Sun, Guo, and Fässler review the function and regulation of integrin-mediated mechanotransduction and discuss how its dysregulation impacts cancer progession.
Abstract
Cells can detect and react to the biophysical properties of the extracellular environment through integrin-based adhesion sites and adapt to the extracellular milieu in a process called mechanotransduction. At these adhesion sites, integrins connect the extracellular matrix (ECM) with the F-actin cytoskeleton and transduce mechanical forces generated by the actin retrograde flow and myosin II to the ECM through mechanosensitive focal adhesion proteins that are collectively termed the “molecular clutch.” The transmission of forces across integrin-based adhesions establishes a mechanical reciprocity between the viscoelasticity of the ECM and the cellular tension. During mechanotransduction, force allosterically alters the functions of mechanosensitive proteins within adhesions to elicit biochemical signals that regulate both rapid responses in cellular mechanics and long-term changes in gene expression. Integrin-mediated mechanotransduction plays important roles in development and tissue homeostasis, and its dysregulation is often associated with diseases.
Introduction
Multicellularity in the metazoan evolved by developing and diversifying genes involved in cell differentiation, cell–cell communication, and cell adhesion (Rokas, 2008). Cell adhesion to the ECM and to neighboring cells allows cells of different lineages to interact at the organ level by facilitating the exchange of biochemical and biophysical information. The ECM of the metazoan is mainly composed of fibrous proteins (e.g., collagens and elastin) that confer the ECM with tensile strength and elasticity, proteoglycans (e.g., perlecan and hyaluronan) that allow interfibrillar slippage under tensile loads and thus confer the ECM with viscosity, and multiadhesive glycoproteins (e.g., fibronectin and laminins) that bind proteoglycans and collagen fibers (Mouw et al., 2014). ECM proteins are recognized by specific cell surface receptors such as integrins, syndecans, CD44, and dystroglycan. ECM receptors induce signaling pathways and facilitate the assembly of different ECM components into sheet-like fibrous structures (basement membranes) or seemingly chaotic meshworks of fibrils and fibers (connective tissue) whose biochemical composition, compliance, and geometric and topographic features in nanometer to micrometer scale vary and correlate with tissue-specific physiological functions (Gasiorowski et al., 2013). The complex biochemical and biophysical characteristics of the ECM contain a wealth of biological information that, in concert with soluble growth factors that are often immobilized within the ECM, exerts a profound impact on many cellular behaviors, including migration, proliferation, and differentiation. To detect and interpret the biological information in the ECM, cells adhere and transduce myosin-generated traction forces to the ECM via integrin-based adhesions and elicit a series of dynamic signaling events that are jointly termed mechanotransduction (Hoffman et al., 2011). Integrin-mediated mechanotransduction commences with force transmission between cells and the ECM (termed mechanotransmission), a process that occurs across the mechanosensitive, integrin-based adhesions. The mechanical load on the adhesion sites leads to force-induced functionalities, such as changes in protein conformation or enzymatic reactions (e.g., kinase activities) that in turn induce biochemical signals (termed mechanosignaling). Finally, the mechanically induced biochemical signals generate appropriate cellular responses that adapt to physiological processes (e.g., polarity, migration, differentiation, and survival) accordingly. In this review, we introduce the main concepts of integrin-mediated mechanotransduction, summarize recent progress on the underlying biophysical principles and the in vivo functional significance, and discuss how the viscoelasticity of the ECM influences integrin-mediated mechanotransduction and how its dysregulation impacts on cancer progression.
Structure of integrin-based adhesions
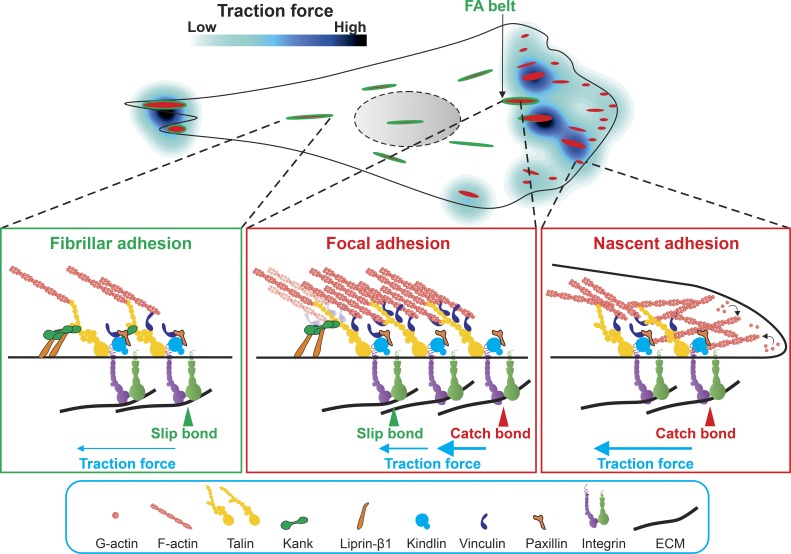
Integrins, which connect the ECM with intracellular actin cytoskeleton and thereby mechanically integrate the extracellular and intracellular compartments, are heterodimeric transmembrane receptors composed of α and β subunits. There are 24 different integrin receptors in mammals, each recognizing a specific set of ECM ligands (Hynes, 2002). Integrin-mediated adhesion starts with conformational changes in the integrin ectodomain (integrin activation) that shift integrins from a low- to high-affinity state for ligand binding (Luo et al., 2007; Su et al., 2016). Kindlin and talin bind integrin cytoplasmic tails (with the exception of β4 integrin tails; de Pereda et al., 2009) and promote integrin activation (Calderwood et al., 2013). Upon ligand binding, integrins recruit numerous proteins to their short cytoplasmic tails, resulting in the assembly of various adhesion structures that differ in their morphology and subcellular localization as well as in their protein composition and mechanical properties (Schiller and Fässler, 2013). The first adhesion structure assembles at the leading edge of cell protrusions by nucleating three to six integrins interspaced by less than 70 nm into short-lived nascent adhesions (NAs; Fig. 1; Cavalcanti-Adam et al., 2007; Hu et al., 2007; Bachir et al., 2014). The mechanism by which integrins first assemble is unclear. Apart from their signaling function, NAs are able to transmit the retrograde pushing forces from the polymerizing branched actin network in membrane protrusions to the ECM via mechanosensitive proteins such as talin and vinculin. The assembly of NAs between lamellipodium and lamellum correlates with the switch from a fast to a slow actin retrograde flow rate caused by the transient coupling of integrins to F-actin (Hu et al., 2007). This coupling is required for the assembly and turnover of NAs (Alexandrova et al., 2008; Thievessen et al., 2013; Swaminathan et al., 2016). In turn, focal adhesion kinase (FAK) in NAs promotes actin dynamics by recruiting Arp2/3, the major actin nucleator in the lamellipodium (Serrels et al., 2007; Swaminathan et al., 2016). β-PIX and the kindlin–paxillin complex in NAs further promote Arp2/3 activity through activating the Rac1 GTPase (Kuo et al., 2011; Theodosiou et al., 2016). Most NAs are disassembled quickly, whereas a few of them grow in size along templates of actomyosin bundles into bigger focal adhesions (FAs) in the lamellum (Fig. 1). FA maturation is a coordinated process requiring further integrin clustering, F-actin bundling, and the reinforcement of the linkages between integrin and actomyosin (Straight et al., 2003; Choi et al., 2008; Iskratsch et al., 2013). Behind the lamellum, the mechanical linkage between FAs and the contractile actomyosin bundles is relaxed and/or released, consequently leading to clathrin-dependent FA disassembly (Ezratty et al., 2009; Yu et al., 2015) or the translocation of β1 integrin–containing adhesions into central, fibrillar adhesions (FBs; Fig. 1; Zamir et al., 2000; Sun et al., 2016). Although the lengths of integrin-based adhesions ranges from below 1 µm in NAs up to 8 µm in the elongated FAs and FBs, the breadth of integrin-based adhesions is usually below 1 µm, corresponding to the scale of the diameter of a single ECM fiber (Gasiorowski et al., 2013; Kim and Wirtz, 2013). Therefore, individual adhesion structures can detect local ECM properties with subcellular precision in a biophysically inhomogeneous 3D environment (Doyle et al., 2015). It is also important to note that there is no simple correlation between the adhesion size and the magnitude of the transmitted force (Oakes et al., 2012; Schiller et al., 2013), as small NAs may also transmit forces that are high enough to drive cell migration (Beningo et al., 2001).
Figure 1.
Model of a migrating cell containing diverse integrin-based adhesion structures that transmit different levels of traction forces. Nascent adhesions (NAs) emerge at the leading edge of cell protrusions by nucleating multiple ligand-bond integrins that have been activated by talin and kindlin. Adhesome proteins such as vinculin are subsequently recruited to adhesion sites via talin in a tension-dependent manner or via paxillin in a tension-independent manner. NAs are dynamically coupled to the polymerizing branched actin network through proteins of the molecular clutch such as talin and vinculin, which convert the retrograde movement of polymerizing branched actin network into a protrusive force at the leading edge membrane and rearward traction force on the ECM. A small number of NAs matures into large focal adhesions (FAs) along actomyosin bundles in the lamella. Within mature FAs, the molecular clutch becomes strongly engaged by F-actin binding to the talin ABS2 and ABS3 sites and vinculin binding to VBS where high traction forces are transmitted across integrins, leading to catch bond formation between integrin and ligand. Behind the lamella, Kank2 is recruited to the FA belt, where it maintains talin in its active integrin-bond state and at the same time diminishes F-actin binding to talin ABS2. Consequently, Kank2 decreases force transmission leading to the slip bond formation between integrin and its ligand and the translocation of FA belt-localized β1 integrins into fibrillar (or central) adhesions. At the rear end of migrating cells, trailing edge FAs may apply such high traction forces that detach the cell rear, probably together with integrin-bound ECM fragments.
