Abstract
RNA quality control of endogenous RNAs is an integral part of eukaryotic gene expression and often relies on exonucleolytic degradation to eliminate dysfunctional transcripts. In parallel, exogenous and selected endogenous RNAs are degraded through RNA silencing, which is a genome defense mechanism used by many eukaryotes. In plants, RNA silencing is triggered by the production of double-stranded RNAs (dsRNAs) by RNA-DEPENDENT RNA POLYMERASEs (RDRs) and proceeds through small interfering (si) RNA-directed, ARGONAUTE (AGO)-mediated cleavage of homologous transcripts. Many studies revealed that plants avert inappropriate PTGS of endogenous coding genes by using RNA surveillance mechanisms as a safeguard to protect their transcriptome. The tug-of-war between RNA surveillance and RNA silencing ensures the appropriate partitioning of endogenous RNA substrates among these degradation pathways. Here we review recent advances on RNA quality control and its role in the suppression of RNA silencing at endogenous genes and discuss the mechanisms underlying the crosstalk among these pathways.
Keywords: Gene silencing, RNA quality control, aberrant RNA, siRNA
INTRODUCTION
RNA silencing is an evolutionarily conserved genome defense mechanism used by many eukaryotic organisms to combat viruses, foreign transgenes and transposable elements (Eamens et al., 2008; Martinez de Alba et al., 2013). Small interfering RNAs (siRNAs) of 21-24 nucleotides (nt) long are central players of RNA silencing. Given the widespread production of endogenous siRNAs, mainly from repeats and transposable elements, in plants’ genomes, a question arises as to how plants avoid inappropriate RNA silencing of endogenous protein-coding genes. The key appears to lie in RNA quality control, a surveillance mechanism that allows the selective elimination of endogenous aberrant RNAs to prevent the dysfunctional transcripts from being translated into nonfunctional proteins (Moore, 2005). RNA silencing and RNA quality control interact and coordinate to ensure the correct partitioning of RNA substrates, and the careful balance between these two pathways is important for maintaining plant transcriptome integrity and, consequently proper plant development. In this review, we summarize the recent advances on RNA surveillance and RNA silencing and discuss the mechanisms underlying the crosstalk among these degradation pathways.
RNA SILENCING PATHWAYS IN PLANTS
Plants employ multiple strategies to constantly defend against various abiotic and biotic stresses from the changing environment. RNA silencing in plants is a nucleotide-sequence-specific gene regulation mechanism that counteracts viral infections, maintains heterochromatin and controls developmental processes (Baulcombe, 2005; Molnar et al., 2011; Bologna and Voinnet, 2014). Two major types of RNA silencing are transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS). Mechanisms of silencing at the TGS level include DNA methylation or histone modifications in the nucleus, and at the PTGS level involve mRNA cleavage or translational repression in the cytoplasm (Voinnet, 2009; Law and Jacobsen, 2010; Matzke and Mosher, 2014). In plants, TGS mainly targets endogenous transposable elements and repetitive DNA to epigenetically repress their transcription, while PTGS functions primarily to eliminate invading RNAs, regulate stress-related genes and genes required for organ patterning or cell-type specification (Ruiz-Ferrer and Voinnet, 2009; Incarbone and Dunoyer, 2013; Pumplin and Voinnet, 2013).
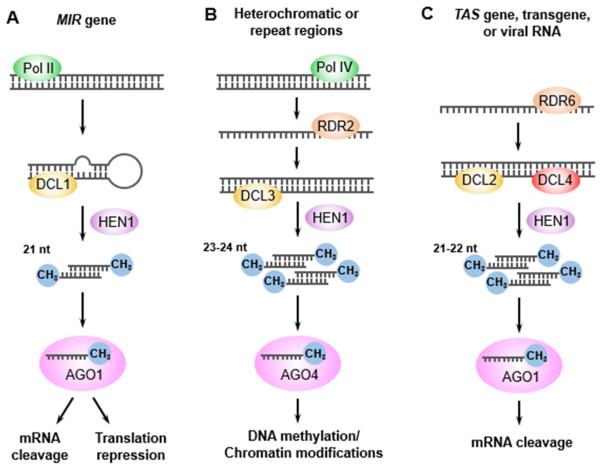
RNA silencing depends on the actions of small RNA molecules of 21-24 nt. Two main classes of small RNAs have been identified in plants: microRNA (miRNA), and small interfering RNA (siRNA) (Poethig et al., 2006; Ramachandran and Chen, 2008; Carthew and Sontheimer, 2009; Chen, 2012). A miRNA is produced as the most predominant species from a Pol II-transcribed, long, single-stranded RNA that forms an imperfectly paired hairpin structure (Xie et al., 2005; Rogers and Chen, 2012). Sequentially processed by the RNase III protein DICER-LIKE 1 (DCL1) with the aid of other RNA-binding cofactor proteins, the precursor releases 21 nt or 22 nt small RNA duplexes (Park et al., 2002; Reinhart et al., 2002). The miRNA duplexes are stabilized by 2′-O-methylation mediated by the methyltransferase HUA ENHANCER 1 (HEN1) (Yu et al., 2005; Yang et al., 2006). After being transported to the cytoplasm, the mature guide strand miRNAs load into AGO1 to form the RNA-Induced Silencing Complex (RISC), which in turn causes mRNA cleavage or translation repression of the target genes (Fig. 1A) (Bartel, 2004; Tang, 2005; Rogers and Chen, 2013).
Figure 1.
