Daley and collaborators show that endogenous Lin28b drives erythroid-dominant fetal hematopoiesis and that decreases in Lin28b activate adult granulocyte-predominant hematopoiesis.
Abstract
For appropriate development, tissue and organ system morphogenesis and maturation must occur in synchrony with the overall developmental requirements of the host. Mistiming of such developmental events often results in disease. The hematopoietic system matures from the fetal state, characterized by robust erythrocytic output that supports prenatal growth in the hypoxic intrauterine environment, to the postnatal state wherein granulocytes predominate to provide innate immunity. Regulation of the developmental timing of these myeloerythroid states is not well understood. In this study, we find that expression of the heterochronic factor Lin28b decreases in common myeloid progenitors during hematopoietic maturation to adulthood in mice. This decrease in Lin28b coincides with accumulation of mature let-7 microRNAs, whose biogenesis is regulated by Lin28 proteins. We find that inhibition of let-7 in the adult hematopoietic system recapitulates fetal erythroid-dominant hematopoiesis. Conversely, deletion of Lin28b or ectopic activation of let-7 microRNAs in the fetal state induces a shift toward adult-like myeloid-dominant output. Furthermore, we identify Hmga2 as an effector of this genetic switch. These studies provide the first detailed analysis of the roles of endogenous Lin28b and let-7 in the timing of hematopoietic states during development.
Hematopoiesis is a tightly controlled process in which stem and progenitor cells generate mature progeny to maintain a dynamic complement of blood cells that together support the physiological needs of oxygen transport, immune competence, and hemostasis. Hematopoietic stem cells (HSCs) produce common myeloid progenitors (CMPs) that commit to either granulocyte–monocyte progenitors (GMPs) or megakaryocyte–erythroid progenitors (MEPs), which further differentiate into mature cells, possibly by acquisition of transcriptional priming that confers ultimate lineage fate (Akashi et al., 2000; Traver et al., 2001; Paul et al., 2015). Situations of stress, such as hemorrhage or systemic inflammation, can enhance erythroid or myeloid output, respectively, while maintaining an adaptable pool of myeloerythroid progenitors (Doulatov et al., 2009; Xiang et al., 2015).
During development from the fetal to adult state, the hematopoietic system undergoes controlled maturation characterized by changes in transcriptional programs (McKinney-Freeman et al., 2012) that promote reduction in HSC self-renewal (Rebel et al., 1996; Bowie et al., 2007), postnatal hemoglobin recruitment (Sankaran et al., 2010), alterations in progenitor priming (Notta et al., 2016), and lymphoid respecification (Ikuta et al., 1990; Ikuta and Weissman, 1991). Efforts to date have identified Sox17 and Polycomb family members as key regulators of this process (He et al., 2011; Mochizuki-Kashio et al., 2011; Xie et al., 2014). These findings illustrate the broader concept of developmental timing of organogenesis, whereby tissues must undergo controlled maturation so that their specialized functions dovetail with organism morphogenesis. Recently, the Lin28b RNA-binding protein has been implicated in the developmental stage–specific regulation of hematopoiesis, with its expression confined to fetal hematopoietic tissues in vivo (Yuan et al., 2012; Copley et al., 2013). Ectopic expression of Lin28b in adult hematopoietic tissues confers fetal-like lymphoid differentiation (Yuan et al., 2012; Zhou et al., 2015), induces fetal globin expression (Lee et al., 2013), and promotes fetal-like self-renewal capabilities in HSCs (Copley et al., 2013). To date, experiments documenting the regulation of developmental hematopoiesis by Lin28b have relied on ectopic overexpression, whereas efforts that probe the effect of loss of endogenous Lin28b are lacking.
LIN28A and LIN28B are RNA-binding proteins that suppress the maturation of the let-7 family of microRNAs and modulate the translation of several mRNAs (Viswanathan et al., 2008; Shyh-Chang and Daley, 2013). Initially identified as regulators of developmental timing in Caenorhabditis elegans (Ambros and Horvitz, 1984; Moss et al., 1997), recent attention has focused on the capacity of LIN28 proteins to participate in the induction of pluripotency in somatic cells (Yu et al., 2007; Hanna et al., 2009) and to contribute to oncogenesis through regulation of let-7 levels (Viswanathan et al., 2009; West et al., 2009; Jiang et al., 2012; Rao et al., 2012; Urbach et al., 2014; Wang et al., 2015; Wu et al., 2015). The mechanisms by which the Lin28–let-7 axis projects its influence on such diverse pathophysiologic processes are beginning to come to light (Shyh-Chang and Daley, 2013). An emerging body of evidence posits a model wherein the presence of LIN28A/B is associated with a pluripotent, dedifferentiated phenotype, whereas accumulation of mature let-7 occurs secondary to LIN28A/B down-regulation to drive terminal differentiation (Viswanathan and Daley, 2010). Given their roles in developmental timing and stem cell function, we hypothesized that the Lin28–let-7 axis regulates myeloerythroid progenitor activity during development.
RESULTS
Maturation of the myeloerythroid compartment
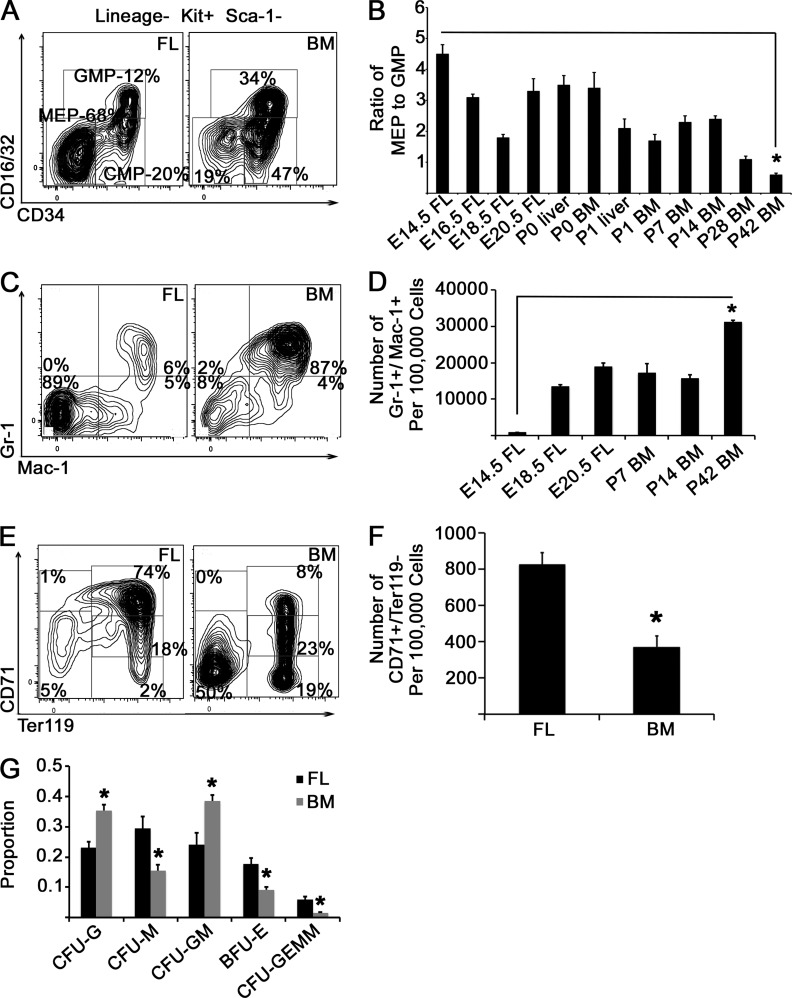
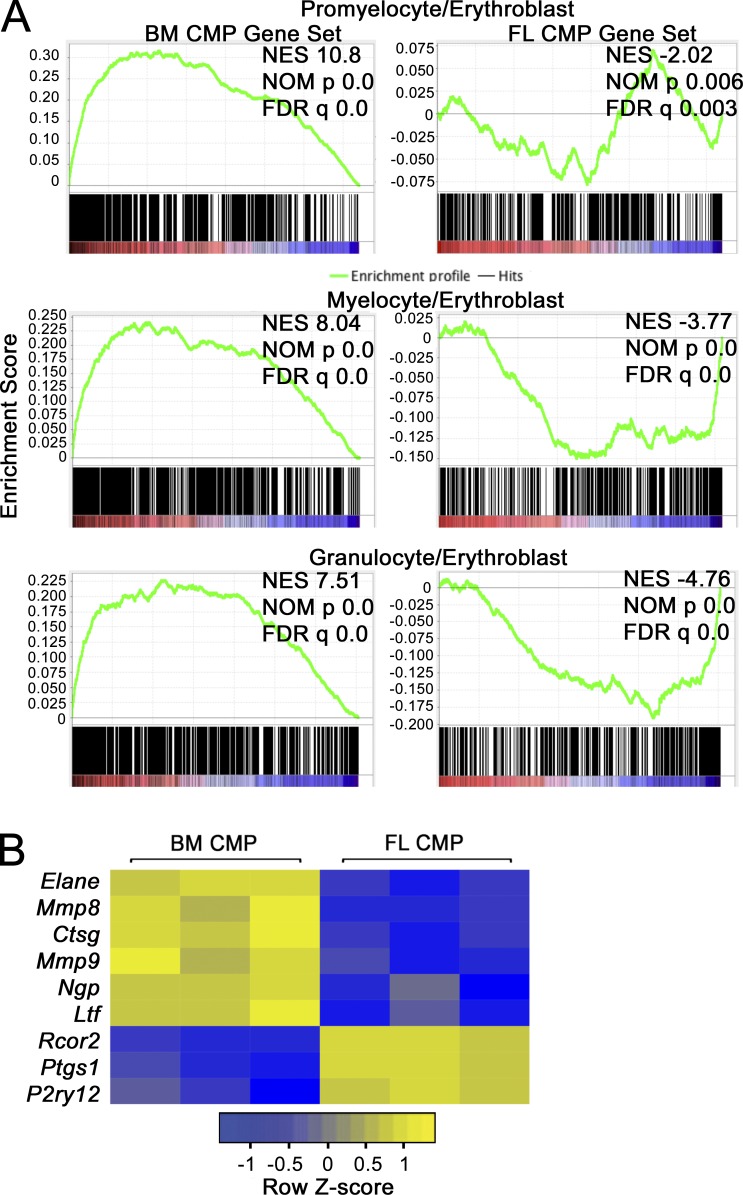
We examined the WT mouse myeloerythroid compartment at various stages of definitive hematopoiesis ranging from the E14.5 fetal liver (FL) to the mature BM, first assessing CMP, MEP, and GMP populations (Fig. 1 A). Compared with adult BM, numbers of MEPs in E14.5 FL were sixfold higher than GMPs, a ratio that switched with time to GMP predominance in adult mice (Fig. 1, A and B). By querying granulocyte populations at different stages (Sakamoto et al., 2000), we observed a progressive increase in mature granulocytes with time, paralleling the increase in GMPs (Fig. 1, C and D). We also delineated stages of erythroid development over time (Socolovsky et al., 2001). Compared with adult BM, we found more Ter119+-maturing erythroid cells in E14.5 FL, as well as a larger CD71+/Ter119− population containing early proerythroblasts (Fig. 1, E and F). Next, we cultured FL and BM mononuclear cells in methylcellulose with myeloerythropoietic growth factors and monitored hematopoietic colony formation. We observed a larger proportion of erythroid colonies in the FL compared with the BM, with more granulocytic colonies arising from BM cells (Fig. 1 G). Finally, as the CMP population is thought to differentiate to either MEPs or GMPs, we analyzed CMP transcriptome profiles by whole RNA sequencing (RNAseq; n = 3 biological replicates for each condition). A comparison with published datasets (Wong et al., 2013; Wu et al., 2014) revealed that transcripts specific to BM CMPs are enriched in myeloid lineage–specific transcripts (relative to erythroblasts), whereas transcripts specific to FL CMPs are enriched in erythroblast-specific transcripts (Fig. 2 A). Consistent with this finding, CMPs from BM contained a significantly higher expression of myeloid-specific genes, including Elane, Mmp8, Ctsg, Mmp9, Ngp, and Ltf, whereas FL CMPs expressed higher levels of the platelet/erythroid factors Rcor2, Ptsg1/Cox1, and P2ry12 (Fig. 2 B). Collectively, these data demonstrate that MEPs and their differentiated progeny predominate in the E14.5 FL compared with BM, in which GMPs and granulocytes predominate, possibly because of transcriptional priming of lineage commitment of CMPs, and describe normal developmental maturation within the myeloerythroid compartment.
