Abstract
The interaction between the cytoskeletal proteins talin and vinculin plays a key role in integrin-mediated cell adhesion and migration. We have determined the crystal structures of two domains from the talin rod spanning residues 482–789. Talin 482–655, which contains a vinculin-binding site (VBS), folds into a five-helix bundle whereas talin 656–789 is a four-helix bundle. We show that the VBS is composed of a hydrophobic surface spanning five turns of helix 4. All the key side chains from the VBS are buried and contribute to the hydrophobic core of the talin 482–655 fold. We demonstrate that the talin 482–655 five-helix bundle represents an inactive conformation, and mutations that disrupt the hydrophobic core or deletion of helix 5 are required to induce an active conformation in which the VBS is exposed. We also report the crystal structure of the N-terminal vinculin head domain in complex with an activated form of talin. Activation of the VBS in talin and the recruitment of vinculin may support the maturation of small integrin/talin complexes into more stable adhesions.
Keywords: focal adhesions, integrin, talin, vinculin
Introduction
Integrins, the principal family of adhesion molecules supporting cellular interactions with the extracellular matrix, are αβ-heterodimers that exist in both high- and low-affinity states (Hynes, 2002). The short integrin αβ cytodomains play a key role in affinity modulation, and more than 20 proteins that interact with these domains, largely the β-subunit, have been identified (Liu et al, 2000). These include talin, α-actinin, filamin, ILK and tensin, which are thought to couple integrins to F-actin and to recruit proteins involved in integrin signalling. The hypothesis that talin is a key player in integrin-mediated adhesion is supported by numerous observations: (i) microinjection of talin antibodies disrupts the cell–extracellular matrix junctions or focal adhesions formed by cells in culture (Nuckolls et al, 1992; Bolton et al, 1997); (ii) downregulation of talin using antisense RNA technology inhibits focal adhesion assembly and also cell spreading (Albiges-Rizo et al, 1995); (iii) disruption of the talin gene compromises focal adhesion assembly in ES cells (Priddle et al, 1998); (iv) gene knockout studies in Caenorhabditis elegans (Cram et al, 2003), Drosophila (Brown et al, 2002) and mouse (Monkley et al, 2000) confirm that talin is essential for integrin-mediated developmental events and (v) talin activates integrins (Tadokoro et al, 2003) by disrupting the interaction between the αβ-subunit cytodomains (Vinogradova et al, 2002; Kim et al, 2003).
The biochemical properties of talin are also consistent with a role in connecting integrins to F-actin. Talin is an elongated (2541 residues; 60 nm) flexible antiparallel dimer, with a small globular head connected to an extended rod (Winkler et al, 1997). The talin head contains an FERM domain (residues 86–400) with binding sites for several β-integrin cytodomains (Calderwood, 2004), as well as the cytodomain of the C-type lectin layilin, which colocalizes with talin in membrane ruffles (Borowsky and Hynes, 1998). The FERM domain also binds to and activates the type 1 gamma isoform of PI4′5′-kinase, which is involved in focal adhesion assembly (Di Paolo et al, 2002; Ling et al, 2002), and the protein tyrosine kinase FAK (Chen et al, 1995), which is important in focal adhesion turnover and cell migration (Ilic et al, 1997). The talin rod, which is dimeric, contains a second lower affinity integrin-binding site (residues 1984–2541) (Xing et al, 2001; Tremuth et al, 2004), a highly conserved C-terminal actin-binding site (residues 2345–2541) (Hemmings et al, 1996; McCann and Craig, 1997) and three binding sites for the cytoskeletal protein vinculin (Hemmings et al, 1996), which in turn has multiple binding partners including F-actin (Jockusch and Rudiger, 1996).
Current data suggest a model in which integrin-mediated adhesion triggers the translocation of a talin/PIP kinase complex to the membrane resulting in localized production of PIP2 (Ling et al, 2002), a molecule previously shown to activate the integrin-binding site(s) in talin (Martel et al, 2001). Talin would then bind, activate and couple integrins to F-actin, and studies using talin1 (−/−) fibroblasts show that talin is required to support the initial weak 2 pN bond between fibronectin/integrin complexes and actomyosin (Jiang et al, 2003), as well as the subsequent assembly of focal adhesions (Giannone et al, 2003). PIP2 also activates the cytoskeletal protein vinculin (Gilmore and Burridge, 1996), which although not required for focal adhesion assembly per se may stabilize the initial integrin/talin/actin complex—vinculin null cells are more motile and have fewer, smaller focal adhesions (Xu et al, 1998).
Recent structural studies have begun to shed light on the above molecular interactions. The crystal structure of part of the talin FERM domain identifies a PTB-like domain that binds to the membrane proximal NPxY motif in the β3-integrin cytodomain (Garcia-Alvarez et al, 2003). Interestingly, the same domain binds PI4′5′-kinase (Barsukov et al, 2003). The crystal structure of vinculin subdomains has also been determined (Bakolitsa et al, 1999; Izard et al, 2004) including a complex of the vinculin head bound to a synthetic talin peptide containing a minimal vinculin-binding site 1 (VBS1) and 3 (Izard et al, 2004; Izard and Vonrhein, 2004). However, to date, there is no structural data on the talin rod. Here we describe the structure of the N-terminal part of the talin rod (residues 482–789), a region that also contains one of the three vinculin-binding sites (VBS1) in talin (Hemmings et al, 1996; Bass et al, 1999). We have identified the residues involved in vinculin binding, which align on the hydrophobic face of an amphipathic helix. These residues are normally buried within the hydrophobic core of a five-helix bundle, and biochemical studies show that activation of VBS1 involves a marked conformational change in the talin rod. Our results provide novel insights into the mechanisms underlying the assembly of the talin/vinculin complex on the cytoplasmic face of integrins.
Results
Structure of the N-terminal talin rod domains
Limited proteolysis with trypsin of talin purified from chicken gizzard identified a resistant 52 kDa fragment spanning residues 482–979. The equivalent boundaries of the murine sequence were cloned and expressed in Escherichia coli, but the purified protein failed to yield crystals. Further limited proteolysis of this protein showed that the N-terminus was stable under a variety of conditions whereas the C-terminus was progressively degraded. A series of truncations at the C-terminus were engineered and expressed in E. coli and high-quality crystals were obtained from constructs encoding residues 482–655 (17 kDa) and 482–789 (30 kDa). The structure of the 17 kDa domain, talin 482–655, was determined to 2.5 Å resolution using two single anomalous diffraction data sets from a platinum derivative and selenomethionine labelled crystals (Supplementary Table 1). Subsequently, the structure of the 30 kDa construct was determined to 2.6 Å using molecular replacement with the 17 kDa model. The talin 482–789 structure is composed of an N-terminal five-helix bundle and a C-terminal four-helix bundle connected by a short loop. The two domains form a flattened elongated Z-shaped structure with dimensions 90 Å × 45 Å × 25 Å (Figure 1).
Figure 1.
