Abstract
Short interfering RNA (siRNA) is widely used for studying post-transcriptional gene silencing and holds great promise as a tool for both identifying function of novel genes and validating drug targets. Two siRNA fragments (siRNA-a and -b), which were designed against different specific areas of coding region of the same target green fluorescent protein (GFP) gene, were used to silence GFP expression in cultured gfp transgenic cells of rice (Oryza sativa L.; OS), cotton (Gossypium hirsutum L.; GH), Fraser fir [Abies fraseri (Pursh) Poir; AF], and Virginia pine (Pinus virginiana Mill.; PV). Differential gene silencing was observed in the bombarded transgenic cells between two siRNAs, and these results were consistent with the inactivation of GFP confirmed by laser scanning microscopy, Northern blot, and siRNA analysis in tested transgenic cell cultures. These data suggest that siRNA-mediated gene inactivation can be the siRNA specific in different plant species. These results indicate that siRNA is a highly specific tool for targeted gene knockdown and for establishing siRNA-mediated gene silencing, which could be a reliable approach for large-scale screening of gene function and drug target validation.
Key words: gene inactivation, gene silencing, green fluorescent protein, short interfering RNAs, transgenic plant cells
Introduction
Post-transcriptional gene silencing (PTGS) is the plant-based silencing of an endogenous gene caused by the introduction of a homologous double-stranded RNA (dsRNA), transgene or virus. In PTGS, the transcript of the silenced gene is synthesized but does not accumulate because it is rapidly degraded (1., 2., 3.). RNA interference (RNAi) is the process whereby dsRNA directly induces the homology-dependent degradation of cognate mRNA (4., 5., 6.). It was first used by researchers studying Caenorhabditis elegans (6). Quelling is the simultaneous silencing or co-suppression of homologous endogenous genes and transgenes that has been observed in fungi. It is functional silencing of chromosomal loci that can be induced by transgenes, which was first reported in Neurospora crassa by the introduction of a transgene (7., 8., 9.). PTGS in plants, RNAi in animals, and quelling in fungi, collectively known as RNA silencing, share many components that are needed to degrade the mRNA homologous to the applied dsRNA (10., 11., 12.).
RNA silencing is thought to be involved in certain developmental or physiological processes in addition to its role in cellular resistance to viral RNA (13., 14., 15.). It has been shown to be effective in a number of organisms including Drosophila (5., 16.), nematodes (6), trypanosomes (7., 14.), mammals (17., 18., 19.) and plants (20., 21., 22., 23.). The dsRNA, either formed intracellularly or delivered exogenously, plays a central role in triggering degradation of the target mRNA (1., 22.). The dsRNA is cleaved into 21–23 nt short interfering RNA (siRNA) that binds to the RNA-induced silencing complex (RISC). RISC is a nuclease complex, composed of proteins and siRNA, which targets and destroys endogenous mRNAs complementary to the siRNA within the complex (24., 25., 26.). The siRNA is amplified by RNA-dependent RNA polymerase (RdRP) to maintain an excess molar ratio over the target mRNA (14., 15.). RdRP provides amplification by several routes, such as replication of long trigger dsRNAs or copying of short siRNAs in a primer-independent manner—siRNA-primed RdRP reaction converts target mRNA into dsRNA, as well as possibly replicating trigger dsRNA (27., 28., 29.). In animal systems, dsRNA is introduced either by injection or by feeding cells in dsRNA-containing medium (5., 6.). In plant systems, PTGS has been studied mostly by transforming plants with dsRNA-forming vectors for the selected gene of Agrobacterium (30., 31., 32., 33.), by bombardment (34), infiltration (35), or by infecting plants with viral vectors that produce dsRNA (26). In addition, Agrobacterium-mediated transient expression was successfully adopted to study PTGS in the intact tissues without generating transgenic plants (36). Most of these methods require vector construction and plant transformation.
The use of RNAi for inhibiting gene expression represents a powerful tool for exploring gene function (37., 38., 39.), identifying and validating new drug targets, and treating diseases (18., 19.). The process of RNAi is mediated by dsRNA, which is cleaved by the enzyme named Dicer into 21- to 23-nt duplexes containing a 2-nt overhang at the 3′ end of each strand. Dicer is a dsRNA-specific endonuclease responsible for processing of the long targeting dsRNA into siRNA (40., 41.). This cleavage requires ATP and RDE-4 (a C. elegans Dicer homolog), a protein containing two dsRNA-binding domains. These intermediates of RNAi and siRNA are double-stranded, and a 2-nt 3′-overhang is present in each sense and antisense strand of siRNA due to the cleavage characteristics of Dicer (39., 42.). The 5′ phosphate group of siRNA is maintained by a specific kinase; the free 3′ hydroxyl group is essential for priming of the subsequent RdRP reaction (43., 44.). These duplexes are incorporated into RISC. Directed by the antisense strand of the duplex, RISC recognizes and cleaves the target mRNA (9., 12.). Although long double-stranded RNAs invoke an interferon response, siRNAs that resemble the products produced by Dicer have been reported to specifically inhibit gene expression in many different mammalian cell lines (18., 19.). It has been shown that even single nucleotide mismatches between the antisense strand of the siRNA and target mRNA can abolish RNAi (19., 40.). In addition, mapping of mRNA cleavage sites has revealed no cleavage sites outside of the region of complementarity (18., 45.). However, the specificity of siRNA at the cellular level remains to be comprehensively studied.
For siRNAs, to be a useful tool in gene knockdown experiments, it is critical that siRNA-mediated post-transcriptional silencing be specific (18., 21.). It is not enough to simply show that a control siRNA with a scrambled nucleotide sequence fails to knock down the protein of interest or produce the same cellular phenotype (4., 21.). Ideally, the siRNA must not cause any effects other than those related to the knockdown of the target gene (18). There are several types of nonspecific effects that siRNA could potentially display (18). In addition to the possibility for cross-hybridization of the antisense strand of the siRNA to different mRNAs, siRNAs could bind in a sequence-dependent manner to various cellular proteins (18). Indeed, antisense oligonucleotides have been shown to bind to many different proteins and cause significant nonspecific effects (18., 21.). It may also be possible for siRNAs to induce common, nonspecific changes in gene expression. These nonspecific or off-target effects could complicate the interpretation of gene knockdown experiments and severely limit the utility of siRNA (17., 18.).
