Abstract
Cellular heterogeneity can emerge from the expression of only one parental allele. However, it has remained controversial whether, or to what degree, random monoallelic expression of autosomal genes (aRME) is mitotically inherited (clonal) or stochastic (dynamic) in somatic cells, particularly in vivo. Here, we used allele-sensitive single-cell RNA-seq on clonal primary mouse fibroblasts and in vivo human CD8+ T-cells to dissect clonal and dynamic monoallelic expression patterns. Dynamic aRME affected a considerable portion of the cells’ transcriptomes, with levels dependent on the cells’ transcriptional activity. Importantly, clonal aRME was detected but was surprisingly scarce (<1% of genes) and affected mainly the most low-expressed genes. Consequently, the overwhelming portion of aRME occurs transiently within individual cells and patterns of aRME are thus primarily scattered throughout somatic cell populations rather than, as previously hypothesized, confined to patches of clonally related cells.
Stochastic cellular processes, such as gene expression1,2, can cause phenotypic variation even in the absence of genetic variation and within equivalent environments3–6. aRME represents an important facet of cellular stochasticity7–9, although its levels and nature have remained contentious. Early microarray studies reported clonally inherited aRME for 5–15% of genes in bulk-population analyses of long-term cultured human10 and mouse11 cells. These data were the basis for several subsequent investigations of histone modifications over promoters and gene bodies of reported clonal aRME genes12, computational inference of clonal aRME in other cell types12, and an exploration of the phenotypic consequences of clonal aRME8. Recently, a study analyzed evolutionary signatures in 4,227 inferred clonal aRME genes13, assuming clonal aRME for nearly 20% of autosomal genes. On the other hand, RNA-seq analyses of clonal somatic cell populations arrived at lower rates (2–3%) of clonal aRME14,15, and single-cell studies suggested that high levels of cellular aRME reflect burst-like transcription from each allele16–18. However, available single-cell data on allelic expression16–18 lacked information on clonality, precluding dissection of clonal and dynamic aRME. Finally, transcriptome-wide studies of clonal aRME in vivo are completely lacking. Therefore, we used single-cell RNA-seq on clonal primary cells to simultaneously investigate clonal and dynamic aRME. Moreover, by analyzing clonal T-cells isolated directly from human blood, we provide the first global analysis of aRME in vivo.
We started by sequencing the transcriptomes of individual mouse primary fibroblasts (n=285, passage 2–4, CAST/EiJxC57BL/6J reciprocal cross, Supplementary Table 1a), either picked randomly or after monoclonal expansions, using Smart-seq219 (Fig. 1a). Seven clones were harvested after 2–7 cellular divisions (4–115 cells). We detected (RPKM>1) on average 10,702 genes in fibroblasts (Supplementary Fig. 1), exhibiting highly correlating expression levels (average Spearman rho=0.85) (Supplementary Fig. 2) and strain-specific single nucleotide polymorphisms (SNPs) in 82% of genes. Using SNPs, we classified the expression of each gene as biallelic, maternal monoallelic, paternal monoallelic, or not detected, in each cell (Online Methods and Supplementary Figs. 3–7).
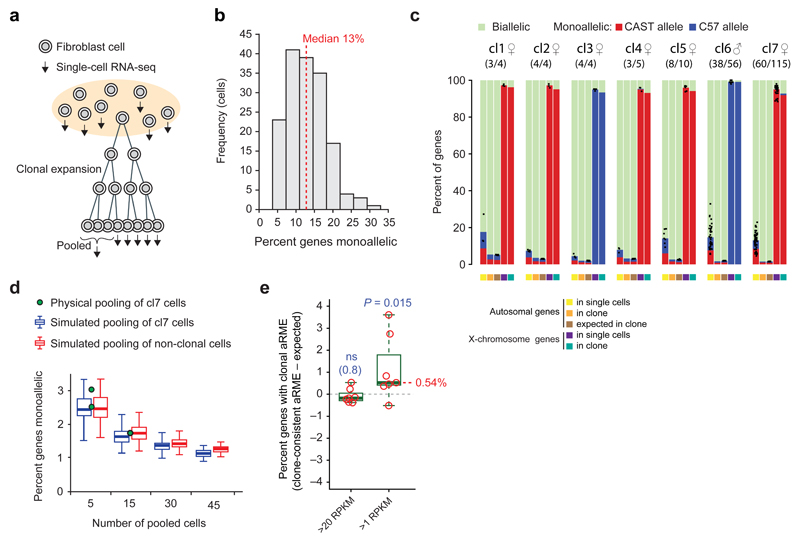
Figure 1. The vast majority of aRME in primary mouse fibroblasts is dynamic.
(a) Schematic overview of the experimental design. (b) Histogram showing percent aRME in randomly picked primary mouse fibroblasts (n=163). (c) Percent autosomal genes with bi- or monoallelic expression in single cells, after aggregating all cells of a clone, and expected levels from randomly pooling allelic calls of the same number of non-clonal cells. Dots represent percent monoallelic genes observed in single cells and bars depict the average. Observed allelic expression on the X-chromosome is shown to the right of each clonal set. Biallelic genes on female X-chromosomes are X-inactivation escapees (Supplementary Fig. 7–9). Sex and number of sequenced cells (out of all cells of the clone) are indicated above. (d) Percent of genes with aRME when sequencing transcriptomes of 5 or 15 pooled clonal cells (dots), and values obtained after in silico pooling of clonal or non-clonal cells shown as boxplots; indicating median (belt), interquartile range (box) and farthest points at maximum 1.5 times the interquartile rage (whiskers). Expression threshold in (b–d): RPKM>20. (e) Percent clonal aRME in the seven clones (circles) (observed minus expected), for genes detected either above 20 or 1 RPKM. The P-value denotes a median significantly greater than zero (one-sided Wilcoxon test). “ns”: not significant.