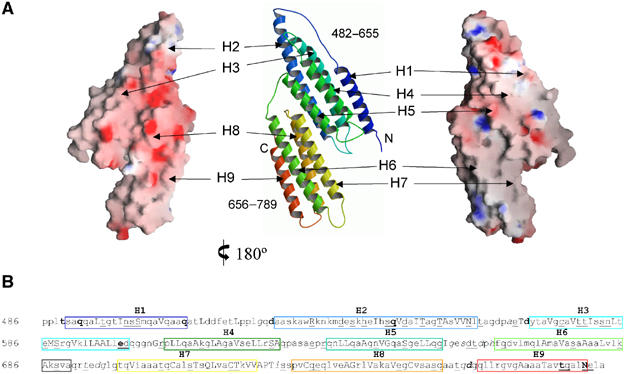
Topology and molecular surfaces of talin 482–789. (A) Ribbon diagram of the talin 482–789 topology (centre) with helices labelled H1–H9 from the N-terminus. Charged molecular surface representations of the same orientation as the ribbon diagram are shown at the far right and the opposing view 180° rotated is shown at the far left. (B) The talin 482–789 sequence annotated with three-dimensional structural features using the program JOY (Mizuguchi et al, 1998). Boxed regions span residues within α-helices and are coloured to match the equivalent helix in the topology diagram above. Key to residue annotation is as follows: solvent inaccessible, upper case; solvent accessible, lower case; positive Φ, italic; hydrogen bond to other side chain, tilde; hydrogen bond to main-chain amide, bold; hydrogen bond to main-chain carbonyl, underline.
Within talin 482–655, helices H2–H5 fold into a right-handed up and down four-helix bundle. Helix H2 is elongated compared to the other helices and bends allowing a 12-residue loop to reach over H3 and connect to H1. The addition of H1 to the H2–H5 fold represents a novel topology, and results in the structure having a triangular rather than square equatorial cross-section. The hydrophobic core is comprised predominantly of leucine, isoleucine and valine side chains. Residues from the hydrophobic faces of helices H3 and H4 form a close association at the centre of the core, and contribute eight side chains each. Helices H1 and H2 sit on opposite sides of H3 and H4 and each contributes five side chains, with the shorter helix H5 forming a more peripheral association and contributing only three side chains to the hydrophobic core. The second domain, talin 656–789, folds into a mirror image of the common up and down four-helix bundle. The first three helices H6–H8 are all approximately of the same length (∼25 residues), while the C-terminal helix H9 is slightly shorter (19 residues). All the helices in the bundle are straight, and there is a small angle between the H6, H7 and the H8, H9 helical hairpins.
The interface between talin 482–655 and talin 656–789 buries 608 Å2 and is formed by hydrophobic patches on helices H2 (Ala552, Thr551, Ala555 and Ser556) and H5 (Leu638, Gly642, Gly645 and Gly649) packing against H8 (Ser728, Val730, Cys731 and Gln734) and H7 (Gln714, Ala 717, Cys718, Val721 and Pro724), respectively. This effectively aligns the helices from the two domains in a parallel fashion resulting in two flat surfaces. A charged molecular surface representation reveals that the surface generated by helices H2, H3 and H8, H9 has an overall negative charge, whereas the H1, H4 and H5–H7 surface is relatively uncharged with hydrophobic patches in the region of H1 and H6, H7 (Figure 1A).
The 17 and 30 kDa crystal forms have residues 482–655 in common, and in each case the five-helix bundle has an identical conformation (r.m.s.d. of 0.93 Å for superposition of 167 Cα atoms). The 17 kDa crystal form has three molecules in the asymmetric unit. Two of these talin 482–655 molecules form a dimer through the association of hydrophobic patches present on helices H2 and H3. The third molecule in the asymmetric unit forms a very similar dimeric arrangement via a crystallographic two-fold axis. The top of the dimer interface is maintained, but the bottom of the interface twists such that the Cys575 residues move 2.1 Å closer to each other and the electron density indicates that they form a disulphide bridge. The 30 kDa crystal form has one molecule in the asymmetric unit and does not form any equivalent dimeric arrangements. The addition of talin 656–789 to the talin 482–655 structure results in an overall negative charge in the area of helices H2 and H3, which would preclude dimer formation.
The primary sequence of the entire talin rod is characterized by a high content of alanine and a low percentage of aromatic residues (McLachlan et al, 1994). Within the structure, there are 57 alanine residues constituting 19% of the total. The alanines cluster within the α-helices but do not contribute significantly to the hydrophobic core, which is predominantly formed by leucine, valine and isoleucine residues. Instead, the alanines lie on the surface or form packing interactions with adjacent helices allowing a close antiparallel alignment (Figure 1B). The talin structure has four aromatic side chains (Phe516, Trp531, Tyr570 and Phe664) and these lie at the top and bottom of the helical bundles, and fill in the gaps between loops at the termini of the helices. The sequence of the remaining ∼1800 residues of the talin rod has very similar characteristics to talin 482–789, indicating that the four helix bundle will most likely be a recurring motif along its length. However, the high alanine content and repetitive nature of the talin amphipathic helices makes it difficult to predict exact N- and C-terminal boundaries for other four-helix bundles within the rod.
Mapping the VBS in talin 482–655
The VBS at the N-terminus of the talin rod, which was mapped using a yeast two-hybrid assay (Bass et al, 1999), spans residues 606–636 (VBS1), and encompasses the length of helix H4 and the H4, H5 loop. The interaction between talin VBS1 and the vinculin head domain (Vh′ residues 1–258) is of high affinity, with a binding constant of ∼39 nM (Bass et al, 2002). In order to define the underlying principles of the VBS1/Vh′ interaction, we synthesized a series of VBS1 peptides containing alanine substitutions. This array was prepared by SPOT synthesis (Frank and Overwin, 1996) on a cellulose membrane and assayed simultaneously for any effects on binding to Vh′ using an ELISA type procedure. A total of 24 peptides were synthesized including single, double, triple and quadruple substitutions by alanine, and also single substitutions to glutamate in circumstances where a change to alanine was too conservative. The results are shown in Figure 2A and are mapped onto the structure of talin 482–655 in Figure 2B. Reductions in binding result from substitution of Leu608, Ala612, Leu615, Val619 and Leu623 on the buried face of helix H4. However, the most marked effects are only observed with triple and quadruple substitutions at these positions (Figure 2A, A1–A5), while double substitutions resulted in more moderate reductions in binding to Vh′. Substitutions that are distributed more widely along H4 (Figure 2A, B1–B4 and B9) show a stronger effect than those where adjacent residues are changed, that is, B7 (Leu608 and Leu609) and B8 (Leu622 and Leu623). Compared to the double alanine substitutions, single glutamate substitutions at Ala612, Val619 and Leu623 showed more striking reductions in binding (Figure 2A, C2, C4 and C5), with little change observed for Leu609, an increase in binding observed for Gly614 and no change for Ser625 (Figure 2A, C1, C3 and C6). Remarkably, double substitutions of surface residues Arg606, Gln610 or Glu621, Arg624 resulted in an increase in Vh′ binding (Figure 2A, B11 and B13) with a similar effect observed for the single substitution of Lys613 (Figure 2A, B12). These data implicate six key hydrophobic residues from the buried face of helix H4 in forming the principal contact with Vh′, but the presence of charged residues on the exposed surface can affect the binding in a negative fashion, probably by attracting a larger water structure to the VBS (Figure 2B).
Figure 2.