To examine the specificity of siRNAs, we used four plant species, rice (Oryza sativa L.; OS), cotton (Gossypium hirsutum L.; GH), Fraser fir [Abies fraseri (Pursh) Poir; AF], and Virginia pine (Pinus virginiana Mill.; PV), which are ideal for gene silencing analysis. Two siRNAs were designed against different regions of the same target gene in the four species. We hypothesized that if siRNAs elicit a specific response, then all of the siRNAs designed against the same target would be expected to produce similar gene expression signatures even though each siRNA has a different nucleotide sequence. At the same time, the green fluorescent protein (gfp) gene was used as the target gene that did not exist in the plant species tested. This test of specificity is more stringent and comprehensive than those reported thus far that followed the knockdown of particular proteins. We have analyzed the effectiveness of RNAi in the four plant species systematically by identifying RNAi lines and by using GFP as a reporter. We found that RNAi lines targeting the same gene generally reduced target transcript levels to a similar extent in the four species, and the maximal degree of reduction of target transcripts appears to be different between two siRNA fragments.
Results
Transgenic cells
To generate transgenic cell lines, 176 cell cultures from each species, which are derived from single embryos, were infected with an Agrobacterium tumefaciens strain containing pBIN-m-gfp5-ER (Figure 1), which carrys a modified kanamycin phosphotransferase gene and a modified GFP protein, with an endoplasmic reticulum targeting sequence. Twenty independent cell lines that were transformed with the pBIN-m-gfp5-ER plasmid (49., 50., 51.) and resistant to kanamycin were generated from each species. After the T-DNA insert was confirmed by both PCR and Southern blot analyses (data not shown) and measurement of growth and cell survival rates at the end of the subculture period (7 days), one transgenic cell line with high growth rate and cell survival rates containing one copy of the T-DNA insert was selected from each species as candidates for siRNA-mediated PTGS experiments. A summary of cell growth rate of all the four transgenic cell lines was demonstrated in Table 1. These transgenic cell lines were transferred weekly into fresh proliferation medium for 10 weeks to produce more cells. No background GFP expression was observed in non-transformed control cell lines.
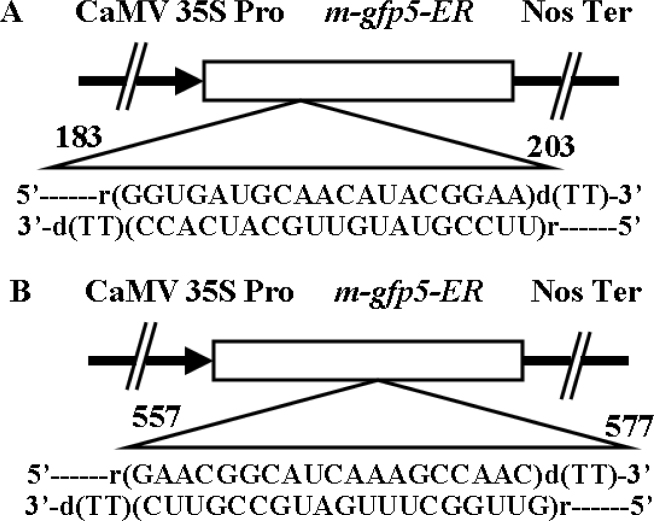
Fig. 1 A and B.
Linear maps of m-gfp5-ER gene indicating the localization of the m-gfp5-ER, a modified GFP protein with an endoplasmic reticulum targeting sequence. CaMV35SPro, the cauliflower mosaic virus 35S promoter; NosTer, the terminator from nopaline synthase gene. Arrows indicate gene translation orientation. The probe used in Northern blot analysis of transgenic cells is the 816-bp fragment of the m-gfp5-ER gene. The sequences of sense and antisense siRNA were indicated immediately below the m-gfp5-ER gene, and the positions of siRNAs were between nucleotides 183 and 203 (siRNA-a) and between nucleotides 557 and 577 (siRNA-b) of the m-gfp5-ER gene.
Table 1.
Fresh and Dry Weight Increases of Transgenic Cell Cultures in Four Plant Species
| Transgenic cell lines | Fresh weight (mg/L/day) |
Dry weight (mg/L/day) |
||
|---|---|---|---|---|
| Week 1 | Week 3 | Week 1 | Week 3 | |
| OS | 9.7±2.1a | 9.2±2.7a | 6.8±2.6a | 6.4±2.4a |
| GH | 9.6±2.4a | 9.1±2.8a | 6.7±2.5a | 6.3±2.2a |
| AF | 7.8±2.1b | 7.2±2.4b | 4.9±2.2b | 4.7±2.5b |
| PV | 7.2±2.2b | 7.1±2.3b | 4.8±2.1b | 4.5±2.2b |
Fresh and dry weight increases (mg fresh weight/liter cell cultures/day) of transgenic cell cultures in four plant species. Data represent the mean ± SD. Values followed by different letters are significantly different (α=0.05) by ANOVA.
siRNA-mediated gfp silencing
Transgenic cells with insertion of m-gfp5-ER reporter gene (Figure 1) were produced using Agrobacterium tumefaciens (Strain GV3850) mediated gene transfer as described in Tang et al. (46). After PCR and Southern blotting analyses (data not shown), transgenic cells with one copy of m-gfp5-ER were selected and used for siRNA-mediated gfp silencing. Gene silencing was observed only after transgenic cells were bombarded with siRNA to silence the mRNA transcripts of the m-gfp5-ER. Silencing of gene expression was monitored by fluorescence and confocal microscopy.