We first characterized aRME using an expression threshold suitable for determining rates of dynamic aRME (RPKM>20; see Online methods), and observed 13% (median) monoallelic expression of autosomal genes in fibroblasts (Fig. 1b). To determine the contribution of clonal and dynamic aRME, we investigated whether the monoallelic expression was the same across clonal cells by pooling cellular allelic calls in silico and determining the percent consistent monoallelic expression over clonal cells (Fig. 1c and Supplementary Fig. 6e). We excluded imprinted genes as well as regions with cell- or clone-specific chromosomal aberrations (Online Methods and Supplementary Fig. 7) – which often appear in cultured cells20. Since dynamic aRME can generate consistent allelic expression patterns in groups of cells by random chance (with probability inversely related to the number of cells), we contrasted the percent allele-consistent aRME in clones with the levels expected by dynamic aRME alone, by in silico pooling of the same number of non-clonal cells (Fig. 1c). This strategy was experimentally validated by physical pooling and joint sequencing of multiple cells from one clone (Fig. 1d). Our data showed that dynamic aRME accounted for nearly all aRME in fibroblasts. Indeed, above the expression-level threshold RPKM>20 we did not detect clonal aRME (P=0.8, one-sided Wilcoxon test), whereas analyses including all expressed genes (RPKM>1) revealed a low frequency of clonal aRME (Fig. 1e), affecting 0.5% of expressed genes (median, P=0.015, one-sided Wilcoxon test). As expected due to X-chromosome inactivation (XCI), X-linked genes had clone-consistent monoallelic expression of a single parental X-chromosome in female cells (Fig. 1c and Supplementary Fig. 7a–c) – thereby serving as an internal positive control for detectability of clonal monoallelic expression; confirming that the scarcity of clonal aRME was not due to insufficient power. Additionally, the X-chromosome data provided the opportunity to explore properties of genes that escape XCI from a single-cell perspective (Supplementary Note 1, Supplementary Figs. 8–9 and Supplementary Table 2).
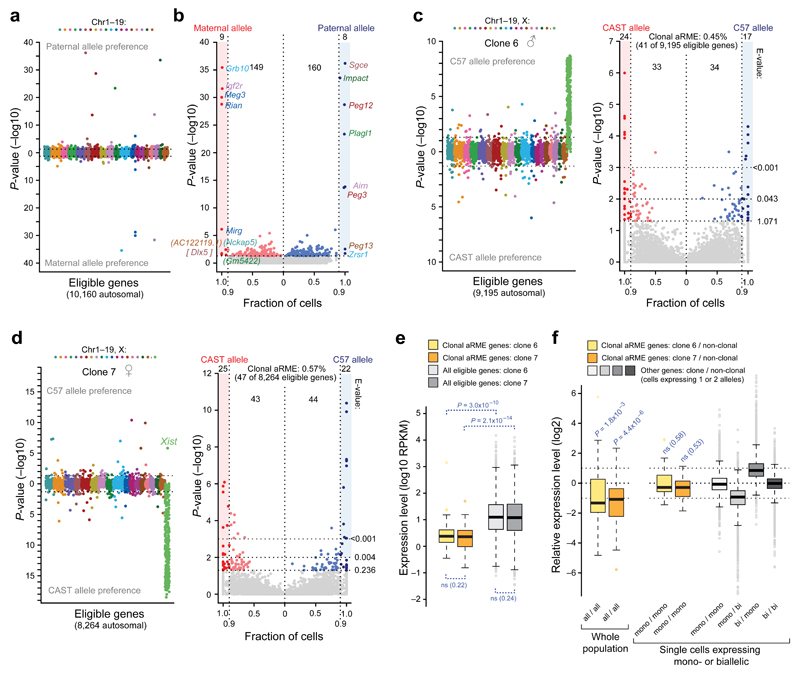
Thus, our data demonstrate that dynamic aRME constitutes the vast majority (>95%) of aRME occurring in primary fibroblasts. Next, to identify individual clonal aRME genes we devised a gene-level test detecting significantly skewed frequencies of monoallelic expression compared to the background expectation of random allelic expression fluctuation (Online Methods and Supplementary Fig. 6f). Genetic biases in allelic expression were observed for ~24% of genes (Supplementary Fig. 10), in line with a recent study21, and were accounted for in the background expectation. We validated our gene-level test on imprinted genes (i.e. parental-specific monoallelic expression), which were reliably identified (Fig. 2a–b and Supplementary Table 3a). As further validation of sensitivity in allele-specific detection using our single-cell assay, we performed deep sequencing on bulk RNA extracted from a fibroblasts culture using stranded mRNA TruSeq, and found that the single-cell data performed at least equally well in correctly identifying known imprinted genes (Supplementary Note 2, Supplementary Table 3b and Supplementary Fig. 9b; including information on allelic expression leakage of the imprinted gene Impact). Next, we applied the gene-level test on the two largest fibroblast clones (having adequate number of cells) and found significant (FDR<5%) clonal aRME for 41 and 47 autosomal genes (0.45 and 0.57% of eligible genes) in the two clones respectively (Fig. 2c–d, Supplementary Table 4 and Supplementary Fig. 11). Again, X-chromosome genes were highly significant for clonal monoallelic expression (Fig. 2d), verifying sensitivity.
Figure 2. Scarce clonal aRME in low-expressed genes.
(a) Genes with parental-specific (imprinted) monoallelic expression, ordered and colored according to chromosomal localization, with –log10 P-values (Fisher’s exact test) on y-axis. (b) Gene scatter with fraction cells having consistent maternal or paternal monoallelic expression (x-axis) plotted against the –log10 P-values (Fisher’s exact test). Genes with P<0.05 and consistent monoallelic in ≥90% of expressing cells were classified parent-of-origin-specific monoallelic, all being known imprinted genes except the novel candidates in brackets (DLX5 is known imprinted in human). (c) Test on clonal aRME (as in (a)) for male primary fibroblast clone 6 (n=38 cells), and scatterplot (as in (b)). E-values denote expected number of false positives above thresholds. (d) Test on clonal aRME for female primary fibroblast clone 7 (n=60 cells) and scatterplot (as in (c)). (e) Expression-level boxplots of clonal aRME (colored) and other genes (gray) in clones 6 and 7. P-values from two-sided Wilcoxon test. (f) Boxplots of relative expression of clonal aRME genes, over whole cell populations (left), or single cells expressing monoallelic (clonal or dynamic aRME) in both clone and non-clonal cells. P-values signify deviation from equal expression (two-sided Wilcoxon test). Additional boxplots (right) show relative expression levels in cells with dynamic aRME and biallelic expression, demonstrating the expected ~1:2 expression-level when transcribing 1 versus 2 alleles. (a–f) Threshold: RPKM>1.