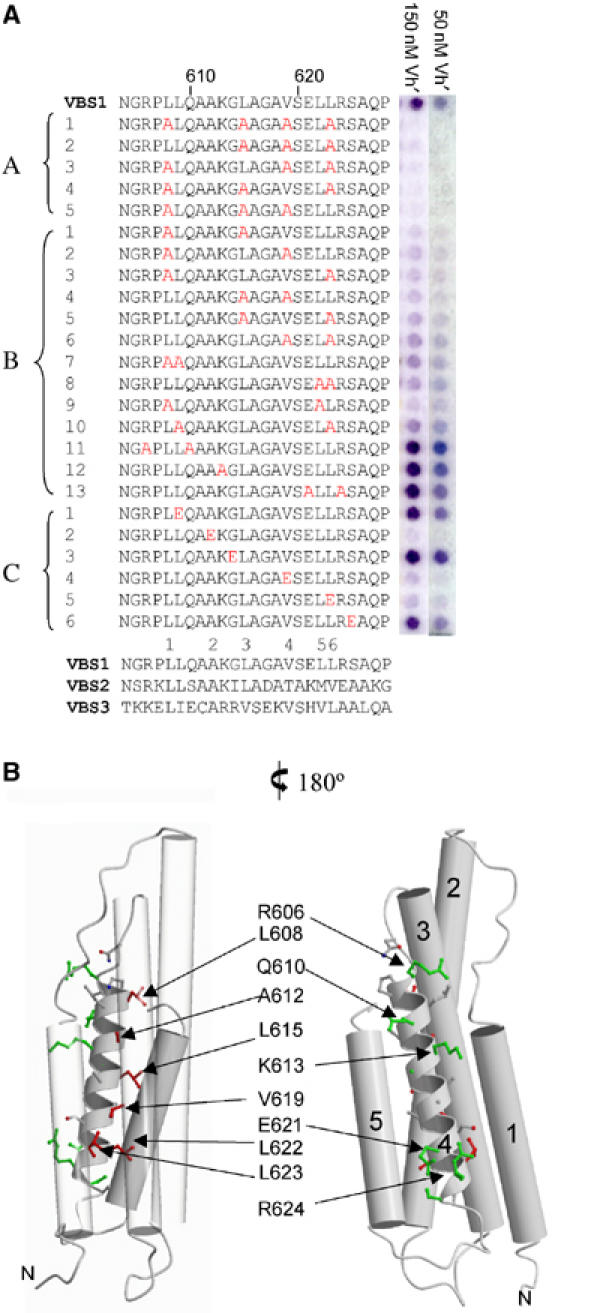
Vinculin head binding to talin peptide spot arrays. (A) Analysis of Vh′ binding to talin VBS1 and a series of 24 VBS1 synthetic peptide 25-mers with alanine and glutamic acid residues substituted into various positions. For each peptide, the substituted positions are highlighted in red. Peptides A1–A5 represent triple and quadruple alanine substitutions. Peptides B1–B13 represent single and double alanine substitutions. Peptides C1–C6 represent single glutamic acid substitutions. The VBS-like peptides were spot-synthesized on a nitrocellulose membrane (Frank and Overwin, 1996) with 0.5 nmol of peptide per spot and bound Vh′ was detected using a polyclonal GST antibody. The blot was assayed with 50 and 150 nM Vh′ domain with each spot shown to the right of the respective peptide sequence. For comparison, the talin rod VBS2 and VBS3 sequences are shown aligned with VBS1 (Bass et al, 1999). (B) Two views of the talin 482–655 structure are illustrated with residues implicated in Vh′ binding from the peptide substitution experiments highlighted. Residues are colour coded red for a reduction in binding and green for those that were equivalent to the wild type or result in an apparent increase in binding. Helices H1, H2, H3 and H5 are shown as cylinders and the VBS1 helix H4 is shown as a helical ribbon. On the left-hand view, helices H1, H2 and H5 are transparent to illustrate the VBS side chains on the buried face of H4.
Talin 482–655 requires activation to bind Vh′
The H4 side chains identified as important in binding to Vh′ are all buried within the core of the talin 482–655 fold (Figure 2B) forming hydrophobic contacts with residues from the other helices, in particular H3. The conformation of talin 482–655 observed in the crystal structures would therefore be predicted to be inactive in terms of Vh′ binding. This notion is reinforced by data from a yeast two-hybrid assay in which it is apparent that the strength of Vh′ binding to VBS1 is critically dependent on the talin construct used as bait (Bass et al, 1999). Thus talin polypeptides spanning talin 498–655, now shown to comprise helices H1–H5, bound to vinculin over 20-fold less well than a truncated construct spanning residues 498–636, which contains only helices H1–H4. Furthermore, a construct spanning residues 606–636 and containing just H4 also bound around five times more strongly than the H1–H5 talin polypeptide. Using NMR spectroscopy we were able to provide a structural explanation for these observations from the yeast two-hybrid screen and also to monitor the talin–vinculin interaction. The [1H,15N]HSQC spectrum of uniformly 15N-labelled talin 482–655 (Figure 3A) has high chemical shift dispersion and uniform cross-peak linewidth, demonstrating a stably folded state of the protein in solution. A shorter construct, talin 482–636, spanning H1–H4 but missing helix H5 yields a spectrum (Figure 3B) containing a population of sharp cross-peaks with low chemical shift dispersion and a population of very broad cross-peaks that have significantly larger chemical shift dispersion (Figure 3B). These spectral characteristics indicate that talin 482–636 contains both unfolded highly mobile regions, corresponding to the sharp signals in the spectrum, and more structured regions giving rise to the broad signals. The signals from the latter regions are broader than those in the spectrum of talin 482–655, suggesting the presence of exchange broadening and hence of an interconversion of the more structured region(s) of talin 482–636 between more than one conformation. Thus the removal of the C-terminal helix clearly destabilizes the helical bundle and leads to the partial unfolding of the protein. This conclusion is supported by its high apparent molecular weight observed using gel filtration chromatography (Figure 4A) described in the following section.
Figure 3.
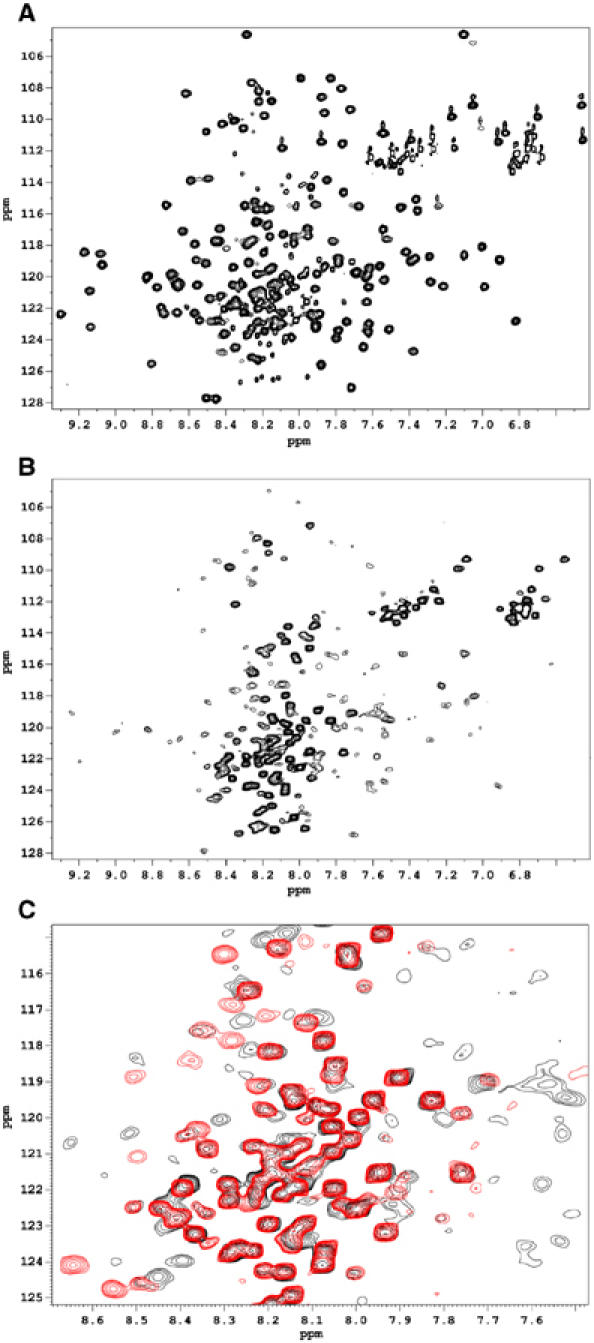
NMR analysis of the talin/vinculin interaction. Two-dimensional [15N/1H]HSQC spectra of uniformly 15N-labelled talin polypeptides. (A) Free talin (482–655). (B) Free talin (482–636). (C) Superposition of the spectra of talin (482–636) free in solution (black) and in the presence of 1.5 molar excess of Vh′ (red).