Talin-based molecular clutch mediates mechanotransmission
Activated integrins are dynamically coupled to the actomyosin system through integrin- and/or F-actin–binding proteins, such as talin and vinculin. These molecules belong to the molecular clutch (see text box) that harnesses the power of the retrograde actin flow generated by the actin polymerization against the leading-edge cell membrane and the contractile actomyosin movements generated by nonmuscle myosin II to pull on FAs to propel the cell body forward (Mitchison and Kirschner, 1988; Chan and Odde, 2008; Giannone et al., 2009; Swaminathan and Waterman, 2016). The spatiotemporal regulation of the clutch engagement determines directional cell migration, for instance, in response to an ECM stiffness gradient in a process called durotaxis (Lo et al., 2000).
The molecular clutch
This concept was originally proposed in 1988 by Tim Mitchison and Marc Kirschner to explain how the actin retrograde flow exerts tension on the substrate (Mitchison and Kirschner, 1988). The molecular clutch generally refers to the mechanical linkage formed by dynamic associations between the ECM-bound integrins and the force-generating actomyosin cytoskeleton. Because integrin cytoplasmic domains do not contain an ABS, the clutch between integrins and actomyosin is mediated by integrin- and/or F-actin-binding proteins such as talin and vinculin. Talin directly interacts with the cytoplasmic domain of activated integrins and F-actin. The connection between integrins and F-actin can be further strengthened by vinculin that binds to talin and F-actin. Other proteins such as kindlin and α-actinin may also contribute to the molecular clutch. In situations where integrin-based adhesions are not connected to actomyosin, the clutch is not engaged, the retrograde actin flow at the cell leading edge is fast, and traction forces are low or not even generated. The engagement of the clutch has two consequences: the kinetic power of the actin retrograde flow and actomyosin contractility are converted into a traction force that pulls on the ECM, and the polymerizing F-actin pushes the resistant plasma membrane at the cell leading edge forward.
The molecular clutch is a highly tunable system that sensitively responds to a wide range of ECM rigidities. On stiff substrates, fast mechanical loading rates on talin induce partial protein unfolding and expose cryptic VBS. Vinculin binding to talin reinforces the clutch by further elevating force transmission. On soft ECM substrates that are easily deformed, the slow mechanical loading rates on talin are not sufficient to induce vinculin-dependent clutch reinforcement, which keeps the transmitted force at a low level.
The molecular clutch is a highly dynamic system. Clutch molecules exhibit fast exchange rates (minute scale) between FAs and their cytosolic pools, indicating that the mechanical linkages within the molecular clutch cannot be static. Furthermore, early study by Odde and colleagues predicted a “frictional slippage” model on stiff substrates caused by constant rupture of the molecular clutch under high mechanical loading rates (Chan and Odde, 2008). Although the probability of clutch rupture is reduced by the vinculin-dependent clutch reinforcement (Elosegui-Artola et al., 2016), multiple slippage interfaces are still observed between different clutch molecules on stiff substrates, indicating that the force is transduced across the molecular clutch via transient linkages between different frictional interfaces in a “slip-stick”–like manner rather than via static connections (Hu et al., 2007).
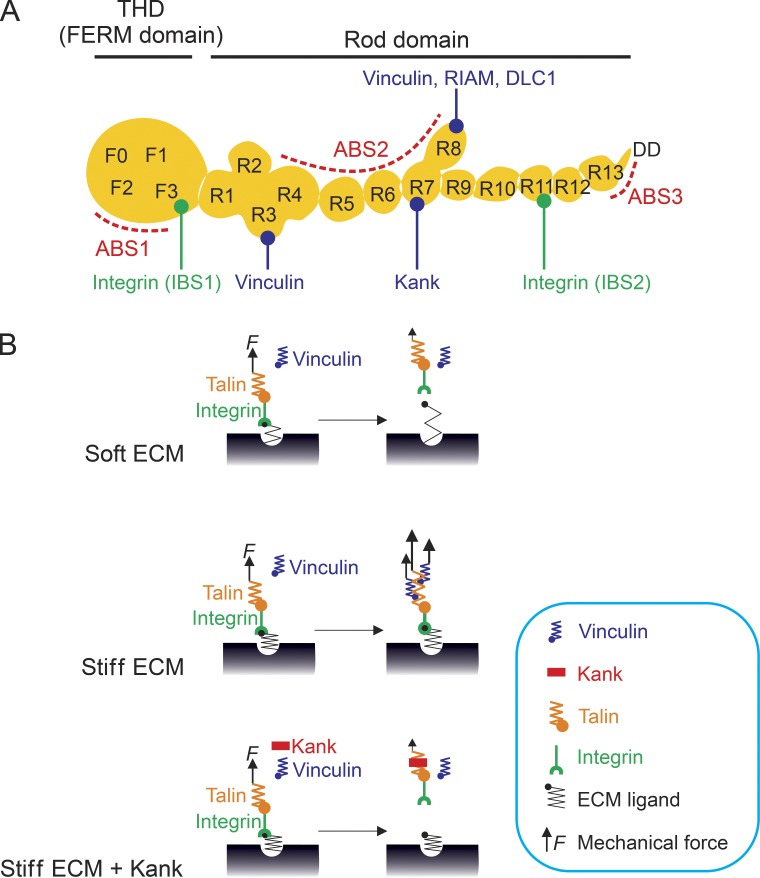
Talin plays a central role among the proteins that constitute the molecular clutch (Fig. 2 A). Mammals have two talin paralogue genes, Talin-1 and Talin-2, which share >80% homology and show different expression patterns. Although Talin-1 is ubiquitously expressed, Talin-2 is mainly expressed in muscle and neuronal cells. Talin consists of an N-terminal FERM (four-point-one, ezrin, radixin, moesin) domain, also called talin head domain (THD), and a long C-terminal rod domain composed of 13 helical bundles (R1–R13) followed by a dimerization motif. The THD directly interacts with the membrane-proximal NPxY motif of β integrin tails, negatively charged lipids in the inner leaflet of the plasma membrane and the cortical F-actin network (actin-binding site 1 [ABS1]). The talin rod domain contains two additional F-actin–binding sites (ABS2 and ABS3), 11 vinculin-binding sites (VBSs), and binding sites for regulatory proteins, including RIAM (Rap1–GTP-interacting adapter molecule), Kank (KN motif and ankyrin repeat domains) family proteins (whose functions are discussed in the following paragraphs), and the Rho GTPase activating protein DLC1 (deleted in liver cancer 1; Calderwood et al., 2013; Sun et al., 2016). Talin adopts an autoinhibited conformation in the cytosol through an intramolecular interaction between the THD and the R9 domain that masks the integrin-binding site of the THD. Kank2 and RIAM interact with the talin rod and release Talin from autoinhibition (Chang et al., 2014; Sun et al., 2016). Negatively charged lipids such as PtdIns(4,5)P2, synthesized by a FA-localized splice variant of phosphatidylinositol-4-phosphate 5-kinase type Iγ (PIPKIγ90) or the actin retrograde flow, may also facilitate talin activation at the plasma membrane (Goksoy et al., 2008; Zhu et al., 2008a; Legate et al., 2011; Comrie et al., 2015).
Figure 2.
Talin-based molecular clutch mediates mechanotransmission. (A) Domain organization of talin. The N-terminal talin head domain (THD) is an atypical FERM domain composed of F0, F1, F2, and F3 subdomains containing an integrin tail–binding site (IBS1). THD is linked to the talin rod domain via a flexible linker of ∼80 amino acids. The talin rod domain contains 13 helix bundles (R1–R13) and a dimerization domain (DD) and a second, underinvestigated integrin tail-binding site (IBS2). The IBS1, IBS2, three actin-binding sites (ABS1–3), two critical vinculin-binding sites (VBSs) in the R3 and R8 domains, and binding sites for RIAM, Kank2, and DLC1 are shown. The remaining VBSs are not depicted. (B) Model depicting the mechanical response of the molecular clutch and integrin–ligand bonds on the soft or rigid ECM in the presence or absence of Kank2. On soft substrates, the slow loading rates fail to induce talin unfolding and vinculin recruitment before the slip bond between integrin and the ligand ruptures under low force. In contrast, on rigid substrate, high loading rates induce vinculin-dependent clutch reinforcement, catch bond formation, and high force transmission. Kank2 interferes with F-actin binding to the talin ABS2, leading to reduced force transmission across talin as well as a diminished activation of VBSs. Consequently, Kank2 abrogates the clutch reinforcement and induces frequent ruptures of the slip bond between integrin and ligand even on rigid substrates.
The engagement between talin and F-actin is likely initiated at the C-terminal ABS3 site. Although the talin ABS3 is not required for cell adhesion, ∼45% of cells fail to assemble FAs when the function of ABS3 is impaired, suggesting an important role of the ABS3 site in initiating force transmission, which may, at least partially, be compensated by unknown mechanisms (Atherton et al., 2015). Subsequent to ABS3 engagement, the R3 domain serves as the first mechanosensor because of its lowest mechanical stability among all domains in the talin rod (Yao et al., 2016). Forces in the piconewton range lead to the stretching of the talin rod and the exposure (and activation) of the cryptic VBS in the R2R3 domain (del Rio et al., 2009; Yao et al., 2014). The subsequent vinculin binding to R2R3 triggers the engagement of talin ABS2 (R4–R8) with F-actin to further increase the force transmission across the talin–integrin complex. The increased force unfolds other helical bundles within the talin rod and exposes more cryptic VBSs. Eventually, vinculin strengthens the talin–F-actin linkages by further recruiting F-actin (Fig. 2 B).
