmt1 and Dnmt3 families of methyltransferases contain the highly conserved C-5 methyltransferase motifs at their C termini, but they show no sequence similarity in their N-terminal regions (5). A number of functional domains have been defined in the N-terminal region of Dnmt1. These include a nuclear localization signal (NLS), several domains involved in targeting the enzyme to replication foci during the S phase, and a CXXC domain (a cysteine-rich Zn2+-binding motif) implicated in binding DNA sequences containing CpG dinucleotides (5). The Dnmt3a and Dnmt3b proteins are very similar in structural organizations. Their N-terminal regulatory domains contain a variable region (≈280 amino acids in Dnmt3a and ≈220 amino acids in Dnmt3b), followed by two conserved regions, a PWWP domain and a cysteine-rich domain that shares homology with a region in ATRX (29, 38). While the ATRX homology domains of Dnmt3a and Dnmt3b have been shown to interact with histone deacetylase 1 and repress transcription in reporter assays (2, 13), the structural basis for the function of de novo methyltransferases remains largely unknown.
The PWWP domain, characterized by the presence of a highly conserved proline-tryptophan-tryptophan-proline motif, was originally identified in a group of proteins related to hepatoma-derived growth factor and in the protein product of the Wolf-Hirschhorn syndrome candidate gene 1 (WHSC1) (17, 35). Subsequent database searches have resulted in the identification of >60 PWWP domain-containing proteins, many of which associate with chromatin (31, 34). The PWWP domain is a moderately conserved region of 100 to 150 amino acids. Studies with crystallography and nuclear magnetic resonance spectroscopy reveal that the N-terminal half of the domain forms a five-stranded β-barrel and the C-terminal half forms a helical bundle (31, 33).
The Dnmt3a and Dnmt3b PWWP domains span 143 and 135 amino acids, respectively, with a conserved SWWP motif. The Dnmt3b PWWP domain has been shown to bind DNA in vitro, probably through a surface area that is enriched with basic residues (31). However, most of these residues are not conserved (31, 33). It remains to be determined, therefore, whether DNA binding is a universal property of the PWWP domain. Although the function of the PWWP domain is still unknown, its significance is underscored by the recent identification of a point mutation (S270P) in the Dnmt3B PWWP domain from two patients with immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome (32), a rare autosomal recessive disorder characterized by hypomethylation of classical satellite DNA (12, 18).
In this study, we provide evidence that the PWWP domains of Dnmt3a and Dnmt3b are involved in functional specialization of these enzymes. We demonstrate that the Dnmt3a and Dnmt3b proteins are concentrated in the major satellite repeats at pericentric heterochromatin. Disruption of their PWWP domains prevents their association with pericentric heterochromatin and abolishes their ability to methylate the major satellite repeats. We also show that the DNMT3b PWWP domain is a nonspecific DNA-binding module, whereas the Dnmt3a PWWP domain has little DNA-binding ability, suggesting that targeting of the Dnmt3 enzymes to pericentric heterochromatin by their PWWP domains is mediated by a mechanism other than direct protein-DNA interactions.
MATERIALS AND METHODS
DNA constructions.
The green fluorescent protein (GFP)-Dnmt3a, GFP-Dnmt3b1, GFP-Dnmt3b2, and GFP-Dnmt3b3 constructs were described previously (7). All GFP fusion constructs containing Dnmt3a or Dnmt3b point mutations or internal deletions were generated with a two-step cloning strategy; cDNA fragments 5′ and 3′ to the mutation or deletion were amplified with PCR and subcloned sequentially into pEGFP-C1 (Clontech). A common forward primer for the 5′ fragments and a common reverse primer for the 3′ fragments were used for these Dnmt3a or Dnmt3b constructs, and their sequences were 5′-AAT GAA TTC CAG CGG CCC CGG GGA C-3′ (Dnmt3a, common forward, the restriction site used for cloning is italic), 5′-TAT GGA TCC TAC TTC AGT TTG CCC CCA-3′ (Dnmt3a, common reverse), 5′-AAT GAA TTC AGA CAG CAG ACA TCT GAA-3′ (Dnmt3b, common forward), and 5′-TCT GGA TCC GAG CTC CCC AGT CCT GGG T-3′ (Dnmt3b, common reverse). The primer pairs that introduced the point mutations or internal deletions (i.e., the reverse primers for the 5′ fragments and the forward primers for the 3′ fragments) were 5′-GTC ATT GTC GAC ACT GCC TCC AAT CAC CAG-3′ and 5′-GGC AGT GTC GAC AAT GAC CTC TCC ATT GTC-3′ (for Dnmt3a:PC→VD), 5′-AAA GGT ACC CGA TGT TTC TGC ACT TCT-3′ and 5′-GCG GGT ACC AGG GCA CCT ATG GGC TGC TGC-3′ (for Dnmt3aΔATRX), 5′-CCA TCC GTC GAC TCA GGC TCA TCG TCG-3′ and 5′-TTG GTG GTC GAC AGG CCG GAG CCG AGC-3′ (for Dnmt3aΔPWWP), 5′-CGG CCT GTC GAC CAG GAG AAG CCC CGA AGT-3′ and 5′-CTC CTG GTC GAC AGG CCG AAT TGT GTC TTG-3′ (for Dnmt3a:WP→ST), 5′-ATT GGT ACC GCT TCC ACC AAT CAC CAA-3′ and 5′-AGC GGT ACC AAT GAT CTC TCT AAC GTC-3′ (for Dnmt3b1:PC→GT), 5′-AGC GGT ACC CCA GAT TGC CCT TGT TGT T-3′ and 5′-AGC GGT ACC ATG GGG TCC TCC GAC GC-3′ (for Dnmt3b1ΔATRX), 5′-TTA GGT ACC TGA TAC TCT GTG CTG T-3′ and 5′-GGT GGT ACC CTG GAA AGC CAC CTC CA-3′ (for Dnmt3b1ΔPWWP), 5′-ATC GGA TCC CTC CTG AGG TCA CCT ATT CCA AAC T-3′ and 5′-GTG GGA TCC GAT CAA GGG CTT CTC CTG-3′ (for Dnmt3b1:VW→RR), 5′-TCA GGA TCC TTG CCA TCA CCA AAC CAC-3′ and 5′-CAA GGA TCC TGA GAT CTC TGC TGA CA-3′ (for Dnmt3b1:S→P), 5′-CCA GGT ACC CCG TTG CAA TTC CAT CAA-3′ and 5′-ATC GGT ACC GCA AAG GTT TAT ATG AGG-3′ (for Dnmt3b1ΔI-IV), 5′-TTA GGT ACC TGA TAC TCT GTG CTG T-3′ and 5′-GAA GGT ACC AGT GGT TAA TAA GTC-3′ (for Dnmt3b1Δ227-360), and 5′-TTA GGT ACC TGA TAC TCT GTG CTG T-3′ and 5′-TGA GGT ACC CAA CAA CAA GGG CAA TCT-3′ (for Dnmt3b1Δ227-429).
