Abstract
Molecular chaperones of the Hsp70 family have diverse functions in cells. They assist the folding of newly synthesized and stress-denatured proteins, as well as the import of proteins into organelles, and the dissociation of aggregated proteins. The well-conserved Hsp70 chaperones are ATP dependent: binding and hydrolysis of ATP regulates their interactions with unfolded polypeptide substrates, and ATPase cycling is necessary for their function. All cellular functions of Hsp70 chaperones use the same mechanism of ATP-driven polypeptide binding and release. The Hsp40 co-chaperones stimulate ATP hydrolysis by Hsp70 and the type 1 Hsp40 proteins are conserved from Escherichia coli to humans. Various nucleotide exchange factors also promote the Hsp70 ATPase cycle. Recent advances have added to our understanding of the Hsp70 mechanism at a molecular level.
Keywords: chaperone, Hsp70, Hsp40, J domain, Hsp110
Hsp70 and the ATPase cycle
Although much of our knowledge of Hsp70 biochemistry was first derived from studies of Escherichia coli DnaK, the outlines of the mechanism appear conserved in the orthologous cytosolic forms in eukaryotes, including human (or mammalian) Hsc70 (gene name HSPA8) and heat shock inducible Hsp70 (HSPA1), and Ssa1 from Saccharomyces cerevisiae (Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002; Daugaard et al. 2007). The Hsp40 family of proteins are also conserved and are essential activators of the Hsp70 chaperones (Mayer and Bukau 2005; Qiu et al. 2006). Particularly in eukaryotes, cytosolic Hsp70 proteins fulfill many tasks. They can interact with nascent polypeptides as they emerge from ribosomes, or bind to mature proteins denatured by stress conditions (Hartl and Hayer-Hartl 2002; Young et al. 2004; Mayer and Bukau 2005). In the cytosol of human and S. cerevisiae cells, a range of Hsp40 and other co-chaperone proteins can recruit Hsp70s for different purposes including degradation by proteasomes, import into mitochondria, conformational switching during signaling events, and disassembly of protein complexes (Young et al. 2003a). Hsp70s also cooperate with other ATP-dependent chaperones including Hsp90 and chaperonins to fold some substrates, and with certain AAA family proteins to dissociate aggregates of misfolded proteins (Hartl and Hayer-Hartl 2002; Young et al. 2004; Mayer and Bukau 2005).
The extensive experimental work on the Hsp70 ATPase cycle has been reviewed elsewhere and is summarized here. The Hsp70 proteins function primarily as monomers, although they can transiently contact regulatory co-chaperone proteins. The invariant domain structure of an Hsp70 consists of an N-terminal nucleotide-binding domain (NBD) having ATPase activity and a C-terminal substrate-binding domain (SBD) joined by a conserved linker. The fundamental ATPase cycle of Hsp70 proteins has been well established. In the ATP-bound state, an Hsp70 has low affinity for substrate; upon conversion to the ADP-bound state, Hsp70 binds substrate with high affinity (Fig. 1). This basic mechanism is used to accomplish the many cellular functions of Hsp70 proteins. Recent progress in understanding the Hsp70 chaperone machine at a mechanistic level will be discussed in this review.
Fig. 1.
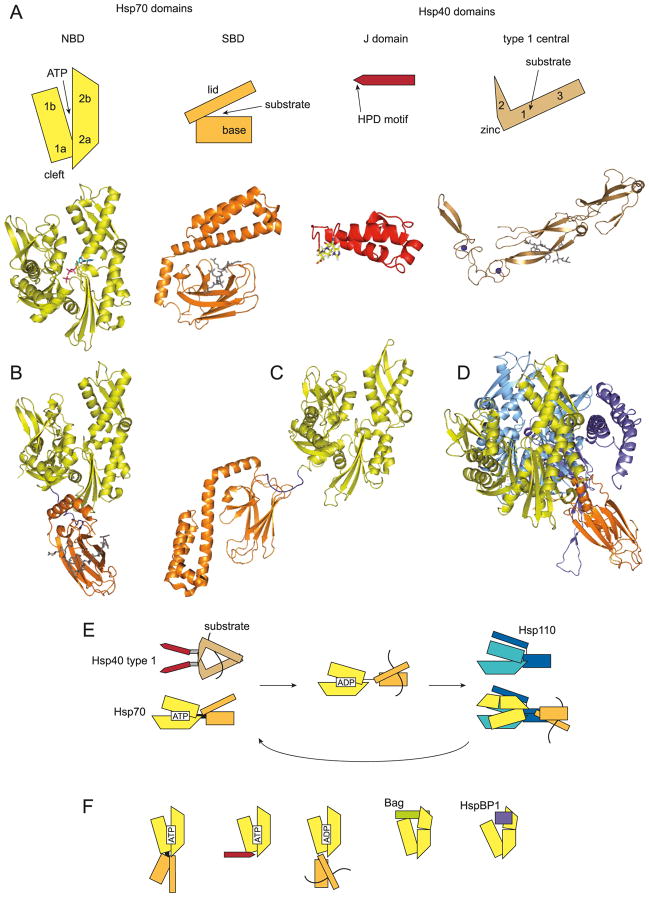
The Hsp70 and Hsp40 type 1 machinery. (A) Schematic of Hsp70 and Hsp40 domains with original structures shown below. The Hsp70 nucleotide-binding domain (NBD) is divided into 2 lobes made up of subdomains 1a, 1b, 2a, and 2b, with ATP (ADP-PO4 in the structure) bound in the opening (3HSC in PDB) (Flaherty et al. 1990). A cleft between 1a and 2a may be a regulatory interaction site. The Hsp70 substrate binding domain (SBD) has a base and a helical lid, which hold polypeptide substrate (dark gray) between them, in a groove in the base (1DKX) (Zhu et al. 1996). The Hsp40 J domain is elongated with the Hsp70-interacting HPD motif (coloured) at one end (1HDJ) (Qian et al. 1996). The central region of Hsp40 type 1 proteins has subdomains 1, 2, and 3 arranged in a hooked structure, with the zinc finger motifs at the angle and the main dimerization site at the end of 3. Substrate (dark gray) is bound by subdomain 2 (1NLT) (Li et al. 2003). (B– D) Structures of Hsp70 two-domain constructs: (B) with the NBD and SBD