Clonal aRME genes were expressed at significantly lower levels than other genes (P<10−9, Wilcoxon test, Fig. 2e), with a median of only ~2 RNA copies per cell (estimated using spike-in RNA). The clonal aRME expression was not dosage compensated, as clonal aRME tended to produce half the expression-level recorded in non-clonal populations (P-values 1.8x10−3 and 4.4x10−6, Wilcoxon test) (Fig. 2f). Furthermore, comparing clonal aRME to monoallelic observations in non-clonal cells (essentially only dynamic aRME) revealed equal transcriptional output (Fig. 2f). Thus, clonal aRME in primary fibroblasts mainly affects low-expressed genes without dosage compensation. Clonal aRME genes were enriched for membrane, extracellular and signaling functions (Supplementary Table 5). Five genes (Mr1, Chn1, Ceacam1, Entpd1 and 5830432E09Rik) had clonal aRME in both clones, either from the same or opposite alleles in the clones (Supplementary Table 4). Although this represents a statistically significant overlap (P<10−3, hypergeometric distribution) it also highlights the scarcity of clonal aRME and that most clonal aRME genes differ between clones.
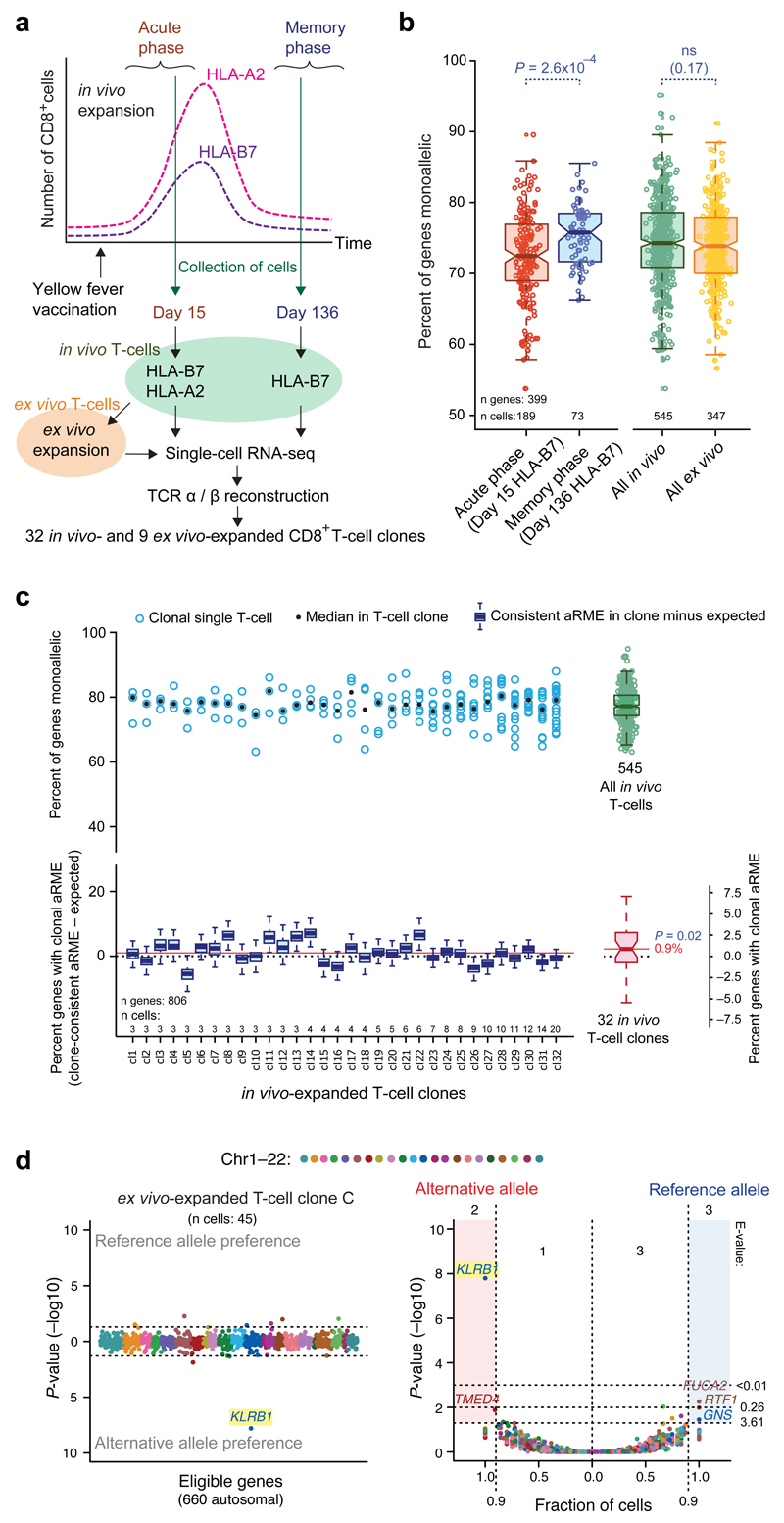
Next, we determined the prevalence of dynamic and clonal aRME in a somatic cell type in vivo for the first time. A male human donor was vaccinated with a yellow fever vaccine (YFV-17D), and blood samples were collected at the acute (day 15) and memory phase (day 136) of the vaccine response (Fig. 3a). We tracked the CD8+ T-cell responses using HLA class I dextramers that identified cells responding to an immunodominant (HLA-A02:01/LLWNGPMAV, “HLA-A2”) and a subdominant (HLA-B07:02:RPIDDRFGL, “HLA-B7”) T-cell epitope22 by fluorescence-activated cell sorting (FACS) (Supplementary Fig. 12). We sequenced the transcriptomes19 of individual in vivo T-cells (n=545 post quality filtering), and reconstructed their rearranged T-cell receptor sequences (TCR-α and TCR-β) (Online Methods). As rearrangements of the two TCR chains result in immense sequence variability23, cells with identically rearrangements were identified as clones (Supplementary Table 1b). We identified 32 in vivo T-cell clones with 3–20 sampled cells each. To identify SNPs, we performed exome sequencing of the donor, and used confirmed SNPs to determine allelic expression in the single T-cells (mean 1,846 genes expressed; 806 allele-informative genes passing SNP filtering). We observed aRME for ~60–85% of expressed genes (RPKM >20) across in vivo T-cells (Fig. 3b and Supplementary Fig. 13). Interestingly, aRME was more prevalent in T-cells collected during the memory phase (P=2.6x10−4, Wilcoxon test) (Fig. 3b), coinciding with decreased transcriptional activity in these cells (Supplementary Fig. 14). Next, we determined the prevalence of clonal aRME by comparing clonally consistent monoallelic expression to that expected from the background of fluctuating allelic expression using in silico pooling. Although the T-cells had high levels of dynamic aRME, clonal aRME was only observed for 0.9% (median) of genes (P=0.02, one-sided Wilcoxon test), demonstrating that clonal aRME is surprisingly scarce also in T-cells in vivo (Fig. 3c). To obtain sufficient number of T-cells per clone for gene-level identification of clonal aRME, we FACS-sorted single HLA-A2-specific T-cells from the same donor into separate culture wells, and clonally expanded cells ex vivo using autologous-antigen-presenting cells and LLWNGPMAV peptide in the presence of IL-2. We collected and sequenced cells from nine clonal expansions (in total 347 T-cells, 29–48 cells per clone). As expected from the in vivo results, the gene-level test identified a small number of clonal aRME genes, most notably a significant (P=1.6x10−8) clonal RME of KLRB1 (Killer Cell Lectin-Like Receptor) in one clone (Fig. 3d) and allelic imbalance in other clones (Supplementary Figs. 15–16 and Supplementary Table 6). Additionally, we noted clone-consistent aRME for CD7 (T-cell antigen) in two clones with opposite alleles expressed. Both these genes are immune related and membrane associated. We conclude that even in T-cells with high levels of monoallelic expression, the vast majority of aRME is dynamic with only sparse clonal aRME.