All other GFP fusion constructs were generated by subcloning the corresponding Dnmt3a or Dnmt3b cDNA fragments into pEGFP-C1. These fragments were generated by PCR with primer pairs 5′-AAT GAA TTC CAG CGG CCC CGG GGA C-3′ and 5′-GAG AGA GTC GAC GCG GAT GGG CTT CCT C-3′ (for Dnmt3a:1-632), 5′-AGG GAA TTC CTA CTA CAT CAG CAA AC-3′ and 5′-AAA GGT ACC CGA TGT TTC TGC ACT TCT-3′ (for Dnmt3a:192-487), 5′-AAT GAA TTC TGT GGA AGA GAA CCA GG-3′ and 5′-AAA GGT ACC CGA TGT TTC TGC ACT TCT-3′ (for Dnmt3a:223-487), 5′-AGG GAA TTC CTA CTA CAT CAG CAA AC-3′ and 5′-GGC GGT ACC CAC ATG TCG GTG TAA-3′ (for Dnmt3a:192-438), 5′-AAT GAA TTC TGT GGA AGA GAA CCA GG-3′ and 5′-GGC GGT ACC CAC ATG TCG GTG TAA-3′ (for Dnmt3a:223-438), 5′-AAT GAA TTC AGA CAG CAG ACA TCT GAA-3′ and 5′-CCA GGT ACC CCG TTG CAA TTC CAT CAA-3′ (for Dnmt3b1:1-594), and 5′-AAT GAA TTC AGA CAG CAG ACA TCT GAA-3′ and 5′-TCA TAT GTC GAC TTG CGG GCA GGA TTG ACG-3′ (for Dnmt3b1:1-670).
The glutathione S-transferase (GST)-Dnmt3a:PWWP and GST-Dnmt3b:PWWP fusion constructs were generated by subcloning cDNA fragments encoding the PWWP domains of Dnmt3a and Dnmt3b, respectively, into pGEX-KG. The primers used to generate the cDNA fragments were 5′-AAT GGA TCC AAA GCA GCC GAC GAT GAG-3′ and 5′-TCT AAG CTT CTG GTG GCT CCA GGC-3′ (for Dnmt3a:PWWP) and 5′-GCA GGA TCC AGA GAT GGA GAC AGC ACA-3′ and 5′-TAT AAG CTT CTG GTT GCT TCT TGT TGG-3′ (for Dnmt3b:PWWP). The GST fusion proteins were expressed in Escherichia coli and purified by affinity chromatography with glutathione beads. The untagged Dnmt3a and Dnmt3b vectors used for stable transfections in ES cells were constructed by subcloning the corresponding cDNAs into pCAG-IRESblast (6).
The Dnmt3b exon 7 targeting vector, in which a 0.6-kb region containing exon 7 was replaced by a phosphoglycerate kinase (PGK)-puromycin cassette flanked with loxP sites, was generated by sequentially subcloning Dnmt3b genomic fragments (arms), the PGK-puromycin cassette flanked with loxP sites, and the PGK-diphtheria toxin fragment A (DTA) cassette into pBluescript II SK. The dnmt3b genomic fragments were generated by PCR with a bacterial artificial chromosome clone (Genome Systems Inc.) as the template and the following pairs of oligonucleotides as primers: 5′-CCA ACG CGT CAT GGA ACC CAC TTT ACC-3′ and 5′-AGA ACG CGT TCA GCC CTT AAG AGC TCT TAC-3′ (for the 5′ arm, 3.1 kb) and 5′-TCT AAG GTC GAC TTT CTG CCA GTG GCT GCA-3′ and 5′-GTC GCG GCC GCA GGT GAC CAA ATC CAT TTA-3′ (for the 3′ arm, 3.0 kb). The identities of all constructs were verified by DNA sequencing.
Antibodies.
The Dnmt3a and Dnmt3b rabbit polyclonal antibodies, which recognize the N-terminal variable regions of Dnmt3a and Dnmt3b, respectively, were described previously (7). The antitubulin monoclonal antibody Ab-1 was obtained from Oncogene Research Products. Peroxidase- and fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G preparations were purchased from Jackson ImmunoResearch Laboratories, Inc.
Cell culture and transfections.
NIH 3T3 cells were maintained as suggested by the American Type Culture Collection. Wild-type J1 and mutant ES cells were maintained as described previously (6). Differentiating ES cells were obtained by culturing them as embryoid bodies for 5 days in suspension in the absence of leukemia inhibitory factor. Cells in embryoid bodies were then trypsinized and plated on gelatin-coated petri dishes. Transient transfections of GFP fusion constructs in NIH 3T3 cells were carried out with the use of Lipofectamine Plus reagent (Invitrogen). Stable transfections of Dnmt3a and Dnmt3b expression vectors in ES cells were performed according to a procedure described previously (6).
Fluorescence analyses.
Transfected and untransfected cells were fixed with 2% paraformaldehyde in phosphate-buffered saline for 5 min at room temperature. If the cells were to be visualized for only GFP, they were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 5 min at room temperature, and their nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). If the cells were to be stained with antibodies, they were permeabilized with a mixture of methanol and acetone (1:1) for 15 min at −20°C. Indirect immunofluorescence analyses with anti-Dnmt3a (1:1,000) or anti-Dnmt3b (1:2,000) were performed as described previously (4). Fluorescence in situ hybridization (FISH) was carried out with the peptide nucleic acid FISH kit (Dako). indocarbocyanine-labeled peptide nucleic acid probes (minor satellite, 5′-ACTCATTGATATACACT-3′; major satellite, 5′-CTTGCCATATTCCACGT-3′) were purchased from Applied Biosystems.
Targeted disruption of the Dnmt3b PWWP domain in ES cells.
The Dnmt3b exon 7 targeting vector was transfected into Dnmt3a−/− Dnmt3b+/− ES cells (28) via electroporation, and transfected cells were selected with puromycin. To screen for homologous recombination, genomic DNA isolated from puromycin-resistant colonies was digested with EcoRV and analyzed by Southern hybridization with an external probe 5′ to the targeting construct. Immunoblotting with anti-Dnmt3b was used to determine whether the wild-type or mutant allele of Dnmt3b was targeted. Deletion of the PGK-puromycin cassette flanked by loxP sites was achieved by cotransfecting pBS185 (an expression vector encoding Cre recombinase) and pcDNA-6 (a vector containing a blasticidin resistance gene) into ES cells, followed by selection with blasticidin.
DNA methylation analysis.
Genomic DNA isolated from various ES cell lines was digested with methylation-sensitive restriction enzymes and analyzed by Southern hybridization with various probes (6, 21). Bisulfite sequencing analysis of the major satellite repeat unit was described previously (6).