RNA-blot hybridization was used to compare the degradation of gfp mRNAs in silencing transgenic cells of the four plant species 48 h after bombardment with siRNA. Figure 2 shows that silencing of gfp expression was correlated with a dramatic decrease in gfp mRNA accumulation in the four species. The gfp transgenic cells not bombarded with siRNA demonstrated no decrease in mRNA levels, which has the same levels of mRNA from transgenic cells bombarded with siRNA at 0 h (data not shown). These results confirm that the siRNA has efficiently induced gfp silencing at the posttranscriptional level and in different plant species tested. Gene silencing in gfp transgenic single living cells was observed with laser scanning microscopy 48 h after particle bombardment (Figure 3). Differential silencing of gfp expression was observed in all the four plant species 48 h after bombardment with siRNA. More than 500 transgenic cells were monitored in individual cell line. Most of the single cells (98%) demonstrated the sequence gene silencing with the same pattern. Compared to siRNA-a, gene silencing mediated by siRNA-b was observed to be more effective in all four species.
Fig. 2.
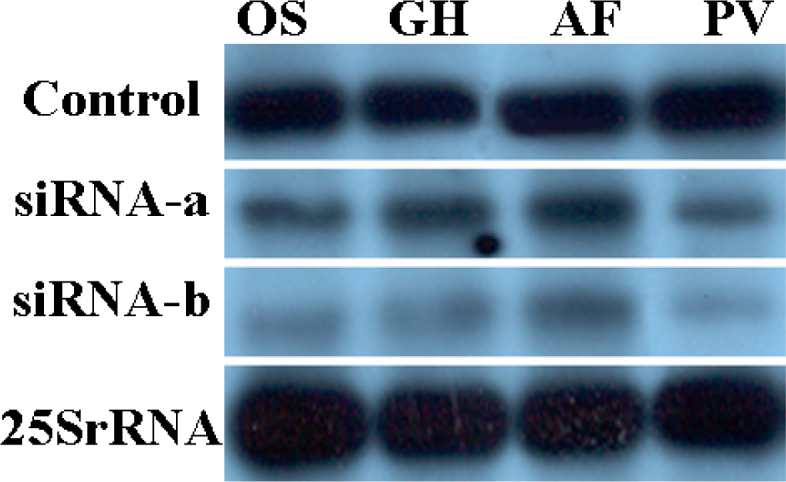
Northern blot analysis of total RNA from transgenic cell lines of rice (OS), cotton (GH), Fraser fir (AF), and Virginia pine (PV). RNA (10 µg) was extracted from transgenic cells bombarded with two siRNAs 48 h after bombardment, and was hybridized at 65°C with the 816-bp m-gfp5-ER probe corresponding to the m-gfp5-ER gene, which was labeled with DIG. The control panel is Virginia pine gfp transgenic cells that did not bombarded with siRNA. The integrity and the amount of RNA applied to each lane were verified by the control of 25S rRNA (lower panel).
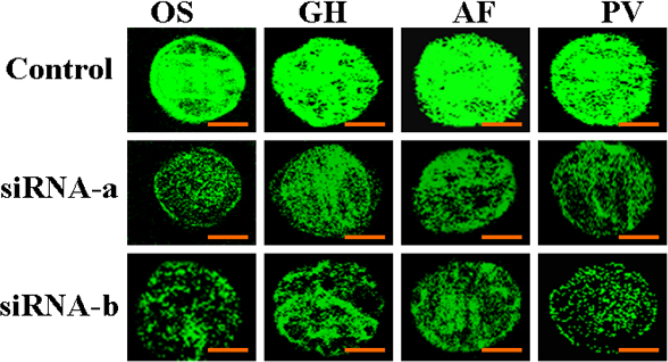
Fig. 3.
Laser scanning microscopy of silenced GFP expression by two siRNAs in transgenic cells 48 h after bombardment with siRNAs in rice (OS), cotton (GH), Fraser fir (AF), and Virginia pine (PV). The control is the GFP expression in Virginia pine gfp transgenic cells not bombarded with siRNAs. GFP fluorescence was decreased more in all transgenic cells after bombardment with siRNA-b than with siRNA-a. No GFP fluorescence was observed in non-transgenic control cells (bars = 0.04 mm).
Time lapse tracking of GFP silencing
For time lapse tracking of GFP silencing, siRNA-mediated gfp silencing was monitored by fluorescence and confocal microscopy in all the four plant species. At the same time, we collected samples for RNA-blot hybridization that were used to compare the degradation of gfp mRNAs in silencing transgenic cells at different times (0, 8, 16, 24, 32, 40, 48, and 56 h) after bombardment with siRNA. Figure 4 shows that silencing of gfp expression was correlated with a dramatic decrease in gfp mRNA accumulation in a time course. The gfp transgenic cells not bombarded with siRNA demonstrated no decrease in mRNA levels, which has the same levels of mRNA from transgenic cells bombarded with siRNA at 0 h (data not shown). Sequence gene silencing was observed in transgenic single living cells at 0, 12, 24, 36, 48, and 60 h after particle bombardment and confocal images were taken by a laser-scanning microscope (Figure 5). High-level silencing of gfp expression was observed 48 h after bombardment with siRNA. Near complete gene silencing was observed 60 h after bombardment with siRNA.
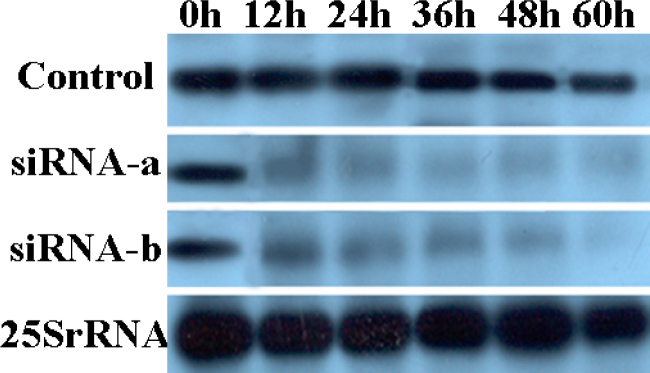
Fig. 4.