Figure 3. Dynamic and clonal aRME in human T-cells.
(a) Schematic representation of the experimental design. (b) Boxplots of percent aRME genes in HLA-B7 restricted T-cells, isolated at day 15 (red) or 136 (blue) after vaccination; and in all in vivo- (green) and ex vivo-expanded (orange) T-cells. Threshold: RPKM>20. (c) Upper panel: Percent aRME in 32 in vivo T-cell clones (circle: per cell, dot: median) and in all in vivo T-cells as a boxplot to the right. Lower panel: Percent clone-consistent aRME (observed minus expected, from sampled in silico pooling) as blue boxplots. The pink boxplot (lower right) and red solid line show median percent clonal aRME estimated over all clones. P-value for clonal aRME above zero according to a one-sided Wilcoxon test. Threshold: RPKM>1. (d) Example of gene-level identification on an ex vivo-expanded T-cell clone (clone C), showing highly significant clonal aRME of KLRB1 (Scatter plots as described in Fig. 2c–d).
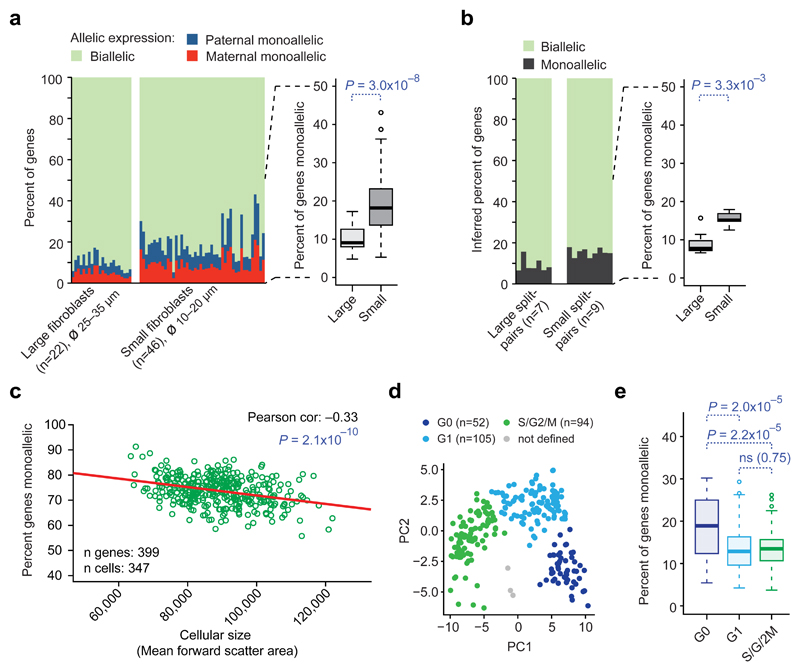
We detected remarkable variation in dynamic aRME across different cell types as well as among equivalently cultured fibroblasts (4–33%, Fig. 1b–c). We therefore pursued determinants of dynamic aRME levels, hypothesizing that increased transcription rates in larger cells24 could result in lowered levels of dynamic aRME. We therefore picked and sequenced large (ødissociated 25–35µm) and small (ødissociated 10–20µm) fibroblasts in culture, and expectedly, large cells contained more polyA+ RNA than small cells (P=5.3x10−9, Wilcoxon test) (Supplementary Figs. 17-18). Importantly, large cells had lower degree aRME than small cells (P=3.0x10−8, Wilcoxon test, median 9.1%, and 17% of genes respectively) (Fig. 4a). This was supported by split-cell control experiments (Online Methods and Supplementary Fig. 19), analytically inferring less monoallelic expression in large (median 7.8% of genes) than in small (median 15% of genes) fibroblasts from the paired allelic calls in split-cell lysates (P=3.3x10−3, Wilcoxon test) (Fig. 4b). Experiments considering intermediate cell sizes, further supported a dependence between size and dynamic aRME (Supplementary Fig. 20). Correlation between cellular size (determined by FACS) and degree dynamic aRME was furthermore observed in T-cells (Pearson correlation=–0.33, P=2.1x10−10, Fig 4c). Across additional mouse in vivo cell types we found that aRME levels varied with the total RNA content (Supplementary Fig. 21). In addition to maintaining overall RNA concentrations24, we found evidence that concentrations for cellular compartments are maintained (Supplementary Tables 7–11), e.g. membrane- and cytosol-related transcripts were elevated along with increased cell size, as recently hypothesized25. Using the fibroblast data, we further investigated how the degree dynamic aRME varies with cell-cycle phase. Using the most-variable cell-cycle genes, we classified the cells into G0, G1 and S/G2/M phases (Supplementary Fig. 22; S/G2/M could not be sub-stratified). Interestingly, levels of aRME were elevated in the “resting” G0 phase (P≤2x10−5) (Fig. 4d–e), in line with elevated aRME in human memory T-cells in vivo (Fig. 3b) that also reside primarily in G026.
Figure 4. Cellular size and cell-cycle phase affect the degree dynamic aRME.
(a) Observed allelic expression in primary fibroblasts of large and small cellular size. P-value: two-sided Wilcoxon test. (b) Percent monoallelic expression in large and small fibroblasts inferred by split-cell analysis. P-value: two-sided Wilcoxon test. (c) Scatter plot and Pearson correlation of percent aRME and cell size (estimated by FACS) in ex vivo-expanded T-cells. (d) Principal component analysis (scatterplot of component 1 and 2) of fibroblasts using the top 100 most-variable cell-cycle genes, with cells colored according to cell-cycle classification. (e) Boxplots of percent aRME in fibroblasts of different cell-cycle phases. P-values: two-sided Wilcoxon test. (a–d) Expression threshold: RPKM>20.