in contact (1YUW) (Jiang et al. 2005); (C) domains separated in the ADP-bound state (2KHO) (Bertelsen et al. 2009); (D) complexed with an Hsp110 co-chaperone (3C7N) (Schuermann et al. 2008). Colours of the domains are as in A., the linker in dark blue, and substrate in dark gray. The orientations of the NBD domains are the same as in A. The NBD of Hsp110 is light blue, the SBD of Hsp110 is indigo. (E) Outline of Hsp70 ATPase cycle. Substrate can be bound by Hsp40 type 1 proteins, which stimulates Hsp70 ATP hydrolysis through their J domains. The J domain of a subunit is connected to the central region by a G/F rich linker (gray bar). Hsp70 in the ATP state has the NBD, SBD and the interdomain linker (black bar) closely packed together, and binds substrate poorly. Hsp70 in the ADP state binds substrate tightly, and Hsp40 dissociates (Hartl and Hayer-Hartl, 2002; Young et al. 2004; Mayer and Bukau, 2005). The domains of Hsp70 in the ADP state may be separate and the linker flexible (black line). Hsp110 has an NBD and SBD structurally related to Hsp70, and binds the Hsp70 NBD to open it and release nucleotide. Hsp70 can then return to the ATP state (Polier et al. 2008; Schuermann et al. 2008). (F) Other modes of interaction of the Hsp70 NBD. The NBD may be in close contact with the interdomain linker and SBD in the ATP state (Liu and Hendrickson, 2007; Swain et al. 2007). The J domain of Hsp40 may interact near the linker binding cleft of the NBD (Jiang et al. 2007). In the ADP state, the SBD may also contact the NBD (Jiang et al. 2005), or rotate separately as shown in B (Swain et al. 2007; Bertelsen et al. 2009). The nucleotide exchange factors (NEF) of the Bag family and HspBP1, like Hsp110, shift NBD subdomain 2b to release nucleotide (Sondermann et al. 2001; Shomura et al. 2005; Andréasson et al. 2008a; Xu et al. 2008).
Crystal structures of Hsp70 domains have provided a first molecular interpretation of this cycle (Flaherty et al. 1990; Zhu et al. 1996; Hartl and Hayer-Hartl 2002; Mayer and Bukau 2005). The NBD has two lobes with a deep opening between them, and nucleotide is bound at the bottom of the opening. Each lobe can be divided into subdomains, 1a and 1b in one and 2a and 2b in the other, with 1a and 2a joined to form the bottom of the nucleotide pocket (Fig. 1A). A cleft surface between 1a and 2a on the other side from the nucleotide pocket may be a regulatory interaction site. Flexibility among the subdomains allows them to shift conformation from ATP to ADP or nucleotide-free states, with conserved amino acids coordinating the nucleotide and supporting catalysis (Flaherty et al. 1990). The SBD also contains two subdomains, a β-sheet base with a hydrophobic groove for polypeptide binding, and an α-helical structure forming a lid over the polypeptide binding site. A short hydrophobic section of a single polypeptide chain in an extended conformation can be bound by the SBD structure (Fig. 1A) (Zhu et al. 1996). To fulfill the ATPase driven cycle, conformational changes in the NBD must be transmitted to the SBD. ATP binding appears to promote flexibility between the base and lid of the SBD, effectively opening up the peptide binding site. Conversely, polypeptide binding in the SBD can also transmit changes to the NBD, increasing the ATP hydrolysis rate (Bukau and Horwich 1998).
Initial work with Escherichia coli DnaK identified two key regulatory co-chaperones. DnaJ increases the ATP hydrolysis rate of DnaK, inducing substrate binding by Hsp70. Another protein, GrpE, acts as a nucleotide exchange factor (NEF) for DnaK, promoting the release of ADP and re-binding of ATP, along with the release of bound substrate. Together, these co-chaperones raise the overall ATPase rate of DnaK (Liberek et al. 1991; Schröder et al. 1993). In addition, DnaJ binds to unfolded polypeptides itself, although it is not an ATPase and does not have an intrinsically regulated substrate binding cycle. It is thought that substrate is bound by DnaJ before and during activation of the DnaK ATP hydrolysis step, with DnaJ then dissociating from the tight DnaK–substrate complex; in effect, substrate is transferred from DnaJ to DnaK upon activation of the DnaK ATPase (Schröder et al. 1993; Szabo et al. 1994; Hartl and Hayer-Hartl 2002; Mayer and Bukau 2005).