Northern blot analysis of total RNA from transgenic cell lines of Virginia pine (PV). RNA (10 µg) was extracted from transgenic cells bombarded with two siRNAs at 0 (transgenic control), 12, 24, 36, 48, and 60 h after bombardment, and was hybridized at 65°C with the 816-bp m-gfp5-ER probe corresponding to the m-gfp5-ER gene, which was labeled with DIG. The control panel is Virginia pine gfp transgenic cells that did not bombarded with siRNA. The integrity and the amount of RNA applied to each lane were verified by the control of 25S rRNA (lower panel).
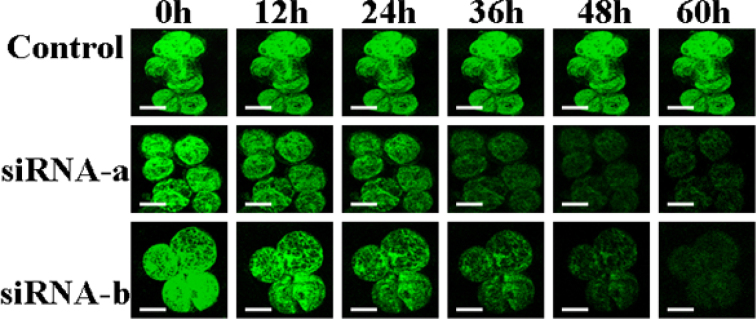
Fig. 5.
Detection of silenced GFP expression by siRNA in transgenic cells and at a time course in Virginia pine (PV). The control is the GFP expression in gfp transgenic cells not bombarded with siRNA. GFP fluorescence in gfp transgenic cells was decreased at 12, 24, 36, 48, and 60 h after bombardment with siRNA. No GFP fluorescence was observed in non-transgenic control cells (bars = 0.04 mm).
The process of gene silencing of more than 2,000 single cells obtained from four transgenic cell lines in the four plant species (one transgenic cell line from each species, 500 transgenic cells per cell line) was monitored by a time-lapse tracking technique. Three-day-old suspensions were passed through a series of sieves with successive 600, 400, 200, and 80 μm pore sizes. Tracking of gene silencing from four different transgenic cell lines has shown the same silencing pathway. An example is shown in Figure 5. No remarkable changes occurred within the first 12 h (Figure 5). Differences in GFP fluorescence appeared from 24 to 36 h after siRNAs were introduced into transgenic cells. Gene silencing in transgenic cells had continued from 12 h onward leading to expand through more than 60 h in all transgenic cell lines. These results confirm that the siRNA has efficiently induced gfp silencing at the posttranscriptional level and at a time course.
Quantitative analyses of gfp silencing
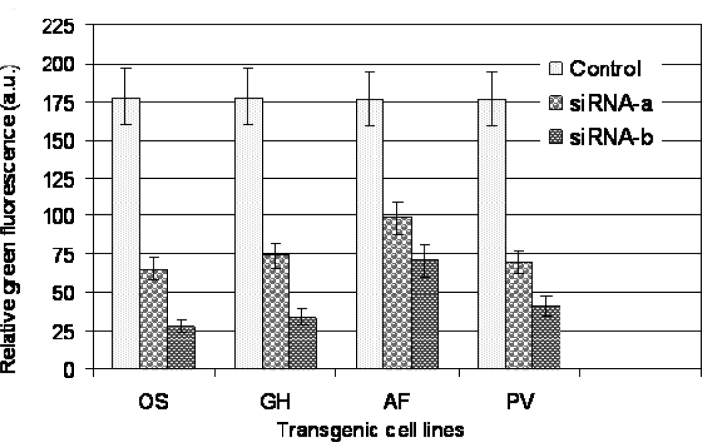
The efficiency of silencing was quantitatively determined from the confocal images taken by an LSM 510 Laser Scanning Microscope (Carl Zeiss Inc., Thornwood, USA) using excitation with the 488-nm Argon laser line and detection of emitted light between 500 and 520 nm in four transgenic cell lines OS, GH, AF, and PV. The similar level of GFP fluorescence was observed in the control cells of four species. The siRNAs used in this study significantly decreased GFP expression in the four transgenic cell lines. Both siRNA-a and siRNA-b are effective in the four species. However, more efficiency was observed in AF transgenic cell line. Compared to siRNA-b, siRNA-a was more effective in the four species (Figure 6).
Fig. 6.
Quantitative analysis of gfp expression silenced by siRNA in transgenic cells in rice (OS), cotton (GH), Fraser fir (AF), and Virginia pine (PV) 48 h after bombardment. GFP fluorescence was expressed as fluorescence intensity (arbitrary unit). Fluorescence of gfp transgenic cells without bombardment with siRNA was also presented as a control. Experiments were repeated three times, and each replicate consisted of 30–50 cells. Values represent the means ± S.D.
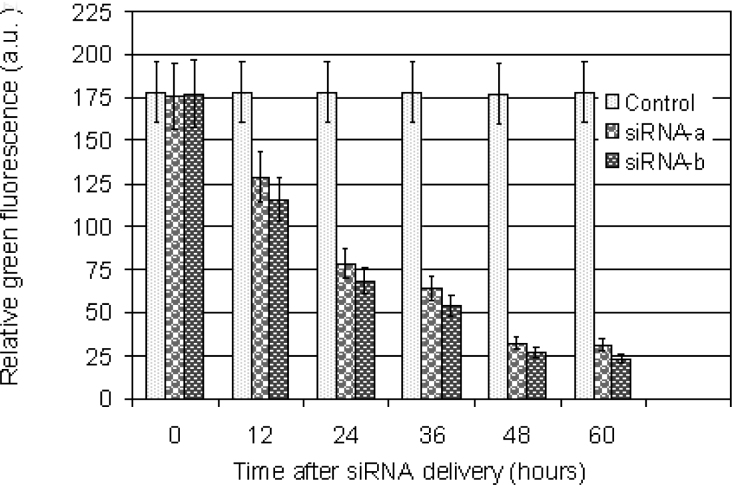
For quantitative time lapse tracking of GFP silencing, transgenic cells were observed over the 60 h culture period. Figure 7 demonstrates the dynamics of gene silencing measured by green fluorescence intensities in transgenic living cells at 0, 12, 24, 36, 48, and 60 h after particle bombardment with siRNA. A higher incidence of silencing was detected 0–48 h after bombardment with siRNA. No silencing was detected from transgenic cells without the bombardment of siRNA and non-transgenic cells (data not shown). These results confirm that the efficiency of gene silencing can be monitored by quantitative analysis of green fluorescence of silencing gfp transgenic cells. It appears that the method described here provides a reliable approach for analyses of gene silencing. To assess the effectiveness of siRNA-mediated gfp silencing in plant cell systems, we monitored gfp silencing over 60 h after transgenic cells were bombarded with siRNA by confocal images, Northern blotting, and measurement of intensities of green fluorescence. The constant results were obtained in our experiments. These results demonstrated that the method presented here could be useful in the investigation of gene silencing using GFP as a visual marker.