Figure 1.
Myeloerythroid development. (A) Whole WT FL mononuclear cells or BM cells were gated on viable, lineage−, c-kit+, and Sca-1− cells, and representative staining patterns for CD16/32 and CD34 are presented. MEPs are defined as lineage−, c-kit+, Sca-1−, CD16/32−, and CD34−; GMPs are defined as lineage−, c-kit+, Sca-1−, CD16/32hi, and CD34+; and CMPs are defined as lineage−, c-kit+, Sca-1−, CD16/32lo, and CD34+. (B) FL or BM cells were isolated at the indicated time points, and the ratio of MEPs to GMPs is presented. *, P < 0.0001 by Student’s t test, comparing E14.5 FL and P42 BM time points. n = 6 animals for each time point over two experiments. (C) BM or FL cells were stained with the indicated antibodies (including erythroid and lymphoid lineage antibodies), and representative flow cytometry plots are presented. (D) FL or adult BM mononuclear cells were isolated at the indicated developmental time points, and the number of Gr-1+/Mac-1+ cells per 100,000 cells is presented. *, P < 0.001 by Student’s t test for E14.5 FL and P42 BM time points. n = 6 animals over two experiments. (E) FL or BM cells were stained with antibodies against Ter119 and CD71, and representative flow cytometry plots are presented. (F) The number of CD71+/Ter119− cells in FL versus adult BM is presented. *, P < 0.0001 by Student’s t test. n = 6 animals over two experiments. (G) Mononuclear cells were cultured in methylcellulose for 14 d in the presence of growth factors (stem cell factor, IL-3, IL-6, and erythropoietin). Colonies were scored by morphology, and the proportion of each colony type in the cultures is presented. *, P < 0.05 for each colony type by Student’s t test. n = 4 adult BM and 6 E14.5 FL over two experiments. Error bars represent SEM. CFU-M, CFU monocyte; CFU-G, CFU granulocyte; CFU-GM, CFU granulocyte–monocyte.
Figure 2.
RNAseq analysis of CMPs. (A) FL and adult BM CMPs (n = 3 biological replicates for each condition isolated over at least two separate sorting experiments) were compared by RNAseq to identify significantly different genes between the two populations at a false discovery rate (FDR) cutoff of <0.001. GSEA was performed to compare adult BM CMP- and FL CMP-specific transcripts to normal adult promyelocyte, myelocyte, granulocyte, and erythroblast transcriptomes. In the top row, a significant positive correlation was observed between adult BM CMP-specific transcripts and promyelocyte-enriched transcripts (left), indicating enrichment of adult BM CMP-specific transcripts among transcripts increased in promyelocytes relative to erythroblasts. A significant negative correlation was observed between FL CMP-specific transcripts (right) and transcripts increased in promyelocytes relative to erythroblasts, indicating correlation with erythroblast-specific transcripts. Normalized enrichment score (NES), nominal p-value (NOM p), and FDR are presented. The same analysis was performed in the middle with myelocyte transcripts and at the bottom with granulocyte transcripts. (B) Heat maps showing expression of erythroid/platelet-specific and granulocyte/monocyte-specific transcripts measured by RNAseq are presented.
Levels of Lin28b decrease, whereas let-7 species increase in myeloid progenitors during development
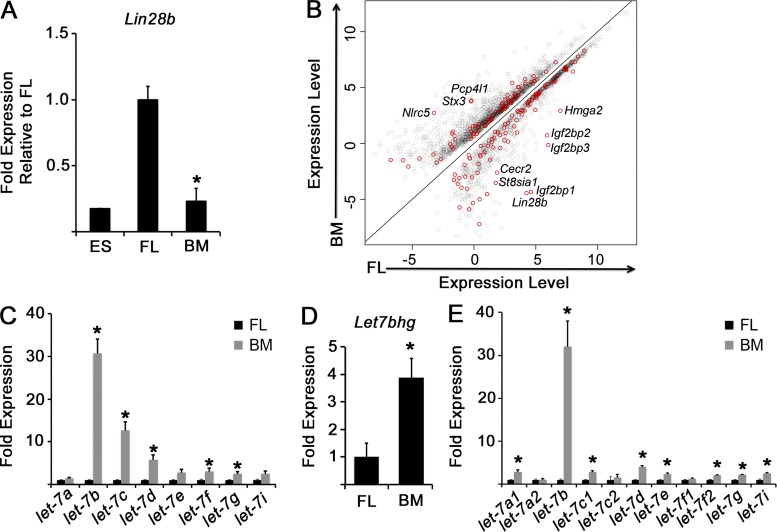
Using RNAseq, we found that a total of 1,081 genes were higher and 1,636 genes were lower in E14.5 FL CMPs compared with BM CMPs at a significance cutoff of 0.001 (Table S1). Lin28b was the second most significantly enriched transcript in the FL, which we confirmed by quantitative PCR (qPCR; Fig. 3 A). We noted enrichment of known let-7 target mRNAs in FL CMPs, suggesting that FL CMPs contain lower overall let-7 levels, possibly as a consequence of relatively higher levels of Lin28b compared with BM CMPs (Fig. 3 B). Therefore, we examined developmental distribution of mature let7 forms and observed significantly higher levels of several mature let-7 microRNAs in adult BM CMPs compared with FL CMPs (Fig. 3 C). Because both transcriptional and posttranscriptional mechanisms govern let-7 levels, we measured levels of pre– and pri–let-7 in FL and BM CMPs, as well as levels of a let-7 host gene. We observed significantly higher levels of several let-7 precursors as well as the host gene for pre–let-7b and pre–let-7c2 in adult BM compared with FL CMPs (Fig. 3, D and E). Together, these data support the notion that let-7 accumulates in CMPs with hematopoietic maturation.
Figure 3.
The Lin28–let-7 axis during myeloerythropoiesis. (A) FL or adult BM CMPs were isolated by FACS. The level of Lin28b mRNA was measured by qPCR in E14.5 FL or BM CMPs, and the levels were compared with mouse embryonic stem (ES) cells by qPCR. *, P < 0.0001 by Student’s t test. n = 6 replicates for FL and BM each over two experiments. An 8.08-fold expression increase in Lin28b was observed in FL CMPs compared with adult BM CMPs; P = 2.34 × 10−15 by RNAseq. (B) Scatter plot of differentially expressed genes between FL and adult BM CMPs as measured by RNAseq. Predicted let-7 targets with significantly different expression between FL and adult BM CMPs at an FDR of 0.001 are indicated in red. Pertinent transcripts are denoted. n = 3 biological replicates over at least two separate sorting experiments for BM and FL. (C–E) Levels of the indicated mature let-7 microRNA family members (C), Let7bhg (D), or let-7 precursors (E) were measured by qPCR in FACS-sorted FL or adult BM CMPs. The results are presented as fold expression compared with FL, normalized to an expression value of 1. *, P < 0.05. n = 6 embryos/mice sorted from three independent experiments. Error bars represent SEM.
Suppression of let-7 activity in the adult confers an FL-like myeloerythropoietic phenotype
Because several mature let-7 species are present at higher levels in adult CMPs relative to FL CMPs, we endeavored to inhibit let-7 activity in the adult and assess effects on myeloerythropoiesis. To this end, we used three complementary tools: (1) ectopic overexpression of LIN28B to impair maturation of let-7 precursors, (2) expression of a pan–let-7 sponge in BM CMPs to functionally sequester mature let-7 species from their endogenous mRNA targets, and (3) use of a mouse model deficient for the polycistronic transcript encoding let-7b (the let-7 form most enriched in BM CMPs compared with FL CMPs) and let-7c2.