The J domain at the N-terminus of E. coli DnaJ is conserved in a large family of proteins in humans and yeast, often referred to as the Hsp40 family, with DnaJ as the founding member. The J domain (Fig. 1A) forms the contact site with Hsp70 proteins, and provides the ATPase-stimulatory function. Following the J domain, DnaJ contains a glycine/phenylalanine-rich (G/F) linker, a central domain containing two zinc-finger motifs (Fig. 1A), and a C-terminal homodimerization domain. Hsp40 proteins preserving this domain architecture are classified as type 1 family members, and include the major cytosolic co-chaperones DJA1 (DnaJA1/Hdj2/dj2/HSDJ) and DJA2 (DnaJA2/dj3/HIRIP4) in humans, and Ydj1 in S. cerevisiae. These eukaryotic Hsp40s also have a conserved cysteine farnesylation site at their C-termini. Type 2 family members have the J domain and G/F linker, but diverge in the rest of the protein, lacking the zinc-finger motifs and having a distinct homodimerization site. The main human type 2 protein in the cytosol is DJB1 (DnaJB1/Hdj1/dj1/Hsp40), normally found at low levels but strongly expressed following heat shock. Sis1 is the equivalent protein in S. cerevisiae, but has functions under normal growth conditions (Cheetham and Caplan 1998; Qiu et al. 2006). The type 3 family members are homologous only in the J domains, and contain various other unrelated domains; they are thought to activate Hsc70 or Ssa1 for specialized functions directed by their additional domains (Young et al. 2003a; Walsh et al. 2004; Sahi and Craig 2007).
The NEF co-chaperones in the human and yeast cytosol are structurally unrelated to E. coli GrpE, perhaps surprisingly given the strong conservation of the Hsp70 and type 1 Hsp40 proteins. Bag domains, conserved within a family of human proteins, were the first of these NEFs identified. The human Bag proteins (numbered from 1 to 5) are divergent outside the Bag domains and seem to have specific roles in cells, particularly in regulation of apoptosis (Höhfeld and Jentsch 1997; Brehmer et al. 2001; Takayama and Reed 2001). Human HspBP1 was initially reported as an inhibitor of Hsc70, but it was next found to have NEF activity, despite having no similarity to Bag domains; its S. cerevisiae ortholog is Fes1 (Kabani et al. 2002). More recently, NEF function has been assigned to the Hsp110 proteins, which are different again from Bag proteins and HspBP1. Intriguingly, the Hsp110s have clear homology with Hsp70s and have domains corresponding to the NBD and SBD, although with additional insertions. The main family member in the cytosol of human cells is Hsp110 (Hsp105α, gene name HSPH1), and the counterpart Sse1 in S. cerevisiae (Dragovic et al. 2006; Raviol et al. 2006; Shaner et al. 2006).
Interdomain mechanism of Hsp70
The coordination between the NBD and SBD of Hsp70 chaperones has been the focus of recent work. A number of early mutagenesis experiments identified residues in both the NBD and SBD of DnaK that were important for interdomain communication, particularly around the nucleotide-binding cleft of the NBD, and both the base and lid subdomains of the SBD (Mayer and Bukau 2005). An allosteric pathway inside the NBD of E. coli DnaK has recently been defined. Upon the binding of ATP, a series of conformation shifts are transmitted through subdomain 1a, which result in specific changes in both the base and lid of the SBD (Mayer et al. 2000; Pellecchia et al. 2000; Fernández-Sáiz et al. 2006; Swain et al. 2006; Vogel et al. 2006a). Intriguingly, mutations in the interdomain linker also disrupted communication (Laufen et al. 1999; Montgomery et al. 1999; Han and Christen 2001; Vogel et al. 2006b), suggesting that the short but hydrophobic and highly conserved segment is a functional unit.
The first high-resolution data on the NBD interaction with SBD was derived from a crystallographic structure of bovine Hsc70 (Fig. 1B) (Jiang et al. 2005). Although the protein used had a truncated SBD lacking part of the lid it, was functionally active; similar constructs with a shortened SBD have been used by many other studies due to technical advantages. In the structure, the lobes of the NBD were open in the nucleotide-free state, and the C-terminus of the protein was packed into the polypeptide binding groove, mimicking a substrate. A conserved surface, near the cleft between the lobes of the NBD, contacted the base and lid fragment of the SBD, with the linker along one side (Fig. 1B and F). By comparison with earlier NBD structures, it was suggested that a shift in linker position in the ATP-bound state would disrupt the interdomain contact, changing the relative positions of the domains (Jiang et al. 2005).
Subsequent work on E. coli DnaK using hydrogen–deuterium (HD) exchange with electrospray ionization mass spectrometry (MS) addressed some of these ideas (Rist et al. 2006). Solvent accessibility suggested that in the ATP state the NBD and linker of the protein were more tightly packed and the SBD more open, compared with the nucleotide-free state. There was evidence of contact between the domains in the nucleotide-free state, but more protection of regions in both domains in the ATP state. Addition of peptide substrate caused shifts in both domains but increased the exposure of the linker (Rist et al. 2006). Similar proposals had been made based on previous spectroscopy data, but this study provided detail on changes at several specific residues.
A later study agreed only in part with the crystallographic structure (Swain et al. 2007). Nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopy of the E. coli DnaK NBD indicated the linker stabilized a closed, ATPase-activated form, by binding to the cleft between the lobes, near where the SBD was docked in the Hsc70 structure. NMR data of DnaK with shortened SBD in the ADP state suggested that both NBD and SBD appeared independent, with no evidence of contact between them. In the ATP state, the stabilization of the NBD and linker extended to parts of the SBD, and hydrogen–deuterium exchange NMR experiments outlined a putative contact site on the SBD base subdomain. Polypeptide binding in turn reduced the stabilization between domains. It was proposed that DnaK cycled between an ATP state in which the domains are tightly docked, and an ADP state where they are separate. The position of the linker in the ATP state could allow substrate binding to promote ATP hydrolysis by the NBD (Swain et al. 2007). However, the precise orientation of the domains could not be assigned. This model is less consistent with that suggested by the crystal structure, but supports previous biophysical work that indicated a compact ATP state for Hsp70.