Fig. 7.
Quantitative analysis of gfp expression silenced by siRNA in transgenic cells in Virginia pine (PV) at 0 (transgenic control), 12, 24, 36, 48, and 60 h after bombardment. GFP fluorescence was expressed as fluorescence intensity (arbitrary unit). Fluorescence of gfp transgenic cells without bombardment with siRNA was also presented as a control. Experiments were repeated three times, and each replicate consisted of 30–50 cells. Values represent the means ± S.D.
Detection of siRNA in silenced transgenic cells
To understand the observation that gfp is silenced only in transgenic cells and to confirm the silencing capacities of siRNA against target genes, we assessed the accumulation of sequence-specific small RNAs. First, we determined whether small RNAs specific for the silencing-inducing locus could be detected. Then we examined the ability of target loci to give rise to target-specific small RNAs. We used a gfp transcript as a probe to detect small RNAs originating from the gfp sequences. The siRNA molecules were detected in samples of the four transgenic cell lines 48 h after bombardment with siRNA (Figure 8). Transgenic line containing a normally expressed gfp transgene but not bombarded with siRNA and non-transgenic cells bombarded with siRNA did not accumulate small gfp RNAs (data not shown). We conclude that small gfp RNAs accumulate only upon silencing of gfp genes in the presence of siRNA and are mainly derived from the gfp transcripts. Therefore, we suggest that siRNAs corresponding to the coding region of gfp gene may direct the formation of small RNAs in transgenic cell lines and can be effective in different plant species.
Fig. 8.
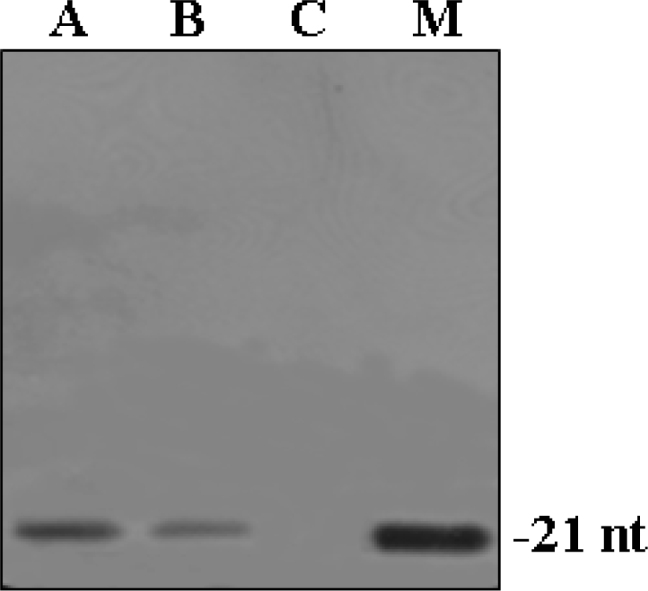
Detection of small RNAs. Low molecular weight RNA fractions were isolated from transgenic cells of Virginia pine, separated on polyacrylamide gels, blotted onto Hybond N+ membranes, and hybridized with 816-bp gfp-coding sequences. The 21-nt siRNA oligomers were used as size controls (size indicated in nucleotides). Each numbered lane contains the low molecular weight RNA fraction of transgenic cells bombarded with siRNA-a and siRNA-b. No specific signal could be detected in transgenic cells without bombardment of siRNAs with the probes. Lane A: transgenic cells bombarded with siRNA-a gave rise to small gfp-specific RNAs of approximately 21 nucleotides. Lane B: transgenic cells bombarded with siRNA-b gave rise to small gfp-specific RNAs of approximately 21 nucleotides. Lane C: gfp transgenic cells without bombardment with siRNA was also presented as a control. Lane M: the 21-nt small gfp-specific RNAs were used as a marker.
Discussion
Gene silencing of endogenous or reporter genes has been described in transgenic plants (52., 53., 54.). In cases of short hairpin RNAs and siRNA-mediated gene inactivation, it was reported that gene silencing can be stable, inducible, and reversible, and this kind of regulation of RNAi has broad applications in the areas of mammalian genetics and molecular therapeutics (18., 19., 55.). However, siRNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells (18., 21.). The gfp gene does not exist in plant cells (56), so the use of gfp transgenic cells as material to study the process of gene silencing and to quantitatively analyze the degree of transgene silencing in a time course is an ideal choice for understanding mechanism of gene silencing. In this study, we observed efficient silencing of gfp genes by the introduction of siRNA in four plant species. It was suggested that the siRNA would be a powerful tool to explore gene function. Although siRNA has been widely used for gene silencing mostly in animal cells and higher plants, this technology has not been reported in conifers. Our results showed that siRNA was effective in different plant species including conifers.
It is suggested that one of the prevalent applications of PTGS is to study the function of gene by degrading the corresponding mRNA (18., 21.). If one can readily induce PTGS in cells, PTGS can be a strong alternative to the widely used methods such as T-DNA insertional mutagenesis (4., 21.). In view of convenience of preparation and delivery of siRNA, siRNA-mediated PTGS technology may be applied to the studies of functional genomics or some other biotechnological researches (18). The observation that target transcript reduction in RNAi lines relative to wild-type varies among targets suggests that each target sequence possesses an inherent degree of susceptibility to RNAi (22., 29.). In Caenorhabditis elegans, strong RNAi effects as assessed by a phenotypic analysis were found to correlate with high expression levels of the targeted genes (6., 14.). RNA gel blot analysis of the genes targeted in our study exhibited strongly reduced target transcript levels in RNAi transgenic cells, suggesting that endogenous transcript accumulation of the targeted gene is the only target-specific determinant of RNAi effectiveness in the four species tested. However, it is reported that factors may affect RNAi effectiveness in a gene-specific manner including sequence composition, spatial and temporal gene expression patterns, and the normal RNA turnover rate of the targeted gene (57., 58., 59.).