RNA Silencing Pathways in Plants
The origination of small RNAs from MIR genes (A), heterochromatic or repeat regions (B), TAS genes, transgenes and viral RNAs (C) requires dsRNA regions generated by transcription of imperfectly matched hairpins, or dsRNAs synthesized by RNA-DEPENDENT RNA POLYMERASEs (RDRs). The dsRNA regions or dsRNAs are processed by DICER-LIKE proteins (DCLs) to give rise to small RNA molecules of 21-24 nt. The small RNAs are methylated by the methyltransferase HUA ENHANCER 1 (HEN1), and one strand from each duplex is loaded into an ARGONAUTE (AGO) protein to guide DNA methylation or histone modifications at the target DNA, or RNA cleavage or translational repression of the target transcripts. The related RNA polymerase (POL), RDR, DCL and AGO proteins required in each pathway and the specific sizes of the small RNAs are indicated.
siRNAs are produced as a population of small RNA species from partially or perfectly paired double-stranded RNAs (dsRNAs) generated by transcription of inverted-repeat genes, convergent transcription of sense-antisense gene pairs or synthesis by RNA-DEPENDENT RNA POLYMERASEs (RDRs) (Kasschau et al., 2007; Kim et al., 2014; Matzke et al., 2015). The biogenesis of canonical heterochromatic siRNAs (hc-siRNA) requires Pol IV to produce singe-stranded RNAs (ssRNAs) that serve as the template for RDR2, which transcribes the ssRNAs into dsRNAs. The dsRNAs are processed by DCL3 into 24 nt hc-siRNAs with 3′ overhangs, which are then methylated by HEN1. hc-siRNAs are primarily loaded into AGO4 to form the RNA-directed DNA methylation (RdDM) effector complex including RNA polymerase V (Pol V), and direct the DNA methylation or histone modification of the targeted DNA repeats and transposon loci at the TGS level (Fig. 1B) (Chapman and Carrington, 2007; Zhang and Zhu, 2011; Castel and Martienssen, 2013; Zhang et al., 2013). siRNAs mediating PTGS mainly include those derived from endogenous aberrant RNAs, exogenous transgene, and viral RNAs. The biogenesis of these siRNAs requires the cellular RDR6 and the RNA stabilizing protein SUPPRESSOR OF GENE SILENCING 3 (SGS3), which transform ssRNAs into dsRNAs. Then the dsRNAs are processed by DICER-LIKE 4 (DCL4) or DCL2 into 21-22 nt siRNAs, which primarily load into AGO1 and mediate the cleavage of target mRNAs (Fig. 1C) (Dalmay et al., 2000; Mourrain et al., 2000; Gasciolli et al., 2005). It has been reported that RDR6 and SGS3 accumulate in siRNA-bodies in the cytoplasm (Kumakura et al., 2009; Jouannet et al., 2012). Trans-acting siRNA (tasiRNA) is one type of endogenous siRNAs functioning at the PTGS level. The biogenesis of tasiRNAs represents an interesting case whereby single-stranded tasiRNA precursors are initially targeted for cleavage by a miRNA, which is usually 22 nt long, and then are converted into dsRNAs by RDR6 to generate 21 nt tasiRNAs (Allen et al., 2005; Vaucheret, 2005). The tasiRNAs regulate protein-coding target mRNAs and play a role in plant development (Peragine et al., 2004; Vazquez et al., 2004).
RNA QUALITY CONTROL AS A SURVEILLANCE MECHANISM TO ELIMINATE ABERRANT RNAS
RNA quality control is a surveillance mechanism in eukaryotes that allows the elimination of selected endogenous aberrant RNAs to guard against defects in gene expression (Chiba and Green, 2009; Schoenberg and Maquat, 2012). The expression of protein-coding genes entails a complicated series of coordinately regulated processes, such as transcription, 5′-end capping, 3′-end cleavage and polyadenylation, splicing, mRNA export from the nucleus, mRNA translation, and eventually mRNA degradation (Moore, 2005). 5′-end capping is the addition of a 7-methyl guanosine cap (m7G-cap) to the 5′ RNA terminus during transcription. The 5′ cap and the cap binding protein complex prevent mRNA degradation by 5′-3′ exonucleases. The formation of the 3′ end includes an endonucleolytic cleavage followed by polyadenylation. The poly(A) tail along with the poly(A) binding proteins helps to protect the mRNA from 3′-5′ exonucleolytic attack (Kahvejian et al., 2001; Mangus et al., 2003). Besides, the 5′-cap and 3′-poly(A)/PABP complexes can interact and form a closed loop to facilitate the initiation of translation and prevent mRNA degradation caused by exonucleases attacking the ends (Kahvejian et al., 2001; Tomek and Wollenhaupt, 2012). During splicing of the primary transcripts, introns are removed and exons are assembled to form the mature mRNAs (Stamm et al., 2005; Kelemen et al., 2013). The multiple steps of mRNA processing should be error free to ensure the eventual production of a functional protein. However, cells routinely make mistakes during these processes. The RNA metabolic processes are subject to quality control and scrutinized at every step. RNA surveillance is employed to discriminate and eliminate dysfunctional RNAs and ensure the production of functional proteins (Isken and Maquat, 2007).
There are three types of mRNA surveillance pathways in eukaryotes, including nonsense-mediated decay (NMD), non-stop decay (NSD) and no-go decay (NGD) (Isken and Maquat, 2007; Chiba and Green, 2009; Garcia et al., 2014). The NMD pathway is responsible for the degradation of mRNAs containing premature termination codons to prevent the production of truncated proteins (Isken and Maquat, 2008; Wen and Brogna, 2008; Xu and Chua, 2011). The NSD pathway mediates the decay of mRNAs lacking translation termination codons, and NGD targets mRNAs with sequence features that cause the stalling of translating ribosomes (Shoemaker and Green, 2012). These RNA decay pathways have not been well studied in plants, but they could function in plants to ensure the proper control of RNA stability and normal cellular functions in the following ways. First, RNA surveillance degrades defective endogenous mRNAs to prevent the production of toxic dysfunctional proteins. Second, RNA surveillance may control the abundance of cellular transcripts and thus protein levels. Third, RNA surveillance may defend against invading exogenous RNAs and maintain transcriptome integrity (Parker and Song, 2004; Garneau et al., 2007; Moreno et al., 2013; Staiger et al., 2013).