Another possibility was raised by the structure of S. cerevisiae Sse1, the Hsp70-related NEF (Liu and Hendrickson 2007). Sse1 can itself bind ATP, and the ATP-bound form with intact NBD and SBD was solved by crystallography. In the structure, the NBD, linker, and SBD were packed against each other. A cleft in the NBD, close to the site suggested by the NMR study above, was opened to bind the linker. Several contacts were observed between the NBD around the cleft and the base subdomain of the SBD where it joins the lid. The orientation of these units was significantly different from that in the Hsc70 ADP state structure (Fig. 1F). Moreover, the lid subdomain was completely separate from the base unlike in previous structures, binding instead along one side of the NBD (Liu and Hendrickson 2007). The large shift in lid position may not be related to the substrate-binding mechanism of canonical Hsp70s. Other experiments, including NMR studies on substrate-bound and free DnaK SBD, suggested more subtle shifts in conformation (Pellecchia et al. 2000; Swain et al. 2006). Instead, as discussed below, the Sse1 lid position appears to act in nucleotide exchange (Polier et al. 2008). The close interaction between the domains of Sse1 in the ATP state was broadly similar to that proposed in the NMR study of DnaK (Fig. 1F), but its resemblance to the ATP state of other Hsp70 proteins remains to be determined. The question of interdomain contacts in the ADP or nucleotide-free states was left open.
Other data have supported a model in which the Hsp70 domains are separate in the ADP state. A crystallographic structure of shortened SBD DnaK from the bacterium Geobacillus kaustophilus in the ADP state was found to have the NBD and SBD completely apart, with the linker extended (Chang et al. 2008). Another group used time-resolved difference infrared (TD-DIR) spectroscopy with photoactivatable caged ATP to probe conformational changes in E. coli DnaK. The binding of ATP to nucleotide-free protein induced significant changes in both the NBD and SBD, but the changes caused by ADP binding were minimal and restricted to the NBD (Moro et al. 2006).
Most recently, NMR was also used to study DnaK from the bacterium Thermus thermophilus. The shortened SBD form had been analyzed in the ATP and ADP states by NMR, and interdomain interactions proposed for both states (Revington et al. 2005). A newer report on the protein in the ADP- and peptide-bound state, including spin-labeling NMR experiments and atomic assignments, found that the NBD and SBD were relatively independent (Figs. 1C and 1E) (Bertelsen et al. 2009). Movement of the domains appeared to be restricted within a range, and collisional interactions between the domains may define the limits of movement. The putative linker-binding cleft in NBD appeared partially closed in the ADP state. It was proposed that in the ATP state, the cleft would open to allow linker binding and docking of the NBD on the SBD (Bertelsen et al. 2009).
A combined model may be outlined in which the Hsp70 domains are alternately closely packed with the linker in the ATP state, then rotated and with the linker displaced in the ADP state. However, the orientation of the domains in the bovine Hsc70 crystallographic structure was outside the range of rotation proposed in the T. thermophilus DnaK study. Although conservation suggests that Hsp70 mechanisms should be universal, it is possible that eukaryotic Hsp70 proteins have different structural states than the bacterial forms. A study of Hsp40 interactions with bovine Hsc70 using NMR also reported significant interdomain contacts within Hsc70 in the ADP state (Jiang et al. 2007). Further study of human and eukaryotic Hsp70 chaperones will be needed to finally resolve this.
Hsp40 type 1 mechanisms
Mechanistic information on the Hsp40 proteins is more limited than for Hsp70s, and complicated by the divergence between types 1, 2, and 3. A number of structural studies of J domains have been reported, by X-ray crystallography and NMR, and supported by mutational analyses. J domains are helical bundles forming an elongated domain, with the absolutely conserved Hsp70 interacting motif His–Pro–Asp (HPD) exposed on a loop at one end (Fig. 1A) (Qian et al. 1996; Cheetham and Caplan 1998; Hennessy et al. 2005; Qiu et al. 2006). There has been recent progress in understanding the activation of Hsp70, and also in substrate binding by type 1 in particular.
The J domain interaction with Hsp70 NBD has been a challenge to study because of its transient nature, and crystallographic structures of the complex have only recently been determined. Auxilin is a type 3 Hsp40 protein with a J domain and separate clathrin binding site, which activates mammalian Hsc70 to dissociate clathrin baskets (Young et al. 2003a). The isolated J domain of bovine auxilin was complexed with the NBD-linker fragment of bovine Hsc70 by engineering a disulfide bond and solved at high resolution in the ADP state. The J domain was bound close to the the proposed linker-binding cleft of the NBD, with the linker alongside (Fig. 1F). This region of the NBD appears key to ATPase activation of Hsp70, either by J domains, or by SBD peptide binding transmitted through the linker. The NBD-linker and J domain complex in the ATP state showed only minor differences, so the actual mechanism of catalytic activation is still unknown, but may involve multiple small shifts at the active site (Jiang et al. 2007).