In the case of T-DNA insertional RNA silencing, the efficiency of gene silencing was influenced by the copy number of the RNAi construct (60., 61., 62.). Single copy RNAi lines are better than those lines carrying multiple copies of the RNAi construct for PTGS analysis. Also, multiple copies of transgenes did not reduce target levels more than single copy of transgenes did (10., 63.). Previous studies demonstrated that RNAi lines targeting endogenous genes can produce a series of mutant phenotypes that vary from weak phenotypes to phenotypes resembling known null mutants of the targeted gene (43., 64., 65.). Chuang and Meyerowitz (33) showed that RNAi constructs driven by a weaker promoter are less effective than those driven by a stronger promoter. Based on our findings, we recommend that functional genomics programs seeking to produce permanent collections of RNAi lines generate single copy transgenic lines in order to maximize RNAi effectiveness and stability. An alternative approach to vary the degree of effect of RNAi may be to express dsRNA from transgene promoters of different strengths (14., 53.).
Although siRNA shows tremendous promise as a tool for targeted gene silencing, its utility will depend on its specificity including the ability to specifically knock down the target gene without interfering with the expression or function of other genes (18., 19.). In cell-based knockdown experiments, siRNA-mediated gene silencing allows for the rapid analysis of the resulting phenotype and thus provides insight into the target gene’s function in a high-throughput manner (21., 66.). Before the discovery of RNAi, antisense oligonucleotides were the primary tools for targeted gene silencing (9., 67.). However, they have been shown to cause significant nonspecific effects. There are also several nonspecific effects that could be induced by siRNAs including: (1) degradation of mRNA other than the target due to cross-hybridization followed by downstream effects, (2) binding to cellular proteins in a sequence-specific manner and all of the downstream transcriptional effects, and (3) translational silencing through miRNA effect (9., 14., 53.). Our approach used a gfp reporter gene that does not exist in plant. It provided advantages for gene silencing research because the nonspecific cellular effects of siRNA can be eliminated. In our study, siRNA only produced specific (on-target) effects, and different siRNAs against the same gene have generated highly similar gene expression signatures.
The optimized siRNA design rules and transfection conditions were useful to generate gene expression signatures for multiple siRNAs directed against different regions of the same target (4., 30.). These experiments were performed for a total of two target regions in our study. Our data indicate a very close qualitative and quantitative correlation between the expression signatures for two siRNAs against the same gene. This correlation implies that, under the optimized conditions, the effects of siRNA are limited to specific target knockdown, and suggests that, when properly designed and used, siRNA does not undergo cross-hybridization. The interactions with cellular proteins may cloud the silencing effect of an agent by limiting its availability for targeting the mRNA and cause nonspecific effects in the cell (18., 19., 42.). Furthermore, the expression signatures generated by siRNA-mediated gene knockdown are unique to each target region, ruling out the possibility that the observed correlation is due to a nonspecific cellular response to RNA.
The biochemistry of RNA-mediated post-transcriptional gene regulation has rapidly advanced through studies on nematodes (6., 9., 16., 27.). While single base-pair mismatches reduce the efficacy of RNAi, they do not permit complete discrimination between two closely related sequences (18., 38.). It would appear that the well-characterized, silenced transgenic cell lines and RNAi sequences described here will be valuable for gaining further insight to maintain gene silencing in these plant species (18., 52.). Indeed, the gene silencing assay system described here provides a versatile and rapid approach to evaluate the role of any candidate protein in gene silencing. In this study, we have demonstrated that siRNA is a highly specific tool for targeted gene knockdown in four plant species. Although a number of questions related to siRNA-mediated gene knockdown remain to be addressed, the observed differences in transgenic cells of siRNAs targeted to different regions of the same mRNA suggest that target accessibility is an important factor governing the siRNA response. Therefore, our data established siRNA-mediated gene silencing as a reliable and valuable approach for effective gene silencing. With the progresses made from genomics research (18., 21., 68.), siRNA-mediated gene silencing would be a powerful tool for gene function and plant functional genomics.
Materials and Methods
Preparation of transgenic cells
Transgenic cells of rice (Oryza sativa L.; OS), cotton (Gossypium hirsutum L.; GH), Fraser fir [Abies fraseri (Pursh) Poir; AF], and Virginia pine (Pinus virginiana Mill.; PV) with insertion of sense m-gfp5-ER reporter gene (Figure 1A) were produced using Agrobacterium tumefaciens (Strain GV3850) mediated gene transfer as described in Tang et al. (46). After PCR, Southern blotting, Northern blotting, cell growth, and green fluorescence analyses (data not shown), one transgenic cell line with one copy of sense m-gfp5-ER, middle fluorescence (150–180 fluorescence intensity), and fast growth (1.2 mg dry weight/liter of cell cultures/day) was selected from transgenic cell lines of rice, cotton, Fraser fir, and Virginia pine, respectively. Four selected gfp transgenic cell lines were used for the study of siRNA-mediated PTGS.