Interestingly, the R1–R3 domains in talin-1 and talin-2 exhibit distinct mechanosensitivities that have consequences for the isoform-specific functions of talins. Unlike the cryptic VBS in the talin-1 R1–R3 domains, the VBS in the equivalent region of talin-2 recruits vinculin in an ABS2- and ABS3-independent manner (Austen et al., 2015). Furthermore, ABS2 and ABS3 in talin play distinct roles in mechanotransmission. Whereas ABS3 is required for high force transmission at the cell periphery, ABS2 plays important roles in sensing differences in ECM stiffness (Atherton et al., 2015; Austen et al., 2015; Kumar et al., 2016). In mouse embryonic fibroblasts, talin seems to be less important for force transmission on soft substrates that have a Young’s elastic modulus below 10 kPa in comparison with stiffer substrates (Elosegui-Artola et al., 2016), suggesting that other integrin- and F-actin-binding proteins such as kindlin (Bledzka et al., 2016), α-actinin (Roca-Cusachs et al., 2013), or the ILK (integrin linked kinase)–PINCH–parvin complex (Wickström et al., 2010) may link integrin to F-actin and contribute to the molecular clutch under different rigidity regimes.
The activation of talin and the engagement of F-actin on talin are tightly coupled. RIAM activates talin and provides an immediate connection between activated talin and F-actin polymerization by recruiting the F-actin stabilizer and elongator VASP (vasodilator-stimulated phosphoprotein; Lafuente et al., 2004; Worth et al., 2010). Such a rapid coupling may be essential for leukocyte β2 integrins to mediate robust cell adhesion and spreading in the presence of shear stress exerted by the blood flow (Klapproth et al., 2015; Su et al., 2015). In contrast to RIAM, Kank family proteins represent a novel type of talin activator that negatively regulates the connections between talin and F-actin. Kank family proteins consist of four members (Kank1–4) in mammals and are evolutionarily conserved in metazoans. They are characterized by a short KN motif at the N terminus followed by several coiled-coil domains in the central part of the protein and five ankyrin repeats at the C-terminal end (Zhu et al., 2008b). Mutations in Kank family genes have been associated with nephrotic syndrome, which can also be caused by compromised integrin-mediated adhesion of podocytes to the glomerular basement membrane or by elevated RhoA signaling (Gee et al., 2015). Kank proteins interact with liprin-β1 through a unique coiled-coil domain and are recruited to a large protein complex containing liprin-β1, liprin-α1, ELKS, and LL5β (Lansbergen et al., 2006; van der Vaart et al., 2013). Interestingly, this multiprotein complex clusters in the proximity of mature FAs, where the complex on the one hand captures the microtubule plus ends by recruiting CLASP proteins and mediates the local exocytosis of metalloprotease MMT-MP1 to promote FA disassembly, and on the other hand recruits KIF21a (kinesin family member 21A) through C-terminal ankyrin repeats of Kank to restrict microtubule outgrowth at the cell periphery (van der Vaart et al., 2013; Stehbens et al., 2014). Recent studies revealed that Kank proteins directly bind to the talin R7 domain through the KN motif and activate Talin at the peripheral rim of FAs termed the FA belt (Bouchet et al., 2016; Sun et al., 2016). The binding of Kank family proteins to the talin R7 occurs in close proximity to the talin ABS2 and interferes with F-actin recruitment to ABS2, resulting in reduced force transmission across talin and ECM-bound integrins, adhesion sliding, β1 integrin translocation into fibrillar adhesion, and a reduction in migration speed (Fig. 2 B; Sun et al., 2016).
The talin-based molecular clutch plays important roles in the 3D organization of FAs. Hundreds of different proteins are recruited to FAs and form a dynamic interaction network, collectively termed the “integrin adhesome” (Winograd-Katz et al., 2014). Despite the molecular complexity, FAs exhibit a 3D, modular, nanometer-scale organization. Different functional modules in FAs are vertically laminated and mechanically linked in agreement with the molecular clutch model. The integrin layer spans ∼20 nm across the plasma membrane, which is followed by the integrin signaling module containing THD, FAK, and paxillin within 20 nm of the plasma membrane, and then a force transduction module containing the talin rod domain, vinculin, and the VASP–zyxin complex that are connected to F-actin (Hu et al., 2007; Kanchanawong et al., 2010). The vertical positioning of vinculin is controlled by its interaction with talin and paxillin during the dynamic transmission and sensing of forces (Plotnikov et al., 2012; Case et al., 2015). In addition to the vertical layering of molecules in FAs, mature FAs assembled on fibronectin (FN) also display a horizontal layering with β3 integrins in FA cores and talin-bound Kank and β1 integrins at the FA belt (Sun et al., 2016; Fig. 1). The lateral segregation of different integrin subtypes may be determined by their differential integrin–ligand bond stabilities under different force regimes. For instance, β1 integrins can assemble adhesion complex without myosin-driven contractility and exhibit fast translocation in FAs, whereas β3 integrins are stationary in FAs and quickly internalized when force is released (Rossier et al., 2012; Schiller et al., 2013; Yu et al., 2015). Consistently, although the β3-enriched FA core is highly engaged with F-actin to transmit high forces, the enrichment of Kank in the FA belt diminishes the engagement of the talin ABS2 with F-actin and hence reduces the force transmission pointing to a force-dependent self-organization of FA structures and a compartmentalized force transmission within FAs (Sun et al., 2016).
ECM viscoelasticity regulates mechanotransmission
Mechanotransmission through the molecular clutch establishes a mechanical equilibrium between the extracellular mechanical resistance and the intracellular contractility and thus is essential for detecting the viscoelasticity of the ECM. Both relaxation and rupture of the force-transducing linkages can terminate mechanotransmission (Schoen et al., 2013; Chen et al., 2015). While relaxation can be caused by the displacement of the ECM ligands when tensile loads induce the deformation or viscous sliding of ECM materials, rupture may stochastically but preferentially occur at the weakest protein–protein interface where the bond lifetime may be regulated by mechanical loads. Whereas most protein–protein interactions exhibit a slip bond behavior in which bond lifetimes are shortened when forces are applied to the bond, some protein–protein interactions are allosterically activated under force, resulting in a prolonged bond lifetime (Thomas, 2008). Such a force-stabilized bond is called a “catch bond.” The α5β1 integrins in their relaxed state bind to the tripeptide RGD (Arg-Gly-Asp) motif in the 10th type III module of fibronectin (FNIII10; Takagi et al., 2003), whereas they additionally engage the synergy site in FNIII9 under tensile load to form a catch bond (Friedland et al., 2009; Kong et al., 2009). Importantly, repeated stretching of the bond between α5β1 integrin and FN, even with a low force amplitude of ∼10 pN, significantly prolongs the bond lifetime in a process called “cyclic mechanical reinforcement” (Kong et al., 2013). Catch bond behaviors have also been documented within the actomyosin system, such as the associations between F-actin/G-actin and between F-actin and myosin II (Guo and Guilford, 2006; Lee et al., 2013). On soft substrates, the slow loading rates on the molecular clutch fail to induce talin unfolding and vinculin recruitment before the slip bond between integrin and the ligand ruptures, whereas on a rigid substrate, high loading rates induce vinculin-dependent clutch reinforcement and catch bond formation (Fig. 2 B). Consequently, both catch bond–dependent adhesion strengthening and vinculin-dependent clutch reinforcement are essential to trigger downstream signaling such as activation of FAK and nuclear translocation of the transcription factor YAP (Friedland et al., 2009; Elosegui-Artola et al., 2016).
Parallel clustering of force-transducing linkages enhances the robustness of mechanotransmission. A theoretical analysis suggests that lateral cross-linking of adjacent force-transducing linkages by adaptor proteins allows the redistribution of the tensile load between parallel bonds within the integrin cluster, increases integrin–ligand rebinding rates, and extends the duration of mechanotransmission (Schoen et al., 2013). The functional significance of integrin clustering has been tested in tumor models, where enhanced β1 integrin clustering induced by the V737N point mutation in the β1 integrin transmembrane domain, but not constitutive β1 integrin activation induced by G429N point mutation in the β1 integrin ectodomain, can bypass the requirement of ECM stiffness for inducing FAK activation and malignant phenotypes in cancer cells cultured on soft substrates (Paszek et al., 2005; Levental et al., 2009). The viscoelasticity of ECM materials also influences mechanotransduction by regulating integrin clustering. In a pure elastic environment, soft ECM negatively modulates integrin rebinding rates and clustering (Qian and Gao, 2010). However, in a viscoelastic environment, where elastic energy can be dissipated through material deformations and translocation, cells counteract the softness by actively remodeling the ECM and increasing local ligand density, which in turn facilitates integrin clustering. Indeed, the spreading and osteoblast differentiation of human mesenchymal stem cells (hMSCs) are inhibited on soft polyacrylamide hydrogel, but not on soft polydimethylsiloxane, a highly viscous material (Trappmann et al., 2012). Furthermore, cell spreading, proliferation, and osteogenic differentiation of hMSCs are increased by stress relaxation on viscoelastic material (Chaudhuri et al., 2015, 2016). Similarly, when hMSCs are cultured on soft networks formed by RGD-functionalized dextran methacrylate fibers, cells recruit nearby fibers by contractile force, thereby increasing local ligand density and consequently promoting cell spreading, FA assembly, and FAK activation (Baker et al., 2015).