Figure 4.
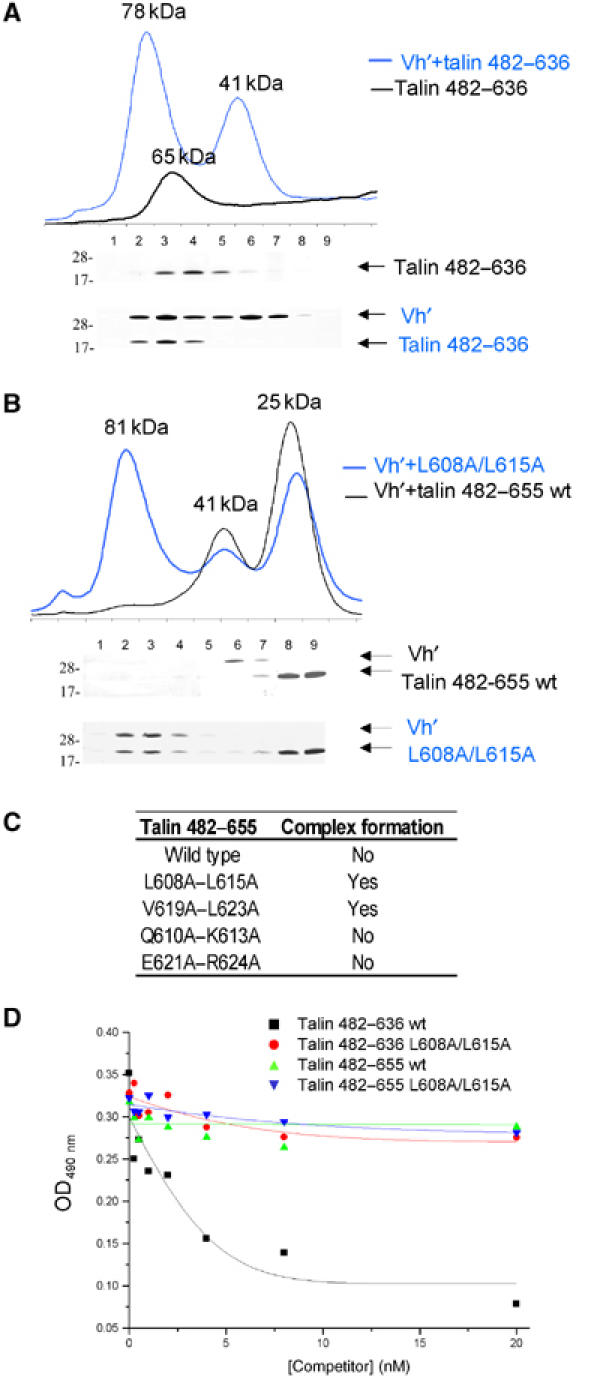
Binding of Vh′ to activated talin mutants. Superdex-75 (10/30) gel filtration chromatography of wild-type and mutant talin polypeptides in the presence/absence of the Vh′ domain. In each case, 0.5 ml fractions were collected and analysed using a Coomassie-stained 4–12% gradient SDS–PAGE gel, which is shown beneath each elution profile. The molecular weight markers of 28 and 17 kDa are indicated. (A) The profile for talin 482–636 is shown as a black line and the mixture with Vh′ as a blue line. Molecular weights as estimated from standards are indicated. An excess of Vh′ is revealed as a 41 kDa peak in the blue profile. (B) Talin 482–655 and a double mutant L608A–L615A complex with Vh′ are shown as black and blue lines, respectively. (C) Tabulated summary of the gel filtration experiments shown in panels A and B and equivalent experiments performed on double alanine mutants V619A–L623A, Q610A–K613A and E621A–R624A where elution profiles are not shown. (D) Binding of GST-Vh′ to microtitre wells coated with the activated talin 482–636 polypeptide. The effects of preincubating GST-Vh′ with increasing concentrations of various talin polypeptides are shown.
The addition of Vh′ has very different effects on the NMR spectra of the two talin fragments. In the case of talin 482–655, only gradual small chemical shift changes were observed for a limited number of cross-peaks over the full range of concentrations used, up to six-fold molar excess of Vh′ (data not shown), indicating a very weak interaction between talin 482–655 and Vh′. With the shorter talin fragment, addition of Vh′ leads to the appearance of a new set of cross-peaks and a decrease in the intensities of the cross-peaks corresponding to the free state of talin 482–636. When the Vh′ concentration exceeds that of talin 482–636, the signals of the free form disappear completely and only the signals of the complex are observed (Figure 3C). This behaviour is characteristic of a high-affinity interaction between the two proteins with slow exchange between the free and bound states. Most of the broad resonances disappear from the spectra of the complex as the overall mobility of talin 482–636 decreases due to the increase of the molecular weight on complex formation. At the same time, a large number of sharp signals with low chemical shift dispersion are observed in the spectrum of the fully formed complex, similar to those observed for the free protein. Indeed, many of these sharp cross-peaks have similar chemical shifts in both the free and the bound states, most likely due to the same regions being unfolded in both cases. However, some of the sharp cross-peaks are observed in significantly different positions in the complex and the free form of talin 482–636 indicating changes in the internal dynamics and/or structure of talin upon complex formation.
Further characterization of the talin VBS1/Vh′ interaction
In order to further characterize binding of Vh′ to talin 482–655 and the truncated mutant 482–636, we used analytical gel filtration. Both talin 482–655 (18 kDa) and Vh′ (29 kDa) eluted as monomers with molecular weights corresponding to 25 and 41 kDa proteins, respectively. Talin 482–636 (16 kDa) eluted at 65 kDa, which concurs with the NMR data indicating that it has a partially disordered or extended conformation. Approximately equimolar mixtures of talin 482–655 and Vh′ showed no indication of complex formation, whereas similar experiments with talin 482–636 and Vh′ resulted in a 78 kDa peak that contained both proteins in a 1:1 complex as revealed by SDS–PAGE (Figure 4A). These data confirm the observations from NMR that talin 482–636 but not talin 482–655 binds tightly to Vh′, consistent with the idea that helix H5 masks VBS1 in the latter construct.
We also analysed Vh′ binding to a series of talin 482–655 and talin 482–636 mutants, the design of which was directed by data from the synthetic peptide-binding experiments. Remarkably, a talin 482–655 double mutant in which the buried Leu608 and Leu615 were substituted with alanine (B1 in Figure 2A) showed a similar ‘gain of function' to the truncated mutant lacking helix H5. The talin 482–655 L608A, L615A mutant alone eluted at the same molecular weight as wild type, but an equimolar mixture of the mutant with Vh′ resulted in an 81 kDa peak that contained both polypeptides (Figure 4B). A similar gain of function was also found with the talin 482–655 double mutant in which the buried Val619 and Leu623 were substituted with alanine (data not shown). In contrast, double mutations of residues on the exposed surface of the VBS1 helix, Q610A and K613A or E621A and R624A, showed the same profiles as wild type, and no complex formation was detected (Figure 4C).