In plants, RNA decay occurs through two general mechanisms: 5′-3′ degradation by XRN exonucleases and 3′-5′ degradation by the multimeric exosome complex (Meyer et al., 2004; Shoemaker and Green, 2012). Both mechanisms involve multiple steps including deadenylation, decapping and exonucleolytic degradation(Chiba and Green, 2009). Deadenylation and decapping are prerequisites for RNA decay and are often considered as rate-limiting steps in the degradation process (Chen and Shyu, 2011). RNA decay is initiated by deadenylation, which is catalyzed by the conserved 3′-5′ poly (A) specific ribonuclease (PARN) and carbon catabolite repressor 4 (CCR4) complex (Dupressoir et al., 2001; Chiba et al., 2004; Reverdatto et al., 2004; Virtanen et al., 2013). After this, the 5′ m7G-cap structure is removed by a set of conserved decapping proteins, including DECAPPING 1 (DCP1), DECAPPING 2 (DCP2), DECAPPING 5 (DCP5), VARICOSE (VCS) and possibly DEA (D/H)-box RNA HELICASE HOMOLOG 1 (DHH1) (Xu et al., 2006; Goeres et al., 2007; Iwasaki et al., 2007; Xu and Chua, 2009). DCP2 hydrolyzes the 5′-m7G-cap, whereas the other proteins probably act in mRNA recognition or facilitate the decapping process (Gunawardana et al., 2008). The deadenylation and decapping proteins localize in cytoplasmic foci called RNA processing bodies (P-bodies), which are sites of RNA turnover (Xu and Chua, 2011; Chen and Shyu, 2013; Maldonado-Bonilla, 2014). After deadenylation and decapping, the aberrant RNAs undergo degradation by 5′-3′ and 3′-5′ exonucleolytic pathways. In plants, 5′-3′ degradation is carried out by three exoribonuclease XRN proteins: the nuclear XRN2 and XRN3 and the cytoplasmic XRN4/EIN5 (Kastenmayer and Green, 2000; Souret et al., 2004; Rymarquis et al., 2011; Nagarajan et al., 2013). It has been reported that XRN4/EIN5 co-localizes with cytoplasmic P-bodies (Kastenmayer and Green, 2000; Weber et al., 2008). The 3′-5′ degradation pathway involves in the exosome complex as well as its co-factors (Schmid and Jensen, 2008; Belostotsky, 2009). In plants, although the exosome has both nuclear and cytoplasmic functions in RNA processing pathways, it seems that the cytoplasmic function is primarily responsible for the degradation of mRNAs (Mitchell et al., 1997). Arabidopsis exosome core subunits contain RIBOSOMAL RNA PROCESSING4 (RRP4), RRP40, RRP41, RRP42, RRP43, RRP45, RRP46, CENTROMERE ENHANCER OF POSITION EFFECT1 SYNTHETIC LETHAL PROTEIN4 (CSL4) and mRNA TRANSPORT REGULATOR3 (MTR3) (Chekanova et al., 2007). Other components that function together with the core subunits are RRP44, RRP6L1, RRP6L2, RRP6L3, HUA ENHANCER2 (HEN2) and MTR4 (Hooker et al., 2007; Zhang et al., 2010; Lange and Gagliardi, 2011; Lange et al., 2011; Lange et al., 2014). Besides, the SKI complex composed of SKI2, SKI3 and SKI8 is also required to tether the exosome to mRNA targets (Anderson and Parker, 1998; Brown et al., 2000; Araki et al., 2001; Orban and Izaurralde, 2005).
RNA SURVEILLANCE SUPPRESSES RNA SILENCING AT ENDOGENOUS GENES
RNA surveillance and RNA silencing are originally considered as exclusive pathways, which are responsible for eliminating aberrant endogenous mRNAs and exogenous RNAs, respectively (Belostotsky, 2004; Chen, 2008). Recently, many studies revealed that the two pathways are actually spatially and functionally linked, and they compete for similar RNA substrates, including not only transgene RNAs but also genome-wide endogenous mRNAs (Belostotsky, 2004; Herr et al., 2006; Gregory et al., 2008; Moreno et al., 2013; Branscheid et al., 2015; Martinez de Alba et al., 2015; Yu et al., 2015; Zhang et al., 2015). The RNA silencing mechanism is activated when RNA surveillance cannot degrade aberrant RNAs in cells– this was revealed by the fact that several players in RNA surveillance act as repressors of RNA silencing (Gazzani et al., 2004; Herr et al., 2006; Gy et al., 2007; Moreno et al., 2013; Lange et al., 2014; Martinez de Alba et al., 2015). How RNA surveillance and RNA silencing interact is worthy of further investigation.
Proper RNA processing is essential for preventing RNA silencing of transgenes
Several factors involved in RNA splicing and 3′-end formation were identified as suppressors of RNA silencing of a transgene in Arabidopsis, including a putative DEAH RNA helicase homologue of the yeast PRP2 RNA splicing cofactor (ESP3), and homologues of mRNA 3′-end formation proteins cleavage stimulation factor CstF64 (ESP1), symplekin/ PTA1 (ESP4), and cleavage polyadenylation specificity factor CPSF100 (ESP5) (Herr et al., 2006; Chen, 2008). Mutants in these genes display enhanced silencing of a transgene, which indicates that proper RNA processing is crucial to prevent RNA silencing in plants. Arabidopsis SmD1b is another factor involved in RNA splicing that participates in the partitioning of transgene-derived aberrant RNAs between RNA surveillance and RNA silencing. Arabidopsis SmD1b encodes an ortholog of the yeast Sm domain-containing protein SmD1, which is a small nuclear ribonucleoprotein of the conserved Smith (Sm) complex (Elvira-Matelot et al., 2016). Besides, it has been reported that truncated and non-polyadenylated mRNAs generated from abortive elongation or improper termination of transgene transcription are subject to RDR6-mediated RNA silencing in Arabidopsis (Luo and Chen, 2007). These results indicate that cellular mRNAs are monitored tightly for their integrity in cells. If mis-processed RNAs (i.e. aberrant RNAs) are not discriminated and eliminated by RNA surveillance mechanisms, they are channeled into the RNA silencing pathway for degradation (Herr et al., 2006). However, it is still unknown whether endogenous mRNAs also undergo RNA silencing in RNA processing mutants.
Decapping and deadenylation required for RNA decay prevent RNA silencing
The 5′-m7G-cap and 3′-poly (A) tail structures are essential features to distinguish a functional mRNA from a dysfunctional mRNA; thus they protect the transcripts from exonucleolytic cleavage as well as ensure their proper translation. The absence of the 5′-cap or 3′-poly (A) tails triggers mRNA degradation. The shortening of the 3′-poly (A) tails is catalyzed by 3′-5′ poly (A) specific ribonuclease PARN as well as the carbon catabolite repressor CCR4 complex. Impairing Arabidopsis PARN and CCR4a has been reported to enhance sense transgene (S)-PTGS, indicating that deadenylation suppresses RNA silencing (Moreno et al., 2013). Decapping of mRNAs involves DCP2, DCP1 and VCS, which are key components of the decapping complex, and generates uncapped transcripts as the substrates for RNA turnover. In Arabidopsis, DCP2, DCP1 and VCS are found to suppress RDR6-dependent transgene PTGS, and more interestingly, these decapping components also prevent the RNA silencing of endogenous protein-coding genes (Thran et al., 2012; Martinez de Alba et al., 2015). In the decapping mutants, hundreds of endogenous mRNAs generate a new class of RDR6-dependent 21-nt RNA quality control siRNAs (rqc-siRNAs) at the genome-wide level. These rqc-siRNAs may be a subset of a large ensemble of endogenous siRNAs whose generation is suppressed by the RNA decay processes. It is proposed that impaired decapping deters the decay of aberrant RNAs, which in turn serve as substrates for PTGS and generate siRNAs. It is also observed that P-bodies and siRNA-bodies are often spatially associated and dynamically interact in the cytoplasm, which might allow the exchange of ribonucleo particle substrates and crosstalk of the two machineries (Moreno et al., 2013; Martinez de Alba et al., 2015). The P-body-localized decapping and deadenylation machinery may limit the unintended entry of dysfunctional RNAs into the siRNA-bodies containing RNA silencing machinery, and thus avoid the generation of rqc-siRNAs and silencing of the target transcripts.