Here we used single-cell transcriptomics to dissect the nature of monoallelic expression in somatic cells. In contrast to previous studies that analyzed bulk-populations of longer-term expanded cell cultures8,10,11,14,15, we show that clonal aRME is very scarce (<1%) in primary and in vivo cells. Indeed >95% of observed cellular aRME was dynamic, with levels correlating with the transcriptional activity of the cells and further reflected from immune-activated responses and cell-cycle phase. These data alter the interpretation of several previous reports on monoallelic expression27–33, as cellular observations of monoallelic expression alone is not indicative of stable allele-level regulation. This also questions the notion of widespread clonal aRME affecting thousands of genes8,10,13,34. The revelation that clonal aRME appear only at miniscule levels in somatic primary and in vivo cells advances our knowledge of allele-specific gene expression and shed light on how variable expressivity and penetrance may emerge from aRME. Thus, in addition to heterogeneity emerging from scarce clonal aRME8, by far the majority of cellular variability in allelic expression occurs randomly throughout somatic cell populations and fluctuates over time.
Experimental procedures
Derivation of primary mouse fibroblasts
Primary fibroblasts were derived from adult CAST/EiJxC57BL/6J or C57BL/6JxCAST/EiJ mice (ethical permit: N343/12, Jordbruksverket) by skinning, mincing and culturing tail explants in fibroblast medium (Supplementary Note 2). For one clone (cl6) the fibroblasts were derived from minced E14.5 skin instead of tails, using the same procedure. After removal of explants, the culture was passaged twice to attain a pure fibroblast culture. Cells from passage 2–4 were dissociated (TrypLE Express, Gibco Ref. 12604-013), diluted to low cell density, and cells were picked by mouth pipetting using a thin glass capillary under 10x or 20x magnification. Cells for RNA-seq were transferred to RNAse-free PCR tubes containing 2µl Smart-seq2 lysis buffer and snap-frozen on dry ice.
Single-cell RNA-sequencing: Reverse transcription and cDNA amplification
Smart-seq2 cDNA libraries were prepared as described earlier19 and outlined in Supplementary Note 2. The cDNA was purified using AMPure XP beads (Beckman Coulter, Ref. A63882) and inspected on an Agilent 2100 Bioanalyzer to determine cDNA concentration and size distribution.
Single-cell RNA-sequencing: Tagmentation and sequencing
Successful cDNA libraries were tagmented, either using the Nextera XT DNA kit (Illumina Cat. FC-131-1024) or using our in-house-generated Tn5 (which produces sequencing libraries of indistinguishable characteristics to those generated using the Nextera XT kit)35. Details on tagmentation and library amplification are available in Supplementary Note 2. Libraries were purified using AMPure XP beads and inspected on an Agilent 2100 Bioanalyser. Sequencing was performed on an Illumina HiSeq2000. All mouse single-cell RNA-seq libraries used in the analyses of aRME are listed in Supplementary Table 1 and deposited to GEO (GSE75659).
Generation of libraries from split-cell lysates
Split-cell libraries were prepared as for single cells, with the following modifications: Single cells were lysed in 4µl lysis buffer instead of 2µl, and the lysate was thoroughly homogenized by pipetting up and down. 2µl of the homogenized lysate was then transferred to two separate tubes, each containing 0.5µl lysis buffer (to facilitate complete release of the lysate), and processed into sequencing libraries. Only split-pairs that generated similar Agilent Bioanalyzer profiles were selected for sequencing.
Exogenous spike-in RNA
Spike-in RNA (ERCC, Ambion, Ref. 4456740) was diluted in 10mM Tris-HCl pH7.5. 0.1µl of diluted (1:109,658, stock:final concentration) ERCC was added to each cell’s RT priming buffer (corresponding to 56,848 ERCC molecules). For T-cell libraries spike-in RNA was diluted 1:1,200,000 and 0.1µl (corresponding to 5,195 ERCC molecules) per reaction was added to the RT priming buffer.
Monoclonal fibroblasts
Single fibroblasts (from passage 2–4) were placed within marked areas in the bottom of gelatinized culture dishes containing fibroblast medium, observing the release of a single cell per dish under microscope. Monoclonally expanding cells were picked after 2–7 cellular divisions. For one clone (cl7), we also picked multi-cell samples of 5 or 15 cells that were jointly lysed and processed into sequencing libraries.
Human subjects for isolation of T-cells
Human volunteers were recruited to participate in an ongoing study examining the longitudinal immune response to the yellow fever vaccine YFV 17D (approved by the Regional Ethical Review Board in Stockholm, Sweden: 2008/1881-31/4, 2013/216-32 and 2014/1890-32). A single subject (ID: YFV2001(male)) was selected for in depth analysis based on being both HLA-A2 and HLA-B7 positive and having T-cell responses against YFV epitopes presented on both HLA types. Peripheral blood was collected at day 15 and day 136 after vaccination. Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation (Lymphoprep, Stem Cell Technologies, Ref. 07801) and stored in liquid nitrogen in 90% FCS and 10% dimethylsulfoxide (DMSO) until sorting.
Single-cell sorting of human CD8+ T-cells
Cryopreserved PBMCs were thawed and re-suspended in cold FACS buffer (PBS, 2% bovine serum, 2mM EDTA). CD8+ T-cells were enriched by negative selection (Miltenyi Human CD8 negative selection kit, Ref. 130-096-495), and were incubated for 15 min with either HLA-B07:02/RPIDDRFGL-dextramers (day 15 and day 136 cells) or HLA-A02:01/LLWNGPMAV-dextramers (day 15 cells) (Immudex, Denmark), followed by incubation with a panel of surface antibodies to detect live CD3+CD8+ T-cells (CD3-Alexa Flour 700 (UCHT1, BD Biosciences), CD8-APC Cy7 (SK1, BD Biosciences), CD4-PE-Cy5 (RPA-T4, Ebiosciences), CD14-Horizon V500 (MΦP9, BD Biosciences), CD19-Horizon V500 (HIB19, BD Biosciences), and Live/Dead Fixable Aqua dead cell stain kit (Invitrogen, Cat. L34957)). After 30 minutes of incubation at 4°C, the cells were washed and resuspended in cold FACS buffer for sorting. The cells were sorted using single-cell mode on a BecktonDickinson ARIA III cell sorter (equipped with a 405nm violet laser, 488nm blue laser, 561nm yellow/green laser, and 633nm red laser) into PCR plates containing 4ul Smart-seq2 lysis buffer.