Normally, mutations disrupting the folded structure of a protein would be expected to result in loss of activity. In this case, substitution of the buried leucines by alanine most likely destabilizes the hydrophobic core H4, H3 interaction, shifting the equilibrium towards an active conformation in which the VBS1 in H4 is exposed. However, the finding that the VBS1 L608A/L615A and V619A/L623A double mutants are still able to bind to Vh′ is surprising given that these residues are implicated in binding of a synthetic VBS1 peptide to Vh′ (Figure 2A). Therefore, we compared the relative abilities of the various talin polypeptides to inhibit binding of GST-Vh′ to talin 482–636 immobilized on plastic microtitre plates (Figure 4D). As expected, preincubation of GST-Vh′ with increasing concentrations of talin 482–636 in solution efficiently inhibited binding of GST-Vh′ to the talin-coated wells (IC50∼3 nM), while talin 482–655 was a much less effective competitor. Similarly, neither talin 482–636 nor 482–655 containing an L608A/L615A double mutation were able to compete effectively with talin 482–636 for binding to Vh′. The results confirm that the buried hydrophobic residues in H4 are important determinants of Vh′ binding, although they also play a role in stabilizing the inactive talin 482–655 five-helix bundle.
Structure of the talin Vh′/talin complex
The complex formed by talin 482–636 and Vh′ described above was purified using preparative gel filtration and crystals grew from the resulting concentrated samples in space group P21212. The structure was determined to 2.0 Å and refined to an Rfactor of 0.229 (Supplementary Table 1). One complex is observed in the asymmetric unit with residues 1–255 modelled for vinculin and residues 605–628 for talin (Figure 5A and Supplementary Figure S2). The structure reveals that the hydrophobic face of the talin VBS helix is indeed buried within the complex and residues identified from the peptide analysis form key interactions. The overall organization of the Vh′/VBS1 structure is topologically equivalent to the Vh′/VBS3 and Vh′/VBS1 synthetic peptide complexes (Izard et al, 2004; Izard and Vonrhein, 2004) and consists of two subdomains. At the N-terminus, four helices (α1–α4) from Vh′ surround residues 605–636 from the talin VBS1. This is connected to the second Vh′ domain comprising the C-terminal four-helix bundle through an elongated helix α4. A remarkable feature of the structure is that the manner in which the Vh′ helices α1–α4 surround VBS1 results in a five-helix architecture, which is identical to the talin 482–655 fold (Figure 5B). Vh′ helices α1, α2, α3 and α4 occupy the positions equivalent to talin H1, H5, H2 and H3, respectively. Key hydrophobic contacts with VBS1 within the core of the talin structure are substituted by similar side chains from the Vh′ helices.
Figure 5.
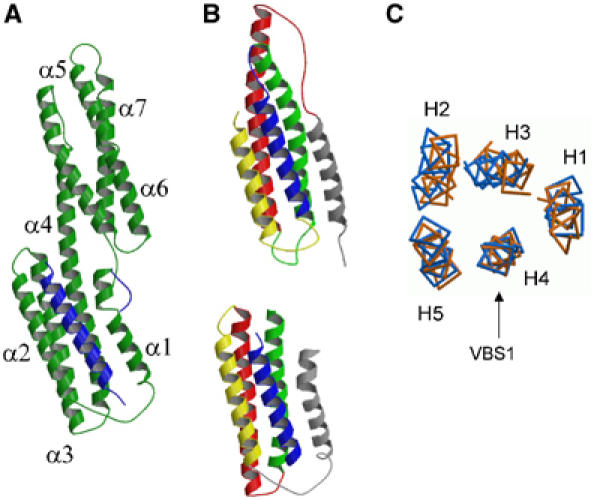
Structure and comparison of Vh′/VBS1 and talin 482–655. (A) Complex crystal structure of Vh′ residues (green) and talin VBS1 (blue). (B) Topological equivalence of the VBS1 helix (blue) within the talin 482–655 structure (top) and the Vh′/VBS1 complex structure (bottom). (C) Cα backbone traces of the superposed talin 482–655 (blue) and the structure of the Vh′/VBS1 complex (orange). The structures were superposed based on 15 Cα atoms spanning the five turns of the VBS1 helix. The helices from talin 482–655 are numbered.
Although residues 605–636 from the VBS1 helix are clearly visible sandwiched between helices α1 and α2 from Vh′, no further density for the remaining talin helices is observed. An SDS–PAGE gel of the washed Vh′/talin 482–636 complex crystals revealed a clear band for Vh′ but no observable band for talin 482–636. Clearly in complex with Vh′, the remaining helices of the activated talin 482–636 are partially disordered and are proteolysed during the crystallization. We carried out a series of tests to examine the proteolytic sensitivity of talin 482–636 and the talin 482–655 L608A/L615A mutant and compared these to wild-type talin 482–655. Figure 6A shows that in the presence of trypsin, wild-type 482–655 is stable whereas talin 482–636 and the talin 482–655 L608A/L615A mutant show increasing sensitivity to proteoylsis with the former being completely degraded after 1 h treatment. Furthermore, Vh′ is sensitive to proteolysis in its uncomplexed form. Addition of talin 482–636, but not talin 482–655, results in a stable Vh′/VBS1 complex with the remaining talin helices degraded. These data indicate that in isolation Vh′ and talin 482–636 both have conformationally dynamic folds whereas, upon binding, a structurally stable complex is formed between Vh′ and residues spanning the VBS1 from talin.
Figure 6.
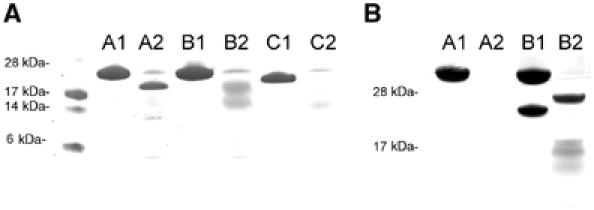
Limited proteolysis of talin and vinculin domains. (A) SDS–PAGE. Lane A1: talin 482–655 wild type untreated; A2: trypsin treated; B1: talin 482–655 L608A/L615A double mutant; B2: trypsin treated; C1: talin 482–636; C2: trypsin treated. Talin 482–655 is resistant to proteolysis, whereas the activated mutant talin 482–655 L608A/L615A and talin 482–636 are increasingly susceptible and most likely exist in an unfolded or a partially unfolded state. The reduction in molecular weight observed in lane A2 results from the cleavage of the talin 482–655 histidine tag as confirmed by N-terminal sequencing. (B) SDS–PAGE. Lane A1: vinculin 1–258 (Vh′) untreated; A2: Vh′ trypsin treated; B1; Vh′, talin 482–636 1:1 mixture; B2: trypsin treated. Vh′ is sensitive to proteolysis and is degraded by trypsin. When Vh′ is incubated with talin 482–636, Vh′ becomes partially protease resistant. The remaining structure of the talin helices is degraded. All proteolysis experiments were carried out using a 1:10 wt/wt trypsin incubated for 1 h at 20°C.