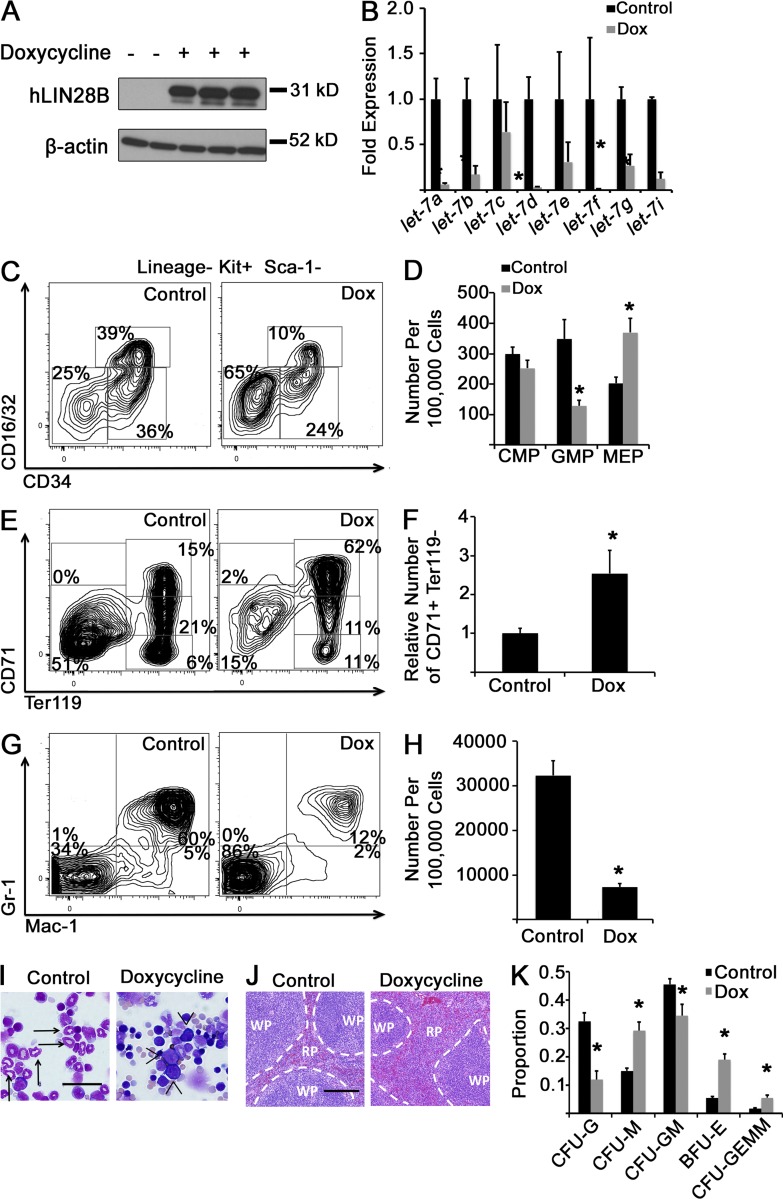
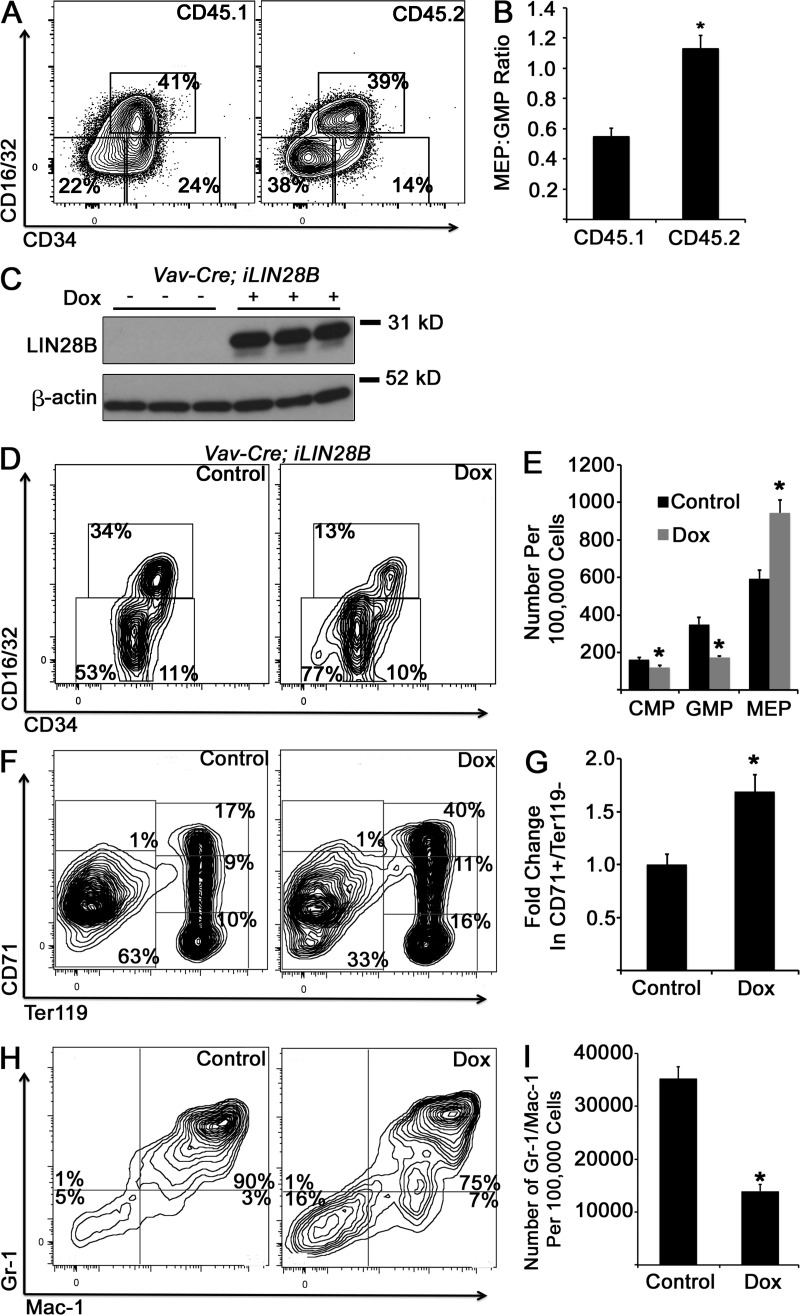
We first used a mouse model of doxycycline-induced human LIN28B expression (iLIN28B mice; Shyh-Chang et al., 2013). 14 d of doxycycline exposure achieved a robust induction of LIN28B in BM cells (Fig. 4 A), suppression of several mature let-7 species (Fig. 4 B), as well as a decrease in GMPs and an FL-like increase in MEPs in the BM (Fig. 4, C and D) compared with unexposed iLIN28B mice. LIN28B-expressing adult hematopoietic tissues were generally more proliferative than control tissues by Ki67 staining (not depicted), but we did not observe obvious differences in overall tissue cellularity or differences in the rates of CMP proliferation or apoptosis as measured by in vivo bromodeoxyuridine incorporation and annexin V staining between iLIN28B and control mice (control 21.9 ± 5.5% compared with iLIN28B 25.9 ± 5.7% bromodeoxyuridine-positive CMPs after a 4-h in vivo pulse [P = 0.63]; control 20.5 ± 3.4% compared with iLIN28B 27.0 ± 4% annexin V–positive CMPs in vivo [P = 0.25]; n = 5 control and iLIN28B mice across three independent experiments for both assays; both analyses by Student’s t test). LIN28B induction resulted in increased numbers of Ter119-positive committed erythroid cells (as well as CD71+/Ter119− early erythroblasts, similar to E14.5 FL) and decreased numbers of mature BM granulocytes compared with controls (Fig. 4, E–H). By morphology, we observed more erythroblasts with fewer mature neutrophils in the BM (Fig. 4 I) and expansion of the splenic red pulp (Fig. 4 J) in doxycycline-exposed TRE-LIN28B mice compared with unexposed mice. We cultured iLIN28B BM cells in methylcellulose with myeloerythropoietic growth factors and observed increased erythroid colony forming activity in LIN28B-induced cells, analogous to WT FL-derived cultures (compare Fig. 4 K with Fig. 1 G). Competitive transplantation of iLIN28B HSCs with WT cells (Fig. 5, A and B) as well as a hematopoietic-specific LIN28B ectopic expression model showed similar effects on myeloid progenitors (Fig. 5, C–I).
Figure 4.
Activation of LIN28B in adult BM. (A) Adult iLIN28B mice were exposed to 1 g/liter doxycycline in drinking water or standard water for 14 d, at which time BM mononuclear cells were isolated, and Western blot analysis for human LIN28B was performed. (B) RNA was isolated from BM CMPs with and without doxycycline (Dox) exposure, and levels of mature let-7 microRNAs were measured by qPCR. For each let-7 form, results are normalized to the control, nondoxycycline-exposed condition. *, P < 0.05 by Student’s t test. n = 4 uninduced and 5 doxycycline-exposed across three experiments. (C) BM mononuclear cells from adult iLIN28B mice with and without 14 d of doxycycline exposure were isolated and immunostained, with representative flow cytometry plots presented. (D) Numbers of myeloid progenitor cells per 100,000 BM mononuclear cells are shown. *, P < 0.01 by Student’s t test. (E) Whole BM cells were analyzed for red blood cell progenitor populations by flow cytometry. Representative plots are shown. (F) Numbers of CD71+/Ter119− cells in doxycycline-exposed iLIN28B animals relative to the control are shown. *, P < 0.02 by Student’s t test. (G) BM mononuclear cells under the indicated conditions were immunostained for Gr-1 and Mac-1. Representative flow cytometry plots are shown. (H) Numbers of Gr-1+/Mac-1+ mature granulocytes per 100,000 BM mononuclear cells under the indicated conditions are presented. *, P < 0.0001 by Student’s t test. (D, F, and H) n = 8 mice for the control condition and 7 mice for the doxycycline-exposed condition across three experiments. (I and J) Morphology of BM cells (I) or splenic tissue (J) from control or doxycycline-treated iLIN28B mice is shown. Images are representative of specimens obtained over at least three independent experiments. (I) Bar, 40 µm. Arrows, neutrophils; arrowheads, erythroblasts. (J) Bar, 200 µm. RP, red pulp; WP, white pulp. Dashed lines indicate borders of these two splenic tissues. (K) BM mononuclear cells from iLIN28B mice exposed to doxycycline for 14 d or unexposed mice were cultured in methylcellulose in the presence of growth factors. After 14 d of culture, colony morphology was scored, and the mean proportions of each colony type are presented. *, P < 0.05 by Student’s t test comparing each colony type. n = 5 control mice and 4 doxycycline-treated mice across three experiments. Error bars represent SEM. CFU-M, CFU monocyte; CFU-G, CFU granulocyte; CFU-GM, CFU granulocyte–monocyte.
Figure 5.
Transplantation of iLIN28B BM into WT recipients and tissue-specific LIN28B induction. (A) Lineage−, c-kit+, Sca-1+, CD150+, and CD48− long-term HSCs were purified by FACS from BM of iLIN28B mice (CD45.2) exposed to doxycycline. Long-term HSCs were cotransplanted with CD45.1 BM cells into lethally irradiated CD45.1 recipient mice. Recipient mice continued to receive doxycycline in their drinking water and were sacrificed 16–20 wk after transplantation, and myeloid progenitors in the BM were analyzed within the CD45.1 and CD45.2 fractions. Representative flow cytometry plots are shown. (B) Ratio of MEP to GMP in the CD45.1 and CD45.2 fractions in mixed chimeric recipient mice is presented. *, P < 0.0001 by Student’s t test. n = 10 mice. (C) Triple transgenic mice carrying alleles for iLIN28B, the reverse tetracycline transactivator (rtta) downstream of a loxp-flanked stop codon cassette, and Vav1-cre for hematopoietic-specific excision of the loxp-flanked stop cassette and hematopoietic-specific expression of rtta and transcription of the iLIN28B transgene in the presence of doxycycline. Exposure of triple transgenic mice to doxycycline (Dox) for 14 d resulted in robust LIN28B induction in the adult BM as assessed by Western blotting. (D and E) Myeloid progenitor profiles were assessed after 14 d of doxycycline exposure in triple transgenic mice compared with unexposed mice. *, P < 0.05 by Student’s t test. (F and G) BM cells were stained for erythroid cells, and numbers of CD71+/Ter119− cells were quantified. *, P = 0.02 by Student’s t test. (H and I) BM cells were stained to assess granulocyte maturation, and numbers of Gr-1+/Mac-1+ cells were quantified. *, P = 0.001 by Student’s t test. (D–I) n = 3 mice across two experiments for each condition. Error bars represent SEM.