Electrophoretic mobility shift assay.
32P-labeled DNA probes (105 cpm per reaction) were incubated at room temperature for 30 min with GST fusion proteins (1 μg per reaction) in phosphate-buffered saline in the absence or presence of sheared salmon sperm DNA (2 to 100 ng per reaction). The samples were electrophoresed on native polyacrylamide gels, and the radioactive signals were detected by autoradiography. The major satellite probe is the 234-bp unit sequence of the major satellite repeats, and the control probes are genomic fragments of similar sizes amplified from various regions of Dnmt3a.
RESULTS
Dnmt3a and Dnmt3b accumulate in the major satellite repeats at pericentric heterochromatin.
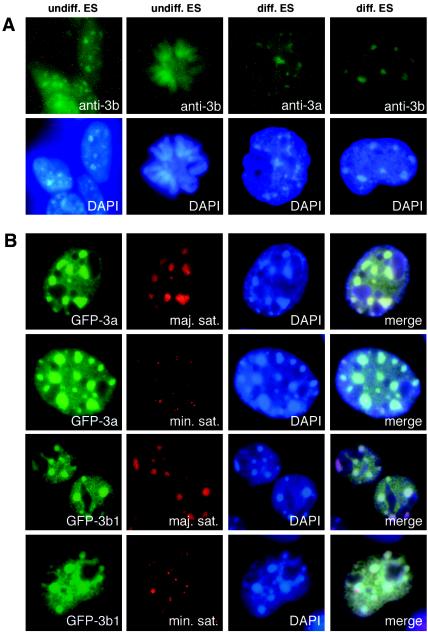
Epitope-tagged Dnmt3a and Dnmt3b proteins have been shown to localize to heterochromatin in transfected cells (2, 7). The subcellular localization of endogenous Dnmt3a and Dnmt3b proteins, however, has not been well characterized, largely because the expression levels of these enzymes are very low in somatic tissues and most cell lines. We carried out indirect immunofluorescence studies in mouse ES cells with specific polyclonal Dnmt3a and Dnmt3b antibodies (7). In undifferentiated ES cells, Dnmt3b displayed nuclear localization with enrichment in large nuclear foci that correspond to DAPI bright spots during interphase and associated with chromosomes during mitosis (Fig. 1A), whereas Dnmt3a was undetectable by immunostaining (data not shown).
FIG. 1.
Dnmt3a and Dnmt3b are enriched in pericentric heterochromatin. (A) Localization of endogenous Dnmt3a and Dnmt3b. Undifferentiated and differentiated ES cells were fixed, permeabilized, and immunostained with rabbit polyclonal anti-Dnmt3a or anti-Dnmt3b, followed by fluorescein isothiocyanate-conjugated goat anti-rabbit secondary antibody. The top panel shows the fluorescein isothiocyanate signals, and the bottom panel shows the nuclei stained with DAPI. (B) Colocalization of Dnmt3a and Dnmt3b with the major satellite repeats. GFP-Dnmt3a and GFP-Dnmt3b1 were individually transfected into NIH 3T3 cells, and the cells were fixed and analyzed by FISH with indocarbocyanine-labeled oligonucleotide probes specific for mouse major or minor satellite repeats. The signals for GFP, indocarbocyanine (Cy3), and DAPI as well as the merged images are shown.
It has been shown that the expression of Dnmt3a and Dnmt3b is upregulated in differentiating ES cells (7). We therefore let ES cells differentiate for 5 days as embryoid bodies and examined the localization of Dnmt3a and Dnmt3b. In these cells, both Dnmt3a and Dnmt3b were readily detectable by immunostaining and exhibited a localization pattern similar to that of Dnmt3b in undifferentiated ES cells (Fig. 1A). These observations demonstrated that endogenous Dnmt3a and Dnmt3b proteins localize in the nuclei and concentrate in heterochromatic regions.
A major component of heterochromatin is various types of repetitive DNA sequences, including the major satellite repeats at the pericentric region and the minor satellite repeats at the centromeric region. To determine whether Dnmt3a and Dnmt3b preferentially localize to these repeats, we expressed GFP-Dnmt3a and GFP-Dnmt3b1 fusion proteins in NIH 3T3 cells and performed fluorescence in situ hybridization (FISH) analysis with indocarbocyanine-labeled DNA probes. Fluorescence microscopy revealed that the major satellite repeats and the large Dnmt3a and Dnmt3b foci show identical distribution patterns and complete colocalization, whereas the minor satellite repeats occupy much smaller nuclear domains, which usually localize at the periphery of the Dnmt3a and Dnmt3b foci (Fig. 1B). These results indicate that Dnmt3a and Dnmt3b accumulate in the major satellite repeats at pericentric heterochromatin.
PWWP domains of Dnmt3a and Dnmt3b are required for these enzymes to localize to the major satellite repeats.
To determine the regions of the Dnmt3a and Dnmt3b proteins that are involved in proper localization, we transfected a series of GFP-Dnmt3a and GFP-Dnmt3b fusion constructs containing deletions or point mutations into NIH 3T3 cells and examined the localizations of the expressed proteins with fluorescence microscopy (Fig. 2). Wild-type Dnmt3a and Dnmt3b1 displayed punctate nuclear localization patterns with two major types of nuclear foci: large foci, which corresponded to pericentric heterochromatin, and small foci, which were distributed throughout the nucleus, excluding the nucleoli. Both types of foci were present in the majority of transfected cells (pattern A, ≈80% for Dnmt3a and ≈60% for Dnmt3b1), and only the small foci were visible in some transfected cells (pattern B, ≈20% for Dnmt3a and ≈40% for Dnmt3b1). It should be noted that the localization patterns of Dnmt3a and Dnmt3b1 were similar but not identical. In most cells, Dnmt3a showed more obvious enrichment in pericentric heterochromatin, whereas Dnmt3b1 showed a stronger nucleoplasmic signal. The Dnmt3b gene encodes multiple variants via alternative splicing of exons 10, 21, and 22. Dnmt3b2 showed the same localization patterns as Dnmt3b1. Dnmt3b3, however, displayed diffuse nucleoplasmic localization patterns with or without accumulation in the major satellite repeats (patterns C and D, respectively), suggesting that the region encoded by exons 21 and 22 is required for Dnmt3b to localize to the small foci.
FIG. 2.