Discussion
The talin 482–655 and talin 656–789 structures both have architectures based on a four-helix bundle. The sequence of the remaining 1760 residues of the talin rod has very similar characteristics to these structures and the four-helix bundle will most likely be a recurring motif along the length of the rod. The high alanine content and repetitive nature of the talin amphipathic helices makes it difficult to predict exact N- and C-terminal boundaries for other five/four-helix bundles within talin. Nevertheless, it is likely that the hydrophobic surfaces of VBS2 and VBS3 will also be buried within a similar architecture observed for VBS1.
Our data reveal that the formation of the complex between the talin rod VBS1 and vinculin requires prior unfolding of the stable talin 482–655 domain. Once released from the talin hydrophobic core, the VBS1 helix is then available to induce the ‘bundle conversion' conformational change within Vh′ thereby displacing the intramolecular interaction with Vt and allowing vinculin to bind actin (Figure 7; Bass et al, 2002). Superimposition of the VBS1 helix from the Vh′/VBS1 complex and talin 482–655 (Figure 5C) reveals that these structures have a very similar architecture. Thus, the environment of talin VBS1 (H4) is the same in the talin 482–655 domain and in the Vh′/VBS1 complex, one set of hydrophobic interactions being replaced by another.
Figure 7.
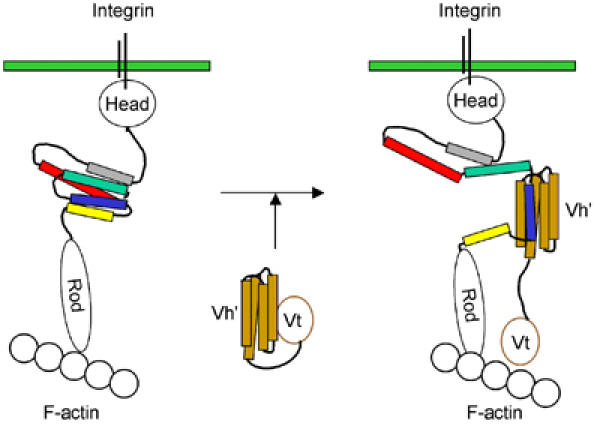
Schematic diagram showing the binding/activation of talin and vinculin. Talin is shown bound to integrins via the talin head domain and to F-actin via the C-terminal region of the talin rod. The structure of residues 482–655 (which contains the vinculin-binding site VBS1) is indicated using the same colour scheme as in Figure 1. In this conformation, talin is unable to bind vinculin because the residues involved in vinculin binding to helix 4 (blue) are buried in the hydrophobic core of the five-helix bundle. In order to bind vinculin, talin must undergo a major conformational change in which VBS1 is released from the core of the talin 482–655 structure. The talin VBS1 helix is then available to activate vinculin by inducing a conformational change within Vh′, which displaces Vt, allowing Vt to bind F-actin. Upon binding Vh′, the talin VBS inserts itself into the Vh′ N-terminal domain converting this four-helix bundle (orange) into a stable five-helix bundle.
There is a striking similarity between the regulation of the talin rod VBS1 and the exchangeable apolipoproteins, which cycle between stable helical bundles and extended membrane-bound forms. Here the hydrophobic lipid-binding surface is also buried in the inactive form within the core of a helical bundle with similar topological elements to talin 482–655 (Supplementary Figure S1). Our data showing that deletion of H5 results in activation of talin VBS1 are also reminiscent of apolipoprotein A/I whereby a 43 residues truncation has been observed to result in helical re-arrangement and conformational change to the active ligand-binding form (Borhani et al, 1997). A truncation mutant of apolipoprotein Ia has been crystallized, and the structure reveals that the helices are intact, but they are fully extended into a horseshoe shape. The authors propose a model in which the hydrophobic interactions that stabilized the helical bundle are replaced by the hydrophobic lipid core of the lipoprotein particle. What happens to the remaining talin 482–655 amphipathic helices when H4 is released and complexed with vinculin remains to be established. The limited proteolysis data provide evidence that the remaining helices do not re-arrange into a stably folded conformation. Furthermore, the 1:1 stoichiometry of binding indicates stabilization by formation of talin dimers, as can occur in activated apolipoproteins A/I in the absence of lipids, is unlikely (Borhani et al, 1997). In this context, it is interesting that talin has been shown to associate with liposomal membranes (Saitoh et al, 1998) raising the possibility that the hydrophobic faces of the amphipathic helices exposed upon talin activation may insert into the plasma membrane.
There are several mechanisms that might lead to the exposure of talin VBS1 in vivo. PIP2 has been implicated in the allosteric activation of the integrin-binding site(s) in talin (Martel et al, 2001) and also in the activation of vinculin (Gilmore and Burridge, 1996). However, to date only the talin head has been shown to bind acidic phospholipids (Niggli et al, 1994). Interestingly, the fibronectin self-association sites important in fibrillogenesis are exposed by mechanical stretch (Erickson, 2002), and stretch has also been shown to increase the capacity of Triton X-100 cytoskeleton preparations to bind FAK, paxillin, and p130Cas, although binding of vinculin was not upregulated (Sawada and Sheetz, 2002). External mechanical force has been shown to induce focal adhesion assembly (Bershadsky et al, 2003), and talin is essential for this process and for the recruitment of vinculin to fibronectin-coated glass beads (Giannone et al, 2003). Laser tweezers studies show that talin is required for the formation of the initial weak 2 pN linkage between a fibronectin/integrin complex and actomyosin (Jiang et al, 2003). Reinforcement of this link by the recruitment of additional components is thought to be essential for focal adhesion assembly. Since talin is clearly subject to the force exerted by actomyosin contraction, it is possible that stretch activates talin VBS1 allowing recruitment of vinculin and the maturation of small focal complexes into larger focal adhesions (Galbraith et al, 2002).
Materials and methods
Protein expression and purification
cDNAs encoding talin polypeptides (482–655, 482–789 and 482–636) were synthesized by PCR using the mouse talin-1 cDNA as template, and cloned into expression vector pET15b (Novagen, Cambridge Bioscience, Cambridge). E. coli BL21 (DE3) or B834 strains (for selenomethionine incorporation) were transformed and cultured in appropriate minimal media. Cells were lysed in 10 mM Tris–HCl pH 8.0, 0.5 M NaCl and 10 mM imidazole. The recombinant polypeptides were extracted by washing with 10 mM Tris–HCl pH 8.0, 0.5 M NaCl and 0.5 M imidazole and dialysed against 10 mM Tris–HCl pH 8.0, 10 mM NaCl, 2 mM EDTA and 2 mM DTT. The polypeptides were then applied to a monoQ ion exchange column (Bio-Rad) and eluted with a 10 mM to 0.5 M NaCl gradient. All mutants prepared were purified using Talon metal affinity resin (Clontech). Glutathione S-transferase (GST) vinculin head (1–258) was expressed and purified as previously described by Bass et al (1999). Recombinant His-tagged vinculin head (1–258) constructs were expressed using the pET15b expression vector and purified as described for the talin polypeptides.