Substrate binding by the Hsp40s has been most well studied in the type 1 proteins, which can typically bind stably to unfolded polypeptides. Early experiments located the substrate-binding site within the central to C-terminal regions, and the zinc-finger like motifs were an initial focus of research. Experiments with E. coli DnaJ and S. cerevisiae Ydj1 suggested that the motifs were important for in vivo function, with different biochemical functions proposed (Szabo et al. 1994; Banecki et al. 1996; Lu and Cyr 1998; Fliss et al. 1999; Johnson and Craig 2000, 2001; Linke et al. 2003). Experiments on purified Ydj1 found that mutation of the zinc finger region did not interfere with substrate binding, assayed by prevention of denatured polypeptide aggregation, but was still involved in the functional interaction with Ssa1 in refolding of the model polypeptide firefly luciferase. The central to C-terminal region was identified as the substrate-binding site (Lu and Cyr 1998). A C-terminal truncation of Ydj1 lacking the dimerization site but having a complete central domain was sufficient to complement Ydj1 in S. cerevisiae cell growth. Furthermore, the Ydj1 N-terminus including the J domain, G/F linker and zinc-finger motifs fused to the divergent C-terminus of Sis1 also supported growth (Johnson and Craig 2001). A chimeric protein having the zinc finger motifs and central region of Ydj1 J domain fused between the J domain and C-terminus of Sis1 could function in activating Ssa1 to refold luciferase in vitro, and had similar substrate-binding properties as wild-type Ydj1 (Fan et al. 2004).
Detailed structural information of the substrate-binding site of Ydj1 was provided by a crystallographic study (Fig. 1A) (Li et al. 2003). Potential substrate peptides of Ydj1 were identified by phage display screening, and one sequence bound as a synthetic peptide with reasonable affinity to Ydj1. The peptide was then crystallized with a Ydj1 fragment containing the zinc finger motifs and central domain, up to the start of the dimerization site. The fragment formed an L-shaped structure with two β-barrel subdomains in the long arm (subdomains 1 and 3), the zinc finger motifs forming the angle at the end of subdomain 1, and the sequence between the motifs forming the short arm or hook (subdomain 2) (Fig. 1A). The substrate was bound in a hydrophobic groove in subdomain 1, on one face of the fragment (Li et al. 2003). Point mutations in the peptide binding site only mildly disrupted activity in vitro and in cells (Li and Sha 2005), and substrate binding may be more distributed than proposed. Recently, a modeling study proposed a consensus sequence of peptides bound by Ydj1, consistent with earlier peptide experiments on E. coli DnaJ (Kota et al. 2009). There may be additional interactions with substrate. Previous data suggested the Ydj1 farnesylation site at the C-terminus was not essential for its function (Fliss et al. 1999; Johnson and Craig 2000). However, live cell experiments suggested that Ydj1 could interact with some substrates (Ste11 kinase and androgen receptor, AR) through the farnesylation, but with other substrates (luciferase) at the peptide binding site in subdomain 1 (Flom et al. 2008; Summers et al. 2009).
A later crystallographic structure of the Ydj1 C-terminal dimerization site suggested the overall shape of the protein (Wu et al. 2005). The dimerization site was formed by extensions of subdomain 3 of the two subunits, joined at an angle. Combined with the structure of the central fragment by modeling, a triangular shape was suggested: two sides made up by subdomains 1 and 3 of each subunit, and the two hooks of subdomain 2 forming the third side (Fig. 1E). A remarkably large space would be framed by the triangle, with the potential to bind an unfolded polypeptide on two separate sites (Wu et al. 2005). Importantly, the model was consistent with a small-angle X-ray scattering (SAXS) analysis of human DJA1 (Borges et al. 2005), which provided the first low-resolution structure of a full-length type 1 Hsp40. A similar triangular form with a gap on the inside was modeled, but with the ends of subdomain 2 more apparently in contact. Next to each subdomain 2 outside the triangle was density consistent with the J domain, suggesting possible interactions between these units (Borges et al. 2005). A following SAXS study suggested a similar overall shape for Ydj1 at low resolution, although with the two J domains extended away from the triangle and possible contacting each other (Fig. 1E). The type 2 Hsp40, Sis1, had a different configuration with the J domains apart (Ramos et al. 2008).
The J domains and substrate-binding domains of type 1 Hsp40s appear to act in coordination, activating both ATP hydrolysis and substrate binding by Hsp70, and transfering substrate from the Hsp40 to Hsp70. This was supported by experiments with E. coli DnaJ and DnaK, in which DnaJ could not efficiently stimulate the ATPase of DnaK mutants defective in substrate binding (Laufen et al. 1999; Mayer et al. 2000). A later study compared DnaJ–DnaK ATPase activation by short and long polypeptides of denatured rhodanese; the longer substrates were most efficient, suggesting that DnaJ and DnaK bind substrate at different sites in a transient ternary complex to effect substrate transfer (Han and Christen 2003). Experiments with S. cerevisiae Ydj1 and Ssa1 showed that the structure of the Hsp40 substrate binding region was important for substrate transfer. Point mutants of the Ydj1 zinc finger motifs reduced binding of a model polypeptide to Ssa1 in live cells, and increased Ydj1 binding. The Ydj1 mutants also could not promote polypeptide refolding by Ssa1 in vitro, although J domain ATPase activation and substrate binding were both normal in the mutants (Fan et al. 2005). In light of the structural models, these mutations likely reduced stability of the angles formed by the zinc fingers, and increased flexibility of subdomain 2 hooks. It is possible that the hooks are important to position the J domains and therefore Hsp70 for efficient substrate transfer. Recent evidence of substrate transfer was reported for the murine endoplasmic reticulum proteins ERdj3, a type 2 Hsp40, and BiP, the lumenal Hsp70; and for murine cytosolic p58/IPK, a type 3 Hsp40, and Hsc70 (Jin et al. 2008; Petrova et al. 2008). Both of these Hsp40 proteins lack the zinc finger motifs, indicating that structural requirements for substrate transfer differ between Hsp40s.