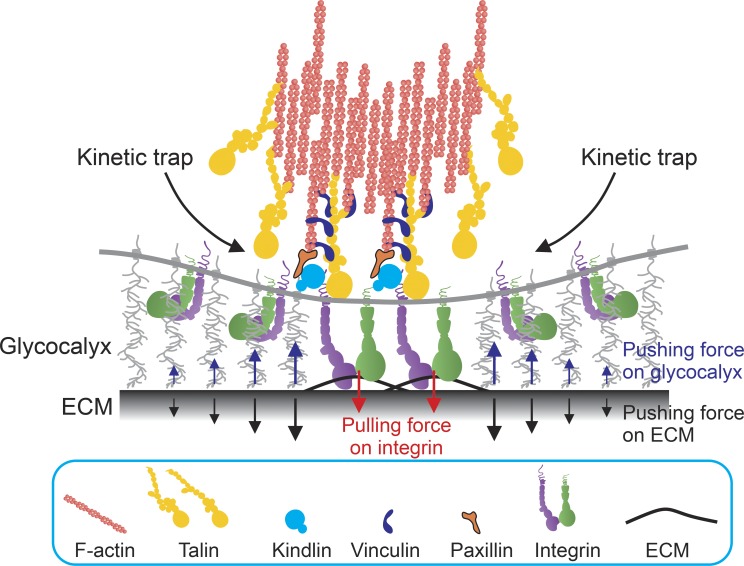
Integrin clustering can also be regulated by the mechanical resistance of the glycocalyx on the cell surface independent of ECM viscoelasticity (Paszek et al., 2009). The glycocalyx is a layer of glycoprotein–polysaccharide complex on the cell surface. Although integrins extend ∼11 nm from the cell surface in their inactive conformations and up to ∼20 nm when they adopt active conformations (Ye et al., 2010; Su et al., 2016), the glycocalyx can extend >100 nm from the cell surface (Hattrup and Gendler, 2008). Although the glycocalyx imposes electrosteric and osmotic repulsion between integrins and the ECM ligands (Bell et al., 1984; Hammer and Tirrell, 1996), local compression of the glycocalyx around integrin–ligand complexes reciprocally imposes a pulling force on integrins to promote integrin extension, clustering, and FA maturation in a kinetic trap-like manner (Fig. 3; Paszek et al., 2009). Importantly, cancer cells and various normal cells such as chondrocytes are armed with a bulky mucin-enriched glycocalyx, which induces a cellular perception of high ECM stiffness even on soft substrate and facilitates cancer cell survival and proliferation in a FAK- and phosphoinositide 3-kinase–dependent manner (Paszek et al., 2014).
Figure 3.
Glycocalyx around integrin–ligand complexes promotes integrin–ligand binding and clustering. The glycocalyx is a layer of glycoprotein–polysaccharide complexes on the cell surface that exerts electrosteric and osmotic repulsion to the ECM. Because the height of the glycocalyx exceeds that of the active integrins, the glycocalyx must be mechanically compressed around integrin–ligand complexes (indicated as blue arrows). Ligand-bound integrins within the compressed glycocalyx reciprocally sense the pulling force that promotes catch-bond formation and mechanotransduction. Glycocalyx-embedded integrin–ligand complexes shorten the distance between the plasma membrane and the ECM, which increases the probability of integrin activation and clustering around existing integrin-based adhesion sites in a kinetic trap-like manner. Talin is immobilized by actomyosin bundles within FAs, where it captures and activates integrins that enter the kinetic trap.
Detection of piconewton force with molecular tension sensors
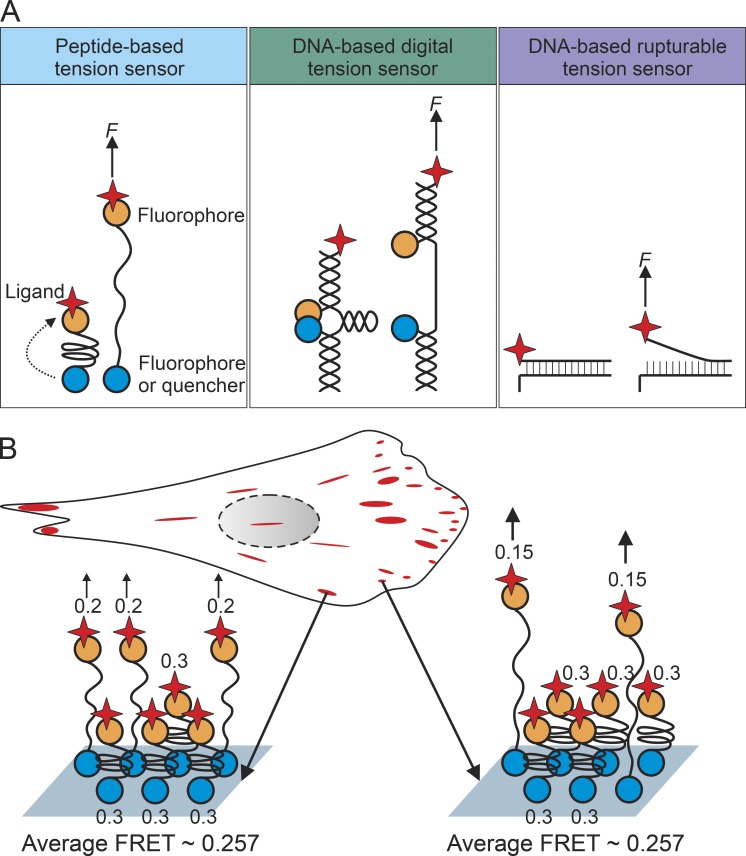
Measuring the exact magnitude of force sensed by each mechanosensitive unit is not a trivial issue (Polacheck and Chen, 2016). Atomic force microscopy and optical/magnetic tweezers can measure mechanical properties with subcellular resolution and in single proteins, but usually outside the physiological context. The development of molecular tension sensors (MTSs) allows measuring the forces sensed by mechanosensitive proteins in living cells with piconewton accuracy. A typical MTS contains a Förster resonance energy transfer (FRET) pair (or fluorophore-quencher pair) connected with a spring-like molecule. Tensile loads from both sides of the MTS increase the distance between the fluorophore pair, leading to either a gradual fluorescence change within a defined force range or a digital fluorescence change at a defined force threshold (Fig. 4 A; Freikamp et al., 2016). Although MTSs provided molecular insights on force transmission, there are considerable controversies between results from different MTS measurements. Using genetically encoded intracellular MTSs with different dynamic ranges, the mean forces sensed by vinculin and talin within mature FAs were shown to be ∼2.5 pN and between 7 and 10 pN, respectively (Grashoff et al., 2010; Austen et al., 2015). Single-molecule tension measurements using an extracellular MTS sensitive to forces below 6 pN revealed that the majority of integrins apply forces between 1 and 5 pN (Morimatsu et al., 2013, 2015). In contrast, DNA-based digital MTSs demonstrated that integrins can apply wider range of forces up to ∼15 pN (Blakely et al., 2014; Zhang et al., 2014). DNA-based tension probes that are irreversibly ruptured by forces above defined thresholds revealed that single integrins transmit forces of up to 40 pN during cell adhesion and even higher forces during FA maturation (Wang and Ha, 2013). These discrepancies may originate from intrinsic technical limitations of the current MTS technology. The bulk measurements of mean forces using FRET- or dequenching-based MTSs lack single-molecule information because of large background signals of tension sensors that are not mechanically engaged (Fig. 4 B). The disadvantage of digital MTSs is their inability to measure forces in a wider dynamic range. Rupturable MTSs on the other hand cannot monitor dynamic changes such as cyclic mechanical reinforcement. Another important issue is to determine mechanical forces in 3D and hence closer to a physiological context. This will probably be resolved in the near future with the help of high-resolution 3D traction force microscopy of cultured cells (Steinwachs et al., 2016) and oil microdroplet force sensors in intact tissue (Campàs et al., 2014).
Figure 4.
Molecular tension sensors are used to measure the forces sensed by mechanosensitive proteins. (A) Models of the different types of molecular tension sensors (MTSs). A typical MTS is composed of a FRET pair (or fluorophore-quencher pair) connected by a spring-like linker that can be extended by force. Three types of MTSs have been invented: the peptide-based MTS uses engineered tension-sensitive peptide as the linker and can be genetically encoded into intracellular proteins, the DNA-based digital MTS unwinds its hairpin structure and changes fluorescence intensity when force is above a defined threshold and is used to determine the lower limits of mechanical stress, and the DNA-based rupturable MTS is irreversibly ruptured by forces above defined thresholds and is used to approach the upper limits of forces transmitted by cell surface receptors. (B) Limitation of the bulk measurement in an MTS experiment. Because of the spatial resolution, current MTS measurements calculate forces as the mean mechanical stress of all MTS probes within each pixel. Such a measurement is strongly influenced by the background of unengaged MTS probes and the variation of forces sensed by each individual MTS probe.
