Abstract
The importance of gene duplication in evolution has long been recognized. Because duplicated genes are prone to diverge in function, gene duplication could plausibly play a role in species differentiation. However, experimental evidence linking gene duplication with speciation is scarce. Here, we show that a hybrid-male sterility gene, Odysseus (OdsH), arose by gene duplication in the Drosophila genome. OdsH has evolved at a very high rate, whereas its most immediate paralog, unc-4, is nearly identical among species in the Drosophila melanogaster subgroup. The disparity in their sequence evolution is echoed by the divergence in their expression patterns in both soma and reproductive tissues. We suggest that duplicated genes that have yet to evolve a stable function at the time of speciation may be candidates for “speciation genes,” which is broadly defined as genes that contribute to differential adaptation between species.
Some genes diverge in function during or soon after speciation, leading to differential adaptation between closely related species and, often, reproductive incompatibilities in their hybrids. Such genes are sometimes referred to as “speciation genes” (1, 2). What makes speciation genes prone to diverge? Gene duplication is an attractive hypothesis because duplicated copies are indeed prone to differentiate into new functional niches (3–7), hence contributing to species differentiation (see Supporting Text, which is published as supporting information on the PNAS web site). However, little empirical evidence has become available despite the many theoretical inferences (5, 7). The hybrid-male sterility gene, Odysseus (OdsH), may fill in the gap (8).
OdsH is responsible for hybrid-male sterility when the Drosophila mauritiana allele is introgressed into an appropriate Drosophila simulans background (9), which carries other D. mauritiana genes that are known to be necessary for this sterility interaction. The genetic result has since been confirmed by transgenic experiments (10), which were also performed in properly controlled backgrounds. OdsH is most closely related to the paired-type homeodomain gene, unc-4, of Caenorhabditis elegans (8, 11) and its mammalian homologues (12, 13). Interestingly, the homeodomain of OdsH from the Drosophila melanogaster subgroup has experienced a 100- to 1,000-fold acceleration in amino acid substitution when compared with the rate of evolution between the unc-4 genes from mouse and C. elegans. The functional homologue of unc-4 has also been reported in Drosophila (14). We refer to this copy as unc-4 in the sense of sequence, as well as expression, conservation.
Materials and Methods
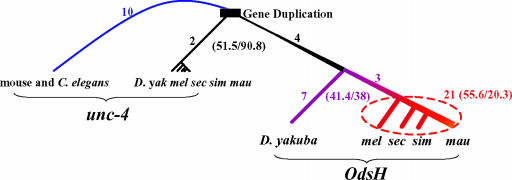
Statistical Analysis. The pairwise distance between extant species was calculated by using the method of Li (15), as implemented in the GCG package. The method, in its intermediate derivation (15), gives A and S between each pair of genes, where A and S are the total number of nonsynonymous and synonymous substitutions, respectively. We decomposed the number of pairwise substitutions for both A and S into branch length by using the method of Fitch and Margoliash (16). The branch length among Drosophila sechellia, D. simulans, and D. mauritiana was resolved first. The branch length from their common ancestor to D. melanogaster was resolved later by using D. sechellia and D. simulans. The next branch length to Drosophila yakuba (purple line in Fig. 2) was resolved by using D. melanogaster and D. simulans. Finally, we used unc-4 sequences to resolve the branch length indicated by the black line in Fig. 2. The choice of species in the resolution does not affect the conclusion of the analysis.
Fig. 2.
Sequence evolution of OdsH and unc-4. The numbers given next to the branches indicate nonsynonymous substitutions only in the homeodomain. Substitutions (nonsynonymous vs. synonymous) on all the homologous sequences aligned in Fig. 1 are given in parentheses. The rate of substitutions is illustrated by the thickness of the lines. yak, D. yakuba; mel, D. melanogaster, sec, D. Sechellia; sim, D. simulans; mau, D. mauritiana.
Whole-Mount in Situ Hybridization. Whole-mount in situ hybridization was carried out by using a digoxigenin-labeled RNA probe, as described (17). Briefly, embryos or tissues were fixed for 15–20 min in 3.7% formaldehyde after dissection in 1× PBS and dehydrated in methanol before being stored at -20°C. The hybridization was carried out at 65°C in the SDS hybridization solution overnight. The samples were then washed with several wash buffers before adding antidigoxigenin alkaline phosphatase. The hybridization results were detected by color reaction.