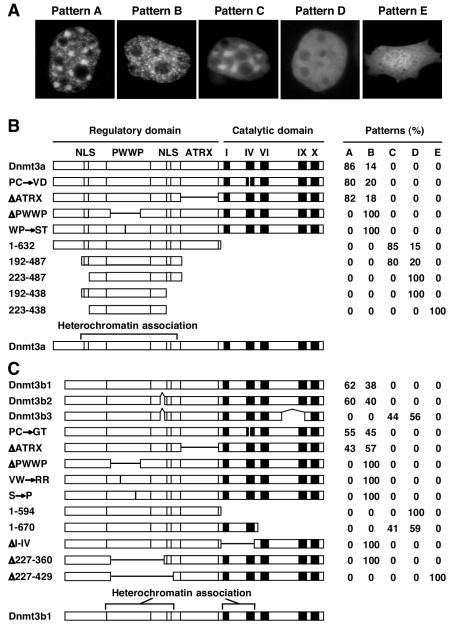
PWWP domain is required for preferential localization of Dnmt3a and Dnmt3b to pericentric heterochromatin. (A) The GFP-Dnmt3a and GFP-Dnmt3b constructs were transfected into NIH 3T3 cells, and the protein localization patterns were analyzed by fluorescence microscopy. The typical localization patterns observed are shown: punctate nuclear localization with accumulation in pericentric heterochromatin (pattern A), punctate nuclear localization without accumulation in pericentric heterochromatin (pattern B), diffuse nuclear localization with accumulation in pericentric heterochromatin (pattern C), diffuse nuclear localization without accumulation in pericentric heterochromatin (pattern D), and diffuse nuclear and cytoplasmic localization (pattern E). (B and C) Schematic diagrams of the GFP-Dnmt3a and GFP-Dnmt3b constructs and quantitation of different localization patterns. In the diagrams, the conserved PWWP and ATRX homology domains, the methyltransferase motifs (I, IV, VI, IX, and X), the putative NLSs, and the sites of alternative splicing are indicated. The positions of deletions and point mutations are indicated by horizontal and vertical thin lines, respectively. At the left, Δ indicates a deletion, → denotes amino acid substitutions, and the numbers are amino acid numbers. The GFP moiety (not shown) was fused to the N termini of the Dnmt3a and Dnmt3b proteins. For each construct, an average of 200 to 300 transfected (green) cells from two separate experiments were counted, and their localization patterns are shown as percentages.
While the reason that each of these proteins displayed more than one localization pattern is unknown, one possibility is that the localization of Dnmt3a and Dnmt3b is regulated during the cell cycle. The catalytic activity of Dnmt3a and Dnmt3b clearly does not play a major role in the localization of these enzymes, as substitutions of the proline-cysteine dipeptide at the catalytic center with other amino acid residues showed no effect on localization (Dnmt3a:PC→VD and Dnmt3b1:PC→GT). Deletion of the ATRX homology domain also did not significantly alter the localization of these proteins (Dnmt3aΔATRX and Dnmt3b1ΔATRX). However, deletion of a large portion of the PWWP domain abolished the ability of Dnmt3a and Dnmt3b to accumulate in large nuclear foci, and all transfected cells showed pattern B (Dnmt3aΔPWWP and Dnmt3b1ΔPWWP). Similar results were obtained when highly conserved residues within the PWWP domain were replaced (Dnmt3a:WP→ST, Dnmt3b1:VW→RR, and Dnmt3b1:S→P). These point mutations are predicted to have profound effect on the overall folding of the PWWP domain structure. Dnmt3a tryptophan 302 and proline 303 are part of the SWWP motif, which is located at the interface of the β-barrel and helical-bundle structures of the PWWP domain (31). Dnmt3b1 valine 236 and tryptophan 237 are located in the middle of β-strand 1, and their side chains form part of the hydrophobic core of the β-barrel (31). Dnmt3b1 serine 277, located in β-strand 4 (31), is equivalent to human DNMT3B serine 270, which was found to be mutated to proline in two patients with ICF syndrome (32). Taken together, our results demonstrated that the PWWP domain is required for Dnmt3a and Dnmt3b to localize to the major satellite repeats at pericentric heterochromatin.
We then tested a number of Dnmt3a and Dnmt3b deletion constructs in order to further map the regions that are responsible for targeting to the major satellite repeats (Fig. 2). Truncation of the C-terminal catalytic domains of Dnmt3a and Dnmt3b resulted in diffuse localization patterns (patterns C and D). For Dnmt3a, deletion of the entire catalytic domain did not affect its ability to associate with the major satellite repeats, as most transfected cells (85%) displayed pattern C (Dnmt3a:1-632). Further truncations at both the N and C termini revealed that the minimal Dnmt3a region required for major satellite localization was from residues 192 to 487 (Dnmt3a:192-487). This region contains, in addition to the PWWP domain, two sequences enriched with basic residues. Deleting either one of these sequences abolished association with the major satellite repeats, but the proteins remained exclusively nuclear (pattern D) (Dnmt3a:223-487 and 192-438). Deleting both sequences, however, resulted in diffuse localization in both the nucleus and the cytoplasm (pattern E) (Dnmt3a:223-438). These results suggested that the two basic sequences are functional NLSs and that they are both required for association with the major satellite repeats.
For Dnmt3b, the N-terminal regulatory domain was not sufficient for major satellite localization, as deletion of the entire catalytic domain abolished accumulation in large nuclear foci and all transfected cells displayed pattern D (Dnmt3b1:1-594). However, a shorter truncation at the C terminus did not affect the ability to accumulate in large nuclear foci, and over 40% of the transfected cells displayed pattern C (Dnmt3b1:1-670), suggesting that the region spanning from motif I to motif IV in the catalytic domain also plays a role in major satellite localization. Indeed, an internal deletion of this region resulted in pattern B in all transfected cells (Dnmt3bΔI-IV). Unlike Dnmt3a, Dnmt3b contains only one putative NLS, which is located between the PWWP and ATRX homology domains. This basic sequence is apparently a functional NLS, as an internal deletion that included the sequence (Dnmt3b1Δ227-429) resulted in both nuclear and cytoplasmic localization (pattern E), whereas a similar deletion that did not include the sequence (Dnmt3b1Δ227-360) showed exclusive nuclear localization (pattern B).
PWWP domains of Dnmt3a and Dnmt3b are required specifically for methylation of the major satellite repeats.