Functional effects of forces on mechanosensitive structures
During mechanotransmission, intra- and extracellular force-bearing mechanosensitive molecules and structures can be stretched into different functional states. These force-induced functional states or functionalities trigger biochemical signals whose quality and quantity are determined by the duration, frequency, and history of each mechanotransmission event. On the level of single proteins, forces induce conformational changes that regulate protein–protein interactions, as shown for FN (Leiss et al., 2008), talin (del Rio et al., 2009), and filamin (Ehrlicher et al., 2011; Rognoni et al., 2012), or alter enzymatic reactions on key signaling molecules. Mechanical stretching allows Src family kinases to phosphorylate p130Cas on multiple tyrosine residues, which subsequently serve as a docking hub for downstream signaling molecules (Tamada et al., 2004; Sawada et al., 2006). Despite the lack of direct experimental proof, molecular simulations suggest that mechanical force may facilitate FAK activation, although the predicted force requirement of >100 pN is terribly high (Zhou et al., 2015). Within the integrin adhesome, the protein interaction network is remodeled in response to mechanical forces (Schiller and Fässler, 2013). A large group of proteins, particularly LIM domain–containing proteins, are recruited to FAs in a force-dependent manner through mechanisms that are still unknown (Schiller et al., 2011). Other system-level changes in the integrin adhesome have been observed during tension release and adhesion turnover, suggesting high plasticity and mechanosensitivity of the adhesome network (Kuo et al., 2011; Horton et al., 2015). On a larger scale, rapid propagation of mechanical stress from integrin adhesion sites along the prestressed cytoskeleton induces the kinase activity of Src at a distance of up to 60 µm away from the site of mechanical stress application and at a speed much higher than achieved by the diffusion or a motor protein-mediated transport of signaling molecules (Na et al., 2008; Wang et al., 2009). Integrins can also mechanically activate growth factor signaling. TGF-βs are growth factors involved in tissue homeostasis, tumor malignancy, and fibrosis (Wells and Discher, 2008). Secreted TGF-β is immobilized on ECM proteins such as fibrillin, proteoglycans, and FN by LAP (latency-associated protein). The integrins αVβ5, αVβ6, and αVβ8 directly bind and pull on LAP, leading to the release of TGF-β, which then activates the heterotetrameric transmembrane TGF-β receptor kinases formed by type II receptors and type I receptors (Buscemi et al., 2011). Intriguingly, type I and II receptors are spatially segregated within FAs and only converge on the less tensed FA belt structure, suggesting an intricate mechanical requirement for optimal TGF-β signaling (Rys et al., 2015).
Cellular responses in mechanotransduction
Mechanotransduction results in rapid changes of the cellular mechanics as well as long-term responses by affecting gene expression, both of which require integrin-dependent RhoA signaling and downstream actin dynamics. Mechanical stretch on integrins activates FAK and Src family kinases and induces immediate cell stiffening through the Rho-GEFs LARG and GEF-H1 (Guilluy et al., 2011). RhoA promotes stress fiber formation through the activation of formins; RhoA also promotes nonmuscle myosin II activity through the activation of Rho-associated coiled-coil containing protein kinase (ROCK). Both RhoA downstream pathways are required for optimal cellular tension and rigidity sensing (Schiller et al., 2013). Remarkably, cells adapt their RhoA signaling in response to ECM rigidity, cell geometry, and cell density to control cell proliferation and differentiation. For examples, high ECM stiffness enhances cell proliferation and induces an invasive phenotype even in nontransformed mammary epithelial cells (Paszek et al., 2005; Aragona et al., 2013). Furthermore, substrate elasticity corresponding to a specific in vivo tissue stiffness predisposes hMSCs to pursue specific cell fates (Engler et al., 2006). hMSCs can be differentiated into multiple mesenchymal lineages in vitro, including adipocytes and osteoblasts that reside in soft and rigid tissue environments, respectively. Inhibition of cellular tension with cytochalasin D or the ROCK inhibitor Y-27632 or by expression of a dominant-negative RhoA promotes adipogenesis on rigid, osteogenic conditions, whereas overexpression of constitutively active ROCK restores osteogenic fate on soft, adipogenic conditions (McBeath et al., 2004; Engler et al., 2006). Most of the effects of ECM stiffness on cell proliferation and differentiation can be attributed to YAP and TAZ (YAP/TAZ), two transcriptional activators that operate in the Hippo pathway to induce the expression of prosurvival genes (Dupont et al., 2011; Aragona et al., 2013; Zanconato et al., 2015). Although incompletely understood, integrin signaling and cellular tension activate YAP/TAZ through a signaling cascade involving FAK, Src, phosphoinositide 3-kinase, and JNK pathways (Codelia et al., 2014; Mohseni et al., 2014; Kim and Gumbiner, 2015; Elbediwy et al., 2016). Besides regulating YAP/TAZ, F-actin polymerization reduces G-actin concentration and releases myocardin-related transcription factors (MRTFs) into the nucleus to activate serum response factor (SRF)–mediated transcription (Olson and Nordheim, 2010). Transcriptional targets of MRTF–SRF significantly overlap with gene signatures related to cancer metastasis, mechanotransduction and YAP/TAZ activation (Esnault et al., 2014). Interestingly, under cyclic stretch, MRTF–SRF activation precedes and facilitates YAP/TAZ activation, suggesting that MRTF–SRF activation is responsible for immediate transcriptional response to mechanical stimulus (Cui et al., 2015).
In addition to the activation of transcription regulators such as YAP, TAZ, and MRTF–SRF, integrin-mediated mechanotransduction also modulates gene expression via the nucleoskeleton. The LINC (linker of the nucleoskeleton and cytoskeleton) complex connects cytoplasmic cytoskeleton with the nuclear lamina through the nuclear transmembrane protein emerin and the inner nuclear protein SUN. This connection enables direct force transmission from integrin-based adhesions to the nucleoskeleton (Isermann and Lammerding, 2013). The consequences of mechanical stress on the nucleus, similar to the mechanical stress in FAs, are changes in protein localization, protein conformation, and complex formation (Poh et al., 2012; Swift et al., 2013; Guilluy et al., 2014; Le et al., 2016). For example, mechanical forces acting on the nucleus during integrin-mediated cell spreading induce nuclear F-actin polymerization, leading to the formation, retention, and activation of the MRTF–SRF complex in the nucleus (Baarlink et al., 2013; Plessner et al., 2015). It has also been demonstrated that chronic cyclic stretches of epidermal stem cells induce remodeling of the nuclear lamina and global rearrangement of chromatin, resulting in epigenetic silencing by polycomb repressive complex 2–mediated trimethylation of Lys27 of histone 3 and global reduction of RNA polymerase II–dependent gene expression (Le et al., 2016). Interestingly, long-term exposure of hMSCs to rigid substrates irreversibly impairs the adaptation of gene expression and adipogenic differentiation on soft substrate, which probably also involves an epigenetic regulation (Yang et al., 2014). The “mechano-epigenetic” regulation may function as a signaling rheostat that memorizes mechanical experiences from the past and thereby predisposes cell fates (Yang et al., 2014). Such a signaling rheostat may play important roles in adult stem cells during tissue homeostasis and repair.
Dysregulated mechanotransduction in cancer
A growing body of knowledge supports the notion that mechanotransduction is also of significance in vivo. In some scenarios, integrin-mediated cell adhesion regulates tissue morphogenesis solely through mechanics. For example, integrin-dependent mesoderm tissue mechanics and intertissue adhesion between the paraxial mesoderm and notochord promote trunk elongation in zebrafish embryos (Dray et al., 2013). Integrin signaling also regulates cell position-dependent proliferation and differentiation through the Hippo pathway in concert with cell–cell adhesion and cell polarity (Hirate et al., 2013; Elbediwy et al., 2016). For a comprehensive review of developmental roles of mechanotransduction in vivo, we refer to a recently published review by Mammoto et al. (2013).
Integrin-mediated mechanotransduction is often dysregulated under pathological conditions such as cancer and tissue degeneration during aging. Tumor progression is typically associated with a pathological increase of tissue stiffness caused by extensive desmoplastic reactions characterized by excessive deposition, cross-linking, and aberrant organization of dense ECM fibers (Malik et al., 2015). In pancreatic ductal adenocarcinoma, which is known for its insensitivity to chemotherapy and immunotherapy, ∼90% of the tumor mass is composed of a thick desmoplastic stroma that maintains an immunosuppressive tumor microenvironment and impedes drug delivery (Heldin et al., 2004; Olive et al., 2009; Provenzano et al., 2012). Cancer-associated fibroblasts (CAFs) represent a key cell population in the fibrotic stroma responsible for ECM deposition. High ECM density and stiffness promote integrin-dependent FAK activation, cell contractility, and subsequent YAP/TAZ activation in cancer epithelial cells, which can be further potentiated by the loss of normal apical–basal polarity (Paszek et al., 2005). At the same time, hyper-activation of YAP/TAZ in CAFs enhances their contractile phenotype (Calvo et al., 2013). The mechanical interactions between the stroma and the cancer epithelial cells act in a self-reinforcing feedback loop to promote tumor progression. This vicious cycle is probably initiated by an oncogene-induced profibrotic JAK–STAT3 signaling pathway and further amplified by aberrant contractility in both cancer cells and CAFs (Sanz-Moreno et al., 2011; Laklai et al., 2016). Although the therapeutic potential of eliminating tumor stroma remains controversial (Olive et al., 2009; Özdemir et al., 2014; Rhim et al., 2014), terminating the vicious cycle of mechanotransduction by inhibiting lysyl oxidase–mediated collagen cross-linking or FAK activity effectively suppresses tumor progression (Levental et al., 2009; Jiang et al., 2016). Impressively, FAK inhibition in pancreatic ductal adenocarcinoma and squamous cell carcinoma was recently shown to decrease immunosuppressive cell populations in tumors and dramatically sensitize tumors to checkpoint immunotherapy and a CD8+ T cell–mediated antitumor response (Serrels et al., 2015; Jiang et al., 2016).
Outlook
Studies in the past 30 years have established the biophysical and biochemical principles of integrin-mediated mechanotransduction. However, there are still a lot of open questions that need to be answered to gain a comprehensive understanding of this process. (1) The recent characterization of the integrin adhesome will allow research efforts to define all mechanosensitive components and delineate their functionalities in response to mechanical stimulus using genetic tools and molecular MTSs. (2) Despite growing evidence pointing to the in vivo importance of an intact and functional mechanotransduction axis, the physiological responses to gradual mechanical changes that occur in tissues during aging remain largely unexplored. (3) It remains a mystery how the low affinity of the talin–integrin tail interaction is able to transmit the sum of all forces originating from all occupied talin–VBS and talin–ABSs. (4) It is also unclear which force regimes are (really) required to disrupt integrin bonds with FN and other integrin ligands. (5) It is unknown how fast ligand-bound integrin classes sense mechanical loads. (6) It will also be important to investigate how the control of force transmission is achieved spatially in cells (FBs and rear FAs). (7) It is still not known whether FA proteins can be damaged by cellular forces, and, if so, how force-damaged proteins are recognized and cleared. (8) More thorough investigations with appropriate animal models are required to elucidate the therapeutic potential of targeting signaling events downstream of mechanotransduction in various diseases and tissue regeneration. Clearly, there is still a lot to do!