siRNA design and biolistic delivery
Based on the rules suggested by Elbashir et al. (4., 40.), the antisense strand of siRNA was targeted against an AA(N)21 sequence at least 100 nt downstream of the start codon. The GC content of the duplexes was kept within the 40%-70% range. In addition to these rules, we used a computer program that maximizes the hybridization specificity of siRNA. All 21-nt sequences against a given coding sequence of gfp gene were generated, which satisfied the suggested rules (4., 40.). Twenty-one-nucleotide RNAs with 3′-dTdT overhangs (Figure 1) were synthesized by QIAGEN Inc. (Valencia, USA). The siRNA sequences that were used to specifically target gfp expression in four transgenic cell lines are: (1) siRNA-a, sense strand 5′-r(GGUGAUGCAACAUACGGAA)d(TT)-3′ (nucleotides 183–203) and antisense strand 3′-d(TT)(CCACUACGUUGUAUGCCUU)r-5′ (Figure 1A); (2) siRNA-b, sense strand 5′-r(GAACGGCAUCAAAGCCAAC)d(TT)-3′ (nucleotides 557–577) and antisense strand 3′-d(TT)(CUUGCCGUAGUUUCGGUUG)r-5′ (Figure 1B). Complementary double-stranded siRNAs with 2-nt overhangs were obtained by spontaneous annealing of mixtures of the antisense and sense oligoribonucleotides at 90°C for 1 min and at room temperature for 1 h. For siRNA-mediated gfp silencing experiments, particle bombardment was used to deliver siRNA to transgenic cells by following the procedure previously described (47). Two μg of siRNA per bombardment was used on gold particles and was delivered at 1,100 psi by using a biolistic PDS-1000 He particle gun (Bio-Rad Laboratories, Hercules, USA), following the manufacturer’s recommendations.
Total RNA extraction and Northern blot analysis
Total RNA was isolated from 1.2 g transgenic cell cultures harvested through 42.5 μm filter papers and ground in liquid nitrogen using a Micro-t-midi total RNA Purification System (Invitrogen life technologies, Carlsbad, USA) following the manufacturer’s protocol. 10 µg RNA was separated by agarose-gel electrophoresis. Electrophoresis and Northern blotting of RNAs were performed as described by Tang and Tian (47). Baked blots were pre-hybridized in 1 M NaCl, 1% SDS, 10% dextran sulphate and 50 µg/mL denatured herring sperm DNA at 64°C, washed with 0.1× SSPE (1× SSPE is 180 mM NaCl, 10 mM NaH2PO4, 1 mM EDTA, pH 6.5), 0.5% SDS at 45°C. Digoxigenin (DIG)-labeling m-gfp5-ER DNA of 816 bp (Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, USA) was used as a hybridization probe. Equal loading of RNA samples was verified on the control of 25S rRNA.
Laser scanning microscopy imaging and quantitative reporter gene silencing assays
GFP expression in transgenic cell cultures was observed with a laser scanning microscope at various times after treatments. For quantitative fluorescence determinations of m-gfp5-ER activity, an LSM 510 Laser Scanning Microscope (Carl Zeiss) using excitation with the 488-nm Argon laser line and detection of emitted light between 500 and 520 nm was used to capture images. The confocal images of m-gfp5-ER expressed cells were created in the Expert Mode. Images were recorded with equal exposure time under non-saturated conditions for randomly chosen gfp expressing transgenic cells. Gene expression was quantified in both control and targeted cells. Fluorescence intensities of different samples were calculated from confocal images with the Zeiss LSM Image Examiner software. The fluorescence level was quantified separately for the whole cell by circumscribing the respective area as a region of interest. Background correction was applied by deducing fluorescence levels in a neighboring non-transgenic cell. Thirty to fifty cells were used for each sample.
Short interfering RNA detection
To detect small RNAs, the procedures described by Hamilton and Baulcombe (20) and Mette et al. (48) were adapted. Transgenic cell cultures were frozen in liquid nitrogen, and total RNA was extracted with Trizol reagent (Invitrogen), according to the manufacturer’s instructions. The lower molecular weight RNAs were recovered from the supernatant as described. For the different samples analyzed, a similar amount of RNA of the lower molecular weight RNA fraction was separated on gel (15% [v/w] polyacrylamide and 7 M urea) and transferred to Hybond N+ membranes (Amersham Biosciences, Piscataway, USA) by electroblotting. DNA oligomers were loaded on the same gels. 32P-labeled probes were synthesized in vitro from a linearized plasmid with an SP6/T7 transcription kit (Roche Diagnostics, Brussels, Belgium) and [α-32P] CTP. The probe was hydrolyzed into fragments of approximately 50 nucleotides. Hybridization and washes were performed as described (20., 48.) at 30°C. Labeled membranes were exposed to a Kodak film (Sigma, St. Louis, USA).
Statistical analyses
Data obtained from different experiments were analyzed by using the General Linear Model procedure of SAS (SAS Institute Inc., Cary, USA), employing ANOVA models. The significant differences between mean values obtained from at least three independent experiments were made with the Least Significant Difference test at 5% level of probability. Each value was presented as means standard errors of the mean, with a minimum of three replicates.
Acknowledgements
The authors are grateful to Dr. C. N. Stewart and Dr. J. Haseloff for providing us with the m-gfp5-ER constructs, to Dr. R. Qu for the facilities used for particle bombardment for part of the work, and to D. Weidner (The Flow Cytometry-Confocal Microscopy Core Facility, the Brody School of Medicine, East Carolina University, NC 27858, USA) for technical assistance with confocal microscopy for imaging and quantitative analysis of green fluorescence. We thank K. S. Rathore for providing us with the cotton and rice calluses.
This work was funded by the East Carolina Christmas Tree Program (2002).