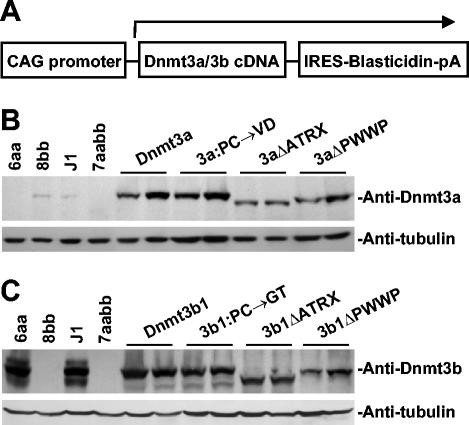
We have recently shown that inactivation of both Dnmt3a and Dnmt3b in ES cells results in progressive loss of global methylation and that introduction of active Dnmt3a and Dnmt3b isoforms back into highly demethylated cells can restore the methylation of most genomic sequences (6, 25). To determine whether the PWWP and ATRX homology domains of Dnmt3a and Dnmt3b play any role in the catalytic activities and specificities of these enzymes, we sought to express Dnmt3a and Dnmt3b proteins that lack these domains (ΔPWWP and ΔATRX, respectively) in late-passage Dnmt3a−/−, Dnmt3b−/− ES cells (7aabb cells) (28) and examine their ability to restore genomic methylation patterns. cDNAs encoding wild-type and mutant Dnmt3a and Dnmt3b were individually subcloned into a plasmid vector in which a CAG promoter drives the expression of a bicistronic transcript that encodes both the intended Dnmt protein and the selection marker, blasticidin S-deaminase (Fig. 3A). These constructs were individually transfected into 7aabb cells, and stable clones were obtained after selection with blasticidin. For each construct, two independent clones were chosen for methylation analysis. Expression of the Dnmt3a and Dnmt3b proteins in these clones was verified by immunoblotting analysis with specific polyclonal antibodies (Fig. 3B and C).
FIG. 3.
Stable expression of Dnmt3a and Dnmt3b proteins in Dnmt3a−/− Dnmt3b−/− ES cells. cDNAs encoding wild-type or mutant Dnmt3a and Dnmt3b proteins were subcloned in a bicistronic expression vector (schematically shown in A), and these constructs were individually electroporated into late-passage (P50) Dnmt3a−/− Dnmt3b−/− double mutant (7aabb) ES cells, which were subsequently selected in blasticidin-containing medium for 7 days. Blasticidin-resistant clones as well as wild-type (J1), Dnmt3a−/− (6aa), Dnmt3b−/− (8bb), and 7aabb ES cells were analyzed with immunoblotting with anti-Dnmt3a (B) or anti-Dnmt3b (C). As a loading control, the same membranes were immunoblotted with antitubulin.
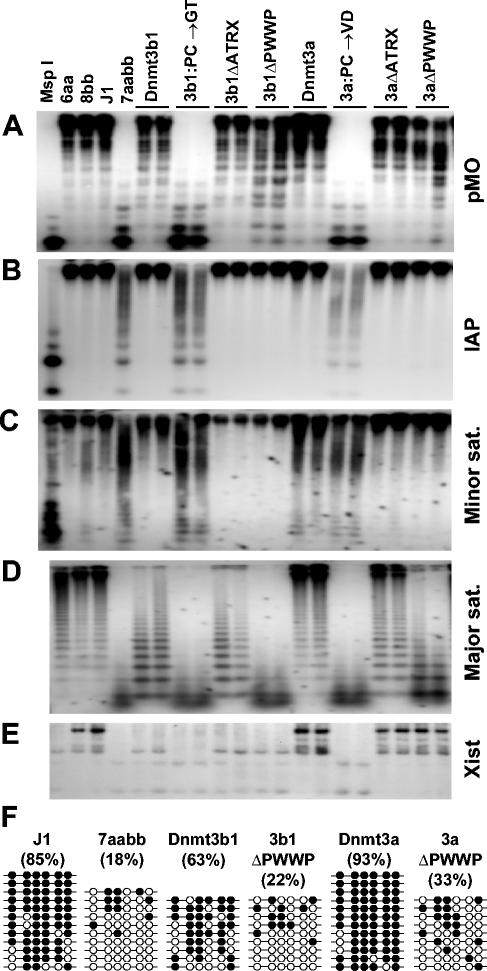
We extracted genomic DNA from the stable clones and control ES cells and examined the methylation status of various repeats and single-copy genes by methylation-sensitive enzyme digestions followed by Southern hybridizations (Fig. 4A to E). The endogenous C-type retrovirus and intracisternal A particle repeats are interspersed in the mouse genome, with about 100 and 1,000 copies per haploid genome, respectively. The major and minor satellite repeats are located in the pericentric and centromeric regions at copy numbers of 700,000 and 50,000 to 100,000, respectively. Xist is a unique gene located on the X chromosome. As reported previously (6), all these sequences were highly methylated in normal ES cells but became severely demethylated in late-passage 7aabb cells. Expression of wild-type Dnmt3a or Dnmt3b1 but not Dnmt3a:PC→VD or Dnmt3b1:PC→GT in 7aabb cells completely or partially restored the methylation patterns.
FIG. 4.
PWWP domain is specifically required for Dnmt3a and Dnmt3b to restore methylation of the major satellite repeats. (A-E) Methylation analysis by Southern hybridization. Genomic DNA from the indicated ES cell lines was digested with HpaII (A-C), Mae II (D), or EcoRV plus HhaI (E) and hybridized to probes for endogenous C-type retrovirus repeats (pMO) (A), the intracisternal A particle repeats (B), the minor satellite repeats (C), the major satellite repeats (D), or the 5′ region of Xist (E). DNA from J1 cells digested with MspI was used as a control for complete digestion. (F) Analysis of the methylation status of the major satellite repeating unit by bisulfite sequencing. Genomic DNA from J1 and 7aabb cells as well as stable cell lines expressing Dnmt3a, Dnmt3aΔPWWP, Dnmt3b1, and Dnmt3b1ΔPWWP was analyzed. The methylation status of six CpG sites from 9 to 12 individual clones is shown schematically (black circles represent methylated sites), and the percentages of methylated CpG sites are indicated in parentheses.
Dnmt3a and Dnmt3b1 exhibited distinct sequence preferences, with Dnmt3a substantially more efficient in methylating the major satellite repeats and the 5′ region of Xist and less efficient in methylating the minor satellite repeats than Dnmt3b1. Deletion of the ATRX homology domains of Dnmt3a and Dnmt3b1 had no effect on the catalytic activities and specificities of these enzymes, as Dnmt3aΔATRX and Dnmt3b1ΔATRX were as efficient as their wild-type counterparts in methylating all the sequences examined. Strikingly, with the deletion of the PWWP domains (Dnmt3aΔPWWP and Dnmt3b1ΔPWWP), the ability of Dnmt3a and Dnmt3b1 to methylate the major satellite repeats was severely compromised, whereas their activities for all the other sequences examined were not significantly affected.
The results were verified by bisulfite sequencing analysis (Fig. 4F). The 234-bp unit sequence of the major satellite repeats contains eight CpG sites. We examined the methylation status of six of these sites (the other two sites were not in the PCR-amplified region). In wild-type ES cells, 85% of the CpG sites were methylated. Only 18% of the sites remained methylated in 7aabb cells. While expression of Dnmt3a and Dnmt3b1 in 7aabb cells restored the methylation levels to 93 and 63%, respectively, expression of Dnmt3aΔPWWP and Dnmt3b1ΔPWWP showed only minimum activities (33 and 22%, respectively). These data showed that the PWWP domains of Dnmt3a and Dnmt3b1 are required for these enzymes to methylate the major satellite repeats de novo.