Ex vivo expansion of human primary T-cells
CD8+ T-cells were isolated by negative selection from thawed cryopreserved PBMCs and were stained with the HLA-A2/LLWNGPMAV dextramer and antibodies as described above. Single live CD8+CD3+HLA-A2 dextramer+ cells were sorted directly into 96 well U-bottom plates containing 2μg/ml peptide (LLWNGPMAV), 20U/ml IL-2, and 50.000 irradiated (40Gy) CD3-depleted autologous PBMCs in complete T-cell media (Supplementary Note 2) and were cultured for 18 days. Every 4–5 days half of the media was replaced with fresh T-cell media containing 50U/ml IL-2 and 2μg/ml peptide, and the wells were visually inspected for proliferation. After 18 days, nine expanded clones were selected for single-cell sorting for RNA-seq based on the presence of sufficient cell numbers (>100) and no evidence of contaminating cells (all live cells were CD8+ and bound the HLA-A2/LLWNGPMAV dextramer). Single-cell RNA-seq libraries were generated as described below for the in vivo studies on YFV-specific CD8+ T-cells.
Single-cell RNA–seq on human T-cells
Smart-seq2 libraries from T-cells were prepared as described for mouse cells, with minor modifications (Supplementary Note 2). All T-cell single-cell RNA-seq libraries used in the analyses are listed in Supplementary Table 1 and deposited to GEO (GSE75659).
Exome sequencing
Exome capture was performed on unsorted PBMCs from donor YFV2001 using the Nextera Exome Sequencing kit (Nextera Rapid Capture Expanded Exome Kit, Illumina, Ref. FC-140-1000).
Computational procedures
Alignment of reads and allelic calling for mouse cells
Reads were aligned using RNA-STAR36 independently towards the mm9 genome assembly (C57 genotype) and an in-house generated CAST mm9 assembly (Supplementary Note 2). We counted C57- and CAST-informative bases for each gene, using the samtools mpileup command. To eliminate false SNP calls, occurring at low frequency, we only conceded the allelic calls for genes with ≥3 allele-informative reads. The allelic expression of each gene in each cell was called “biallelic”, “reference monoallelic”, “alternative monoallelic” or “not detected” (Supplementary Fig. 6a). A gene was called monoallelically expressed in a cell if one allele contributed ≥98% of the genotype-informative bases. Thus we allowed for up to 2% of reads to be aligning to the other allele without calling it “biallelic”, in order to tolerate a small degree of PCR and sequencing errors known to affect high-expressed genes in particular16; considerably more stringent than previous studies RNA-seq studies14,15,37,38. We demonstrated that our criteria (≥98% of the reads from one allele, and ≥3 allele-informative reads) were robust and resulted in accurate calls through analyses of genes on the X chromosome in male cells (i.e. cells with only one parental X chromosome) (Supplementary Fig. 3a–b). We verified that SNPs within the same genes and cells provided coherent allelic information (Supplementary Fig. 5). Fibroblasts expressed on average 10,702 genes. The number of allele-informative autosomal genes, eligible in the gene-level test for fibroblasts, was 8,264–9,195 genes, and the number of allele-informative genes with mean RPKM >20 in fibroblasts was 4,514.
Expression levels
We calculated RPKM values (reads per kilobase and million mapped reads) using rpkmforgenes39 version 7 Feb 2014 with uniquely mapped reads; Ensembl annotation (from UCSC genome browser, last modification date 11 April 2012) for mouse, and hg19 for human; length compensation for unmappable positions40 and the settings -fulltranscript -mRNAnorm -rmnameoverlap and -bothendsceil.
Percent monoallelically expressed genes
Percent monoallelically expressed genes for each cell was calculated as the number of genes with monoallelic calls divided by the total number of genes with informative (biallelic or monoallelic) calls in the cell multiplied by 100, including genes with mean expressions above 1 or 20 RPKM over the group of cells considered (thresholds stated in figure legends). The 20 RPKM threshold was applied when estimating total aRME in fibroblast cells, since at this threshold the false negatives and false positives monoallelic calls are in balance both in previous experiments16 and in the split-cell experiments on the mouse fibroblasts. The 1 RPKM threshold was applied when performing tests on clonal aRME, since clonal aRME genes were expressed at these low levels.
Analyses of allelic calls in split-cell experiments
We inferred monoallelic expression and allelic dropouts in a cell before its lysate were split into two fractions and tubes using the analytical model we recently developed16. In brief, we compared allelic calls for each gene in the paired libraries and determined the fraction of genes for which the splitted cells gave coherent calls, and the fraction of genes with different calls (Supplementary Note 2). We introduced equations that model the observed allelic calls and account for allelic losses (e.g. from biallelic to monoallelic or not detected; from monoallelic to not-detected) as detailed in fig. S26 in Deng et al.16. We solved the equations to find the fraction of monoallelic expression (x), the allelic losses (z) and the fraction biallelic expression (y). From these variables, we calculated the fraction of expressed genes that were monoallelic as x/(x+y) (Supplementary Fig. 19a–b). Since z would depend on the starting number of transcripts, we inferred x, y, z for different expression levels (i.e. RPKM bins) and we plotted the inferred variables as a function of expression level (Supplementary Fig. 19c–d). Importantly, the analytical model solves the allelic dropouts and monoallelic expression per expression bin, and it is a threshold-independent method that can account for varying dropout levels depending on RNA contents of cells prior to lysis and splits. Therefore the approach allows for varying allelic dropouts in the analyses of each split-cell pair, and we observed that smaller fibroblasts obtained slightly higher dropout levels than larger fibroblasts (Supplementary Fig. 19c–d).
Corrections for chromosomal aneuploidies
We surveyed the allelic calls along chromosomes to detect large-scale chromosomal aberrations, including aneuploidies. These were identified as chromosomes, or parts of chromosomes, where a large number of genes were deviating in allelic expression in favor of CAST or C57 genotypes. Chromosomes were flagged if abs(monoallelic calls CAST – monoallelic calls C57) > 50% and were then visually inspected. Supplementary Note 2 provides details on flagged cells and chromosomes.
Inferring number of RNA molecules per cell from spike-ins
To determine the total number of polyA+ molecules per cell (used for Supplementary Fig. 21), we divided the number of added ERCC spike-in molecules by the total RPKM for spike-ins in each a sample and multiplied by the total RPKM for genes (Supplementary Note 2).