References
- 1.Schweizer P.J. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 2000;24:895–903. doi: 10.1046/j.1365-313x.2000.00941.x. [DOI] [PubMed] [Google Scholar]
- 2.Kanno T.S. Post-transcriptional gene silencing in cultured rice cells. Plant Cell Physiol. 2000;41:321–326. doi: 10.1093/pcp/41.3.321. [DOI] [PubMed] [Google Scholar]
- 3.Akashi H.M. Suppression of gene expression by RNA interference in cultured plant cells. Antisense Nucleic Acid Drug Dev. 2001;11:359–367. doi: 10.1089/108729001753411326. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir S.M. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamore P.D. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 6.Fire A. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Cogoni C., Macino G. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 2000;10:638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 8.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 9.Vaucheret H. Post-transcriptional gene silencing in plants. J. Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- 10.Vance V., Vaucheret H. RNA silencing in plants—defense and counter defense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 11.Sijen T. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 12.Sharp P.A. RNA interference. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 13.Mourrain P. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 14.Matzke M. RNA: guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 15.Kooter J.M. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- 16.Zamore P.D. RNA interference: listening to the sound of silence. Nat. Struct. Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 17.Semizarov D. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scacheri P.C. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S. Inducible, reversible, and stable RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 21.Vanithatani R. Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells. Proc. Natl. Acad. Sci. USA. 2003;100:9632–9636. doi: 10.1073/pnas.1733874100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klahre U. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton A.J. A transgene with repeated DNA causes high frequency, post-transcriptional suppression of ACC-oxidase gene expression in tomato. Plant J. 1998;15:737–746. doi: 10.1046/j.1365-313X.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- 24.Baulcombe D.C. Gene silencing: RNA makes RNA makes no protein. Curr. Biol. 1999;9:R599–R601. doi: 10.1016/s0960-9822(99)80383-2. [DOI] [PubMed] [Google Scholar]
- 25.Béclin C. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 26.Dalmay T. An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 27.Nishikura K. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell. 2001;107:415–418. doi: 10.1016/s0092-8674(01)00581-5. [DOI] [PubMed] [Google Scholar]
- 28.Papaefthimiou I. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001;29:2395–2400. doi: 10.1093/nar/29.11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipardi C. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- 30.Van Houdt H. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J. 1997;12:379–392. [Google Scholar]
- 31.Van Houdt H. Both sense and antisense RNAs are targets for the sense transgene-induced posttranscriptional silencing mechanism. Mol. Gen. Genet. 2000;263:995–1002. doi: 10.1007/pl00008700. [DOI] [PubMed] [Google Scholar]
- 32.Waterhouse P.M. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuang C.F., Meyerowitz E.M. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweizer P. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 2000;24:895–903. doi: 10.1046/j.1365-313x.2000.00941.x. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 36.Lisa K. Silencing on the spot induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;126:930–938. doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz M.T. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabarek J.B. RNA interference by production of short hairpin dsRNA in ES cells, their differentiated derivatives, and in somatic cell lines. BioTechniques. 2003;34:734–744. doi: 10.2144/03344st02. [DOI] [PubMed] [Google Scholar]
- 39.Hutvagner G., Zamore P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 40.Elbashir S.M. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 41.Vaistij F.E. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sijen T. RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions. Plant Cell. 1996;8:2277–2294. doi: 10.1105/tpc.8.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bender J. A vicious cycle: RNA silencing and DNA methylation in plants. Cell. 2001;106:129–132. doi: 10.1016/s0092-8674(01)00441-x. [DOI] [PubMed] [Google Scholar]
- 44.Braunstein T.H. Specific degradation of 3′ regions of GUS mRNA in post-transcriptionally silenced tobacco lines may be related to 5′−3′ spreading of silencing. RNA. 2002;8:1034–1044. doi: 10.1017/s1355838202026080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoutjesdijk P.A. hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol. 2002;129:1723–1731. doi: 10.1104/pp.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang W. Regeneration of transgenic loblolly pine (Pinus taeda L.) from zygotic embryos transformed with Agrobacterium tumefaciens. Planta. 2001;213:981–989. doi: 10.1007/s004250100566. [DOI] [PubMed] [Google Scholar]
- 47.Tang W., Tian Y.C. Transgenic loblolly pine (Pinus taeda L.) plants expressing a modified delta-endotoxin gene of Bacillus thuringiensis with enhanced resistance to Dendrolimus punctatus Walker and Crypyothelea formosicola Staud. J. Exp. Bot. 2003;54:835–844. doi: 10.1093/jxb/erg071. [DOI] [PubMed] [Google Scholar]
- 48.Mette M.F. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haseloff J. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart C.N. The utility of green fluorescent protein in transgenic plants. Plant Cell Rep. 2001;20:376–382. doi: 10.1007/s002990100346. [DOI] [PubMed] [Google Scholar]
- 51.Scott A. Model system for plant cell biology: GFP imaging in living onion epidermal cells. BioTechniques. 1999;26:1125–1132. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
- 52.Van Houdt H. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 2003;131:245–253. doi: 10.1104/pp.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ling X., Li F. Silencing of antiapoptotic survivin gene by multiple approaches of RNA interference technology. BioTechniques. 2004;36:450–460. doi: 10.2144/04363RR01. [DOI] [PubMed] [Google Scholar]
- 54.Depicker A. Post-transcriptional reporter transgene silencing in transgenic tobacco. In: Grierson D., editor. Mechanisms and Applications of Gene Silencing. Nottingham University Press; Nottingham, UK: 1996. pp. 71–84. [Google Scholar]
- 55.Smith N.A. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- 56.Millwood R.J. Instrumentation and methodology for quantifying GFP fluorescence in intact plant organs. BioTechniques. 2003;34:638–643. doi: 10.2144/03343pf02. [DOI] [PubMed] [Google Scholar]
- 57.De Buck S. The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 1999;20:295–304. doi: 10.1046/j.1365-313x.1999.t01-1-00602.x. [DOI] [PubMed] [Google Scholar]
- 58.Jones L. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones A.L. De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 1998;17:6385–6393. doi: 10.1093/emboj/17.21.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han Y., Grierson D. The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol. Genet. Genomics. 2002;267:629–635. doi: 10.1007/s00438-002-0696-z. [DOI] [PubMed] [Google Scholar]
- 61.Sanders M. An active role for endogenous β-1,3-glucanase genes in transgene-mediated co-suppression in tobacco. EMBO J. 2002;21:5824–5832. doi: 10.1093/emboj/cdf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas C.L. Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 2001;25:417–425. doi: 10.1046/j.1365-313x.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- 63.Wesley S.V. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 64.Wang M.B. Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA. 2001;7:16–28. doi: 10.1017/s1355838201001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wassenegger M. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 66.Hammond S.M. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–295. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 67.Ingelbrecht I. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc. Natl. Acad. Sci. USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang W. Functional genomics of wood quality and properties. Geno. Prot. Bioinfo. 2003;1:263–278. doi: 10.1016/S1672-0229(03)01032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]