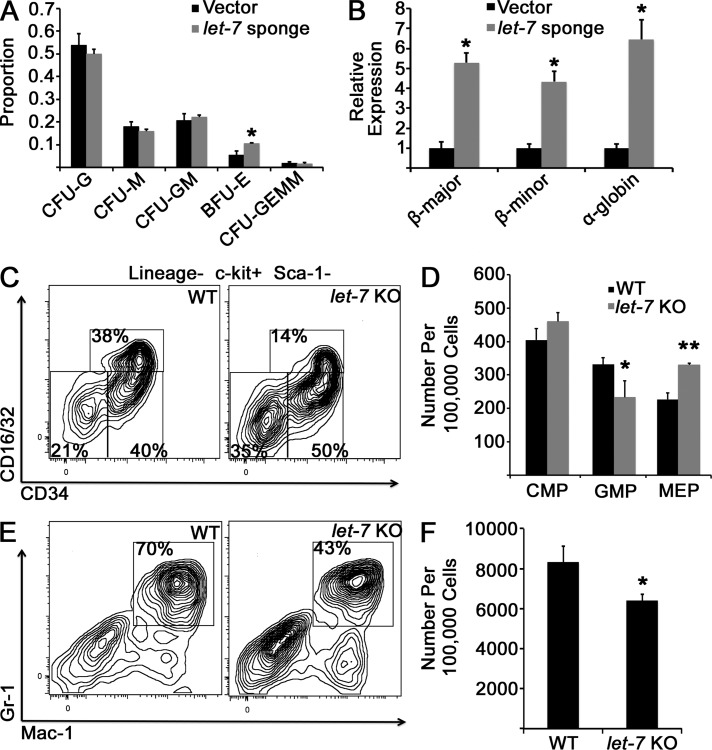
Our second method of interfering with let-7 function in adult myeloerythropoiesis was introduction of a let-7 sponge construct in sorted BM CMPs (Kumar et al., 2008), followed by culture of the sponge- or empty vector–transduced BM CMPs in methylcellulose in the presence of growth factors. Sequestration of let-7 resulted in significantly increased proportions of burst-forming unit erythroid (BFU-E) colonies and mRNA levels of mouse globin genes compared with control cells in culture (Fig. 6, A and B). Third, we analyzed the myeloerythroid progenitor compartment of a strain of mice bearing a deletion of the let-7b and let-7c2 polycistronic host gene (let-7 KO) and observed a trend toward decreased GMPs and increased MEPs (Fig. 6, C and D), as well as fewer BM granulocytes compared with WT isogenic controls (Fig. 6, E and F), but not a significant effect on erythroid maturation or distribution of myeloerythroid colony formation in vitro (not depicted), potentially because of compensatory effects of other let-7 species in this model.
Figure 6.
Inhibition of let-7 in adult myeloerythropoiesis. (A) Purified WT adult BM CMPs were retrovirally transduced with an empty vector or a let-7 sponge construct and cultured in methylcellulose for 14 d, when colonies were scored by morphology. Colony distributions are presented. *, P = 0.03 by Student’s t test. n = 5 replicates for vector and sponge constructs across three independent experiments. CFU-M, CFU monocyte; CFU-G, CFU granulocyte; CFU-GM, CFU granulocyte–monocyte. (B) RNA was isolated from cultures after 14 d, and levels of mouse globin genes were measured by qPCR. *, P < 0.01 by Student’s t test. n = 3 replicates across three experiments. (C) Adult mice with the locus encoding let-7b1 and let-7c2–deleted (let-7 KO) or WT C57 littermates were euthanized, BM cells were stained with the indicated antibodies, and myeloid progenitor populations were analyzed by flow cytometry. (D) Numbers of myeloid progenitor cells are presented. *, P = 0.14; **, P = 0.009 by Student’s t test for each comparison. n = 3 WT and let-7 KO mice analyzed; results are representative of two experiments performed (a separate experiment was performed with mice of strain 129 with similar results). (E) BM from the indicated mice was isolated and stained with the indicated antibodies to evaluate granulocyte populations. (F) Numbers of Gr-1+/Mac-1+ mature granulocytes are presented. *, P = 0.09 by Student’s t test. n = 3 for each genotype; the phenotype is verified in an independent strain in an independent experiment. Error bars represent SEM.
Activation of let-7g in the FL drives an adult BM-like myeloerythroid phenotype
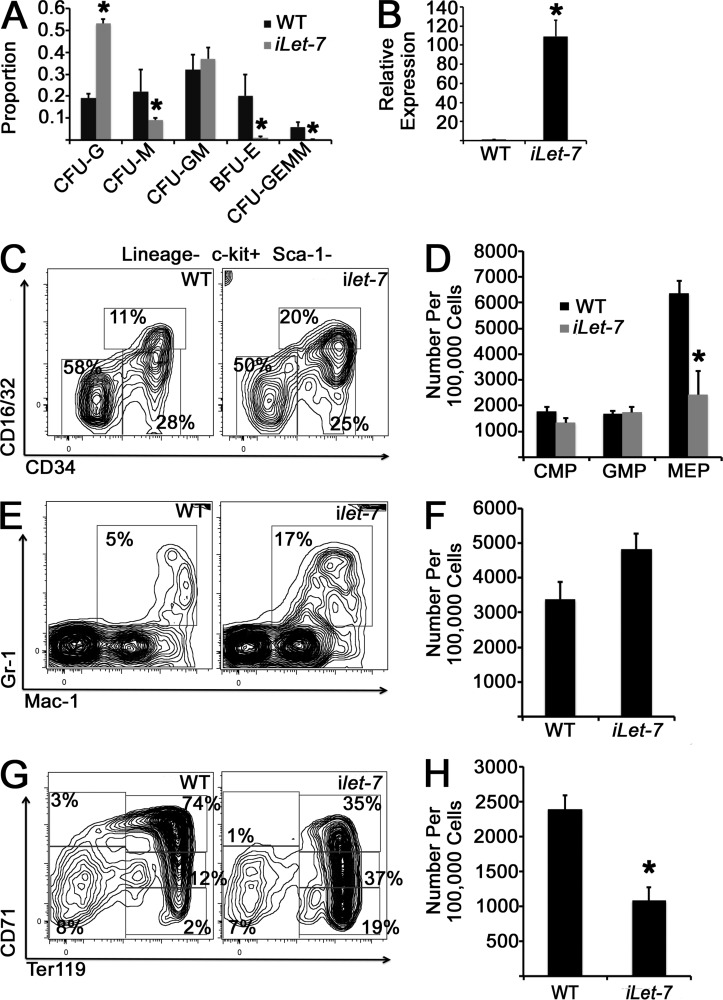
If let-7 function specifies adult BM myeloerythropoiesis, we hypothesized that mistimed ectopic activation of let-7 in the FL would result in an aberrant adult-like myeloerythroid phenotype. To this end, we used a mouse strain wherein expression of a Lin28-resistant chimeric form of let-7g (let-7S21L) is induced by doxycycline exposure (iLet-7 mouse; Zhu et al., 2011) and assessed myeloerythroid colony formation in E14.5 FL-derived cells from iLet-7 embryos and controls after 2–4 d of in utero induction before harvesting of embryos. Induction of let-7S21L resulted in an increased proportion of CFU granulocyte colonies at the expense of BFU-E and CFU-GEMM (granulocyte, erythrocyte, monocyte, and megakaryocyte) colonies (Fig. 7 A). To assess effects of ectopic let-7 in vivo, we induced females bearing mixed litters of iLet-7 embryos and littermate controls at E10.5 with doxycycline in the drinking water, which resulted in robust let-7g expression in the FL when harvested at E14.5 (Fig. 7 B). Although many iLet-7 embryos died, analysis of myeloid progenitors in viable embryos at E14.5 revealed depletion of MEPs (Fig. 7, C and D), a trend toward increased FL granulocytes (Fig. 7, E and F), and diminished numbers of CD71+/Ter119− early erythroid progenitors with an adult-like pattern of erythroid differentiation (Fig. 7, G and H).
Figure 7.
Induction of let-7 in FL hematopoietic cells. (A) Pregnant females carrying iLet-7 embryos and littermate controls were exposed to doxycycline for 2–4 d before harvesting at E14.5. FL mononuclear cells were cultured in methylcellulose for 14 d in the presence of doxycycline, when colony morphology was scored. Proportions of each colony type are presented. *, P < 0.05 by Student’s t test for each colony type comparison. n = 8 control and 4 iLet-7 embryos across three experiments. CFU-M, CFU monocyte; CFU-G, CFU granulocyte; CFU-GM, CFU granulocyte–monocyte. (B) let-7g induction in the FL at E14.5 after 4-d maternal doxycycline exposure as measured by qPCR. *, P < 0.0001 by Student’s t test. n = 7 WT and 2 iLet-7 embryos from two independent experiments. (C and D) FL cells from the indicated genotypes were stained with the indicated antibodies, and myeloid progenitor populations were analyzed by flow cytometry. Representative histograms are shown. (E and F) FL cells were stained with the indicated antibodies to assess mature granulocyte populations. P = 0.05 by Student’s t test for control and iLet-7 comparison of Gr1+/Mac-1+ cells. (G and H) Whole FL cells were stained with anti-CD71 and anti-Ter119 antibodies to analyze red blood cell progenitor populations by flow cytometry. (C–H) *, P < 0.05 comparing CD71+/Ter119− cells. A total of 10 control and 8 iLet-7 embryos were evaluated, obtained from three independent litters. Error bars represent SEM.
Endogenous Lin28b deficiency alters the myeloid progenitor pool in the FL
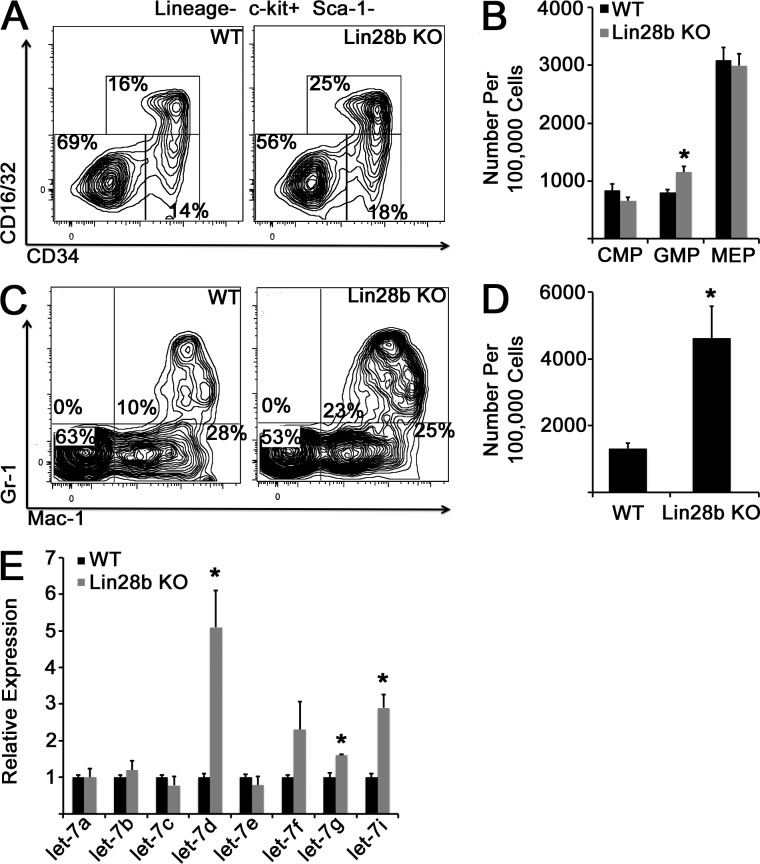
Because Lin28b decreases during myeloerythroid maturation and ectopic LIN28B drives an FL-like myeloerythroid phenotype in BM CMPs, we examined the progenitor pool in Lin28b-deficient E14.5 FLs and observed an increased number of GMPs in Lin28b-deficient embryos compared with littermate controls (Fig. 8, A and B). Although we did not observe a significant phenotype related to erythroid maturation (not depicted), we did find increased numbers of granulocytes in the FLs of Lin28b-deficient embryos compared with WT littermates (Fig. 8, C and D). Culture of FL mononuclear cells in methylcellulose with growth factors did not demonstrate a significant difference in colony formation between genotypes (not depicted). Importantly, we did not observe a compensatory up-regulation of Lin28a in Lin28b-deficient FLs (not depicted). Consistent with a fetal-specific role for Lin28b in myelopoiesis, complete blood counts in adult Lin28b-deficient mice were similar to control mice, with the exception of a mild anemia in the null mice (not depicted). We hypothesized that the incomplete induction of an adult BM-like myelopoietic phenotype in Lin28b-deficient embryos might be caused by a relative paucity of precursor let-7 species in FL CMPs compared with adult BM CMPs (Fig. 3 C). Therefore, we assessed the ability of Lin28b-deficient E14.5 CMPs to form mature let-7 microRNAs. In the absence of Lin28b, we observed an increase in a subset of mature let-7 forms in FL CMPs (let-7d, let-7g, and let-7i); however, levels of let-7 were still lower than those observed during physiological myeloerythroid maturation (compare Fig. 8 E with Fig. 3 C). Thus, it is likely that the consequences of loss of Lin28b in the FL are mitigated by an inability to generate adult BM-like levels of let-7 microRNAs, possibly caused by low levels of basal transcription of let-7 precursors.