5′-3′ and 3′-5′ exonucleolytic degradation suppresses RNA silencing
In RNA quality control, upon deadenylation and decapping, the aberrant RNAs undergo degradation by 5′-3′ and 3′-5′ exonucleolytic digestion. Either impairing 5′-3′ degradation or 3′-5′ degradation alone could enhance PTGS. Disruption of both 5′-3′ and 3′-5′ degradation more dramatically enhances PTGS.
5′ to 3′ degradation of decapped RNA by XRN proteins
In Arabidopsis, 5′-3′ degradation involves three exonuclease XRN proteins, including the nuclear XRN2 and XRN3 and the cytoplasmic XRN4/EIN5 (Kastenmayer and Green, 2000; Souret et al., 2004; Rymarquis et al., 2011; Nagarajan et al., 2013). XRN4/EIN5 exhibits ribonuclease activity and specifically degrades uncapped mRNAs (Murota et al., 2011). Previous studies showed that XRN4/EIN5 deficiency triggers the PTGS of transgenes and certain endogenous genes, including genes that share sequence identity with the transgenes. (Belostotsky, 2004; Gazzani et al., 2004; Hayashi et al., 2012). It is also observed that hundreds of loci mapping to a number of different gene-rich locations generated clusters of 21 nt siRNAs in the xrn4/ein5 mutant (Gregory et al., 2008). These siRNAs derive from both sense and antisense strands of the transcripts, suggesting that they are processed from RDR-dependent dsRNAs. The siRNA-generating mRNAs accumulate in uncapped forms in the absence of XRN4/EIN5, indicating that XRN4/EIN5 eliminates uncapped transcripts to prevent them from being channeled into the PTGS pathway (Chen, 2008; Gregory et al., 2008). Arabidopsis FIERY1, XRN2 and XRN3 were also identified as endogenous RNA silencing suppressors (Gy et al., 2007). The 3′ (2′), 5′-bisphosphate nucleotidase/ inositol polyphosphate 1-phosphatase FIERY1 is one of the six Arabidopsis orthologues of yeast Hal2, which positively regulates the 5′-3′ exonucleases XRN1 and RAT1 (Quintero et al., 1996; Xiong et al., 2001; Xiong et al., 2004). Arabidopsis FIERY1 is a positive regulator of the exonuclease XRN proteins, and the fiery1 mutant mimics an xrn2 xrn3 xrn4 triple mutant (Hirsch et al., 2011). Like XRN4/EIN5, XRN2 and XRN3 act as endogenous S-PTGS suppressors and FIERY1 inhibits PTGS by positively regulating these XRNs (Gy et al., 2007; Yu et al., 2015). It is worth noting that XRN2 and XRN3 are nuclearly localized, while XRN4/EIN5 is a cytoplasmic protein (Kastenmayer and Green, 2000). While individual mutations in XRN2, XRN3 and XRN4/EIN5 all stimulate PTGS of sense transgenes, they target different endogenous substrate RNAs: XRN4/EIN5 degrades uncapped cytoplasmic RNAs, such as the 3′ fragments generated by miRNA-mediated cleavage, whereas XRN2 and XRN3 target excised miRNA loops derived from the miRNA biogenesis pathway in the nucleus (Gy et al., 2007).
3′-5′ degradation by the exosome and its co-factors
Compromising 3′-5′ RNA degradation provokes the entry of transgenes into PTGS as well. The exosome complex and associated co-factors are responsible for the 3′-5′ degradation of endogenous mRNA substrates (Chekanova et al., 2007; Schmid and Jensen, 2008). Mutations in the exosome core subunits RRP4 and RRP41, or exosome co-factors RRP44A, RRP6L1 and HEN2 were found to enhance transgene S-PTGS in Arabidopsis (Moreno et al., 2013; Lange et al., 2014; Hematy et al., 2016). RRP44A and RRP6L1 are essential for the 3′-5′ exonuclease catalytic activity of the core exosome (Chekanova et al., 2007; Shin and Chekanova, 2014). HEN2 is an RNA helicase responsible for the degradation of polyadenylated nuclear exosome substrates (Lange et al., 2014). The zinc-finger protein SOP1 was identified as a novel co-factor of the exosome and a mutation in SOP1 was found to enhance PTGS as well. SOP1 co-localizes with HEN2 in nucleoplasmic speckles and is required for the degradation of a selective subset of nuclear exosome targets (Hematy et al., 2016). Once the activity of the exosome is impaired, aberrant transcripts accumulate and trigger the PTGS pathway. A mutation in the exosome core subunit RRP45B/CER7 was identified from a mutagenesis screen for plant cuticular wax biosynthesis (Hooker et al., 2007; Lam et al., 2012). Abundant small RNAs from endogenous loci were found to accumulate in rrp45b/cer7, and the biogenesis of these small RNAs requires RDR1, RDR6, SGS3, SILENCING DEFECTIVE5 (SDE5), DCL4, HEN1 and AGO1, most of which are components of the tasiRNA biosynthetic pathway (Lam et al., 2015). Although the authors defined these small RNAs as tasiRNAs, it is more likely that they resemble rqc-siRNAs, which are generated due to the derepression of PTGS upon impairment of RNA decay (Lam et al., 2015). All these exosome components including RRP4, RRP41, RRP45B/CER7, RRP44A, RRP6L1, HEN2 and SOP1 are predominantly nuclearly localized (Chekanova et al., 2007; Zhang et al., 2010; Moreno et al., 2013; Lange et al., 2014; Hematy et al., 2016), indicating that nuclear RNAs are also instrumental for the S-PTGS pathway, which is in agreement with the existence of both cytoplasmic and nuclear PTGS (Hoffer et al., 2011; Le Masson et al., 2012). Besides, the DExH-box helicase SKI2, the tetratricopeptide repeat protein SKI3 and the WD-40 (beta-transducin) repeats protein SKI8 were found to mediate the degradation of 5′-fragments generated by miRNA-guided RISC cleavage in Arabidopsis (Branscheid et al., 2015). Moreover, the ski3 single mutant also provokes the entry of non-silenced transgenes into the S-PTGS pathway (Yu et al., 2015). SKI2, SKI3 and SKI8 form a heterotetrameric complex in yeast, and mediate RNA decay through unwinding and threading transcripts into the exosome complex in the cytoplasm (Brown et al., 2000; Synowsky and Heck, 2008). Disruption of this process impairs the exosome degradation pathway and triggers RNA silencing.