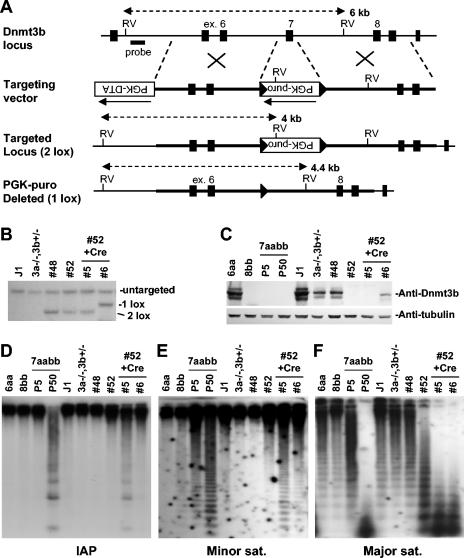
To confirm the results from the rescue experiments, we sought to delete exon 7 of Dnmt3b from the wild-type allele in Dnmt3a−/− Dnmt3b+/− ES cells (28) by gene targeting. Exon 7, which encodes the amino acid residues that form the β-barrel structure of the PWWP domain, consists of 159 bp, and therefore, its removal would not cause a frameshift of the downstream coding sequence. We generated a targeting vector in which a 0.6-kb genomic region containing exon 7 was replaced with a PGK-puromycin cassette with flanking loxP sites (PGK-puromycin in the opposite orientation from Dnmt3b transcription) (Fig. 5A). We expected that homologous recombination on the wild-type Dnmt3b allele would result in Dnmt3b protein products that lack 53 amino acids in the PWWP domain or, if the Dnmt3b transcripts were disrupted, eliminate Dnmt3b protein products.
FIG. 5.
Targeted deletion of Dnmt3b exon 7 results in demethylation of the major satellite repeats. (A) Strategy for targeted deletion of Dnmt3b exon 7. The top line shows the Dnmt3b genomic structure with exons represented by vertical bars. The targeting vector (second line) was constructed by replacing exon 7 with a PGK-puromycin cassette flanked by loxP sites (shown as triangles). A PGK-DTA cassette was introduced for negative selection to increase the targeting frequency. The third line represents the mutant locus resulting from homologous recombination between the targeting vector and the wild-type locus (2 lox). The fourth line represents the targeted locus with the PGK-puromycin cassette deleted by Cre (1 lox). (B) Southern analysis of the genotype of ES cell lines. Genomic DNA was digested with EcoRV and hybridized to a 5′ external probe, as shown in A. The 6.0-kb untargeted allele and the 4.0-kb (2 lox) and 4.4-kb (1 lox) targeted alleles are indicated. (C) Lysates from the indicated cell lines were immunoblotted with anti-Dnmt3b (top) and antitubulin (bottom) antibodies. (D-F) Genomic DNA from the indicated ES cell lines was digested with HpaII (D and E) or Mae II (F) and hybridized to probes for the intracisternal A particle repeats (D), the minor satellite repeats (E), or the major satellite repeats (F).
Southern hybridization showed that homologous recombination occurred in two (numbers 48 and 52) of the 80 clones that we screened (Fig. 5B). Immunoblotting analysis revealed that Dnmt3b protein products were not altered in clone 48 compared to the parental Dnmt3a−/− Dnmt3b+/− cells, whereas they were undetectable in clone 52 (Fig. 5C). We therefore concluded that the mutant Dnmt3b allele was targeted in clone 48 and the wild-type allele was targeted in clone 52. Cells from clone 52 were then cotransfected with expression vectors encoding the Cre recombinase and blasticidin S deaminase, and individual clones were obtained following selection with blasticidin. As expected, slightly smaller Dnmt3b protein products were detected in cells that had undergone Cre-mediated deletion of the PGK-puromycin cassette, although the protein levels were lower than those in the parental Dnmt3a−/−, Dnmt3b+/− cells (Fig. 5B and C, subclone 6). In cells in which deletion of the PGK-puromycin cassette did not occur, Dnmt3b proteins remained undetectable (Fig. 5B and C, subclone 5), and these cells would predictably behave like Dnmt3a−/−Dnmt3b−/− cells. Indeed, progressive demethylation of repetitive sequences occurred in these cells during the 3-week course of selection and clonal expansion after transfection (Fig. 5D to F, compare subclone 5 and parental clone 52). With the deletion of the PGK-puromycin cassette, expression of Dnmt3b proteins that lack a functional PWWP domain largely prevented demethylation of the intracisternal A particle and minor satellite repeats but not the major satellite repeats (Fig. 5D to F, subclone 6). These results were consistent with those from the rescue experiments and provided genetic evidence for involvement of the PWWP domain in maintaining the methylation of the major satellite repeats as well.
PWWP domains of Dnmt3a and Dnmt3b have different DNA-binding abilities.
Our finding that an intact PWWP domain is essential for Dnmt3a or Dnmt3b to be targeted to the major satellite repeats raises the possibility that the PWWP domain interacts with a component of pericentric heterochromatin. Since the Dnmt3b PWWP domain has been shown to bind DNA in vitro (31), we asked whether the PWWP domains of Dnmt3a and Dnmt3b preferentially bind specific sequences in the major satellite repeats. We generated GST fusion proteins containing the entire PWWP domain of Dnmt3a or Dnmt3b (31) (Fig. 6A) and compared their abilities to bind the 234-bp unit sequence of the major satellite repeats and random genomic sequences (as controls). The electrophoretic mobility shift assay showed that the Dnmt3b PWWP domain efficiently bound all the DNA probes tested, consistent with a previous report (31). To our surprise, the Dnmt3a PWWP domain, despite its high homology to the Dnmt3b PWWP domain, showed only minimal DNA-binding abilities (Fig. 6B and data not shown). These results indicated that DNA binding is probably not a property shared by all PWWP domains.
FIG. 6.
PWWP domains of Dnmt3a and Dnmt3b have different DNA-binding abilities. (A) GST fusion proteins stained with Coomassie blue. (B) Electrophoretic mobility shift assay. 32P-labeled DNA probes corresponding to the 234-bp unit sequence of the major satellite repeats or similar-sized random genomic sequences (control) were incubated with phosphate-buffered saline (PBS) or GST fusion proteins (1 μg), and the reactions were analyzed by native acrylamide gel electrophoresis and autoradiography. (C) GST-Dnmt3b:PWWP (1 μg) was used in the presence of increasing amounts (0, 2, 10, 30, and 100 ng) of sheared salmon sperm DNA.
To determine whether the Dnmt3b PWWP domain binds DNA sequences with different affinities, we carried out competition experiments by adding various amounts (2 to 100 ng per reaction) of sheared salmon sperm DNA to the reactions. Salmon sperm DNA inhibited the binding of all the DNA probes to GST-Dnmt3b:PWWP with similar efficiencies (Fig. 6C and data not shown), suggesting that the Dnmt3b PWWP domain is a nonspecific DNA-binding module. It is therefore unlikely that protein-DNA interactions mediated by the PWWP domains play a major role in directing Dnmt3a and Dnmt3b to pericentric heterochromatin.