Figure 8.
Deficiency of Lin28b in the FL. (A) WT or Lin28b KO embryos were isolated at E14.5, and FL cells were stained with the indicated antibodies. (B) The number of myeloid progenitor populations is shown. *, P = 0.007 by Student’s t test. n = 7 WT and 6 Lin28-deficient embryos across three experiments. (C) FL cells of the indicated genotypes were isolated, and granulocyte populations were analyzed by flow cytometry. (D) Numbers of Gr-1+/Mac-1+ granulocytes per 100,000 FL mononuclear cells are shown. *, P = 0.03 by Student’s t test. n = 7 WT and 5 Lin28b-deficient embryos across three experiments. (E) Mature let-7 species were measured in FL CMP cells from E14.5 embryos of the indicated genotypes. *, P < 0.05 by Student’s t test. n = 3 replicates for each genotype from independent litters. Error bars represent SEM.
Transcriptional programs underlying Lin28b/let-7 control of myeloerythropoiesis
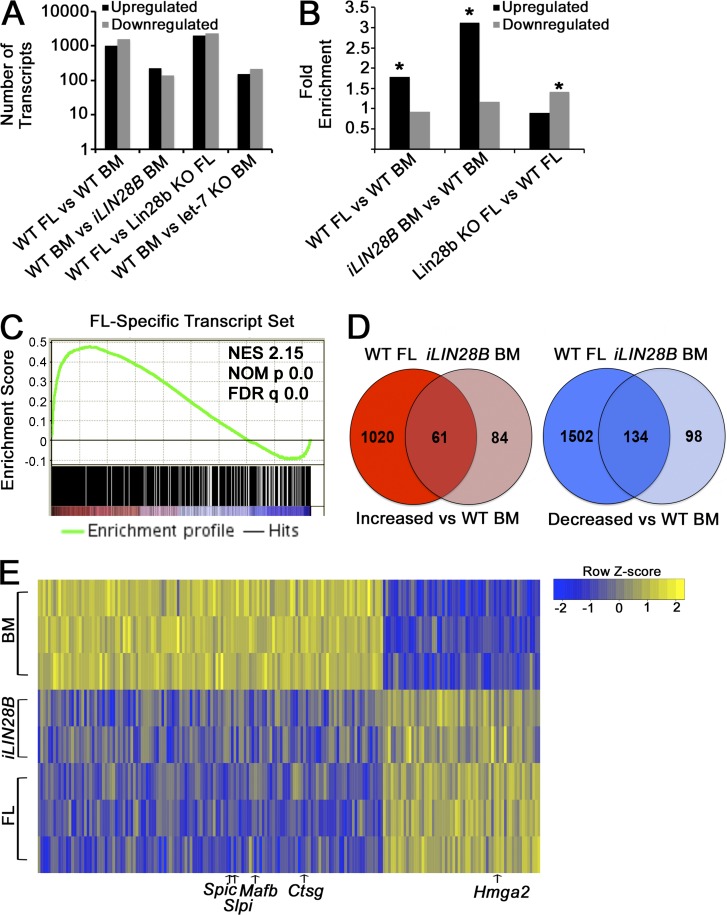
We performed RNAseq on CMPs isolated from WT FL, WT adult BM, Lin28b-deficient FL, and iLIN28B adult BM. The numbers of differentially expressed genes between relevant comparisons are shown in Fig. 9 A. We observed significant enrichment of let-7 target genes in three comparisons (Fig. 9 B), in patterns consistent with the relative let-7 abundance across development described above (Fig. 3 C). Gene set enrichment analysis (GSEA) revealed that mRNAs affected by LIN28B induction in adult BM CMPs had a highly significant coordinate pattern of regulation with those altered during normal development from FL to BM myeloerythropoiesis (Fig. 9 C). As such, we compared the sets of mRNAs altered in FL CMPs and iLIN28B BM CMPs relative to WT BM CMPs and noted a significant degree of overlap (Fig. 9 D). On the list of commonly regulated mRNAs, we found several markers of mature neutrophils (Ctsg and Slpi) and monocytes (Mafb and Spic) that were more abundant in WT BM compared with iLIN28B BM and WT FLs (Fig. 9 E). Among the commonly enriched transcripts in FL CMPs and iLIN28B BM CMPs, 16/61 (26%) had predicted let-7 binding sites, whereas among the transcripts that were commonly lower, only 5/134 (4%) were predicted let-7 targets. Interestingly, LIN28B induction resulted in a 4.58-fold increased expression of embryonic globin Hbb-y relative to WT BM CMPs.
Figure 9.
Analysis of mRNA expression by RNAseq. (A) Raw numbers of differentially expressed mRNAs for the CMP comparisons indicated are shown (FDR cutoff for significance is 0.001). (B) Fold enrichment of let-7 target gene frequency in the indicated pools of transcripts compared with the overall frequency of let-7 targets among all analyzed mRNAs is shown. *, P < 0.001; hypergeometric p-values compared with the expected frequency of let-7 targets based on the frequency of targets among all expressed mRNAs. (C) GSEA demonstrating significant positive enrichment of the FL CMP transcriptional program in iLIN28B CMPs. Transcripts enriched in iLIN28B adult BM CMPs compared with adult BM CMPs were compared with the set of transcripts significantly enriched in WT FL CMPs compared with WT adult BM CMPs. NES, normalized enrichment score; NOM p, nominal p-value. (D) Venn diagram analysis showing overlap of transcripts up-regulated in FL CMPs and iLIN28B CMPs compared with WT BM CMPs (left), as well as down-regulated transcripts for these comparisons (right). The p-value for overlap of up-regulated transcripts is <2.2 × 10−16 (expected number of 16.22); the p-value for overlap of down-regulated transcripts is <2.2 × 10−16 (expected number of 39.15). Analysis was performed with Fisher’s exact test. (E) Heat map demonstrating relative expression levels of the transcripts commonly up-regulated or down-regulated in iLIN28B CMPs and WT FL CMPs compared with WT BM CMPs. Pertinent transcripts are denoted.
LIN28B effect on multipotent progenitors (MPPs)
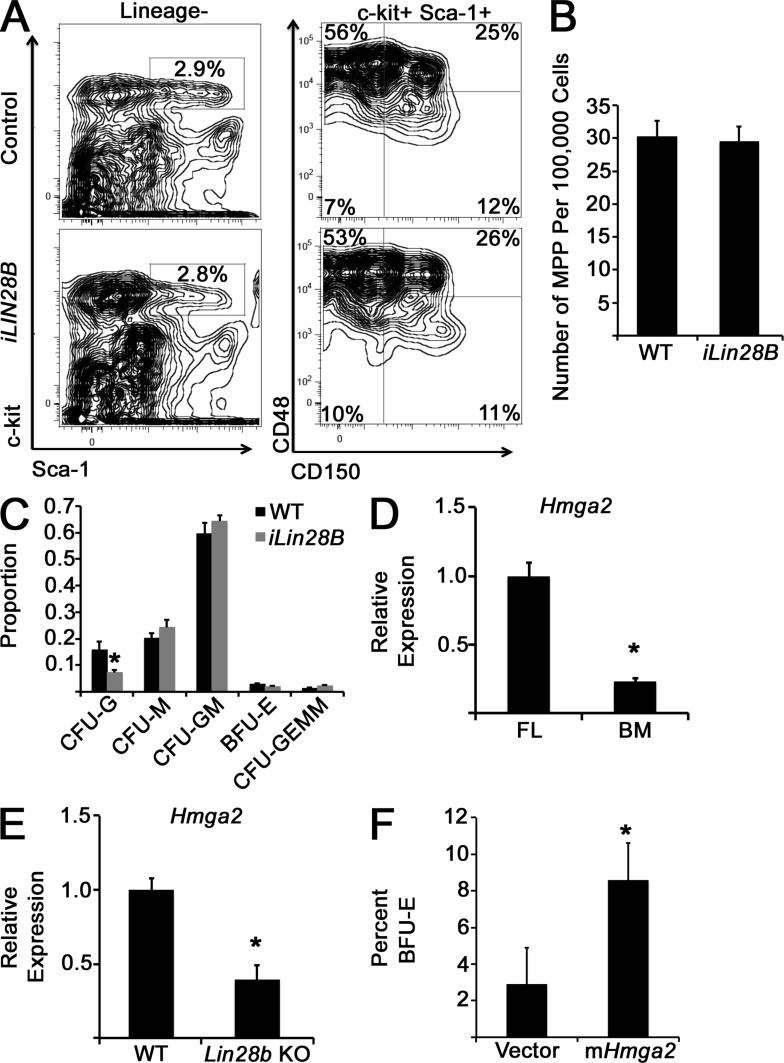
As Lin28/let-7 appears to regulate myeloerythropoiesis at the level of the CMP, possibly via transcriptional priming, we assessed the myeloerythroid differentiation potential of MPPs—which give rise to CMPs in hematopoietic ontogeny—in iLIN28B and control mice. Within the LSK (Lineage−, Sca-1+, c-kit+) compartment, we resolved MPP and long-term and short-term HSC populations, defining MPPs as CD150− and CD48−/low (Fig. 10 A; Oguro et al., 2013). We did not observe differences in MPP frequency between iLIN28B and control BM (Fig. 10 B). MPPs were sorted and cultured in methylcellulose in the presence of myelopoietic cytokines, and colonies were scored by morphology after 10–14 d. We did not observe differences in colony distribution, save for restraint of full CFU granulocyte potential by ectopic LIN28B expression, and no effect on erythroid differentiation (Fig. 10 C), consistent with an effect on myeloerythroid differentiation mediated predominantly at the CMP level.