Quantitative RT-PCR. Total RNA of whole flies or relevant tissues was extracted with the TRIzol reagent according to the manufacturer's instructions. A 1–2.5 μg aliquot was used as template for cDNA synthesis, employing the SuperScript First-Strand Synthesis system and oligo(dT) primers. Specific primers of OdsH, unc-4, and rp49 were used in the quantitative RT-PCRs with 1× SYBR Green PCR Master Mix. All reactions were performed in triplicate. Real-time quantitative PCR analysis was carried out according to the manufacture's instructions (Applied Biosystems).
Results
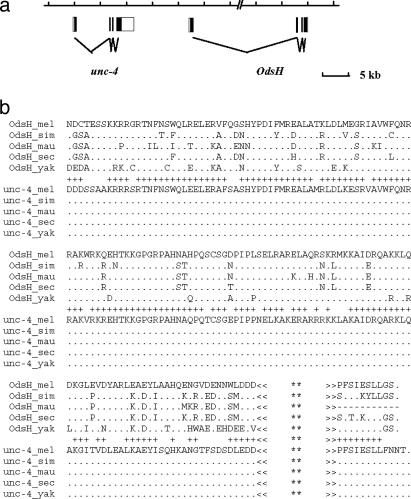
OdsH and unc-4 are 13-kb apart in tandem, as shown schematically in Fig. 1a. They preserve the intron and exon structure but are very divergent in both the N terminus (including the entire exon 1) and the C terminus (most of the exon 4). At the extreme C terminus, the coding region contains a small conserved domain with 8 aa shared between OdsH and unc4 (Fig. 1b). In C. elegans, this domain is responsible for protein–protein interactions (18). Noncoding portion of the gene provides little clue of homology. We also cannot determine whether the exons 1 of the two genes are homologous. If they are homologous, they have diverged greatly since gene duplication.
Fig. 1.
Structure (a) and sequences (b) of OdsH and unc-4. Only regions where the two genes are mutually alignable are shown and analyzed. A plus symbol between the two groups indicates where at least one of the OdsH sequences shares the same amino acid with unc-4. The C terminus, as shown, contains a shared protein–protein interaction domain. **, There are 74 residues for OdsH and 210 residues for unc-4.
It is striking that, within the coding region where unc-4 and OdsH can be unambiguously aligned (Fig. 1b), there is not a single amino acid substitution in unc-4 among the five species in the D. melanogaster subgroup, whereas OdsH has experienced numerous changes. This extreme asymmetry in the sequence evolution between duplicated genes suggests that their functional evolution may also be asymmetric. The level of divergence among the sequences of Fig. 1b is given in Table 1. The divergence time between OdsH and unc-4 is estimated to be four times that of the divergence between D. yakuba and D. melanogaster on the basis of the KS values of OdsH. The duplication might have occurred in the genus Drosophila, likely in the lineage of the subgenus Sophophora after it split from the Drosophila subgenus. The recent genomic information on Anopheles and Drosophila pseudoobscura is consistent with this estimation. Anopheles has only one copy of unc-4-like gene, whereas D. pseudoobscura has both unc-4 and OdsH.
Table 1. Nonsynonymous (Ka) and synonymous (Ks) substitutions in OdsH and unc-4 of the five species of Drosophila.
| Species pair | KA ± SD | KS ± SD | KA/KS |
|---|---|---|---|
| OdsH vs. unc-4* | 0.313 | 1.629 | 0.192 |
| Between unc-4 | |||
| yak (-mel, -sim, -sec, -mau) | 0.003 | 0.122 | 0.025 |
| mel (-sim, -sec, -mau) | 0 | 0.036 | 0 |
| (sim, sec, mau) | 0 | 0.012 | 0 |
| Between OdsH | |||
| yak-mel | 0.158 ± 0.035 | 0.317 ± 0.077 | 0.498 |
| yak-sec | 0.214 ± 0.041 | 0.409 ± 0.108 | 0.523 |
| yak-sim | 0.229 ± 0.043 | 0.425 ± 0.112 | 0.539 |
| yak-mau | 0.244 ± 0.046 | 0.412 ± 0.115 | 0.592 |
| mel-sec | 0.113 ± 0.027 | 0.157 ± 0.066 | 0.720 |
| mel-sim | 0.126 ± 0.029 | 0.194 ± 0.071 | 0.649 |
| mel-mau | 0.151 ± 0.032 | 0.186 ± 0.073 | 0.812 |
| sec-sim | 0.026 ± 0.016 | 0.041 ± 0.025 | 0.634 |
| sec-mau | 0.051 ± 0.021 | 0.040 ± 0.032 | 1.275 |
| sim-mau | 0.061 ± 0.023 | 0.024 ± 0.023 | 2.542 |
See text for the statistical tests. yak, D. yakuba; mel, D. melanogaster; sim, D. simulans; mau, D. mauritiana; sec, D. sechellia.