DISCUSSION
Methylated cytosines are not randomly distributed in the mammalian genome. Rather, some genomic sequences (e.g., heterochromatic DNA) are heavily methylated, while others, such as CpG islands, are almost methylation-free. Yet little is known about the molecular mechanisms by which DNA methylation patterns are generated. Since the Dnmt1 and Dnmt3 families of methyltransferases do not appear to have any sequence specificity beyond CpG dinucleotides (10, 29, 41), chromatin-based mechanisms have been proposed to explain how DNA methyltransferases find their target sequences in the genome (3). One possibility is that accessory factors interact with certain domains present in DNA methyltransferases and target these enzymes to specific genomic sequences or chromatin structures. In this study, we have provided evidence that the PWWP domains of Dnmt3a and Dnmt3b play an important role in targeting these enzymes to pericentric heterochromatin. We show that the Dnmt3a and Dnmt3b proteins with their PWWP domains disrupted failed to accumulate in pericentric heterochromatin, resulting in their inability to methylate the major satellite repeats. The observation that these mutant proteins efficiently methylate other genomic sequences indicates that the PWWP domains are not required for enzymatic activities but are involved in functional specialization.
While Dnmt3a and Dnmt3b display similar localization patterns, Dnmt3a appears to have a stronger tendency to localize to pericentric heterochromatin. We noticed that enrichment of Dnmt3a in DAPI bright spots is more obvious and is observed in more cells than Dnmt3b. Consistent with the localization data, Dnmt3a is more efficient than Dnmt3b in methylating the major satellite repeats. By contrast, Dnmt3b shows more diffuse distribution in the nucleoplasm, raising the possibility that Dnmt3b may have a wider spectrum of substrates than Dnmt3a. Different domains of the Dnmt3 proteins may be involved in targeting these enzymes to various chromosomal regions.
Although the Dnmt3b PWWP domain is capable of binding DNA in vitro (31), DNA binding does not appear to be the major determinant for the specificity of the PWWP domain in targeting Dnmt3a and Dnmt3b to pericentric heterochromatin. We show that the Dnmt3b PWWP domain is a nonspecific DNA-binding module and the Dnmt3a PWWP domain, surprisingly, has only minimal DNA-binding ability. Sequence alignment reveals that 5 of the 19 basic residues in the Dnmt3b PWWP domain are not present in the Dnmt3a PWWP domain (31), which may account for the differences in DNA-binding abilities displayed by these domains.
Given that many of the basic residues believed to be involved in DNA binding are not conserved in other members of the PWWP domain family (31, 33), it is likely that the PWWP domain has an intrinsic property other than mediating DNA binding. Indeed, the PWWP domain has been proposed to mediate protein-protein interactions (34). Consistent with this hypothesis, the β-barrel structure of the PWWP domain is remarkably similar to those of several protein-protein interaction modules, including the Tudor domain and the chromodomain (26). Recent studies have demonstrated that DNA methylation is closely linked to and regulated by other epigenetic mechanisms, such as histone modification and chromatin-remodeling systems (23). Inactivation of the histone methyltransferases Suv39h1 and Suv39h2 in mouse ES cells has been shown to cause demethylation of the major satellite repeats but not other repetitive sequences (20). Lsh, a member of the SNF2 family of chromatin-remodeling proteins, has been shown to accumulate in pericentric heterochromatin and play a crucial role in CpG methylation of various repetitive sequences, including the major satellite repeats (9, 40). Thus, an interesting possibility is that the PWWP domains of Dnmt3a and Dnmt3b interact with one or more components of the histone modification and chromatin-remodeling systems that are involved in the establishment and/or maintenance of the structure of pericentric heterochromatin. Alternatively, the high-order structure of pericentric heterochromatin may provide binding sites for the PWWP domains of Dnmt3a and Dnmt3b, and methylation of pericentric DNA may in turn reinforce the stability of the heterochromatin structure.
It should be noted that the PWWP domains of Dnmt3a and Dnmt3b are required but not sufficient for pericentric heterochromatin targeting. Deletion analyses indicate that the pericentric heterochromatin targeting domain of Dnmt3a consists of the PWWP domain and the immediate upstream ≈90 amino acids and downstream ≈70 amino acids. A notable feature of the PWWP domain-flanking regions is that each contains a stretch of lysine- and arginine-rich sequence. While either one of these basic sequences is able to confer nuclear localization, both of them are required for pericentric heterochromatin targeting, suggesting that these sequences have other functions in addition to acting as NLSs. The minimum region responsible for targeting Dnmt3b to pericentric heterochromatin remains to be determined. We show that in addition to the PWWP domain and an NLS, a region spanning from motif I to motif IV in the catalytic domain is required. This region has recently been shown to be required for Dnmt3a and Dnmt3b homo- and heterodimerization (Y. Ueda, M. Okano, C. Williams, T. Chen, K. Georgopoulos, and E. Li, submitted for publication). It is therefore possible that dimerization is required for Dnmt3b to localize to pericentric heterochromatin. Taken together, our results suggest that the PWWP domains of Dnmt3a and Dnmt3b act cooperatively with other functional domains to target these enzymes to pericentric heterochromatin.
Methylation of heterochromatic DNA is involved in maintaining chromosomal stability. Aberrant changes of genomic methylation patterns are associated with various human disorders, most notably the ICF syndrome and cancer. ICF syndrome is a rare genetic disease characterized by demethylation of classical satellites 2 and 3 and chromosomal abnormalities in the pericentric region in lymphocytes (12, 18). While the majority of ICF mutations identified so far occur in the catalytic domain of DNMT3B (15, 28, 37, 39), a missense mutation (S270P) was recently identified in the PWWP domain (32). Serine 270 is conserved in human and mouse Dnmt3a and Dnmt3b. Substitution of this residue with a proline would predictably disrupt one of the β-strands that form the β-barrel structure of the PWWP domain. Indeed, introduction of the equivalent mutation (S277P) into mouse Dnmt3b prevents the accumulation of the protein in pericentric heterochromatin. We therefore conclude that failure to target DNMT3B to pericentric region is the underlying defect of the S270P mutation. In cancer cells, overexpression of DNMTs is frequently observed, yet the overall level of DNA methylation in these cells is usually lower than normal, largely due to loss of methylation in heterochromatic DNA, including classical satellites (19). Hypomethylation has been shown to be able to induce tumor formation in mice, possibly by promoting chromosomal instability (11, 14). While the cause of hypomethylation in cancer cells is not clear, defects in targeting DNMTs to proper chromosomal regions may be a contributing factor.