Figure 10.
Assessment of LIN28B function in MPPs and investigation of Hmga2 as a Lin28b–let-7 target regulating myeloerythropoiesis. (A) Representative flow cytometry plots showing gating strategy to assess MPP populations in control and iLIN28B mice. (B) Numbers of MPP in the indicated mice are shown. P = 0.82 by Student’s t test. n = 5 mice for each condition across three independent experiments. (C) MPPs were isolated by FACS and cultured in methylcellulose in the presence of myelopoietic cytokines for 10–14 d, when colony morphology was scored. Colony distributions are presented. *, P = 0.04 by Student’s t test. n = 5 mice for each condition across three independent experiments. CFU-M, CFU monocyte; CFU-G, CFU granulocyte; CFU-GM, CFU granulocyte–monocyte. (D and E) Levels of Hmga2 mRNA were measured by qPCR and compared in FL versus adult BM CMPs (D) or Lin28b-deficient compared with littermate control FL CMPs (E). (D) *, P = 0.002 by Student’s t test. n = 3 in each group sorted across two experiments. (E) *, P = 0.007. n = 4 WT and 3 Lin28b-deficient samples from three separate litters. (F) WT adult BM CMPs were transduced with the indicated retroviruses and cultured in methylcellulose in the presence of growth factors for 14 d, when colony morphology was scored. The percentages of BFU-E are indicated. *, P < 0.05 by Student’s t test. n = 3 from three independent experiments. Error bars represent SEM.
Hmga2 as an effector of Lin28b–let-7–controlled myeloerythropoiesis
By RNAseq, the let-7 target gene Hmga2 (Copley et al., 2013) was one of the most significantly increased mRNAs in both iLIN28B CMPs and FL CMPs compared with adult BM CMPs (Fig. 9 E). We confirmed this differential expression by qPCR (Fig. 10 D). Moreover, we found lower levels of Hmga2 in Lin28b-deficient FL CMPs (Fig. 10 E). Because Hmga2 is a well-known hematopoietic regulator (Ikeda et al., 2011; Lam et al., 2014), we isolated adult BM CMPs and ectopically expressed a mouse Hmga2 transgene or a control vector and cultured the CMPs in methylcellulose with hematopoietic growth factors for 14 d. Expression of ectopic Hmga2 resulted in an increased proportion of BFU-E colonies relative to control cultures after 14 d (Fig. 10 F), paralleling an FL-like phenotype and indicating that Hmga2 is a mediator of the effect of the Lin28–let-7 axis on hematopoietic maturation.
DISCUSSION
Hematopoiesis is a tightly regulated but adaptable process in which production of terminally differentiated cells responds to pathophysiologic stimuli such as hypoxia, hemorrhage, infection, or inflammation (Orkin and Zon, 2008; Paulson et al., 2011; Takizawa et al., 2012). Basal myeloerythroid output is poised to meet specific developmental stage–specific demands, with erythroid production predominant in the FL to support growth under hypoxic conditions and granulocyte production favored in the adult BM to provide innate immunity and tissue repair capability. Candidate regulators of hematopoietic developmental maturation have begun to emerge (He et al., 2011; Mochizuki-Kashio et al., 2011; Xie et al., 2014). In this study, we document developmental maturation of myeloerythroid progenitors and link this mechanistically to regulation by the Lin28–let-7–Hmga2 axis, which is known to control developmental timing from worms to humans (Ambros and Horvitz, 1984; Yuan et al., 2012; Copley et al., 2013; Shyh-Chang and Daley, 2013).
Lin28 genes function in vertebrates to specify timing of kidney and germ cell development (Ambros and Horvitz, 1984; Moss et al., 1997; West et al., 2009; Urbach et al., 2014), as well as in tissue regeneration (Shyh-Chang et al., 2013) and muscle and nerve cell differentiation (Polesskaya et al., 2007; Yang et al., 2015). Recently, two studies have implicated Lin28 in the timing of hematopoietic phenotypes during development (Yuan et al., 2012; Copley et al., 2013). Specifically, ectopic expression of Lin28 in adult BM cells confers fetal-like HSC self-renewal properties and lymphopoietic phenotypes, indicating that ectopic Lin28 expression may be sufficient to recapitulate prenatal hematopoietic programs (Yuan et al., 2012; Copley et al., 2013). Both of these studies were based on transcriptional analyses showing predominance of let-7 in adult and Lin28 in fetal hematopoietic cells (Yuan et al., 2012; Copley et al., 2013). Our study extends this paradigm to the myeloerythroid system. However, in contrast to these prior studies based exclusively on ectopic overexpression of Lin28, we have dissected the functionality of the Lin28–let-7 axis in myeloerythroid differentiation by manipulating the timing of this developmental switch through both gain-of-function and loss-of-function studies of endogenous alleles in both adult and fetal scenarios, thereby confirming the role of endogenous Lin28 in regulating normal hematopoietic development. Furthermore, our findings provide a paradigm in which the function of this axis or other candidate regulators of developmental timing can be interrogated in other systems.
We observed strong reduction of Lin28b levels and reciprocal recruitment of several mature let-7 species during developmental maturation of CMPs from fetus to adult, as well as coordinate modulation of several let-7–regulated mRNAs with developmental changes in endogenous Lin28b and let-7 expression. Mature let-7 activation in the adult seemed to occur as a result of both the down-regulation of Lin28b—which would be expected to permit full processing of let-7 precursors to mature forms—as well as transcriptional activation of precursor let-7, indicative of two levels of let-7 regulation. Therefore, to investigate the roles of Lin28 and let-7 in early myeloerythroid differentiation, we used the complementary strategies of inhibition of let-7 function in adult tissues and inappropriate let-7 activation in the fetal state.
In the FL, wherein erythroid output predominates, mistimed activation of let-7 activity inappropriately drives granulocyte accumulation at the expense of erythroid activity. This observation agrees with previous studies that let-7 expression increases during terminal granulocyte differentiation (Wong et al., 2014) and that disruption of Lin28 expression can affect granulopoiesis (Wang et al., 2015). These results also parallel studies from the leukemia literature wherein let-7 activity has been shown to promote myeloblast differentiation (Viswanathan et al., 2009; Emmrich et al., 2013; Pelosi et al., 2013). Consistent with both transcriptional and posttranscriptional mechanisms of let-7 regulation, Lin28b deficiency alone did not fully recapitulate an adult-like myeloerythroid phenotype in the FL. In the setting of low levels of endogenous precursor let-7 transcription, there appears to be scant let-7 precursors present for Lin28b to impair. Therefore, the blunted phenotype of Lin28b-deficient FLs is likely caused by a low degree of mature let-7 accumulation, compared with that which occurs during normal myeloerythroid maturation as a consequence of the dual effects of Lin28b depletion and pre/pri–let-7 transactivation. Very little is known regarding regulators of precursor let-7 transcription, with one study implicating HOXA10 as a transactivator of a human let-7–containing tricistron (Emmrich et al., 2014). It is likely that enhanced understanding of the regulation of coordinated Lin28b transcriptional repression and let-7 precursor transcriptional activation will provide further insight into developmental hematopoiesis. Given the number of mRNAs deregulated by Lin28b deficiency in the FL, it could also be speculated that Lin28b may play a significant role in regulating mRNA stability, as has been previously reported (Polesskaya et al., 2007).
We used three methods of let-7 inhibition in adult BM myeloerythroid progenitors: ectopic/mistimed expression of LIN28B, a novel mouse model of precursor let-7b1/c2 deficiency, and a let-7 sponge construct. Although prior studies have suggested that ectopic Lin28 expression in adult hematopoiesis can drive fetal-like hematopoietic output (Yuan et al., 2012; Copley et al., 2013), our findings extend this observation to the myeloerythroid system. Furthermore, our results mechanistically attribute this effect to Lin28 inhibition of let-7 maturation, given the similar effects on myeloerythropoiesis provoked by modulating let-7 activity directly using our let-7 KO mice and let-7 sponge construct. Importantly, we observed that inhibition of let-7 directly in FACS-sorted adult BM CMPs impacted production of differentiated progeny. Together, these findings advance a mechanism by which Lin28b works at least in part via a linear axis involving control of let-7 microRNA maturation and let-7 target genes to specify CMP fate and cell output in the FL and adult BM. Conversely, we observed a very subtle effect of ectopic activation of LIN28B in FACS-sorted MPPs. Together with our RNAseq data, these findings suggest that transcriptional priming at the level of the CMP specifies myeloerythroid differentiation across development. Indeed, human and mouse myeloid progenitor populations possess intrinsic priming in transcriptional programs and ultimate lineage commitment at the single-cell level (Paul et al., 2015; Notta et al., 2016). Given the changes in progenitor lineage priming that occur across development (Notta et al., 2016), we propose that as heterochronic regulators of myeloerythropoiesis, Lin28b/let-7 may play a central role in this process, a hypothesis that deserves further investigation. Finally, our genetic and transplant strategies for hematopoietic-specific LIN28B activation demonstrate that this effect is autonomous within the hematopoietic system.
In WT FL and adult BM, we observed varying patterns of erythroid cell differentiation from the earliest stages of MEP fate commitment through reticulocyte formation. We observed that mistimed LIN28B expression in the adult or let-7 expression in the FL could recapitulate fetal or adult erythroid states, respectively, at inappropriate developmental times, effectively generating erythroid heterochronic mutants. It has been shown that let-7 microRNAs are enriched in adult human erythroid cells compared with umbilical cord cells (Noh et al., 2009), implicating the Lin28–let-7 axis in human developmental erythropoiesis as well. Indeed, these studies have been extended to demonstrate that the Lin28–let-7 axis acts as a regulator of hemoglobin switching (Lee et al., 2013). Further study is necessary to delineate potential roles for let-7 microRNAs in sequential phases of erythroid precursor differentiation.
Our RNAseq data show that CMPs from FL and adult BM differentially express transcripts characteristic of erythroid and granulocytic differentiation, in agreement with priming for production of appropriately differentiated progeny, as described previously (Pronk et al., 2007; Notta et al., 2016). Our data suggest that this priming may be regulated at least in part by Lin28 and let-7 given the coordinate patterns of gene expression we observed between iLIN28B adult BM and normal FL CMPs. We identified Hmga2 as one of the most highly up-regulated mRNAs in FLs and iLIN28B adult BM CMPs compared with WT adult BM CMPs, consistent with a role as a candidate effector of Lin28b/let-7–driven regulation of CMP output. Hmga2 has been previously implicated as a regulator of myeloid progenitor cell function (Lam et al., 2014), and consistent with our results, its ectopic expression can drive adult BM and extramedullary erythropoiesis (Ikeda et al., 2011). Interestingly, this effect parallels a study wherein insertional activation of HMGA2 by a gene therapy lentivirus in a human β-thalassemia patient resulted in autonomous erythropoiesis and transfusional independence (Cavazzana-Calvo et al., 2010). Further supportive of an intersection with Lin28–let-7 function, Hmga2 previously has been shown to regulate developmental timing in neural stem cells (Nishino et al., 2008).