Identification of genes with imprinted or strain-biased allelic expression
We translated the CAST and C57 calls to maternal and paternal calls, according to the parental genotypes. P-values for imprinted allelic expression were calculated using Fisher’s exact test on the frequency of monoallelic expression in a parental-specific manner compared to the overall call frequencies (Supplementary Fig. 6c). To consider a gene imprinted, and for inclusion in Supplementary Table 3a, the criteria were coherent monoallelic parental-specific calls in ≥90% of the cells and P<0.05 for the paternal bias in monoallelic expressions. We further performed imprinting analyses using bulk RNA sequencing on fibroblasts (described in Supplementary Note 2). We identified genes with strain-biased allelic expression using the observed frequencies C57 or CAST monoallelic in cells (Supplementary Fig. 6b), and we evaluated the frequency deviation from P=0.5 using a binominal test (Supplementary Fig. 6d). Previously known imprinted (genes in the GeneImprint repository) and novel imprinted genes were filtered out before strain analyses.
Analysis of clonal random monoallelic expression
Clone-consistent random monoallelic expression was investigated using two approaches. To estimate the percentage of genes with clonal aRME in mouse primary fibroblasts or human in vivo T-cells we compared the observed frequency of clone-consistent monoallelic calls over the cells of each clone separately to the expected clone-consistent aRME deriving from dynamic aRME through “in silico pooling” of cellular allelic calls (Supplementary Fig. 6e). Observed clone-consistent monoallelic expression was computed as the percentage of detected genes with allele-consistent monoallelic in the clonal cells. The expected background frequency of clone-consistent monoallelic expression (being consistent monoallelic over cells by random chance alone, due to transcriptional bursting and/or technical dropout of RNA species) was computed through in silico pooling of non-clonal cells and/or allelic calls. We evaluated the background frequencies derived from in silico pooling of either random cells or randomized allelic calls and both approaches yielded similar background estimates. For analyses presented in the study, we used randomized allelic calls rather than cells, since pooling of randomized calls conserve possible clone-specific patterns of expressed genes. In each round (500 random samplings), we replaced informative allelic calls per gene (i.e. calls assigned as biallelic, reference-monoallelic, or alternative-monoallelic) in the cells of each clone, with a random informative call with probability according to the frequency at which the different calls occurred in the non-clonal cell population (Supplementary Fig. 6b). The clonal aRME (reported in Supplementary Fig. 6e, Fig. 1e and Fig. 3c) was defined as the observed percent of genes with clone-consistent monoallelic expression minus the median percent genes with clone-consistent monoallelic expression in 500 random samplings.
Gene-level test to identify genes with clonal aRME
To identify individual genes with clonal aRME in fibroblasts and ex vivo T-cells we applied a gene-level test (Supplementary Fig. 6f), assessing the statistical significance of skew in allelic call frequencies in a clone compared to the overall call frequencies in non-clonal cells (i.e. taking genetic effects into account). We classified genes as subjected to clonal aRME based on: i) Fisher’s exact test P<0.05 (one-sided test for either over-representation of reference-monoallelic calls or over-representation of alternative-monoallelic calls) and ii) required consistent monoallelic calls in ≥90% of the cells with informative calls for the particular gene and clone. The rationale for requiring 90% of cells having clone-consistent calls was based on analyses of imprinted genes (see the shape of the distribution in Fig. 2b). Imprinted genes were filtered out before all clonal aRME analyses. To account for multiple testing, we computed the expected number of false positives (E-values) using randomly sampled non-clonal cells (1000 randomizations per clone).
Identification of T-cell in vivo clones
T-cell clones were identified based on cDNA sequences corresponding to the TCR-alpha (TCRα) and TCR-beta (TCRβ) chains using the MiTCR platform41, and a list of putative TCRα and TCRβ sequences was identified for each single-cell RNA-seq library. Reads were filtered to remove unproductive sequences corresponding to erroneous calls or PCR artifacts (as annotated in the MiTCR output). A secondary pipeline was developed in which publicly available sequences in the NCBI and Ensemble gene repositories of TCRα- and TCRβ-related gene components were used as reference to screen for reads aligning to these regions. Reads were aligned to this reference, assembled using Velvet42, and submitted to the International ImMunoGeneTics (IMGT) TCR sequence identifying platform43. The resulting hits were compared with the MiTCR screen to validate the individual sequences. In most cases either one or both of the TCR chains could be identified with both methods, and only cells with a high degree of certainty, presenting both the TCRα and TCRβ sequences, were considered for analysis of their clonal identity. Since both TCR chains are generated as separate recombination events it is extremely rare that two distinct T-cells will arise bearing identical both TCRα and TCRβ chains at the nucleotide level23. Cells with identical TCRα and TCRβ sequences were consequently classified as clones. Clones with at least three cells were considered for further analysis of clonal aRME, resulting in 32 individual in vivo T-cell clones.
Identification of heterozygous SNPs in human donor T-cells
To identify high-confidence SNPs in the male donor, we considered only heterozygous bases supported in the exome data and present in dbSNP (build 138) reference database. The criteria for inclusion were: ≥3 reads of each SNP variant in the exome data and ≤50-fold difference between the two, and that both SNP bases were validated by the single-cell RNA-seq data (informative call for the SNP in ≥1% of the cells). To filter out SNPs expressed with strong genetic bias, imprinted genes, and possible remaining false-positive SNPs, we discarded SNPs with a detected bias for one base (reference or alternative) >70% over all cells (Supplementary Fig. 3c), and all X-chromosome genes. This resulted in the inclusion of 1,010 confirmed autosomal SNPs expressed in the human donor’s T-cells, corresponding to 806 unique genes post filtering. For genes with multiple SNPs we used the most informative SNP (i.e. the SNP with highest number of reads). T-cells expressed (RPKM>1) on average 1,846 genes, and the number of expressed genes varied between the acute and memory phase (Supplementary Fig. 14a). The number of eligible allele-informative genes in ex vivo-expanded T-cell clones ranged between 370 and 660 (Supplementary Fig. 15), and the number of allele-informative genes with mean RPKM >20 was 399.
Allelic calling for human cells
For the analysis of allelic expression in T-cells, we used the same criteria and analyses as for fibroblasts (but using human reference/alternative SNPs rather than mouse CAST/C57 SNPs), considering genes with a mean expression of at least 1 or 20 RPKM over the group of T-cells considered, as stated in the figure legends.