Average over the five species
Whereas unc-4 is strongly conserved, OdsH shows an interesting trend in its sequence evolution. The KA/KS ratio (KA being the number of nonsynonymous changes per nonsynonymous site, and KS being the number of synonymous changes per synonymous site) has increased appreciably in the lineages leading to D. melanogaster and its three sibling species. The ratio is highest between D. mauritiana and D. simulans, in which the effect of OdsH on hybrid-male sterility is manifested. To examine the trend statistically, we assign the number of nonsynonymous (A) and synonymous (S) substitutions to different parts of the phylogenetic tree of Fig. 2. Along the branch between the extant unc-4 genes and the common ancestor of D. melanogaster and D. yakuba, indicated by the thin black line in Fig. 2, the A/S ratio is 51.5:90.8. From D. yakuba to the ancestor of the sibling species of D. melanogaster (purple line in Fig. 2), the ratio is 41.4:38.0, and among the D. melanogaster sibling species (solid red line), the A/S ratio becomes 55.6:20.3. All three ratios are significantly different from one another by the G or χ2 test.
Another interesting comparison is the relative evolutionary rate between duplicated genes, especially immediately after their duplication. To make this comparison, we used the unc-4 gene of C. elegans and mouse as outgroup. Given the long divergence, we can only compare the homeodomain because it is the only alignable portion among C. elegans, mouse, and Drosophila. Since the duplication, only two amino acid changes can be unambiguously assigned to the unc-4 branch, whereas 35 (4 + 7 + 3 + 21) changes have occurred along the OdsH branches (Fig. 2).
Note the delayed divergence of OdsH from unc-4. Fig. 2 shows that the accelerated divergence is observed mostly in the last ≈1 million years, after a long period of “quiescence” since duplication. This recent episode of rapid sequence evolution is coupled with the different expression patterns of OdsH among extant sibling species (Fig. 3). The observation contrasts with the widely held view that duplicated genes diverge immediately after duplication (4, 5, 7, 19).
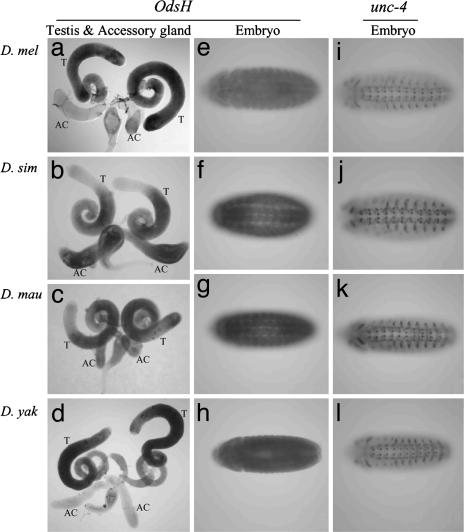
Fig. 3.
Gene expression in the four Drosophila species. (a–d) OdsH expression in male reproductive tissues. T, testis; AC, accessory gland. (e–h) OdsH expression in stage 14 embryos. (i–l) unc-4 expression in stage 14 embryos. D. mau, D. mauritiana; D. mel, D. melanogaster; D. sim, D. simulans; D. yak, D. yakuba.
To study the divergence of OdsH among the sibling species, we surveyed the expression patterns by whole-mount in situ hybridization (Fig. 3). OdsH is expressed in the male reproductive tissues. The expression patterns are variable among the four sibling species, such that in D. melanogaster and D. yakuba, we observed relatively strong expression in the testis but not in the accessory gland (Fig. 3 a and d). In D. simulans, the expression is relatively low at the apical region of the testis, but it is high in the accessory gland (Fig. 3b). In D. mauritiana, the expression in the accessory gland is weaker than expression in the testis (Fig. 3c).