Data collection, structure determination and refinement
Crystals of talin 482–655 were grown in 0.1 M NH4Ac pH 4.6, 0.2 M NH4SO4 and 30% PEG 2KMME at 20°C. The crystals belonged to the space group P3221 with cell dimensions a=95.7 Å, b=95.7 Å, c=115.2 Å, α=90, β=90 and γ=120. The asymmetric unit contained three molecules with a 51% solvent content. Data were collected at the European Synchrotron Radiation Facility (ESRF), beamline 14-4 (λ=0.979). Data sets were collected for native as well as selenomethionine and diiodobis(ethylenediamine) diplatinum(II)nitrate (PIP) derivatized crystals (Supplementary Table 1). Data sets were reduced using DENZO and scaled using Scalepack (HKL Research Inc.). The sites of selenium and platinum atoms were determined using SOLVE (Terwilliger and Berendzen, 1999). Phases were combined and the quality of the resulting map was significantly improved by solvent flattening using the program DM (CCP4 suite). The model of talin 482–655 was built using XtalView (McRee, 1999) and refinement carried out using REFMAC5 (Murshudov et al, 1999). Procheck revealed that 90.3% of the residues lie in most favoured regions, 6.0% in additionally allowed regions and 3.7% in generously allowed regions. The final Rcryst and Rfree for all data between 30 and 2.5 Å were 0.241 and 0.299, respectively. Crystals of talin 482–789 were grown in 0.1 M sodium citrate pH 5.6, 20% PEG 6K and 0.1 M MgCl2 at 20°C. The crystals belonged to the space group R32 with cell dimensions a=105.8 Å, b=105.8 Å, c=173.8 Å, α=90, β=90 and γ=120. The asymmetric unit contained one talin 482–789 molecule with 52% solvent content. Data were collected at the ESRF, beamline 14-1 (λ=0.934). The structure was solved by molecular replacement using AmoRe (CCP4 suite) with the talin 482–655 structure used as the search model. The highest peak in the cross-rotation function gave the correct orientation of the monomer. The translation function gave a strong solution in space group R32 with an Rcryst of 0.433. After rigid body refinement of the model, the 2FoFc and FoFc maps revealed the additional electron density encompassing talin 656–789 helices. The map was further improved by solvent flattening using DM. The model was constructed and refined as previously described with a resultant Rcryst of 0.256 for all data between 20 and 2.6 Å and an Rfree of 0.280. The main-chain torsion angles of 87.5% of the residues lie within most favoured regions, 8% in additional favoured regions and 4.5% in generously allowed regions.
The complex of talin 482–636 with Vh′ was purified using gel filtration chromatography (Hiload Superdex 75). The complex was concentrated to 7.5 mg/ml. Crystals were grown in 30% PEG 5000, 0.1 M MES pH 6.5 and 0.2 M ammonium sulphate at 20°C. The crystals belonged to the space group P21212 with cell dimensions a=51.64 Å, b=71.42 Å, c=96.09 Å, α=90, β=90 and γ=90. Data were collected at the ESRF, beamline 14-4, using an ADSC CCD detector (λ=0.972). The structure was solved by molecular replacement using AmoRe (CCP4 suite). The structure of the Vh′/VBS3 peptide (unpublished data) was used as a search model. The highest peak in the cross-rotation function gave the correct orientation of the monomer. The translation function gave a solution with Rfactor=0.501, which after rigid body fitting dropped to 0.396. The model was built using XtalView (McRee, 1999) and refinement was carried out using REFMAC (CCP4). The model was refined to final Rfactor=0.229 for all data between 30 and 2.0 Å and Rfree=0.286. The main-chain torsion angles of 94.4% of the residues lie within most favoured regions, 4% in additional favoured regions and 1.6% in generously allowed regions.
Vinculin head binding to the peptide spot array
VBS peptides (25-mers) were spot-synthesized on a nitrocellulose membrane (Frank and Overwin, 1996) with 0.5 nmol of peptide per spot. Membranes were blocked in Tris-buffered saline (TBS, 50 mM Tris–HCl, pH 7.0) with 1% fetal bovine serum overnight. Membranes were overlaid for 1 h with GST-Vh′ (50 or 150 nM) in TBS with 1% BSA. Bound Vh′ was detected using a polyclonal GST antibody (Sigma) and alkaline phosphatase (AP)-coupled anti-rabbit Ig (Sigma). Between incubations, membranes were washed three times for 5 min with TBS-T (0.05% Tween 20). Filters were then incubated with 0.24 mg/ml BCIP, 0.3 mg/ml MTT and 5 mM MgCl2 in citrate-buffered saline (CBS, 50 mM sodium citrate, pH 7.0) for up to 30 min and colour development was stopped by washing membranes in PBS.
NMR and gel filtration
Recombinant mouse talin 482–636, 482–655 and vinculin 1–258 were prepared as described previously. 15N/1H-labelled talin constructs were obtained by expression in a 1 l culture of M9 minimal medium supplemented with 1 g of 15N-labelled ammonium chloride (CK gas products Ltd, UK) and 4 g/l of glucose. All NMR samples were prepared by dialysis against 20 mM potassium phosphate buffer containing 200 mM NaCl, 1 mM EDTA and 5 mM DTT at pH 6.5. As an internal lock, 10% D2O (v/v) was added. For the talin 482–655 titration, a 0.06 mM vinculin head solution was added stepwise to 0.2 mM 15N-labelled talin. The talin/vinculin molar ratios tested were 1:0.75, 1:1.5, 1:3 and 1:6. The 1:1.5 talin 482–636/vinculin head solution of Figure 1C was obtained by mixing equal volumes of a 0.3 mM 15N-labelled talin solution with a 0.2 mM vinculin head solution. The NMR experiments were carried out on a Bruker DRX600 spectrometer at 25°C. The NMR data were processed using XWINNMR and plotted using XWINPLOT. Analytical gel filtration chromatography of recombinant talin polypeptides and vinculin head (1–258) was performed using a Superdex-75 column (Pharmacia) at 4°C. The column was pre-equilibrated and run in 20 mM Tris (pH 8.0), 200 mM NaCl and 2 mM DTT at a flow rate of 0.8 ml/min.
ELISA competition experiments
NUNC immunoplates (F96-maxisorp) were incubated overnight at 4°C with 50 μl of talin 482–636 polypeptide (50 nM) in 50 mM NaHCO3 pH 9.6. The wells were washed twice with TBS containing 0.1% Tween 20 and excess protein-binding sites were blocked with 50 μl of 5% BSA in the above buffer at 37°C for 1 h. The blocking buffer was removed, the wells washed twice and then incubated with GST-vinculin (5 nM) in the presence of varying concentrations of talin polypeptides in TBS-T for 2 h at 37°C. The wells were washed three times with TBS-T, and 50 μl of a rabbit polyclonal anti-GST coupled to HRP (Santa Cruz; 1:5000) in 1% BSA TBS-T was added to each well. Plates were then incubated at room temperature for 1 h, washed three times with TBS-T and finally 50 μl of OPD substrate in citrate-phosphate buffer was added and left for 10 min for the colour to develop. The reaction was stopped by adding 50 μl of 8 M sulphuric acid, and the absorbance of the wells was recorded at 490 nm using a Biotek plate reader.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Table 1
Acknowledgments
The work was supported by a BBSRC SMART grant and by the Wellcome Trust. We are grateful to Richard Hynes (MIT, Cambridge, MA, USA) for the mouse talin1 cDNA and to Clive R Bagshaw for many helpful discussions. We also thank S Daenicke for expert technical assistance in the peptide array synthesis. We also acknowledge the support of the staff from the ESRF beamline ID14-4. Dr Jonas Emsley is a Wellcome Trust Funded Fellow.
Accession numbers Atomic coordinates have been deposited in the Protein Data Bank under accession codes 1SJ7, 1SJ8 and 1TO1 for talin 482–655, talin 482–789 and Vh′/VBS1, respectively.