GO analyses of genes correlating with cellular size and mRNA content
GO analyses (described in detail in Supplementary Note 2, results in Supplementary Table 7-11) were performed using DAVID44,45, and genes expressed in fibroblasts (genes with mean expression >1 RPKM) were used as background set.
Cell-cycle classification on fibroblast cells
The cell-cycle phase of individual fibroblasts cells was identified by regressing the cell-cycle genes identified by Whitfield46. A fit was performed using the R function glmgam.fit(), as previously described by Brennecke et al.47. The 100 cell-cycle genes presenting the greatest variability were used for principal component analysis whereby three phases of the cell cycle could be separated.
Supplementary Material
Acknowledgments
We thank Daniel Edsgärd for assistance in handling sequence data and members of the Sandberg lab for their input. J.M. was supported by a Human Frontiers Science Program Long-Term Fellowship (LT-000231/2011-L) and the work was supported by grants from the Swedish Research Council, the European Research Council (648842), the Swedish Foundation for Strategic Research, the Swedish Cancer Society, the Karolinska Institute, Tobias Stiftelsen, the Strategic Research Programme in Stem Cells and Regenerative Medicine at Karolinska Institutet (StratRegen), Knut och Alice Wallenbergs Stiftelse and Torsten Söderbergs Stiftelse.
Footnotes
URLs
GeneImprint (http://www.geneimprint.com/ )
Accession codes
Gene Expression Omnibus: GSE75659 and Sequence Read Archive: SRP066963.
Author contributions
B.R. designed mouse experiments, derived and sequenced mouse cells, performed computational experiments, prepared figures and tables and wrote the manuscript; J.E.M. designed human experiments, performed FACS, sequenced human T-cells and analyzed TCR sequences; D.R. performed computational experiments and prepared figures; Q.D. designed mouse experiments and derived mouse cells; P.J. performed cell-cycle classification; J.M. designed human experiments and performed FACS. J.F. designed human experiments; R.S. designed mouse experiments, supervised the work and wrote the manuscript.
Code Availability
Python and R code used in the analysis of allelic expression will be available on https://github.com/RickardSandberg/Reinius_et_al_Nature_Genetics_2016
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Suter DM, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 3.Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression: implications for haploinsufficiency. Proc Natl Acad Sci USA. 1998;95:15641–15646. doi: 10.1073/pnas.95.26.15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 7.Eckersley-Maslin MA, Spector DL. Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet. 2014;30:237–244. doi: 10.1016/j.tig.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chess A. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet. 2012;13:421–428. doi: 10.1038/nrg3239. [DOI] [PubMed] [Google Scholar]
- 9.Reinius B, Sandberg R. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat Rev Genet. 2015;16:653–664. doi: 10.1038/nrg3888. [DOI] [PubMed] [Google Scholar]
- 10.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 11.Zwemer LM, et al. Autosomal monoallelic expression in the mouse. Genome Biol. 2012;13:R10. doi: 10.1186/gb-2012-13-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nag A, et al. Chromatin signature of widespread monoallelic expression. Elife. 2013;2:e01256. doi: 10.7554/eLife.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savova V, et al. Genes with monoallelic expression contribute disproportionately to genetic diversity in humans. Nat Genet. 2016 doi: 10.1038/ng.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckersley-Maslin MA, et al. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Developmental Cell. 2014;28:351–365. doi: 10.1016/j.devcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gendrel A-V, et al. Developmental Dynamics and Disease Potential of Random Monoallelic Gene Expression. Developmental Cell. 2014;28:366–380. doi: 10.1016/j.devcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 17.Marinov GK, et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borel C, et al. Biased allelic expression in human primary fibroblast single cells. Am J Hum Genet. 2015;96:70–80. doi: 10.1016/j.ajhg.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 20.Baker DEC, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 21.Pinter SF, et al. Allelic Imbalance Is a Prevalent and Tissue-Specific Feature of the Mouse Transcriptome. Genetics. 2015;200:537–549. doi: 10.1534/genetics.115.176263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blom K, et al. Temporal dynamics of the primary human T cell response to yellow fever virus 17D as it matures from an effector- to a memory-type response. J Immunol. 2013;190:2150–2158. doi: 10.4049/jimmunol.1202234. [DOI] [PubMed] [Google Scholar]
- 23.Paul WE. Fundamental Immunology. Lippincott Williams & Wilkins; 2012. [Google Scholar]
- 24.Padovan-Merhar O, et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 26.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Ohlsson R, et al. Random monoallelic expression of the imprinted IGF2 and H19 genes in the absence of discriminative parental marks. Dev Genes Evol. 1999;209:113–119. doi: 10.1007/s004270050233. [DOI] [PubMed] [Google Scholar]
- 28.Miyanari Y, Torres-Padilla M-E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- 29.Faddah DA, et al. Single-cell analysis reveals that expression of nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell Stem Cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filipczyk A, et al. Biallelic expression of nanog protein in mouse embryonic stem cells. Cell Stem Cell. 2013;13:12–13. doi: 10.1016/j.stem.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Bix M. Independent and Epigenetic Regulation of the Interleukin-4 Alleles in CD4+ T Cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- 32.Nutt SL, et al. Independent regulation of the two Pax5 alleles during B-cell development. Nat Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- 33.Holländer GA, et al. Monoallelic expression of the interleukin-2 locus. Science. 1998;279:2118–2121. doi: 10.1126/science.279.5359.2118. [DOI] [PubMed] [Google Scholar]
- 34.Savova V, Patsenker J, Vigneau S, Gimelbrant AA. dbMAE: the database of autosomal monoallelic expression. Nucleic Acids Res. 2016;44:D753–6. doi: 10.1093/nar/gkv1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picelli S, et al. Tn5 transposase and tagmentation procedures for massively-scaled sequencing projects. Genome Res. 2014 doi: 10.1101/gr.177881.114. gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffries AR, et al. Stochastic Choice of Allelic Expression in Human Neural Stem Cells. STEM CELLS. 2012;30:1938–1947. doi: 10.1002/stem.1155. [DOI] [PubMed] [Google Scholar]
- 38.Li SM, et al. Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS ONE. 2012;7:e31751. doi: 10.1371/journal.pone.0031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsköld D, Wang ET, Burge CB, Sandberg R. An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storvall H, Ramsköld D, Sandberg R. Efficient and comprehensive representation of uniqueness for next-generation sequencing by minimum unique length analyses. PLoS ONE. 2013;8:e53822. doi: 10.1371/journal.pone.0053822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolotin DA, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10:813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 42.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefranc M-P, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–12. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.