5′-3′ and 3′-5′ bidirectional degradation
While impairment of either the 5′-3′ or the 3′-5′ degradation pathway has a modest effect in triggering RNA silencing of endogenous genes, simultaneously impairing both degradation pathways triggers endogenous RNA silencing more dramatically (Zhang et al., 2015). This is in agreement with the morphological phenotypes observed in xrn4 or ski2 single mutants and the xrn4 ski2 double mutant. The single mutants exhibit minor development defects (Olmedo et al., 2006), whereas the xrn4 ski2 double mutant displays much severe phenotypes including lethality for severe alleles at the embryo stage (Zhang et al., 2015). Upon impairing bidirectional RNA turnover, a large number of 21-22 nt siRNAs were generated from 441 protein-coding transcripts, including the 5′ cleavage fragments of some miRNA targets. These siRNAs were termed coding transcript-derived siRNAs (ct-siRNAs), and they require RDR6, SGS3, DCL2 and DCL4 for biogenesis and are partially dependent on AGO1 for function (Zhang et al., 2015). It is highly possible that the ct-siRNAs reported here are similar to the RDR6-dependent rqc-siRNAs found in the decapping mutants (Martinez de Alba et al., 2015). Actually, the morphological phenotypes of xrn4 ski2 resemble those of dcp2 or vcs mutants (Martinez de Alba et al., 2015; Zhang et al., 2015). So compromising both 5′-3′ and 3′-5′ RNA decay activities mimics compromising decapping activity; this may indicate that decapping is required for the degradation from both 5′ and 3′ ends.
Core components of nonsense-mediated decay (NMD) are endogenous RNA silencing suppressors
In plants, the specialized RNA decay pathway NMD is activated by the presence of premature termination codons (Chiba and Green, 2009). The NMD machinery consists of three core components, UP FRAMESHIFT1 (UPF1), UPF2 and UPF3, which are recruited to defective transcripts and direct the degradation of these aberrant RNAs either through decapping or deadenylation pathways followed by exonucleolytic decay (Lejeune et al., 2003; Yoine et al., 2006; Kerenyi et al., 2008; Chiba and Green, 2009). Impairing Arabidopsis UPF1 and UPF3 has been reported to enhance sense transgene (S)-PTGS, indicating that both of them are endogenous PTGS suppressors (Moreno et al., 2013). Besides, it was also observed that UPF1 colocalized with both P-body and siRNA-body markers, suggesting that UPF1 has dual roles in RNA surveillance and RNA silencing (Moreno et al., 2013). It is highly possible that endogenous rqc-siRNAs or ctsiRNAs are also present in the mutants of NMD core components, however further investigation is needed.
VIRUS INFECTION TRIGGERS RNA SILENCING AT ENDOGENOUS GENES
RNA silencing is a major antiviral defense mechanism employed by plants and other eukaryotes (Waterhouse, 2006; Ding and Voinnet, 2007; van Mierlo et al., 2011; Wang et al., 2012). Recently it was found that activation of antiviral RNA silencing is accompanied by the production of an abundant class of siRNAs mapped to the exon regions of more than 1000 endogenous genes as well as rRNAs upon viral infection in Arabidopsis (Cao et al., 2014). This novel class of siRNAs is predominantly 21 nt in length, and was designated as virus-activated siRNAs (vasiRNAs). The biogenesis of vasiRNA requires RDR1 and DCL4, and vasiRNAs direct the silencing of target host genes through AGO2 (Cao et al., 2014). The fact that viral infection triggers the production of siRNAs from many endogenous genes suggests that viruses might compromise host’s RNA surveillance system, or that infected plants deliberately suppress RNA surveillance as a means to generate vasiRNAs to combat viruses. Although vasiRNAs, ct-siRNAs and rqc-siRNAs are all derived from endogenous genes, their biogenesis and function involve different RDR and AGO proteina (Martinez de Alba et al., 2015; Zhang et al., 2015). Interestingly, loss of function in XRN4/EIN5 enhances the biogenesis of vasiRNAs and viral resistance, but does not alter the abundance of viral siRNAs, indicating that the RNA silencing of endogenous genes directed by vasiRNAs is independent of anti-viral RNA silencing, and the accumulation of vasiRNAs plays a role in plant virus resistance (Cao et al., 2014). The detailed mechanism of how the endogenous transcripts switch to producing vasiRNAs upon viral infection and the role of vasiRNAs in anti-viral immunity awaits further study.
CONCLUSIONS AND PERSPECTIVE
The last decade has seen a significant increase in our knowledge of understanding how plants elaborately avoid inappropriate RNA silencing of endogenous protein-coding genes. A considerable amount of research has been performed and the results revealed that RNA surveillance is the key to preventing RNA silencing at endogenous genes (Belostotsky, 2004; Herr et al., 2006; Gregory et al., 2008; Moreno et al., 2013; Branscheid et al., 2015; Martinez de Alba et al., 2015; Yu et al., 2015; Zhang et al., 2015). Many players involved in proper RNA processing and RNA decay have been found to suppress RNA silencing (summarized in Table 1), demonstrating the importance of the strict control of the coordination of RNA surveillance and RNA silencing in plants. RNA quality control eliminates dysfunctional transcripts based on their structural features, whereas RNA silencing degrades both aberrant transcripts and their homologous functional transcripts since it relies on the complementarity between the siRNA and its targets (Molnar et al., 2011; Schoenberg and Maquat, 2012). So the expression of endogenous genes is primarily regulated by RNA surveillance, rather than RNA silencing, which might be deleterious for plants. The findings that players in RNA quality control suppress RNA silencing demonstrate that RNA surveillance is the first layer of plant defense against defective nucleic acids. Base on the existing results, one model is that aberrant RNAs are channeled into RNA silencing pathways and become substrates for RDR and Dicer proteins when RNA surveillance is impaired, either due to the mutation of important RNA surveillance components or the saturation of the surveillance machinery by over-accumulated aberrant transcripts (Fig. 2). The tug-of-war between RNA surveillance and RNA silencing ensures the appropriate partitioning of endogenous RNA substrates among these degradation pathways. The fact that P-bodies and siRNA-bodies are spatially associated and functionally linked also supports this model.