Our data posit that the Lin28–let-7 axis acts as a heterochronic switch within the hematopoietic system. The roles of Lin28/let-7 as heterochronic regulators appear to be relevant in pathological contexts as well. In cancer, Lin28 acts as an oncofetal factor, whereby developmentally mistimed Lin28 expression provokes aberrant induction of fetal-like cell dedifferentiation and proliferation in certain tumors (Jiang et al., 2012; Rao et al., 2012; Nguyen et al., 2014; Urbach et al., 2014; Wang et al., 2015).
In sum, we propose that the highly conserved Lin28–let-7–Hmga2 signaling axis serves as a heterochronic genetic regulatory system that matches the myeloerythroid phenotype to the appropriate developmental state to meet physiological needs. We speculate that this paradigm of heterochronic regulation is applicable not only to the hematopoietic system, but also to other mammalian developmental processes.
MATERIALS AND METHODS
Mice
iLIN28B mice, Lin28b-deficient mice, and iLet-7 mice that carry a Lin28-resistant chimeric form of let-7g consisting of the stem loop structure from let-7g fused to the loop of the LIN28-resistant miR-21 have been described previously (Zhu et al., 2011; Shinoda et al., 2013). For doxycycline-inducible expression of LIN28B or let-7g, transgenic mice were crossed to the B6.Cg-Gt(ROSA)26Sortm1(rtTA*M2)Jae/J strain (006965; The Jackson Laboratory; Hochedlinger et al., 2005). Experiments using WT adult mice and embryos used the C57BL/6J strain. All animal procedures were performed according to animal care guidelines approved by the Institutional Animal Care and Use Committee. To generate let-7 KO mice, a mouse genomic clone containing the let-7b and let-7c-2 isoforms was obtained from a genomic P1-derived artificial chromosome (PAC) library (RPCI-21 129s6/SvEvTAC). A gene replacement vector was constructed by recombineering essentially as previously described (Rodriguez et al., 2007). The final vector contained the MC1-DTA–negative selection cassette and the loxP-PGK-EM7-NeobpA-loxP–positive selection cassette. Targeted AB1 embryonic stem cell clones were identified by Southern blots and then used for blastocyst injections. Male chimeras were mated to 129S5 isogenic-CMV-Cre mice to remove the selection cassette. PCR sequencing was then used to confirm the presence of the 801-bp genomic deletion of let-7b and let-7c-2. The deletion breakpoints are as follows: 5′-GACCTCAAGAAGCCACAACATC-(810-bp deletion)-TACAACCTACTGCCTTCCCTGA-3′. Heterozygous mice were intercrossed to obtain homozygous mutant animals. Mutant animals used in these studies were backcrossed at least six generations into a C57BL/6 genetic background. This strain was also analyzed in separate experiments on the 129-strain background.
Flow cytometry and FACS
Cells were isolated from BM by flushing marrow cavities. Cells were isolated from FL from timed pregnancies. Cells were analyzed on a Fortessa flow cytometer or sorted on a FACS Aria (BD). The following anti–mouse fluorescence-conjugated antibodies were used: anti-B220, CD3, Ter119, Mac-1, Gr-1 conjugated to Pacific blue as a lineage cocktail, anti–c-kit–APC-Cy7, anti-CD34–FITC, and anti-CD16/32–PerCp-Cy5.5 (eBioscience and Affymetrix); anti–Sca-1–PE-Cy7; anti-Ter119–APC and anti–Gr-1–PE; anti–Mac-1–PE-Cy5 and anti-CD150–APC (BioLegend); and anti–Mac-1–AF700, anti-CD48–PE, and anti-CD71–FITC (BD). Cells were counterstained for viability with Sytox blue or propidium iodide (Thermo Fisher Scientific). Viability for cell quantification was determined using a parallel tube stained with Sytox blue.
Annexin V staining
BM mononuclear cells were isolated and immunostained with surface markers per the flow cytometry methods in the previous paragraph to delineate progenitor populations. They were then bound with annexin V APCs (eBioscience and Affymetrix) according to the manufacturer’s instructions.
In vivo bromodeoxyuridine incorporation
Adult mice were injected intraperitoneally with 2 mg bromodeoxyuridine. 4 h later, BM cells were isolated and immunostained with surface markers to delineate progenitor populations, after which they were fixed and permeabilized for intracellular staining of incorporated bromodeoxyuridine with a kit from BD, according to the manufacturer’s instructions.
DNA constructs
The let-7 sponge construct was provided in pMSCV-puro and was a gift from P. Sharp (Massachusetts Institute of Technology, Boston, MA; plasmid 29766; Addgene; Kumar et al., 2008). pcDNA 3.1 WT mouse Hmga2 was a gift from D. Bartel (Massachusetts Institute of Technology, Boston, MA; plasmid 14789; Addgene; Mayr et al., 2007). The WT mouse Hmga2 insert was subcloned into pBabe-puro for retrovirus packaging.
Western blotting
The following antibodies were used for Western blotting: rabbit anti-LIN28B (4196; Cell Signaling Technology) and goat anti-actin (sc-1616; Santa Cruz Biotechnology, Inc.).
qPCR
RNA was isolated using TRIzol reagent (Thermo Fisher Scientific), and cDNA was synthesized using the miScript II kit (QIAGEN). Target cDNAs were detected using a SYBR green detection kit (Thermo Fisher Scientific) and gene-specific primers.
Cytology and microscopy
FL and BM mononuclear cells were stained with Wright-Giemsa and May-Grunwald stains (Sigma-Aldrich), and images were photographed.
Retroviral transduction of myeloid progenitors
Myeloid progenitors were isolated by FACS. Ecotropic retrovirus was generated by cotransfection of the appropriate retroviral vector with an ecotropic packaging plasmid. Myeloid progenitors were exposed to retroviral supernatant for 48 h in the presence of 100 ng/ml stem cell factor, 10 ng/ml IL-3, and 10 ng/ml IL-6 (all from R&D Systems), as well as 5 µg/ml polybrene (Sigma-Aldrich).
Methylcellulose culture
BM or FL mononuclear cells were resuspended and plated in methylcellulose medium according to the manufacturer’s protocol (M3434; STEMCELL Technologies). In some experiments, cells were cultured in the presence of 1 µg/ml doxycycline (Sigma-Aldrich) or 2 µg/ml puromycin (Thermo Fisher Scientific).
RNAseq and data analysis
For reverse transcription and library preparation, we used a SMARTer Ultra Low RNA kit (V3) for sequencing and a Low Input Library Prep kit (Takara Bio Inc.) according to the manufacturer’s protocol. The RNAseq libraries were sequenced on the HiSeq 2500 platform (Illumina) yielding 82 million raw reads per sample on average (Table S1). From the raw reads, adapter sequences were removed using a Cutadapt tool (Martin, 2011) allowing some mismatches (the specific parameters we used were −n 2 −e 0.02 −o 15). The cleaned reads were aligned to the mouse genome/transcriptome (10 mm and corresponding University of California, Santa Cruz, gene model) using TopHat2 software (Kim et al., 2013). The mean mapping rate was 81%, and this yields 67 million aligned reads per sample on average (Table S1). The RNAseq raw data and analyzed data have been deposited in NCBI’s Gene Expression Omnibus (GEO) database (Edgar et al., 2002) and are accessible through GEO Series accession no. GSE72521.
To estimate expression levels for each gene, aligned reads per gene were counted using an htseq-count tool (Anders et al., 2015). Counts were then normalized and converted to counts per million and reads per kilobase per million values for further analysis using an edgeR package (Robinson et al., 2010). Differentially expressed genes between any two groups in this study were identified according to the edgeR’s guidelines. Low-level expressed genes, which did not have counts per million values over 1 in at least three samples, were filtered out. Dispersions were estimated, and an exact test was performed. Most computational experiments were conducted on the Orchestra High Performance Compute Cluster at Harvard Medical School. For GSEA analysis, publicly available datasets for normal erythroblasts (GEO accession no. GSM995533), promyelocytes (GEO accession no. GSM1174469), myelocytes (GEO accession no. GSM1174470), and granulocytes (GEO accession no. GSM1174471) were used (Wong et al., 2013; Wu et al., 2014).
Complete blood count analysis
Under isoflurane anesthesia, mice were retroorbitally bled, and complete blood counts were obtained on a hematology system (Hemavet 950FS; Drew Scientific).
Immunohistochemistry
This was done at the Dana-Farber/Harvard Cancer Center Specialized Histopathology Core.
BM transplantation
BM mononuclear cells from age- and gender-matched male or female mice were directly retroorbitally injected (3 × 106 CD45.1 cells/recipient with 150 long-term HSCs [lineage−, c-kit+, Sca-1+, CD48−, and CD150+]) into anesthetized, lethally irradiated (950 rads) congenic CD45.1 recipients. Recipients received trimethoprim-sulfamethoxazole in their drinking water for 4 wk after transplantation to prevent infection.
Statistics
Statistics were done with Student’s t test unless otherwise indicated.
Online supplemental material
Table S1 shows RNAseq data comparing transcript expression between E14.5 FL and adult BM CMPs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151912/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joanna Han, Anna Zhao, and the Harvard Medical School Research Computing Team for RNAseq data management.
This work is supported by grants from the US National Institutes of Health: National Institute for Diabetes and Digestive and Kidney Diseases (R24-DK092760 and R24-DK49216), the National Heart, Lung, and Blood Institute (NHBLI Progenitor Cell Biology Consortium grant UO1-HL100001), and the National Institute of General Medical Sciences (RO1GM107536; PN2 EY018244 and P30DK036836 to A.J. Wagers). G.Q. Daley is an associate member of the Broad Institute and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research. R.G. Rowe was supported by NHLBI (5T32HL007574-34). L.D. Wang was supported by a Damon Runyon-Sohn Foundation Pediatric Cancer Research fellowship (DSRG 02-12), an Alex’s Lemonade Stand Foundation for Childhood Cancer Young Investigator award (31582), and a St. Baldrick’s Foundation PALS Scholar award (243625).
The authors declare no competing financial interests.
Author contributions: R.G. Rowe and L.D. Wang designed the research, performed experiments, analyzed data, and wrote the manuscript. S. Coma performed experiments, analyzed data, and performed mouse husbandry. A. Rodriguez generated let-7 KO mice. P.T. Nguyen, S. Ross, and P. Sousa performed experiments and mouse husbandry. R. Mathieu and D.S. Pearson performed experiments. A. Han analyzed RNAseq data. A.J. Wagers and G.Q. Daley supervised experiments, designed the research, and wrote the manuscript.