In addition to DNMT3B, several genes encoding PWWP domain proteins have been linked to human diseases. Deletions and translocations involving WHSC1 are associated with Wolf-Hirschhorn syndrome and multiple myeloma (34). Haploinsufficiency of nuclear receptor-binding SET domain 1 (NSD1), due to intragenic mutations or submicroscopic deletions, causes Sotos syndrome (36). Mutations in the mismatch repair gene MSH6 predispose to hereditary non-poliposis colorectal cancer (27). The protein products of these genes are involved in processes intimately linked to chromatin, such as transcriptional regulation and DNA repair. Many other PWWP domain family members have also been shown to associate with chromatin, although their functions are poorly understood (31, 34). Thus, chromatin binding might be a universal function of PWWP domains. Consistent with their possible involvement in the regulation of chromatin structure and function, PWWP domain proteins are present in eukaryotic organisms ranging from yeasts to mammals but not in prokaryotes (31, 34). Since PWWP domains show substantial structural variations (33), they may have distinct specificities and various functions.
Acknowledgments
This work was supported by grants CA82389 and GM52106 from the National Institutes of Health (to E.L.). T.C. was a recipient of a long-term fellowship from the Human Frontier Science Program.
We thank Yi Zhang for stimulating discussions.
REFERENCES
- 1.Aoki, A., I. Suetake, J. Miyagawa, T. Fujio, T. Chijiwa, H. Sasaki, and S. Tajima. 2001. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 29:3506-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 3.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T., F. M. Boisvert, D. P. Bazett-Jones, and S. Richard. 1999. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell 10:3015-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, T., and E. Li. 2004. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 60:55-89. [DOI] [PubMed] [Google Scholar]
- 6.Chen, T., Y. Ueda, J. E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, T., Y. Ueda, S. Xie, and E. Li. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277:38746-38754. [DOI] [PubMed] [Google Scholar]
- 8.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 9.Dennis, K., T. Fan, T. Geiman, Q. Yan, and K. Muegge. 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15:2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodge, J., B. H. Ramsahoye, Z. G. Wo, M. Okano, and E. Li. 2002. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene 289:41-48. [DOI] [PubMed] [Google Scholar]
- 11.Eden, A., F. Gaudet, A. Waghmare, and R. Jaenisch. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:455. [DOI] [PubMed] [Google Scholar]
- 12.Ehrlich, M. 2003. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 109:17-28. [DOI] [PubMed] [Google Scholar]
- 13.Fuks, F., W. A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudet, F., J. G. Hodgson, A. Eden, L. Jackson-Grusby, J. Dausman, J. W. Gray, H. Leonhardt, and R. Jaenisch. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489-492. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, R. S., C. Wijmenga, P. Luo, A. M. Stanek, T. K. Canfield, C. M. Weemaes, and S. M. Gartler. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96:14412-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 17.Izumoto, Y., T. Kuroda, H. Harada, T. Kishimoto, and H. Nakamura. 1997. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 238:26-32. [DOI] [PubMed] [Google Scholar]
- 18.Jeanpierre, M., C. Turleau, A. Aurias, M. Prieur, F. Ledeist, A. Fischer, and E. Viegas-Pequignot. 1993. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 2:731-735. [DOI] [PubMed] [Google Scholar]
- 19.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 20.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 21.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 22.Leonhardt, H., A. W. Page, H.-U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 23.Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3:662-673. [DOI] [PubMed] [Google Scholar]
- 24.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 25.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer-Stroh, S., N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber, and C. P. Ponting. 2003. The Tudor domain ‘royal family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28:69-74. [DOI] [PubMed] [Google Scholar]
- 27.Miyaki, M., M. Konishi, K. Tanaka, R. Kikuchi-Yanoshita, M. Muraoka, M. Yasuno, T. Igari, M. Koike, M. Chiba, and T. Mori. 1997. Germline mutation of MSH6 as the cause of hereditary nonpoliposis colorectal cancer. Nat. Genet. 17:271-272. [DOI] [PubMed] [Google Scholar]
- 28.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 29.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 30.Pradhan, S., A. Bacolla, R. D. Wells, and R. J. Roberts. 1999. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274:33002-33010. [DOI] [PubMed] [Google Scholar]
- 31.Qiu, C., K. Sawada, X. Zhang, and X. Cheng. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 9:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirohzu, H., T. Kubota, A. Kumazawa, T. Sado, T. Chijiwa, K. Inagaki, I. Suetake, S. Tajima, K. Wakui, Y. Miki, M. Hayashi, Y. Fukushima, and H. Sasaki. 2002. Three novel DNMT3B mutations in Japanese patients with ICF syndrome. Am. J. Med. Genet. 112:31-37. [DOI] [PubMed] [Google Scholar]
- 33.Slater, L. M., M. D. Allen, and M. Bycroft. 2003. Structural variation in PWWP domains. J. Mol. Biol. 330:571-576. [DOI] [PubMed] [Google Scholar]
- 34.Stec, I., S. B. Nagl, G. J. van Ommen, and J. T. den Dunnen. 2000. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473:1-5. [DOI] [PubMed] [Google Scholar]
- 35.Stec, I., T. J. Wright, G. J. van Ommen, P. A. de Boer, A. van Haeringen, A. F. Moorman, M. R. Altherr, and J. T. den Dunnen. 1998. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4; 14) multiple myeloma. Hum. Mol. Genet 7:1071-1082. [DOI] [PubMed] [Google Scholar]
- 36.Visser, R., and N. Matsumoto. 2003. Genetics of Sotos syndrome. Curr. Opin. Pediatr. 15:598-606. [DOI] [PubMed] [Google Scholar]
- 37.Wijmenga, C., R. S. Hansen, G. Gimelli, E. J. Bjorck, E. G. Davies, D. Valentine, B. H. Belohradsky, J. J. van Dongen, D. F. Smeets, L. P. van den Heuvel, J. A. Luyten, E. Strengman, C. Weemaes, and P. L. Pearson. 2000. Genetic variation in ICF syndrome: evidence for genetic heterogeneity. Hum. Mutat. 16:509-517. [DOI] [PubMed] [Google Scholar]
- 38.Xie, S., Z. Wang, M. Okano, M. Nogami, Y. Li, W. W. He, K. Okumura, and E. Li. 1999. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 236:87-95. [DOI] [PubMed] [Google Scholar]
- 39.Xu, G. L., T. H. Bestor, D. Bourc'his, C. L. Hsieh, N. Tommerup, M. Bugge, M. Hulten, X. Qu, J. J. Russo, and E. Viegas-Pequignot. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187-191. [DOI] [PubMed] [Google Scholar]
- 40.Yan, Q., E. Cho, S. Lockett, and K. Muegge. 2003. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell. Biol. 23:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoder, J. A., N. S. Soman, G. L. Verdine, and T. H. Bestor. 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270:385-395. [DOI] [PubMed] [Google Scholar]