Table 1.
RNA Surveillance Factors Known to Suppress RNA Silencing
| Protein | Name | Gene accession |
Function | References |
|---|---|---|---|---|
| ESP1 | ENHANCED SILENCING PHENOTYPE 1 |
AT1g73840 | 3′ end processing | (Herr et al., 2006) |
| ESP3 | ENHANCED SILENCING PHENOTYPE 3 |
AT1g32490 | RNA splicing | (Herr et al., 2006) |
| ESP4 | ENHANCED SILENCING PHENOTYPE 4 |
AT5g01400 | 3′ end processing | (Herr et al., 2006) |
| ESP5 | ENHANCED SILENCING PHENOTYPE 5 |
AT5g23880 | 3′ end processing | (Herr et al., 2006) |
| SMD1B | SM DOMAIN- CONTAINING PROTEIN 1B |
AT4g02840 | RNA splicing | (Elvira-Matelot et al., 2016) |
| PARN | POLY(A) RIBONUCLEASE | AT1g55870 | Deadenylation | (Moreno et al., 2013) |
| CCR4 | CARBON CATABOLITE REPRESSOR4 |
AT3g58560 | Deadenylation | (Moreno et al., 2013) |
| DCP1 | DECAPPING1 | AT1g08370 | Decapping complex formation |
(Martinez de Alba et al., 2015) |
| DCP2 | DECAPPING2/TRIDENT | AT5g13570 | Decapping complex formation; Hydrolyze the 5′- m7G-cap |
(Thran et al., 2012; Martinez de Alba et al., 2015) |
| VCS | VARICOSE | AT3g13300 | Decapping complex formation |
(Martinez de Alba et al., 2015) |
| XRN2 | EXORIBONUCLEASE2 | AT5g42540 | 5′-3′ exonucleolytic cleavage |
(Gy et al., 2007) |
| XRN3 | EXORIBONUCLEASE3 | AT1g75660 | 5′-3′ exonucleolytic cleavage |
(Gy et al., 2007) |
| XRN4/EIN5 | EXORIBONUCLEASE4 /ETHYLENEINSENSITIVE5 |
AT1g54490 | 5′-3′ exonucleolytic cleavage |
(Gazzani et al., 2004; Souret et al., 2004; Rymarquis et al., 2011; Hayashi et al., 2012) |
| FIERY1 | FIERY1/FRY1/SAL1 | AT5g63680 | Maintaining function of exoribonucleases |
(Gy et al., 2007) |
| RRP4 | RIBOSOMAL RNA PROCESSING4 |
AT1g03360 | Member of core exosome complex |
(Moreno et al., 2013) |
| RRP41 | RIBOSOMAL RNA PROCESSING41 |
AT3g61620 | Member of core exosome complex |
(Moreno et al., 2013) |
| RRP45B/CE R7 |
RIBOSOMAL RNA PROCESSING45B/ ECERIFERUM7 |
AT3g60500 | Member of core exosome complex |
(Lam et al., 2015) |
| RRP44A | RIBOSOMAL RNA PROCESSING44A |
AT2g17510 | Exosome- associated factors; 3′ hydrolytic exonuclease and endonuclease activity |
(Moreno et al., 2013) |
| RRP6L1 | RIBOSOMAL RNA PROCESSING6L1 |
AT1g54440 | Exosome- associated factors; 3′ hydrolytic exonuclease |
(Moreno et al., 2013) |
| HEN2 | HUA ENHANCER2 | AT2g06990 | RNA helicase, degrade polyadenylated nuclear exosome substrates |
(Lange et al., 2014) |
| SOP1 | SUPPRESSOR OF PAS2-1 | AT1g21580 | Zinc-finger protein; exosome co-factor |
(Hematy et al., 2016) |
| SKI2 | SUPERKILLER2 | AT3g49690 | DExH-box helicase, member of exosome cofactor Ski complex |
(Branscheid et al., 2015) |
| SKI3 | SUPERKILLER3 | AT1g76630 | Member of exosome cofactor Ski complex |
(Branscheid et al., 2015; Yu et al., 2015) |
| SKI8 | SUPERKILLER8 | AT4g29830 | Member of exosome cofactor Ski complex |
(Branscheid et al., 2015) |
| UPF1 | UP FRAMESHIFT1 | AT5g47010 | Nonsense- mediated decay |
(Moreno et al., 2013) |
| UPF3 | UP FRAMESHIFT3 | AT1g33980 | Nonsense- mediated decay |
(Moreno et al., 2013) |
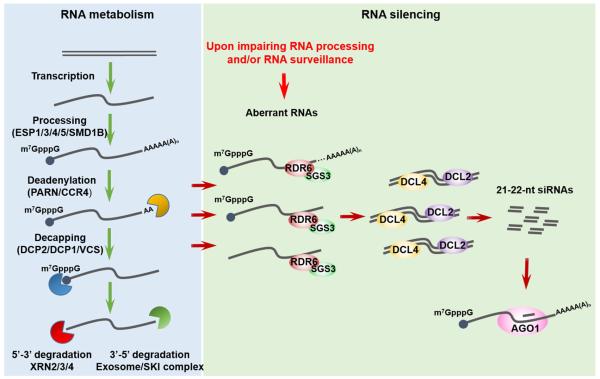
Figure 2.