The first human/mammalian type 1 Hsp40 characterized was DJA1. Immuno-depletion of DJA1 from rabbit reticulocyte lysate suggested it functioned in mitochondrial import of a precursor protein, pre-ornithine transcarbamylase, from the cytosol. Purified DJA1 was also able to activate Hsc70 (Terada et al. 1997). DJA1 also associated with Hsc70 on nascent polypeptides of the cystic fibrosis transmembrane regulator (CFTR) (Meacham et al. 1999). Later work found that DJA1 could function in vitro with Hsc70 and the chaperone Hsp90 in progesterone receptor (PR) folding (Cintron and Toft 2006). A second type 1 Hsp40, DJA2, was reported at levels similar to DJA1, and also functioned with purified Hsc70 (Terada and Mori 2000). A third type 1 protein, DJA4, was most highly expressed in heart and testes of mice and may have a tissue-specific function (Hafizur et al. 2004). Interestingly, knockout mice lacking DJA1 were viable; however, males had greatly reduced fertility — gross defects in spermatogenesis were traced to overactive signaling by AR, a known substrate of the cytosolic Hsp40–Hsp70 system. Thus, DJA2 and DJA4 were unable to substitute entirely for DJA1, indicating some distinction between their biological and biochemical properties (Terada et al. 2005).
More recently, the three mammalian type 1 proteins were identified in complexes with a mitochondrial precursor protein during Hsc70-dependent import involving the Tom70 receptor (Young et al. 2003b). Consistent with the Ydj1 data, substrate binding by the DJA proteins was localized to the central to C-terminal regions, in co-precipitation experiments. Truncation mutants of the DJAs lacking the J domains acted as inhibitors of import in vitro, and stimulation of ATP hydrolysis by purified Hsc70 was similar between the DJAs. However, while DJA2 could efficiently promote refolding of firefly luciferase by Hsc70, DJA1 was much less active; DJA4 was intermediate in this activity. Conversely, DJA1 appeared somewhat more active than DJA2 in promoting luciferase folding in cultured cells (Bhangoo et al. 2007). The substrate binding patterns and Hsc70 ATPase stimulation of DJA1 and DJA2 were then studied more closely. A chimera having the J domain of DJA1 fused to the central to C-terminal region of DJA2 was active in both substrate binding and Hsc70 ATPase stimulation, but like DJA1 was unable to support luciferase refolding (Tzankov et al. 2008). Thus, the J domain, G/F linker and subdomain 2 within DJA1 and DJA2 may have to function together, suggesting interactions between the structural units. There may be partial specialization of DJA1 and DJA2 for different ranges of polypeptide folding intermediates.
Nucleotide exchange factors
GrpE, the NEF of E. coli DnaK, is essential for the normal function of DnaK, together with DnaJ (Hartl and Hayer-Hartl 2002; Mayer and Bukau 2005). As observed in a crystallographic structure (Harrison et al. 1997), GrpE is a homodimer that binds to the NBD domain of DnaK at the tips of subdomains 1b and 2b. The result is an opening of the NBD lobes, particularly a rotation of subdomain 2b, to weaken the interactions with bound nucleotide. Dissociation of GrpE would allow binding of ATP, which would be at much higher concentrations in the cell than ADP, effectively causing nucleotide exchange (Harrison et al. 1997).
The C-terminal domain of human/mammalian Bag1 contains its NEF activity for Hsc70, and is conserved within the Bag family of proteins. A crystallographic structure of this domain complexed with the NBD of human Hsc70 was solved (Sondermann et al. 2001), and revealed a mechanism parallel to that of GrpE with DnaK. The monomeric α-helical Bag domain is very different from the α/β structure of GrpE, and binds Hsc70 at different sites on subdomains 1b and 2b. However, the result is an almost identical opening of the lobes (Fig. 1F), indicating a similar mechanism of nucleotide exchange (Sondermann et al. 2001). Recent work on human Bag2 interaction with Hsc70 suggested that it has a variation of this mechanism (Xu et al. 2008). The Bag2 NEF domain formed a symmetrical homodimer and bound the NBD in a different angle from Bag1, as determined by crystallography. Subdomain 2b of the NBD was shifted directly outwards by Bag2 binding, accomplishing the same result. The interaction was supported by NMR data, which further suggested that Bag2 can also bind polypeptide substrate at its Hsc70 binding site, a novel function for a Bag domain (Xu et al. 2008). How this acts in the chaperone cycle of Hsc70 remains to be established.
Another crystallographic study outlined an alternative mechanism for the human NEF HspBP1 and its S. cerevisiae homolog Fes1 (Shomura et al. 2005). The core NEF domain of HspBP1 was complexed with lobe 2 (subdomains 2a and 2b) of the Hsp70 NBD, and was found to bind to the inside surface of the nucleotide binding pocket. HspBP1 binding caused subdomain 2b to bend away from subdomain 2a, but also appeared to clash sterically with lobe 1 (Fig. 1F). In the complete NBD, HspBP1 increased sensitivity to proteolysis between the lobes, suggesting that the NBD was more dramatically twisted open to release nucleotide (Shomura et al. 2005).