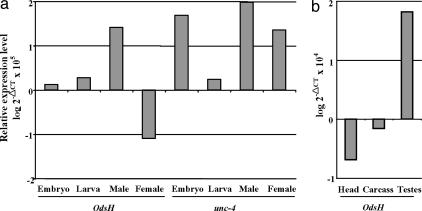
In C. elegans, unc-4 controls the motor-neuron differentiation (18). The published study on Drosophila unc-4 gene has revealed its expression in specific postmitotic neurons (14). As shown in Fig. 3, unc-4 is expressed (segment-wise) in a subset of ectodermal and neural cells in the CNS. This expression pattern is conserved across divergent species, from D. melanogaster, D. yakuba (Fig. 3 i–l), and Drosophila ananassae, to D. pseudoobscura (data not shown). In contrast, there is no detectable expression of OdsH in D. melanogaster or D. yakuba embryos (Fig. 3 e and h). The ubiquitous expression of OdsH observed in the embryos of D. simulans and D. mauritiana does not show specific neuronal patterns (Fig. 3 f and g). We further analyzed the expression levels of OdsH and unc-4 in embryos, larvae, adult males, and females of D. melanogaster by quantitative RT-PCR (Fig. 4a). Consistent with the in situ hybridization results, OdsH is most strongly expressed in adult males in D. melanogaster, whereas only a few-fold differences can be detected between males and females in D. mauritiana. The expression of OdsH in different tissues of the adult males was also assayed, and the result shows that the OdsH expression in males is mainly in the testes (Fig. 4b).
Fig. 4.
Expression levels of OdsH and unc-4 in D. melanogaster, as quantified by real-time RT-PCR. All values are normalized against an internal control (rp-49), and the relative expression level is shown in the log scale. (a) Expression in different stages and sexes. The reference point of zero on the y axis is approximately the average expression of OdsH across the four measurements. (b) Expression of OdsH in different tissues in males.
Discussion
Overall, OdsH has been evolving rapidly in both expression and DNA sequence. Presumably, the duplication of unc-4 has permitted it to evolve into a new functional niche. A recent experiment by sequence-dependent gene knockout has shown that OdsH functions in promoting the fertility of very young males (10). This new functional role of OdsH has been evolving away from the unc-4 expression and toward a testicular function. The OdsH-induced hybrid-male sterility might be the consequence of such a shift in expression. We should note that other genetic factors from both species have been shown to be involved in this sterility interaction (9), making it more complex than the simplest two-locus Dobzhansky–Muller mechanism (20).
Without gene duplication, orthologous genes between the diverging species usually have to assume comparable functions, unless the environments have shifted drastically. However, if gene duplication occurred shortly before speciation, the diverging species would each inherit a duplicated gene whose functions are still prone to differentiate. The undifferentiated and redundant function is expected to evolve along different trajectories in the two diverging species. Although the connection between gene duplication and reproductive isolation has been suggested (4), our model is quite different in that it emphasizes functional divergence and differential adaptation (1).
The conventional view on gene duplication is that one of the two duplicated genes retains the original function, allowing the other to evolve a new one [neo-functionalization (3, 21)]. Recent reports have suggested an alternative hypothesis that the ancestral functions are split between the duplicated genes [subfunctionalization (4, 6, 7)]. The duplicated pair of unc-4 and OdsH appears to have evolved in the former mode.
OdsH is also a “dispensable” gene because its deletion reduces male fertility only partially and only when males are <4 days old (10). The concept of dispensability is important because it makes a distinction between physiological assay and evolutionary interpretation. Dispensability indicates merely that a gene is not needed for normal viability and fertility under general conditions, without implying its fitness effect in the evolutionary context. Many other rapidly evolving genes that bear the signature of positive selection are also dispensable. The examples include the Acp26Aa and desaturase-2 genes in Drosophila and the glycophorins in human (22–24). OdsH is another example of a dispensable gene playing a significant role in evolution. In fact, such genes, being less constrained in evolution, may be more prone to diverge in function during speciation. It is the divergence in function, not the physiological importance of the gene function, that contributes to species differentiation.