Footnotes
Abbreviations used:
- BFU-E
- burst-forming unit erythroid
- CMP
- common myeloid progenitor
- FDR
- false discovery rate
- FL
- fetal liver
- GMP
- granulocyte–monocyte progenitor
- GSEA
- gene set enrichment analysis
- HSC
- hematopoietic stem cell
- MEP
- megakaryocyte–erythroid progenitor
- MPP
- multipotent progenitor
- qPCR
- quantitative PCR
References
- Akashi K., Traver D., Miyamoto T., and Weissman I.L.. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Ambros V., and Horvitz H.R.. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 226:409–416. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie M.B., Kent D.G., Dykstra B., McKnight K.D., McCaffrey L., Hoodless P.A., and Eaves C.J.. 2007. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc. Natl. Acad. Sci. USA. 104:5878–5882. 10.1073/pnas.0700460104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K., et al. 2010. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 467:318–322. 10.1038/nature09328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley M.R., Babovic S., Benz C., Knapp D.J., Beer P.A., Kent D.G., Wohrer S., Treloar D.Q., Day C., Rowe K., et al. 2013. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 15:916–925. 10.1038/ncb2783 [DOI] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Rice K.L., Howell L., Zelent A., Licht J.D., and Dick J.E.. 2009. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 23:2076–2087. 10.1101/gad.1788109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S., Katsman-Kuipers J.E., Henke K., Khatib M.E., Jammal R., Engeland F., Dasci F., Zwaan C.M., den Boer M.L., Verboon L., et al. 2013. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia. 28:1022–1032. 10.1038/leu.2013.357 [DOI] [PubMed] [Google Scholar]
- Emmrich S., Rasche M., Schöning J., Reimer C., Keihani S., Maroz A., Xie Y., Li Z., Schambach A., Reinhardt D., and Klusmann J.H.. 2014. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 28:858–874. 10.1101/gad.233791.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., and Jaenisch R.. 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 462:595–601. 10.1038/nature08592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Kim I., Lim M.S., and Morrison S.J.. 2011. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 25:1613–1627. 10.1101/gad.2052911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K., Yamada Y., Beard C., and Jaenisch R.. 2005. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 121:465–477. 10.1016/j.cell.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Mason P.J., and Bessler M.. 2011. 3′UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood. 117:5860–5869. 10.1182/blood-2011-02-334425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta K., and Weissman I.L.. 1991. The junctional modifications of a T cell receptor gamma chain are determined at the level of thymic precursors. J. Exp. Med. 174:1279–1282. 10.1084/jem.174.5.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta K., Kina T., MacNeil I., Uchida N., Peault B., Chien Y.H., and Weissman I.L.. 1990. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 62:863–874. 10.1016/0092-8674(90)90262-D [DOI] [PubMed] [Google Scholar]
- Jiang X., Huang H., Li Z., Li Y., Wang X., Gurbuxani S., Chen P., He C., You D., Zhang S., et al. 2012. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 22:524–535. 10.1016/j.ccr.2012.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.S., Erkeland S.J., Pester R.E., Chen C.Y., Ebert M.S., Sharp P.A., and Jacks T.. 2008. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA. 105:3903–3908. 10.1073/pnas.0712321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K., Muselman A., Du R., Harada Y., Scholl A.G., Yan M., Matsuura S., Weng S., Harada H., and Zhang D.E.. 2014. Hmga2 is a direct target gene of RUNX1 and regulates expansion of myeloid progenitors in mice. Blood. 124:2203–2212. 10.1182/blood-2014-02-554543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.T., de Vasconcellos J.F., Yuan J., Byrnes C., Noh S.J., Meier E.R., Kim K.S., Rabel A., Kaushal M., Muljo S.A., and Miller J.L.. 2013. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood. 122:1034–1041. 10.1182/blood-2012-12-472308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17:10–12. http://dx.doi.org/ 10.14806/ej.17.1.200 [DOI]
- Mayr C., Hemann M.T., and Bartel D.P.. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 315:1576–1579. 10.1126/science.1137999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney-Freeman S., Cahan P., Li H., Lacadie S.A., Huang H.T., Curran M., Loewer S., Naveiras O., Kathrein K.L., Konantz M., et al. 2012. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 11:701–714. 10.1016/j.stem.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki-Kashio M., Mishima Y., Miyagi S., Negishi M., Saraya A., Konuma T., Shinga J., Koseki H., and Iwama A.. 2011. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 118:6553–6561. 10.1182/blood-2011-03-340554 [DOI] [PubMed] [Google Scholar]
- Moss E.G., Lee R.C., and Ambros V.. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 88:637–646. 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- Nguyen L.H., Robinton D.A., Seligson M.T., Wu L., Li L., Rakheja D., Comerford S.A., Ramezani S., Sun X., Parikh M.S., et al. 2014. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell. 26:248–261. 10.1016/j.ccr.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K., and Morrison S.J.. 2008. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 135:227–239. 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh S.J., Miller S.H., Lee Y.T., Goh S.H., Marincola F.M., Stroncek D.F., Reed C., Wang E., and Miller J.L.. 2009. Let-7 microRNAs are developmentally regulated in circulating human erythroid cells. J. Transl. Med. 7:98 10.1186/1479-5876-7-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., Zandi S., Takayama N., Dobson S., Gan O.I., Wilson G., Kaufmann K.B., McLeod J., Laurenti E., Dunant C.F., et al. 2016. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 351:aab2116 10.1126/science.aab2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Ding L., and Morrison S.J.. 2013. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 13:102–116. 10.1016/j.stem.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., and Zon L.I.. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644. 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A., et al. 2015. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 163:1663–1677. 10.1016/j.cell.2015.11.013 [DOI] [PubMed] [Google Scholar]
- Paulson R.F., Shi L., and Wu D.C.. 2011. Stress erythropoiesis: new signals and new stress progenitor cells. Curr. Opin. Hematol. 18:139–145. 10.1097/MOH.0b013e32834521c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi A., Careccia S., Lulli V., Romania P., Marziali G., Testa U., Lavorgna S., Lo-Coco F., Petti M.C., Calabretta B., et al. 2013. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 32:3648–3654. 10.1038/onc.2012.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., and Harel-Bellan A.. 2007. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21:1125–1138. 10.1101/gad.415007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., and Bryder D.. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442. 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Rao S., Lee S.Y., Gutierrez A., Perrigoue J., Thapa R.J., Tu Z., Jeffers J.R., Rhodes M., Anderson S., Oravecz T., et al. 2012. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 120:3764–3773. 10.1182/blood-2012-03-415349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel V.I., Miller C.L., Eaves C.J., and Lansdorp P.M.. 1996. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood. 87:3500–3507. [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611. 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Ueno H., Sonoda K., Hisatomi T., Shimizu K., Ohashi H., and Inomata H.. 2000. Blockade of TGF-β by in vivo gene transfer of a soluble TGF-β type II receptor in the muscle inhibits corneal opacification, edema and angiogenesis. Gene Ther. 7:1915–1924. 10.1038/sj.gt.3301320 [DOI] [PubMed] [Google Scholar]
- Sankaran V.G., Xu J., and Orkin S.H.. 2010. Advances in the understanding of haemoglobin switching. Br. J. Haematol. 149:181–194. 10.1111/j.1365-2141.2010.08105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda G., Shyh-Chang N., Soysa T.Y., Zhu H., Seligson M.T., Shah S.P., Abo-Sido N., Yabuuchi A., Hagan J.P., Gregory R.I., et al. 2013. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells. 31:1563–1573. 10.1002/stem.1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., and Daley G.Q.. 2013. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 12:395–406. 10.1016/j.stem.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M.T., Tsanov K.M., Nguyen L., Asara J.M., Cantley L.C., and Daley G.Q.. 2013. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 155:778–792. 10.1016/j.cell.2013.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., and Lodish H.F.. 2001. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood. 98:3261–3273. 10.1182/blood.V98.12.3261 [DOI] [PubMed] [Google Scholar]
- Takizawa H., Boettcher S., and Manz M.G.. 2012. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 119:2991–3002. 10.1182/blood-2011-12-380113 [DOI] [PubMed] [Google Scholar]
- Traver D., Miyamoto T., Christensen J., Iwasaki-Arai J., Akashi K., and Weissman I.L.. 2001. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood. 98:627–635. 10.1182/blood.V98.3.627 [DOI] [PubMed] [Google Scholar]
- Urbach A., Yermalovich A., Zhang J., Spina C.S., Zhu H., Perez-Atayde A.R., Shukrun R., Charlton J., Sebire N., Mifsud W., et al. 2014. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 28:971–982. 10.1101/gad.237149.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S.R., and Daley G.Q.. 2010. Lin28: A microRNA regulator with a macro role. Cell. 140:445–449. 10.1016/j.cell.2010.02.007 [DOI] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., and Gregory R.I.. 2008. Selective blockade of microRNA processing by Lin28. Science. 320:97–100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S.R., Powers J.T., Einhorn W., Hoshida Y., Ng T.L., Toffanin S., O’Sullivan M., Lu J., Phillips L.A., Lockhart V.L., et al. 2009. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41:843–848. 10.1038/ng.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.D., Rao T.N., Rowe R.G., Nguyen P.T., Sullivan J.L., Pearson D.S., Doulatov S., Wu L., Lindsley R.C., Zhu H., et al. 2015. The role of Lin28b in myeloid and mast cell differentiation and mast cell malignancy. Leukemia. 29:1320–1330. 10.1038/leu.2015.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Viswanathan S.R., Yabuuchi A., Cunniff K., Takeuchi A., Park I.H., Sero J.E., Zhu H., Perez-Atayde A., Frazier A.L., et al. 2009. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 460:909–913. 10.1038/nature08210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K., et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 154:583–595. 10.1016/j.cell.2013.06.052 [DOI] [PubMed] [Google Scholar]
- Wong J.J., Ritchie W., Gao D., Lau K.A., Gonzalez M., Choudhary A., Taft R.J., Rasko J.E., and Holst J.. 2014. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J. Hematol. Oncol. 7:42 10.1186/1756-8722-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Nguyen L.H., Zhou K., de Soysa T.Y., Li L., Miller J.B., Tian J., Locker J., Zhang S., Shinoda G., et al. 2015. Precise let-7 expression levels balance organ regeneration against tumor suppression. eLife. 4:e09431 10.7554/eLife.09431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Morrissey C.S., Keller C.A., Mishra T., Pimkin M., Blobel G.A., Weiss M.J., and Hardison R.C.. 2014. Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 24:1945–1962. 10.1101/gr.164830.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Wu D.C., Chen Y., and Paulson R.F.. 2015. In vitro culture of stress erythroid progenitors identifies distinct progenitor populations and analogous human progenitors. Blood. 125:1803–1812. 10.1182/blood-2014-07-591453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Xu J., Hsu J.H., Nguyen M., Fujiwara Y., Peng C., and Orkin S.H.. 2014. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 14:68–80. 10.1016/j.stem.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Yang S.L., Herrlinger S., Liang C., Dzieciatkowska M., Hansen K.C., Desai R., Nagy A., Niswander L., Moss E.G., and Chen J.F.. 2015. Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development. 142:1616–1627. 10.1242/dev.120543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318:1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Yuan J., Nguyen C.K., Liu X., Kanellopoulou C., and Muljo S.A.. 2012. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 335:1195–1200. 10.1126/science.1216557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Li Y.S., Bandi S.R., Tang L., Shinton S.A., Hayakawa K., and Hardy R.R.. 2015. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J. Exp. Med. 212:569–580. 10.1084/jem.20141510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shyh-Chang N., Segrè A.V., Shinoda G., Shah S.P., Einhorn W.S., Takeuchi A., Engreitz J.M., Hagan J.P., Kharas M.G., et al. MAGIC Investigators . 2011. The Lin28/let-7 axis regulates glucose metabolism. Cell. 147:81–94. 10.1016/j.cell.2011.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.