The Hsp110 NEF mechanism has recently been analyzed by crystallography (Polier et al. 2008; Schuermann et al. 2008). In one structure (Polier et al. 2008), a fragment of S. cerevisiae Sse1 lacking a flexible internal loop was studied in complex with the NBD of human Hsp70; the interaction was confirmed as functional. The NBD domains from the respective proteins bound each other face to face, making multiple contacts on the surfaces of both lobes. The base and lid subdomains of the Sse1 SBD were also separated, with the base in contact with the Sse1 NBD but the lid binding across both NBDs. These interactions induced a rotation in subdomain 2b of the Hsp70 NBD, opening the nucleotide cleft somewhat wider than observed for Bag1 (Figs. 1D and 1F). Experiments in live yeast suggested that both the NBD and lid subdomain of Sse1 were important for NEF activity with its biological partner Ssa1 (Polier et al. 2008). Another structure was reported of Sse1 complexed with bovine Hsc70 having a complete NBD and shortened SBD (Schuermann et al. 2008). The interaction face between the NBDs, and the orientation of the Sse1 lid subdomain across the complex, was confirmed. In addition, the SBDs of both proteins could be compared (Fig. 1D). Unlike the split SBD of Sse1, the SBD of Hsc70 was separate as a unit from its NBD and drastically rotated from the position in the Hsc70 ADP-bound state. The NBD of Sse1 contacted both the NBD and SBD of Hsc70. The overall configuration of the complex was also supported by electron microscopy three-dimensional reconstruction, suggesting that the Hsc70 SBD had limited flexibility (Schuermann et al. 2008). Thus, Hsp110 may more directly regulate the Hsp70 chaperone cycle by affecting interdomain coordination.
The proposed Hsp110 mechanism has also been extended by HD MS experiments (Andréasson et al. 2008a, 2008b). ATP was found to stabilize the NBD of S. cerevisiae Sse1 and was necessary for its function. Interestingly, unlike canonical Hsp70s, nucleotide did not induce large conformational changes between NBD and SBD. Solvent accessibility of the Sse1 complex with Ssa1 NBD indicated the same contact sites as observed in the crystallographic structures, and provided independent evidence that the Sse1 SBD lid subdomain bound the Ssa1 NBD. Because there was no evidence of changes in the Sse1 lid in response to ATP or ADP, its main function is probably to contact Ssa1 rather than to close on a polypeptide substrate in the SBD (Andréasson et al. 2008a, 2008b).
Although the NEFs are not essential for the function of mammalian Hsc70 or S. cerevisiae Ssa1, they can boost chaperone activity. This has been most clearly shown for human Bag1, but also for human Hsp110 and S. cerevisiae Sse1 (Höhfeld and Jentsch 1997; Terada and Mori 2000; Brehmer et al. 2001; Gassler et al. 2001; Dragovic et al. 2006; Raviol et al. 2006; Shaner et al. 2006; Tzankov et al. 2008). In addition, there have been reports that Hsp110 and Sse1 can bind to polypeptides themselves to suppress aggregation, but not mediate refolding (Oh et al. 1997; Goeckeler et al. 2002). How this activity contributes to Hsp70-mediated refolding is not clear, as the Bag1 NEF domain is unable to bind substrates but was as efficient as Hsp110 in increasing luciferase refolding by Hsc70 and DJA2 (Tzankov et al. 2008). It is possible that Hsp110 substrate binding, like that of Bag2, assists with a subset of substrate polypeptides or folding intermediates.
Questions
A picture is emerging of the Hsp70 chaperone machine. A key mechanism appears to be the movement of the Hsp70 domains relative to one another during the ATPase cycle, aided by Hsp40 and NEF co-chaperones. The exact configuration of Hsp70 domains at each step of the cycle has to be definitively established. The next important question is how the various domains of Hsp40, Hsp70, and possibly Hsp110 move relative to one another during the Hsp70 cycle, particularly the different substrate binding sites. There is potential for such coordinated movements to cause significant conformational changes in a polypeptide substrate, although more passive mechanisms of binding and release are also possible. With molecular insights into such mechanisms, the larger question of how these interactions assist polypeptide folding, and the many other cellular functions of Hsp70s, may then be addressed.
Acknowledgments
J.C.Y. is supported by the Canada Research Chair in Molecular Chaperones, Tier II, and Canadian Institutes of Health Research operating grant MOP-68825.
Footnotes
This paper is one of a selection of papers published in this special issue entitled “Canadian Society of Biochemistry, Molecular & Cellular Biology 52nd Annual Meeting — Protein Folding: Principles and Diseases” and has undergone the Journal’s usual peer review process.