Our results reveal a connection between species differentiation and gene duplication. Duplicated genes that are in the process of evolving into new functions at the time of species separation are likely to contribute to species differentiation. Many of the differentiated genes between recently diverged species may contribute to hybrid incompatibility, sexual isolation, or morphological differences. Molecular analyses of additional speciation genes may shed further light on the genic basis of speciation that has been elusive for many decades (1).
Supplementary Material
Acknowledgments
We thank Justin Fay for helping with part of D. yakuba sequencing and Yu-Ping Poh for sequence analysis. This work was supported by grants from the National Institutes of Health and the National Science Foundation (to C.-I.W.) and the National Science Council of Taiwan (to C.-T.T. and S.-C.T.). N.H.P. is an Investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. 634187, 645582, 645586, 634205, 645592, 634211, 645596, and 645598).
References
- 1.Wu, C.-I. (2001) J. Evol. Biol. 14, 851-865. [Google Scholar]
- 2.Wu, C.-I. & Ting, C.-T. (2004) Nat. Rev. Genet. 5, 114-122. [DOI] [PubMed] [Google Scholar]
- 3.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).
- 4.Lynch, M. & Conery, J. C. (2000) Science 290, 1151-1154. [DOI] [PubMed] [Google Scholar]
- 5.Li, W.-H. (1985) in Population Genetics and Molecular Evolution, eds. Ohta, T. & Aoki, K. (Springer, Berlin), pp. 333-352.
- 6.Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151, 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch, M., O'Hely, M. & Walsh, B. (2001) Genetics 159, 1789-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting, C.-T., Tsaur, S. C., Wu, M.-L. & Wu, C.-I. (1998) Science 282, 1501-1504. [DOI] [PubMed] [Google Scholar]
- 9.Perez, D. P. & Wu, C.-I. (1995) Genetics 140, 201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun, S., Ting, C.-T. & Wu, C.-I. (2004) Science 305, 81-83. [DOI] [PubMed] [Google Scholar]
- 11.Miller, D. M., Shen, M. M., Shamu, C. E., Burglin, T. R., Ruvkun, G., Dubois, M. L., Ghee, M. & Wilson, L. (1992) Nature 355, 841-845. [DOI] [PubMed] [Google Scholar]
- 12.Mansouri, A., Yokota, Y., Wehr, R., Copeland, N. G., Jenkins, N. A. & Gruss, P. (1997) Dev. Dyn. 210, 53-65. [DOI] [PubMed] [Google Scholar]
- 13.Saito, T., Lo, L., Anderson, D. J. & Mikoshiba, K. (1996) Dev. Biol. 180, 143-155. [DOI] [PubMed] [Google Scholar]
- 14.Tabuchi, K., Yoshikawa, S., Okabe, M., Sawamoto, K. & Okano, H. (1998) Neurosci. Lett. 257, 49-52. [DOI] [PubMed] [Google Scholar]
- 15.Li, W.-H. (1993) J. Mol. Evol. 36, 96-99. [DOI] [PubMed] [Google Scholar]
- 16.Fitch, W. M. & Margoliash, E. (1967) Biochem. Genet. 1, 65-71. [DOI] [PubMed] [Google Scholar]
- 17.Patel, N. H. (1996) in A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis, ed. Kreig, P. A. (Wiley, New York), pp. 357-369.
- 18.Winnier, A. R., Meir, J. Y., Ross, J. M., Tavernarakis, N., Driscoll, M., Ishihara, T., Katsura, I. & Miller, D. M., III, (1999) Genes Dev. 13, 2774-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei, M. & Roychoudhury, A. K. (1973) Am. Nat. 107, 362-372. [Google Scholar]
- 20.Wu, C.-I. & Palopoli, M. F. (1994) Annu. Rev. Genet. 28, 283-308. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, A. L. (1994) Mol. Biol. Evol. 11, 417-425. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi, A., Tsaur, S. C., Coyne, J. A. & Wu, C.-I. (2001) Proc. Natl. Acad. Sci. USA 98, 3920-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herndon, L. A. & Wolfner, M. F. (1995) Proc. Natl. Acad. Sci USA 92, 10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, H.-Y., Tang, H., Shen, C. & Wu, C.-I. (2003) Mol. Biol. Evol. 20, 1795-1804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.