Acknowledgments
The authors are grateful for the support of the European Research Council (grant agreement no. 322652), the Deutsche Forschungsgesellschaft (SFB863), and the Max-Planck-Gesellschaft.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ABS
- actin-binding site
- CAF
- cancer-associated fibroblast
- FA
- focal adhesion
- FB
- fibrillar adhesion
- FN
- fibronectin
- FRET
- Förster resonance energy transfer
- hMSC
- human mesenchymal stem cell
- MRTF
- myocardin-related transcription factor
- MTS
- molecular tension sensor
- NA
- nascent adhesion
- ROCK
- Rho-associated coiled-coil containing protein kinase
- SRF
- serum response factor
- THD
- talin head domain
- VBS
- vinculin-binding site
References
- Alexandrova A.Y., Arnold K., Schaub S., Vasiliev J.M., Meister J.J., Bershadsky A.D., and Verkhovsky A.B.. 2008. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS One. 3:e3234 10.1371/journal.pone.0003234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., and Piccolo S.. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 154:1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Atherton P., Stutchbury B., Wang D.Y., Jethwa D., Tsang R., Meiler-Rodriguez E., Wang P., Bate N., Zent R., Barsukov I.L., et al. 2015. Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 6:10038 10.1038/ncomms10038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen K., Ringer P., Mehlich A., Chrostek-Grashoff A., Kluger C., Klingner C., Sabass B., Zent R., Rief M., and Grashoff C.. 2015. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17:1597–1606. 10.1038/ncb3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarlink C., Wang H., and Grosse R.. 2013. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 340:864–867. 10.1126/science.1235038 [DOI] [PubMed] [Google Scholar]
- Bachir A.I., Zareno J., Moissoglu K., Plow E.F., Gratton E., and Horwitz A.R.. 2014. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr. Biol. 24:1845–1853. 10.1016/j.cub.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.M., Trappmann B., Wang W.Y., Sakar M.S., Kim I.L., Shenoy V.B., Burdick J.A., and Chen C.S.. 2015. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat. Mater. 14:1262–1268. 10.1038/nmat4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G.I., Dembo M., and Bongrand P.. 1984. Cell adhesion. Competition between nonspecific repulsion and specific bonding. Biophys. J. 45:1051–1064. 10.1016/S0006-3495(84)84252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo K.A., Dembo M., Kaverina I., Small J.V., and Wang Y.L.. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153:881–888. 10.1083/jcb.153.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely B.L., Dumelin C.E., Trappmann B., McGregor L.M., Choi C.K., Anthony P.C., Duesterberg V.K., Baker B.M., Block S.M., Liu D.R., and Chen C.S.. 2014. A DNA-based molecular probe for optically reporting cellular traction forces. Nat. Methods. 11:1229–1232. 10.1038/nmeth.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledzka K., Bialkowska K., Sossey-Alaoui K., Vaynberg J., Pluskota E., Qin J., and Plow E.F.. 2016. Kindlin-2 directly binds actin and regulates integrin outside-in signaling. J. Cell Biol. 213:97–108. 10.1083/jcb.201501006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet B.P., Gough R.E., Ammon Y.C., van de Willige D., Post H., Jacquemet G., Altelaar A.M., Heck A.J., Goult B.T., and Akhmanova A.. 2016. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife. 5:e18124 10.7554/eLife.18124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi L., Ramonet D., Klingberg F., Formey A., Smith-Clerc J., Meister J.J., and Hinz B.. 2011. The single-molecule mechanics of the latent TGF-β1 complex. Curr. Biol. 21:2046–2054. 10.1016/j.cub.2011.11.037 [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Campbell I.D., and Critchley D.R.. 2013. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14:503–517. 10.1038/nrm3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F., Ege N., Grande-Garcia A., Hooper S., Jenkins R.P., Chaudhry S.I., Harrington K., Williamson P., Moeendarbary E., Charras G., and Sahai E.. 2013. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15:637–646. 10.1038/ncb2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O., Mammoto T., Hasso S., Sperling R.A., O’Connell D., Bischof A.G., Maas R., Weitz D.A., Mahadevan L., and Ingber D.E.. 2014. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods. 11:183–189. 10.1038/nmeth.2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L.B., Baird M.A., Shtengel G., Campbell S.L., Hess H.F., Davidson M.W., and Waterman C.M.. 2015. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17:880–892. 10.1038/ncb3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti-Adam E.A., Volberg T., Micoulet A., Kessler H., Geiger B., and Spatz J.P.. 2007. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 92:2964–2974. 10.1529/biophysj.106.089730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.E., and Odde D.J.. 2008. Traction dynamics of filopodia on compliant substrates. Science. 322:1687–1691. 10.1126/science.1163595 [DOI] [PubMed] [Google Scholar]
- Chang Y.C., Zhang H., Franco-Barraza J., Brennan M.L., Patel T., Cukierman E., and Wu J.. 2014. Structural and mechanistic insights into the recruitment of talin by RIAM in integrin signaling. Structure. 22:1810–1820. 10.1016/j.str.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O., Gu L., Darnell M., Klumpers D., Bencherif S.A., Weaver J.C., Huebsch N., and Mooney D.J.. 2015. Substrate stress relaxation regulates cell spreading. Nat. Commun. 6:6364 10.1038/ncomms7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S.A., Weaver J.C., Huebsch N., Lee H.P., Lippens E., Duda G.N., and Mooney D.J.. 2016. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 15:326–334. 10.1038/nmat4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Ji B., and Gao H.. 2015. Modeling active mechanosensing in cell-matrix interactions. Annu. Rev. Biophys. 44:1–32. 10.1146/annurev-biophys-051013-023102 [DOI] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., and Horwitz A.R.. 2008. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050. 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codelia V.A., Sun G., and Irvine K.D.. 2014. Regulation of YAP by mechanical strain through Jnk and Hippo signaling. Curr. Biol. 24:2012–2017. 10.1016/j.cub.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie W.A., Babich A., and Burkhardt J.K.. 2015. F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. J. Cell Biol. 208:475–491. 10.1083/jcb.201406121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Hameed F.M., Yang B., Lee K., Pan C.Q., Park S., and Sheetz M.. 2015. Cyclic stretching of soft substrates induces spreading and growth. Nat. Commun. 6:6333 10.1038/ncomms7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J.M., and Sheetz M.P.. 2009. Stretching single talin rod molecules activates vinculin binding. Science. 323:638–641. 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pereda J.M., Ortega E., Alonso-García N., Gómez-Hernández M., and Sonnenberg A.. 2009. Advances and perspectives of the architecture of hemidesmosomes: lessons from structural biology. Cell Adhes. Migr. 3:361–364. 10.4161/cam.3.4.9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A.D., Carvajal N., Jin A., Matsumoto K., and Yamada K.M.. 2015. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 6:8720 10.1038/ncomms9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray N., Lawton A., Nandi A., Jülich D., Emonet T., and Holley S.A.. 2013. Cell-fibronectin interactions propel vertebrate trunk elongation via tissue mechanics. Curr. Biol. 23:1335–1341. 10.1016/j.cub.2013.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Ehrlicher A.J., Nakamura F., Hartwig J.H., Weitz D.A., and Stossel T.P.. 2011. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 478:260–263. 10.1038/nature10430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwy A., Vincent-Mistiaen Z.I., Spencer-Dene B., Stone R.K., Boeing S., Wculek S.K., Cordero J., Tan E.H., Ridgway R., Brunton V.G., et al. 2016. Integrin signalling regulates YAP and TAZ to control skin homeostasis. Development. 143:1674–1687. 10.1242/dev.133728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A., Oria R., Chen Y., Kosmalska A., Pérez-González C., Castro N., Zhu C., Trepat X., and Roca-Cusachs P.. 2016. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18:540–548. 10.1038/ncb3336 [DOI] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., and Discher D.E.. 2006. Matrix elasticity directs stem cell lineage specification. Cell. 126:677–689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Esnault C., Stewart A., Gualdrini F., East P., Horswell S., Matthews N., and Treisman R.. 2014. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 28:943–958. 10.1101/gad.239327.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty E.J., Bertaux C., Marcantonio E.E., and Gundersen G.G.. 2009. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J. Cell Biol. 187:733–747. 10.1083/jcb.200904054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freikamp A., Mehlich A., Klingner C., and Grashoff C.. 2016. Investigating piconewton forces in cells by FRET-based molecular force microscopy. J. Struct. Biol. 10.1016/j.jsb.2016.03.011 [DOI] [PubMed] [Google Scholar]
- Friedland J.C., Lee M.H., and Boettiger D.. 2009. Mechanically activated integrin switch controls alpha5beta1 function. Science. 323:642–644. 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- Gasiorowski J.Z., Murphy C.J., and Nealey P.F.. 2013. Biophysical cues and cell behavior: the big impact of little things. Annu. Rev. Biomed. Eng. 15:155–176. 10.1146/annurev-bioeng-071811-150021 [DOI] [PubMed] [Google Scholar]
- Gee H.Y., Zhang F., Ashraf S., Kohl S., Sadowski C.E., Vega-Warner V., Zhou W., Lovric S., Fang H., Nettleton M., et al. 2015. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J. Clin. Invest. 125:2375–2384. 10.1172/JCI79504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Mège R.M., and Thoumine O.. 2009. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 19:475–486. 10.1016/j.tcb.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Goksoy E., Ma Y.Q., Wang X., Kong X., Perera D., Plow E.F., and Qin J.. 2008. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell. 31:124–133. 10.1016/j.molcel.2008.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., and Schwartz M.A.. 2010. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 466:263–266. 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C., Swaminathan V., Garcia-Mata R., O’Brien E.T., Superfine R., and Burridge K.. 2011. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat. Cell Biol. 13:722–727. 10.1038/ncb2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C., Osborne L.D., Van Landeghem L., Sharek L., Superfine R., Garcia-Mata R., and Burridge K.. 2014. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 16:376–381. 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., and Guilford W.H.. 2006. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc. Natl. Acad. Sci. USA. 103:9844–9849. 10.1073/pnas.0601255103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer D.A., and Tirrell M.. 1996. Biological adhesion at interfaces. Annu. Rev. Mater. Sci. 26:651–691. 10.1146/annurev.ms.26.080196.003251 [DOI] [Google Scholar]