References
- Andréasson C, Fiaux J, Rampelt H, Druffel-Augustin S, Bukau B. Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc Natl Acad Sci USA. 2008a;105(43):16519–16524. doi: 10.1073/pnas.0804187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008b;283(14):8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- Banecki B, Liberek K, Wall D, Wawrzynów A, Georgopoulos C, Bertoli E, et al. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J Biol Chem. 1996;271(25):14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106(21):8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo MK, Tzankov S, Fan ACY, Dejgaard K, Thomas DY, Young JC. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol Biol Cell. 2007;18(9):3414–3428. doi: 10.1091/mbc.E07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges JC, Fischer H, Craievich AF, Ramos CH. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J Biol Chem. 2005;280(14):13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- Brehmer D, Rüdiger S, Gässler CS, Klostermeier D, Packschies L, Reinstein J, et al. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8(5):427–432. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92(3):351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chang YW, Sun YJ, Wang C, Hsiao CD. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J Biol Chem. 2008;283(22):15502–15511. doi: 10.1074/jbc.M708992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3(1):28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintron NS, Toft D. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J Biol Chem. 2006;281(36):26235–26244. doi: 10.1074/jbc.M605417200. [DOI] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581(19):3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell. 2004;15(2):761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem. 2005;280(1):695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- Fernández-Sáiz V, Moro F, Arizmendi JM, Acebrón SP, Muga A. Ionic contacts at DnaK substrate binding domain involved in the allosteric regulation of lid dynamics. J Biol Chem. 2006;281(11):7479–7488. doi: 10.1074/jbc.M512744200. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346(6285):623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Fliss AE, Rao J, Melville MW, Cheetham ME, Caplan AJ. Domain requirements of DnaJ-like (Hsp40) molecular chaperones in the activation of a steroid hormone receptor. J Biol Chem. 1999;274(48):34045–34052. doi: 10.1074/jbc.274.48.34045. [DOI] [PubMed] [Google Scholar]
- Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol Biol Cell. 2008;19(12):5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler CS, Wiederkehr T, Brehmer D, Bukau B, Mayer MP. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J Biol Chem. 2001;276(35):32538–32544. doi: 10.1074/jbc.M105328200. [DOI] [PubMed] [Google Scholar]
- Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol Biol Cell. 2002;13(8):2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafizur RM, Yano M, Gotoh T, Mori M, Terada K. Modulation of chaperone activities of Hsp70 and Hsp70-2 by a mammalian DnaJ/Hsp40 homolog, DjA4. J Biochem. 2004;135(2):193–200. doi: 10.1093/jb/mvh023. [DOI] [PubMed] [Google Scholar]
- Han W, Christen P. Mutations in the interdomain linker region of DnaK abolish the chaperone action of the DnaK/DnaJ/GrpE system. FEBS Lett. 2001;497(1):55–58. doi: 10.1016/S0014-5793(01)02435-8. [DOI] [PubMed] [Google Scholar]
- Han W, Christen P. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem. 2003;278(21):19038–19043. doi: 10.1074/jbc.M300756200. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276(5311):431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14(7):1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16(20):6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20(4):513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28(3):422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Awad W, Petrova K, Hendershot LM. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 2008;27(21):2873–2882. doi: 10.1038/emboj.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Craig EA. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol Cell Biol. 2000;20(9):3027–3036. doi: 10.1128/MCB.20.9.3027-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Craig EA. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J Cell Biol. 2001;152(4):851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531(2):339–342. doi: 10.1016/S0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- Kota P, Summers DW, Ren HY, Cyr DM, Dokholyan NV. Identification of a consensus motif in substrates bound by a Type I Hsp40. Proc Natl Acad Sci USA. 2009;106(27):11073–11078. doi: 10.1073/pnas.0900746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96(10):5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sha B. Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem J. 2005;386(3):453–460. doi: 10.1042/BJ20041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11(12):1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88(7):2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Wolfram T, Bussemer J, Jakob U. The roles of the two zinc binding sites in DnaJ. J Biol Chem. 2003;278(45):44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131(1):106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273(10):5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Schröder H, Rüdiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 2000;7(7):586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18(6):1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286(3):915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- Moro F, Fernández-Sáiz V, Muga A. The allosteric transition in DnaK probed by infrared difference spectroscopy. Concerted ATP-induced rearrangement of the substrate binding domain. Protein Sci. 2006;15(2):223–233. doi: 10.1110/ps.051732706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272(50):31636–31640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Montgomery DL, Stevens SY, Vander Kooi CW, Feng HP, Gierasch LM, Zuiderweg ERP. Structural insights into substrate binding by the molecular chaperone DnaK. Nat Struct Biol. 2000;7(4):298–303. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27(21):2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133(6):1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Qian YQ, Patel D, Hartl FU, McColl DJ. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol. 1996;260(2):224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos CH, Oliveira CL, Fan CY, Torriani IL, Cyr DM. Conserved central domains control the quaternary structure of type I and type II Hsp40 molecular chaperones. J Mol Biol. 2008;383(1):155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revington M, Zhang Y, Yip GN, Kurochkin AV, Zuiderweg ER. NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J Mol Biol. 2005;349(1):163–183. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J Biol Chem. 2006;281(24):16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA. 2007;104(17):7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12(11):4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31(2):232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45(50):15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, et al. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17(3):367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291(5508):1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Summers DW, Douglas PM, Ren HY, Cyr DM. The type I Hsp40 Ydj1 utilizes a farnesyl moiety and zinc finger-like region to suppress prion toxicity. J Biol Chem. 2009;284(6):3628–3639. doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JF, Schulz EG, Gierasch LM. Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J Biol Chem. 2006;281(3):1605–1611. doi: 10.1074/jbc.M509356200. [DOI] [PubMed] [Google Scholar]
- Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26(1):27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91(22):10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3(10):E237–E241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J Biol Chem. 2000;275(32):24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- Terada K, Kanazawa M, Bukau B, Mori M. The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J Cell Biol. 1997;139(5):1089–1095. doi: 10.1083/jcb.139.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Yomogida K, Imai T, Kiyonari H, Takeda N, Kadomatsu T, et al. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO J. 2005;24(3):611–622. doi: 10.1038/sj.emboj.7600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzankov S, Wong MJH, Shi K, Nassif C, Young JC. Functional divergence between co-chaperones of Hsc70. J Biol Chem. 2008;283(40):27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006a;21(3):359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006b;281(50):38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- Walsh P, Bursać D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5(6):567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li J, Jin Z, Fu Z, Sha B. The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40. J Mol Biol. 2005;346(4):1005–1011. doi: 10.1016/j.jmb.2004.12.040. [DOI] [PubMed] [Google Scholar]
- Xu Z, Page RC, Gomes MM, Kohli E, Nix JC, Herr AB, et al. Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat Struct Mol Biol. 2008;15(12):1309–1317. doi: 10.1038/nsmb.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003a;28(10):541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003b;112(1):41–50. doi: 10.1016/S0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5(10):781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]