References
- Albiges-Rizo C, Frachet P, Block MR (1995) Down regulation of talin alters cell adhesion and the processing of the α5β1 integrin. J Cell Sci 108: 3317–3329 [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, dePereda JM, Bagshaw CR, Critchley DR, Liddington RC (1999) Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99: 603–613 [DOI] [PubMed] [Google Scholar]
- Barsukov IL, Prescot A, Bate N, Patel B, Floyd DN, Bhanji N, Bagshaw CR, Letinic K, Di Paolo G, De Camilli P, Roberts GC, Critchley DR (2003) Phosphatidylinositol phosphate kinase type 1gamma and beta1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J Biol Chem 278: 31202–31209 [DOI] [PubMed] [Google Scholar]
- Bass MD, Patel B, Barsukov IG, Fillingham IJ, Mason R, Smith BJ, Bagshaw CR, Critchley DR (2002) Further characterization of the interaction between the cytoskeletal proteins talin and vinculin. Biochem J 362: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass MD, Smith BJ, Prigent SA, Critchley DR (1999) Talin contains three similar vinculin-binding sites predicted to form an amphipathic helix. Biochem J 341: 257–263 [PMC free article] [PubMed] [Google Scholar]
- Bershadsky AD, Balaban NQ, Geiger B (2003) Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol 19: 677–695 [DOI] [PubMed] [Google Scholar]
- Bolton SJ, Barry ST, Mosley H, Patel B, Jockusch BM, Wilkinson JM, Critchley DR (1997) Monoclonal antibodies recognizing the N- and C- terminal regions of talin disrupt actin stress fibers when microinjected into human fibroblasts. Cell Motil Cytoskeleton 36: 363–376 [DOI] [PubMed] [Google Scholar]
- Borhani DW, Rogers DP, Engler JA, Brouillette CG (1997) Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci USA 94: 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky ML, Hynes RO (1998) Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J Cell Biol 143: 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NH, Gregory SL, Rickoll WL, Fessler LI, Prout M, White RA, Fristrom JW (2002) Talin is essential for integrin function in Drosophila. Dev Cell 3: 569–579 [DOI] [PubMed] [Google Scholar]
- Calderwood DA (2004) Integrin activation. J Cell Sci 117: 657–666 [DOI] [PubMed] [Google Scholar]
- Chen HC, Appeddu PA, Parsons JT, Hildebrand JD, Schaller MD, Guan JL (1995) Interaction of focal adhesion kinase with the cytoskeletal protein talin. J Biol Chem 270: 16995–16999 [DOI] [PubMed] [Google Scholar]
- Cram EJ, Clark SG, Schwarzbauer JE (2003) Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J Cell Sci 116: 3871–3878 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De Camilli P (2002) Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420: 85–89 [DOI] [PubMed] [Google Scholar]
- Erickson HP (2002) Stretching fibronectin. J Muscle Res Cell Motil 23: 575–580 [DOI] [PubMed] [Google Scholar]
- Frank R, Overwin H (1996) SPOT synthesis. Epitope anlysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol 66: 149–169 [DOI] [PubMed] [Google Scholar]
- Galbraith CG, Yamada KM, Sheetz MP (2002) The relationship between force and focal complex development. J Cell Biol 159: 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC (2003) Structural determinants of integrin recognition by talin. Mol Cell 11: 49–58 [DOI] [PubMed] [Google Scholar]
- Giannone G, Jiang G, Sutton DH, Critchley DR, Sheetz MP (2003) Talin1 is critical for force-dependent reinforcement of initial integrin–cytoskeleton bonds but not tyrosine kinase activation. J Cell Biol 163: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K (1996) Regulation of vinculin binding to talin and actin by phosphatidylinositol-4-5-bisphosphate. Nature 381: 531–535 [DOI] [PubMed] [Google Scholar]
- Hemmings L, Rees DJG, Ohanian V, Bolton SJ, Gilmore AP, Patel B, Priddle H, Trevithick JE, Hynes RO, Critchley DR (1996) Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci 109: 2715–2726 [DOI] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Ilic D, Damsky CH, Yamamoto T (1997) Focal adhesion kinase: at the crossroads of signal transduction. J Cell Sci 110: 401–407 [DOI] [PubMed] [Google Scholar]
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR (2004) Vinculin activation by talin through helical bundle conversion. Nature 427: 171–175 [DOI] [PubMed] [Google Scholar]
- Izard T, Vonrhein C (2004) Structural basis for amplifying vinculin activation by talin. J Biol Chem, (in press) [DOI] [PubMed] [Google Scholar]
- Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP (2003) Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424: 334–337 [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Rudiger M (1996) Crosstalk between cell adhesion molecules: vinculin as a paradigm for regulation by conformation. Trends Cell Biol 6: 311–315 [DOI] [PubMed] [Google Scholar]
- Kim M, Carman CV, Springer TA (2003) Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301: 1720–1725 [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420: 89–93 [DOI] [PubMed] [Google Scholar]
- Liu S, Calderwood DA, Ginsburg MH (2000) Integrin cytoplasmic domain-binding proteins. J Cell Sci 113: 3563–3571 [DOI] [PubMed] [Google Scholar]
- Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, Albiges-Rizo C (2001) Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J Biol Chem 276: 21217–21227 [DOI] [PubMed] [Google Scholar]
- Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP (1998) JOY: protein sequence-structure representation and analysis. Bioinformatics 14: 617–623 [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW (1997) The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA 94: 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AD, Stewart M, Hynes RO, Rees DJG (1994) Analysis of repeated motifs in the talin rod. J Mol Biol 235: 1278–1290 [DOI] [PubMed] [Google Scholar]
- Mcree DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Monkley SJ, Zho X-H, Kinston SJ, Giblett SM, Hemmings L, Priddle H, Brown JE, Pritchard CA, Critchley DR, Fassler R (2000) Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev Dyn 219: 560–574 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Niggli V, Kaufmann S, Goldmann WH, Weber T, Isenberg G (1994) Identification of functional domains in the cytoskeletal protein talin. Eur J Biochem 224: 951–957 [DOI] [PubMed] [Google Scholar]
- Nuckolls GH, Romer LH, Burridge K (1992) Microinjection of antibodies against talin inhibits the spreading and migration of fibroblasts. J Cell Sci 102: 753–762 [DOI] [PubMed] [Google Scholar]
- Priddle H, Hemmings L, Monkley S, Woods A, Patel B, Sutton D, Dunn GA, Zicha D, Critchley DR (1998) Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated ES cells. J Cell Biol 142: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh A, Takiguchi K, Tanaka Y, Hotani H (1998) Opening-up of liposomal membranes by talin. Proc Natl Acad Sci USA 95: 1026–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Sheetz MP (2002) Force transduction by Triton cytoskeletons. J Cell Biol 156: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA (2003) Talin binding to integrin beta tails: a final common step in integrin activation. Science 302: 103–106 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremuth L, Kreis S, Melchior C, Hoebeke J, Ronde P, Plancon S, Takeda K, Kieffer N (2004) A fluorescence cell biology approach to map the second integrin-binding site of talin to a 130-amino acid sequence within the rod domain. J Biol Chem 279: 22258–22266 [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Hass TA, Plow EF, Qin J (2002) Structural mechanism of integrin αIIbβ3 ‘inside out' activation as regulated by its cytoplasmic face. Cell 110: 587–597 [DOI] [PubMed] [Google Scholar]
- Winkler JH, Lunsdorf H, Jockusch BM (1997) Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur J Biochem 243: 430–436 [DOI] [PubMed] [Google Scholar]
- Xing B, Jedsadayanmata A, Lam SC (2001) Localization of an integrin binding site to the C terminus of talin. J Biol Chem 276: 44373–44378 [DOI] [PubMed] [Google Scholar]
- Xu W, Baribault H, Adamson ED (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125: 327–337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Table 1