- Hattrup C.L., and Gendler S.J.. 2008. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 70:431–457. 10.1146/annurev.physiol.70.113006.100659 [DOI] [PubMed] [Google Scholar]
- Heldin C.H., Rubin K., Pietras K., and Ostman A.. 2004. High interstitial fluid pressure—an obstacle in cancer therapy. Nat. Rev. Cancer. 4:806–813. 10.1038/nrc1456 [DOI] [PubMed] [Google Scholar]
- Hirate Y., Hirahara S., Inoue K., Suzuki A., Alarcon V.B., Akimoto K., Hirai T., Hara T., Adachi M., Chida K., et al. 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23:1181–1194. 10.1016/j.cub.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B.D., Grashoff C., and Schwartz M.A.. 2011. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 475:316–323. 10.1038/nature10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton E.R., Byron A., Askari J.A., Ng D.H., Millon-Frémillon A., Robertson J., Koper E.J., Paul N.R., Warwood S., Knight D., et al. 2015. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 17:1577–1587. 10.1038/ncb3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Ji L., Applegate K.T., Danuser G., and Waterman-Storer C.M.. 2007. Differential transmission of actin motion within focal adhesions. Science. 315:111–115. 10.1126/science.1135085 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Isermann P., and Lammerding J.. 2013. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 23:R1113–R1121. 10.1016/j.cub.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Yu C.H., Mathur A., Liu S., Stévenin V., Dwyer J., Hone J., Ehler E., and Sheetz M.. 2013. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell. 27:545–559. 10.1016/j.devcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Hegde S., Knolhoff B.L., Zhu Y., Herndon J.M., Meyer M.A., Nywening T.M., Hawkins W.G., Shapiro I.M., Weaver D.T., et al. 2016. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 22:851–860. 10.1038/nm.4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., and Wirtz D.. 2013. Focal adhesion size uniquely predicts cell migration. FASEB J. 27:1351–1361. 10.1096/fj.12-220160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.G., and Gumbiner B.M.. 2015. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210:503–515. 10.1083/jcb.201501025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapproth S., Sperandio M., Pinheiro E.M., Prünster M., Soehnlein O., Gertler F.B., Fässler R., and Moser M.. 2015. Loss of the Rap1 effector RIAM results in leukocyte adhesion deficiency due to impaired β2 integrin function in mice. Blood. 126:2704–2712. 10.1182/blood-2015-05-647453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F., García A.J., Mould A.P., Humphries M.J., and Zhu C.. 2009. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185:1275–1284. 10.1083/jcb.200810002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F., Li Z., Parks W.M., Dumbauld D.W., García A.J., Mould A.P., Humphries M.J., and Zhu C.. 2013. Cyclic mechanical reinforcement of integrin-ligand interactions. Mol. Cell. 49:1060–1068. 10.1016/j.molcel.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Ouyang M., Van den Dries K., McGhee E.J., Tanaka K., Anderson M.D., Groisman A., Goult B.T., Anderson K.I., and Schwartz M.A.. 2016. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J. Cell Biol. 213:371–383. (published erratum J. Cell Biol. 2016. 214:231). 10.1083/jcb.201510012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J.C., Han X., Hsiao C.T., Yates J.R. III, and Waterman C.M.. 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13:383–393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E.M., van Puijenbroek A.A., Krause M., Carman C.V., Freeman G.J., Berezovskaya A., Constantine E., Springer T.A., Gertler F.B., and Boussiotis V.A.. 2004. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 7:585–595. 10.1016/j.devcel.2004.07.021 [DOI] [PubMed] [Google Scholar]
- Laklai H., Miroshnikova Y.A., Pickup M.W., Collisson E.A., Kim G.E., Barrett A.S., Hill R.C., Lakins J.N., Schlaepfer D.D., Mouw J.K., et al. 2016. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 22:497–505. 10.1038/nm.4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A.B., Grosveld F., and Akhmanova A.. 2006. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell. 11:21–32. 10.1016/j.devcel.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Le H.Q., Ghatak S., Yeung C.Y., Tellkamp F., Günschmann C., Dieterich C., Yeroslaviz A., Habermann B., Pombo A., Niessen C.M., and Wickström S.A.. 2016. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18:864–875. 10.1038/ncb3387 [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Lou J., Wen K.K., McKane M., Eskin S.G., Ono S., Chien S., Rubenstein P.A., Zhu C., and McIntire L.V.. 2013. Actin depolymerization under force is governed by lysine 113:glutamic acid 195-mediated catch-slip bonds. Proc. Natl. Acad. Sci. USA. 110:5022–5027. 10.1073/pnas.1218407110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K.R., Takahashi S., Bonakdar N., Fabry B., Boettiger D., Zent R., and Fässler R.. 2011. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 30:4539–4553. 10.1038/emboj.2011.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiss M., Beckmann K., Girós A., Costell M., and Fässler R.. 2008. The role of integrin binding sites in fibronectin matrix assembly in vivo. Curr. Opin. Cell Biol. 20:502–507. 10.1016/j.ceb.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W., et al. 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139:891–906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.M., Wang H.B., Dembo M., and Wang Y.L.. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152. 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.H., Carman C.V., and Springer T.A.. 2007. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25:619–647. 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R., Lelkes P.I., and Cukierman E.. 2015. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33:230–236. 10.1016/j.tibtech.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T., Mammoto A., and Ingber D.E.. 2013. Mechanobiology and developmental control. Annu. Rev. Cell Dev. Biol. 29:27–61. 10.1146/annurev-cellbio-101512-122340 [DOI] [PubMed] [Google Scholar]
- McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., and Chen C.S.. 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 6:483–495. 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1988. Cytoskeletal dynamics and nerve growth. Neuron. 1:761–772. 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- Mohseni M., Sun J., Lau A., Curtis S., Goldsmith J., Fox V.L., Wei C., Frazier M., Samson O., Wong K.K., et al. 2014. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat. Cell Biol. 16:108–117. 10.1038/ncb2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M., Mekhdjian A.H., Adhikari A.S., and Dunn A.R.. 2013. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 13:3985–3989. 10.1021/nl4005145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M., Mekhdjian A.H., Chang A.C., Tan S.J., and Dunn A.R.. 2015. Visualizing the interior architecture of focal adhesions with high-resolution traction maps. Nano Lett. 15:2220–2228. 10.1021/nl5047335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw J.K., Ou G., and Weaver V.M.. 2014. Extracellular matrix assembly: a multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 15:771–785. 10.1038/nrm3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S., Collin O., Chowdhury F., Tay B., Ouyang M., Wang Y., and Wang N.. 2008. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc. Natl. Acad. Sci. USA. 105:6626–6631. 10.1073/pnas.0711704105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes P.W., Beckham Y., Stricker J., and Gardel M.L.. 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196:363–374. 10.1083/jcb.201107042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive K.P., Jacobetz M.A., Davidson C.J., Gopinathan A., McIntyre D., Honess D., Madhu B., Goldgraben M.A., Caldwell M.E., Allard D., et al. 2009. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 324:1457–1461. 10.1126/science.1171362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., and Nordheim A.. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353–365. 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdemir B.C., Pentcheva-Hoang T., Carstens J.L., Zheng X., Wu C.C., Simpson T.R., Laklai H., Sugimoto H., Kahlert C., Novitskiy S.V., et al. 2014. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 25:719–734. (published erratum appears in Cancer Cell 2015. 28:831-833) 10.1016/j.ccr.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M.J., Zahir N., Johnson K.R., Lakins J.N., Rozenberg G.I., Gefen A., Reinhart-King C.A., Margulies S.S., Dembo M., Boettiger D., et al. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8:241–254. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Paszek M.J., Boettiger D., Weaver V.M., and Hammer D.A.. 2009. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLOS Comput. Biol. 5:e1000604 10.1371/journal.pcbi.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M.J., DuFort C.C., Rossier O., Bainer R., Mouw J.K., Godula K., Hudak J.E., Lakins J.N., Wijekoon A.C., Cassereau L., et al. 2014. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 511:319–325. 10.1038/nature13535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessner M., Melak M., Chinchilla P., Baarlink C., and Grosse R.. 2015. Nuclear F-actin formation and reorganization upon cell spreading. J. Biol. Chem. 290:11209–11216. 10.1074/jbc.M114.627166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov S.V., Pasapera A.M., Sabass B., and Waterman C.M.. 2012. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 151:1513–1527. 10.1016/j.cell.2012.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh Y.C., Shevtsov S.P., Chowdhury F., Wu D.C., Na S., Dundr M., and Wang N.. 2012. Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nat. Commun. 3:866 10.1038/ncomms1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck W.J., and Chen C.S.. 2016. Measuring cell-generated forces: a guide to the available tools. Nat. Methods. 13:415–423. 10.1038/nmeth.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P.P., Cuevas C., Chang A.E., Goel V.K., Von Hoff D.D., and Hingorani S.R.. 2012. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 21:418–429. 10.1016/j.ccr.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., and Gao H.. 2010. Soft matrices suppress cooperative behaviors among receptor-ligand bonds in cell adhesion. PLoS One. 5:e12342 10.1371/journal.pone.0012342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim A.D., Oberstein P.E., Thomas D.H., Mirek E.T., Palermo C.F., Sastra S.A., Dekleva E.N., Saunders T., Becerra C.P., Tattersall I.W., et al. 2014. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 25:735–747. 10.1016/j.ccr.2014.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., del Rio A., Puklin-Faucher E., Gauthier N.C., Biais N., and Sheetz M.P.. 2013. Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc. Natl. Acad. Sci. USA. 110:E1361–E1370. 10.1073/pnas.1220723110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni L., Stigler J., Pelz B., Ylänne J., and Rief M.. 2012. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc. Natl. Acad. Sci. USA. 109:19679–19684. 10.1073/pnas.1211274109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. 2008. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu. Rev. Genet. 42:235–251. 10.1146/annurev.genet.42.110807.091513 [DOI] [PubMed] [Google Scholar]