RNA Surveillance as a Key to Suppressing RNA Silencing at Endogenous Genes
Impairing RNA processing or RNA decay, such as mRNA 3′ end formation, splicing, deadenylation, decapping, 5′-3′ or 3′-5′ exonucleolytic degradation, can generate aberrant RNAs that are channeled into RNA silencing through the activities of RNA-DEPENDENT RNA POLYMERASE 6 (RDR6), DICER-LIKE 4 (DCL4) or DCL2,) and ARGONAUTE1 (AGO1). The tug-of-war between RNA surveillance and RNA silencing ensures the appropriate partitioning of endogenous RNA substrates among these degradation pathways. mRNAs with dotted lines before the polyA tail indicate that they are aberrant forms resulting from defects in splicing or polyadenylation.
An outstanding question is what determines whether an endogenous transcript undergo RNA silencing upon impairment of RNA quality control. Even when both 5′-3′ and 3′-5′ degradation pathways are impaired, only a group of hundreds of RNAs enters PTGS. One model is that levels of aberrant RNAs have to reach a silencing threshold to initiate RNA silencing. In this model, endogenous genes expressed at high levels are prone to be channeled into RNA silencing, whereas the relatively lowly expressed genes are not. However, efforts to find this correlation failed- transcripts undergoing RNA silencing are not expressed at distinctively higher levels compared to other genes (Martinez de Alba et al., 2015). Identification of the common features or specific functional categories of the genes that are prone to enter PTGS is worthwhile. Another area that warrants further research is how, and to what extent, plants modulate the production of siRNAs from endogenous protein-coding transcripts. The finding that plants produce siRNAs from endogenous genes upon viral infection to facilitate anti-viral immunity raises the possibility that plants may utilize siRNAs from endogenous protein-coding genes in responses to various stresses or even in developmental regulation. Understanding the molecular and cell biological mechanisms governing the crosstalk between RNA surveillance and RNA silencing and the regulation of this crosstalk in plant development and stress responses will be of interest in plant biology.
ACKNOWLEDGMENTS
We are thankful to anonymous reviewers for their valuable suggestions to improve this article.
FUNDING
Research in the Chen laboratory is supported by grants from the National Institutes of Health (GM061146), National Science Foundation (IOS-1340001), National Science Foundation of China (91440105), and Guangdong Innovation Research Team Fund (2014ZT05S078).
Footnotes
No conflict of interest declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing. Trends Biochem. Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Belostotsky D. mRNA turnover meets RNA interference. Mol. Cell. 2004;16:498–500. doi: 10.1016/j.molcel.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Belostotsky D. Exosome complex and pervasive transcription in eukaryotic genomes. Curr. Opin. Cell Biol. 2009;21:352–358. doi: 10.1016/j.ceb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O, Brodersen P. SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 2015;43:10975–10988. doi: 10.1093/nar/gkv1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal-On A, Zhou C, Li Y, Ding SW. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:14613–14618. doi: 10.1073/pnas.1407131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. Deadenylation and P-bodies. Adv. Exp. Med. Biol. 2013;768:183–195. doi: 10.1007/978-1-4614-5107-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A silencing safeguard: links between RNA silencing and mRNA processing in Arabidopsis. Dev. Cell. 2008;14:811–812. doi: 10.1016/j.devcel.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs in development - insights from plants. Curr. Opin. Genet. Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Green PJ. mRNA Degradation Machinery in Plants. J. Plant Biol. 2009;52:114–124. [Google Scholar]
- Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene. 2004;328:95–102. doi: 10.1016/j.gene.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4− and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens A, Wang MB, Smith NA, Waterhouse PM. RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol. 2008;147:456–468. doi: 10.1104/pp.108.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira-Matelot E, Bardou F, Ariel F, Jauvion V, Bouteiller N, Le Masson I, Cao J, Crespi MD, Vaucheret H. The nuclear ribonucleoprotein SmD1 interplays with splicing, RNA quality control and post-transcriptional gene silencing in Arabidopsis. Plant Cell. 2016 doi: 10.1105/tpc.15.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Garcia S, Voinnet O. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe. 2014;16:391–402. doi: 10.1016/j.chom.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Gunawardana D, Cheng HC, Gayler KR. Identification of functional domains in Arabidopsis thaliana mRNA decapping enzyme (AtDcp2) Nucleic Acids Res. 2008;36:203–216. doi: 10.1093/nar/gkm1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nanba C, Saito M, Kondo M, Takeda A, Watanabe Y, Nishimura M. Loss of XRN4 function can trigger cosuppression in a sequence-dependent manner. Plant Cell Physiol. 2012;53:1310–1321. doi: 10.1093/pcp/pcs078. [DOI] [PubMed] [Google Scholar]
- Hematy K, Bellec Y, Podicheti R, Bouteiller N, Anne P, Morineau C, Haslam RP, Beaudoin F, Napier JA, Mockaitis K, et al. The Zinc-Finger Protein SOP1 Is Required for a Subset of the Nuclear Exosome Functions in Arabidopsis. PLoS Genet. 2016;12:e1005817. doi: 10.1371/journal.pgen.1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Molnar A, Jones A, Baulcombe DC. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:14994–15001. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Misson J, Crisp PA, David P, Bayle V, Estavillo GM, Javot H, Chiarenza S, Mallory AC, Maizel A, et al. A novel fry1 allele reveals the existence of a mutant phenotype unrelated to 5′->3′ exoribonuclease (XRN) activities in Arabidopsis thaliana roots. PLoS One. 2011;6:e16724. doi: 10.1371/journal.pone.0016724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, Wagner N, Voelker T. Posttranscriptional gene silencing in nuclei. Proc. Natl. Acad. Sci. USA. 2011;108:409–414. doi: 10.1073/pnas.1009805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker TS, Lam P, Zheng H, Kunst L. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell. 2007;19:904–913. doi: 10.1105/tpc.106.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incarbone M, Dunoyer P. RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 2013;18:382–392. doi: 10.1016/j.tplants.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Takeda A, Motose H, Watanabe Y. Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 2007;581:2455–2459. doi: 10.1016/j.febslet.2007.04.051. [DOI] [PubMed] [Google Scholar]
- Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012;31:1704–1713. doi: 10.1038/emboj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Maizel A, Chen X. Traffic into silence: endomembranes and post-transcriptional RNA silencing. EMBO J. 2014;33:968–980. doi: 10.1002/embj.201387262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Lam P, Zhao L, Eveleigh N, Yu Y, Chen X, Kunst L. The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. 2015;167:323–336. doi: 10.1104/pp.114.252825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Zhao L, McFarlane HE, Aiga M, Lam V, Hooker TS, Kunst L. RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 2012;159:1385–1395. doi: 10.1104/pp.112.199646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Gagliardi D. The exosome and 3′-5′ RNA degradation in plants. Adv. Exp. Med. Biol. 2011;702:50–62. doi: 10.1007/978-1-4419-7841-7_5. [DOI] [PubMed] [Google Scholar]
- Lange H, Sement FM, Gagliardi D. MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 2011;68:51–63. doi: 10.1111/j.1365-313X.2011.04675.x. [DOI] [PubMed] [Google Scholar]
- Lange H, Zuber H, Sement FM, Chicher J, Kuhn L, Hammann P, Brunaud V, Berard C, Bouteiller N, Balzergue S, et al. The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 2014;10:e1004564. doi: 10.1371/journal.pgen.1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Masson I, Jauvion V, Bouteiller N, Rivard M, Elmayan T, Vaucheret H. Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell. 2012;24:3603–3612. doi: 10.1105/tpc.112.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Luo Z, Chen Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19:943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD. Composition and function of P bodies in Arabidopsis thaliana. Front. Plant Sci. 2014;5:201. doi: 10.3389/fpls.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Alba AE, Elvira-Matelot E, Vaucheret H. Gene silencing in plants: a diversity of pathways. Biochim. Biophys. Acta. 2013;1829:1300–1308. doi: 10.1016/j.bbagrm.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Martinez de Alba AE, Moreno AB, Gabriel M, Mallory AC, Christ A, Bounon R, Balzergue S, Aubourg S, Gautheret D, Crespi MD, et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015;43:2902–2913. doi: 10.1093/nar/gkv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015;66:243–267. doi: 10.1146/annurev-arplant-043014-114633. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Molnar A, Melnyk C, Baulcombe DC. Silencing signals in plants: a long journey for small RNAs. Genome Biol. 2011;12:215. doi: 10.1186/gb-2010-11-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From Birth to Death: The Complex Lives of Eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Moreno AB, Martinez de Alba AE, Bardou F, Crespi MD, Vaucheret H, Maizel A, Mallory AC. Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 2013;41:4699–4708. doi: 10.1093/nar/gkt152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Murota K, Hagiwara-Komoda Y, Komoda K, Onouchi H, Ishikawa M, Naito S. Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from Arabidopsis callus cultures. Plant Cell Physiol. 2011;52:1443–1453. doi: 10.1093/pcp/pcr080. [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5′-->3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta. 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. ETHYLENE-INSENSITIVE5 encodes a 5′-->3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS, Peragine A, Yoshikawa M, Hunter C, Willmann M, Wu G. The function of RNAi in plant development. Cold Spring Harb. Symp. Quant. Biol. 2006;71:165–170. doi: 10.1101/sqb.2006.71.030. [DOI] [PubMed] [Google Scholar]
- Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Garciadeblas B, Rodriguez-Navarro A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′), 5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;8:529–537. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13:368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA. 2004;10:1200–1214. doi: 10.1261/rna.7540204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. microRNA biogenesis and turnover in plants. Cold Spring Harb. Symp. Quant. Biol. 2012;77:183–194. doi: 10.1101/sqb.2013.77.014530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, Green PJ. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA. 2011;17:501–511. doi: 10.1261/rna.2467911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Chekanova JA. Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet. 2014;10:e1004612. doi: 10.1371/journal.pgen.1004612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Synowsky SA, Heck AJ. The yeast Ski complex is a hetero-tetramer. Protein Sci. 2008;17:119–125. doi: 10.1110/ps.073155908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Thran M, Link K, Sonnewald U. The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 2012;72:368–377. doi: 10.1111/j.1365-313X.2012.05066.x. [DOI] [PubMed] [Google Scholar]
- Tomek W, Wollenhaupt K. The "closed loop model" in controlling mRNA translation during development. Anim. Reprod. Sci. 2012;134:2–8. doi: 10.1016/j.anireprosci.2012.08.005. [DOI] [PubMed] [Google Scholar]
- van Mierlo JT, van Cleef KW, van Rij RP. Defense and counterdefense in the RNAi-based antiviral immune system in insects. Methods Mol. Biol. 2011;721:3–22. doi: 10.1007/978-1-61779-037-9_1. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. MicroRNA-dependent trans-acting siRNA production. Sci. STKE. 2005;2005:pe43. doi: 10.1126/stke.3002005pe43. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Virtanen A, Henriksson N, Nilsson P, Nissbeck M. Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit. Rev. Biochem. Mol. Biol. 2013;48:192–209. doi: 10.3109/10409238.2013.771132. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang MB, Masuta C, Smith NA, Shimura H. RNA silencing and plant viral diseases. Mol. Plant Microbe Interact. 2012;25:1275–1285. doi: 10.1094/MPMI-04-12-0093-CR. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM. Defense and counterdefense in the plant world. Nat. Genet. 2006;38:138–139. doi: 10.1038/ng0206-138. [DOI] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008;56:517–530. doi: 10.1111/j.1365-313X.2008.03623.x. [DOI] [PubMed] [Google Scholar]
- Wen J, Brogna S. Nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2008;36:514–516. doi: 10.1042/BST0360514. [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Huang R, Zhu JK. A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J. 2004;40:536–545. doi: 10.1111/j.1365-313X.2004.02225.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. Processing bodies and plant development. Curr. Opin. Plant Biol. 2011;14:88–93. doi: 10.1016/j.pbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- Yu A, Saudemont B, Bouteiller N, Elvira-Matelot E, Lepere G, Parent JS, Morel JB, Cao J, Elmayan T, Vaucheret H. Second-Site Mutagenesis of a Hypomorphic argonaute1 Allele Identifies SUPERKILLER3 as an Endogenous Suppressor of Transgene Posttranscriptional Gene Silencing. Plant Physiol. 2015;169:1266–1274. doi: 10.1104/pp.15.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a Crucial Step in Plant microRNA Biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He X, Zhu JK. RNA-directed DNA methylation in plants: Where to start? RNA Biol. 2013;10:1593–1596. doi: 10.4161/rna.26312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Murphy C, Sieburth LE. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc. Natl. Acad. Sci. USA. 2010;107:15981–15985. doi: 10.1073/pnas.1007060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Y, Liu X, Hong X, Xu Y, Zhu P, Shen Y, Wu H, Ji Y, Wen X, et al. Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015;348:120–123. doi: 10.1126/science.aaa2618. [DOI] [PubMed] [Google Scholar]