- Rossier O., Octeau V., Sibarita J.B., Leduc C., Tessier B., Nair D., Gatterdam V., Destaing O., Albigès-Rizo C., Tampé R., et al. 2012. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 14:1057–1067. 10.1038/ncb2588 [DOI] [PubMed] [Google Scholar]
- Rys J.P., DuFort C.C., Monteiro D.A., Baird M.A., Oses-Prieto J.A., Chand S., Burlingame A.L., Davidson M.W., and Alliston T.N.. 2015. Discrete spatial organization of TGFβ receptors couples receptor multimerization and signaling to cellular tension. eLife. 4:e09300 10.7554/eLife.09300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gaggioli C., Yeo M., Albrengues J., Wallberg F., Viros A., Hooper S., Mitter R., Féral C.C., Cook M., et al. 2011. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 20:229–245. 10.1016/j.ccr.2011.06.018 [DOI] [PubMed] [Google Scholar]
- Sawada Y., Tamada M., Dubin-Thaler B.J., Cherniavskaya O., Sakai R., Tanaka S., and Sheetz M.P.. 2006. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 127:1015–1026. 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., and Fässler R.. 2013. Mechanosensitivity and compositional dynamics of cell-matrix adhesions. EMBO Rep. 14:509–519. 10.1038/embor.2013.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Friedel C.C., Boulegue C., and Fässler R.. 2011. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12:259–266. 10.1038/embor.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Hermann M.R., Polleux J., Vignaud T., Zanivan S., Friedel C.C., Sun Z., Raducanu A., Gottschalk K.E., Théry M., et al. 2013. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15:625–636. 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Schoen I., Pruitt B.L., and Vogel V.. 2013. The yin-yang of rigidity sensing: how forces and mechanical properties regulate the cellular response to materials. Annu. Rev. Mater. Res. 43:589–618. 10.1146/annurev-matsci-062910-100407 [DOI] [Google Scholar]
- Serrels A., Lund T., Serrels B., Byron A., McPherson R.C., von Kriegsheim A., Gómez-Cuadrado L., Canel M., Muir M., Ring J.E., et al. 2015. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell. 163:160–173. 10.1016/j.cell.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrels B., Serrels A., Brunton V.G., Holt M., McLean G.W., Gray C.H., Jones G.E., and Frame M.C.. 2007. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat. Cell Biol. 9:1046–1056. 10.1038/ncb1626 [DOI] [PubMed] [Google Scholar]
- Stehbens S.J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T.. 2014. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16:561–573. 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwachs J., Metzner C., Skodzek K., Lang N., Thievessen I., Mark C., Münster S., Aifantis K.E., and Fabry B.. 2016. Three-dimensional force microscopy of cells in biopolymer networks. Nat. Methods. 13:171–176. 10.1038/nmeth.3685 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., and Mitchison T.J.. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747. 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Su W., Wynne J., Pinheiro E.M., Strazza M., Mor A., Montenont E., Berger J., Paul D.S., Bergmeier W., Gertler F.B., and Philips M.R.. 2015. Rap1 and its effector RIAM are required for lymphocyte trafficking. Blood. 126:2695–2703. 10.1182/blood-2015-05-644104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Xia W., Li J., Walz T., Humphries M.J., Vestweber D., Cabañas C., Lu C., and Springer T.A.. 2016. Relating conformation to function in integrin α5β1. Proc. Natl. Acad. Sci. USA. 113:E3872–E3881. 10.1073/pnas.1605074113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Tseng H.Y., Tan S., Senger F., Kurzawa L., Dedden D., Mizuno N., Wasik A.A., Thery M., Dunn A.R., and Fässler R.. 2016. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 18:941–953. 10.1038/ncb3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V., and Waterman C.M.. 2016. The molecular clutch model for mechanotransduction evolves. Nat. Cell Biol. 18:459–461. 10.1038/ncb3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V., Fischer R.S., and Waterman C.M.. 2016. The FAK-Arp2/3 interaction promotes leading edge advance and haptosensing by coupling nascent adhesions to lamellipodia actin. Mol. Biol. Cell. 27:1085–1100. 10.1091/mbc.E15-08-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M., et al. 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341:1240104 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J., Strokovich K., Springer T.A., and Walz T.. 2003. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 22:4607–4615. 10.1093/emboj/cdg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M., Sheetz M.P., and Sawada Y.. 2004. Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell. 7:709–718. 10.1016/j.devcel.2004.08.021 [DOI] [PubMed] [Google Scholar]
- Theodosiou M., Widmaier M., Böttcher R.T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Müller D.J., Zent R., and Fässler R.. 2016. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 5:e10130 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thievessen I., Thompson P.M., Berlemont S., Plevock K.M., Plotnikov S.V., Zemljic-Harpf A., Ross R.S., Davidson M.W., Danuser G., Campbell S.L., and Waterman C.M.. 2013. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202:163–177. 10.1083/jcb.201303129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. 2008. Catch bonds in adhesion. Annu. Rev. Biomed. Eng. 10:39–57. 10.1146/annurev.bioeng.10.061807.160427 [DOI] [PubMed] [Google Scholar]
- Trappmann B., Gautrot J.E., Connelly J.T., Strange D.G., Li Y., Oyen M.L., Cohen Stuart M.A., Boehm H., Li B., Vogel V., et al. 2012. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 11:642–649. 10.1038/nmat3339 [DOI] [PubMed] [Google Scholar]
- van der Vaart B., van Riel W.E., Doodhi H., Kevenaar J.T., Katrukha E.A., Gumy L., Bouchet B.P., Grigoriev I., Spangler S.A., Yu K.L., et al. 2013. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell. 27:145–160. 10.1016/j.devcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Wang X., and Ha T.. 2013. Defining single molecular forces required to activate integrin and notch signaling. Science. 340:991–994. 10.1126/science.1231041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Tytell J.D., and Ingber D.E.. 2009. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10:75–82. 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- Wells R.G., and Discher D.E.. 2008. Matrix elasticity, cytoskeletal tension, and TGF-beta: the insoluble and soluble meet. Sci. Signal. 1:pe13 10.1126/stke.110pe13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström S.A., Lange A., Montanez E., and Fässler R.. 2010. The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J. 29:281–291. 10.1038/emboj.2009.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd-Katz S.E., Fässler R., Geiger B., and Legate K.R.. 2014. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15:273–288. 10.1038/nrm3769 [DOI] [PubMed] [Google Scholar]
- Worth D.C., Hodivala-Dilke K., Robinson S.D., King S.J., Morton P.E., Gertler F.B., Humphries M.J., and Parsons M.. 2010. Alpha v beta3 integrin spatially regulates VASP and RIAM to control adhesion dynamics and migration. J. Cell Biol. 189:369–383. 10.1083/jcb.200912014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Tibbitt M.W., Basta L., and Anseth K.S.. 2014. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 13:645–652. 10.1038/nmat3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Goult B.T., Chen H., Cong P., Sheetz M.P., and Yan J.. 2014. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4:4610 10.1038/srep04610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Goult B.T., Klapholz B., Hu X., Toseland C.P., Guo Y., Cong P., Sheetz M.P., and Yan J.. 2016. The mechanical response of talin. Nat. Commun. 7:11966 10.1038/ncomms11966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A.A., McLean M.A., Sligar S.G., Taylor K.A., and Ginsberg M.H.. 2010. Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188:157–173. 10.1083/jcb.200908045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Rafiq N.B., Cao F., Zhou Y., Krishnasamy A., Biswas K.H., Ravasio A., Chen Z., Wang Y.H., Kawauchi K., et al. 2015. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat. Commun. 6:8672 10.1038/ncomms9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir E., Katz M., Posen Y., Erez N., Yamada K.M., Katz B.Z., Lin S., Lin D.C., Bershadsky A., Kam Z., and Geiger B.. 2000. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat. Cell Biol. 2:191–196. 10.1038/35008607 [DOI] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., and Piccolo S.. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17:1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ge C., Zhu C., and Salaita K.. 2014. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 5:5167 10.1038/ncomms6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Aponte-Santamaría C., Sturm S., Bullerjahn J.T., Bronowska A., and Gräter F.. 2015. Mechanism of focal adhesion kinase mechanosensing. PLOS Comput. Biol. 11:e1004593 10.1371/journal.pcbi.1004593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., and Springer T.A.. 2008a Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861. 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Kakinuma N., Wang Y., and Kiyama R.. 2008b Kank proteins: a new family of ankyrin-repeat domain-containing proteins. Biochim. Biophys. Acta. 1780:128–133. 10.1016/j.bbagen.2007.09.017 [DOI] [PubMed] [Google Scholar]