Abstract
According to current models for hematopoiesis, lymphoid-primed multi-potent progenitors (LMPPs; Lin−Sca-1+c-Kit+CD34+Flt3hi) and common myeloid progenitors (CMPs; Lin−Sca-1+c-Kit+CD34+CD41hi) establish an early branch point for separate lineage commitment pathways from hematopoietic stem cells, with the notable exception that both pathways are proposed to generate all myeloid innate immune cell types through the same myeloid-restricted pre-granulocyte-macrophage progenitor (pre-GM; Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD150−CD105−). By single cell transcriptome profiling of pre-GMs we identify distinct myeloid differentiation pathways: a Gata1-expressing pathway generates mast cells, eosinophils, megakaryocytes and erythroid cells, and a Gata1-negative pathway that generates monocytes, neutrophils and lymphocytes. These results identify an early hematopoietic lineage bifurcation, separating the myeloid lineages prior to their segregation from other hematopoietic lineage potentials.
The cell types of the myeloid innate immune system, neutrophils, macrophages, eosinophils, mast cells and basophils, each make a unique contribution to immunity against bacteria and parasites1. All of these cell types are generated from multi-potent hematopoietic stem cells (HSCs) in a process controlled by a complex network of transcription factors2 and extrinsic signals3. Knowledge of the cellular pathways by which hematopoietic cell types are generated, and how their production is regulated by infectious organisms and cytokines, is critical to understanding how the immune system functions, and how its dysfunction results in immune cell deficiency or overproduction under pathological conditions4, 5. Notably, current models for hematopoiesis commonly imply that all myeloid cell types develop from the same granulocyte-macrophage progenitor populations (pre-GM and GMP (defined as Lin−Sca-1−c-Kit+CD41−FcγRII/III+))2, 6, 7, 8, 9, which according to the CMP-LMPP hematopoietic model are produced by both the CMP (which is able to generate myeloid cells, megakaryocytes and erythroid cells, but lacks lymphoid lineage potentials10, 11) and LMPP branch (which can generate myeloid and lymphoid cell types, but, lacks megakaryocyte-erythroid lineage potentials12). Previous studies have shown that GMPs possess combined monocyte-macrophage and neutrophil potential at the clonal level6, 10. However, granulocytes also include functionally distinct mast cells, basophils and eosinophils, each specified through the action of critical transcription factors13, and the presence of these potentials in GMPs has so far not been systematically analyzed. It therefore remains an open question if the LMPP and CMP pathways generate the same complement of myeloid cell types.
We here use global gene profiling of single pre-GMs to identify molecularly distinct pre-GM subsets defined by differential expression of Gata1 and Flt3, encoding the GATA-1 transcription factor and the Fms-like tyrosine kinase like 3 receptor, respectively. Gata1 is of particular interest, as, in addition to its critical role in megakaryocyte and erythrocyte development14, 15, GATA-1 is expressed in eosinophils, basophils and mast cells (but not monocytes-macrophages and neutrophils), and is important for their differentiation16, 17, 18. By generating Gata1-EGFP reporter mice we were able to fractionate myeloid progenitors based on their Gata1 expression, and identify sub-populations that harbored distinct myeloid lineage potentials: Gata1-EGFP+ (henceforth GE+) pre-GMs and GE+ GMPs gave rise to mast cells and eosinophils, but lacked monocyte-macrophage and had little or no neutrophil potential. Conversely, Gata1-EGFP− (GE−) pre-GMs and GE− GMPs (as well as LMPPs) generated monocytes and neutrophils, but no mast cells and few eosinophils. Notably, monocyte-neutrophil and mast cell-eosinophil potentials segregated prior to separation from other blood lineages, as GE+ pre-GMs generated megakaryocytes and erythroid cells, whereas LMPPs contained robust B- and T-lineage potential.
Our findings therefore identify an early blood lineage fate decision segregating lymphoid-macrophage-neutrophil potential from mast cell-eosinophil-megakaryocytic-erythroid potential, establishing separate myeloid-erythroid and myelo-lymphoid differentiation pathways, and generating two distinct myeloid-restricted progenitor subsets, instead of a common GMP harboring all monocyte-macrophage and granulocyte potentials.
Results
Single cell RNA sequencing identifies pre-GM heterogeneity
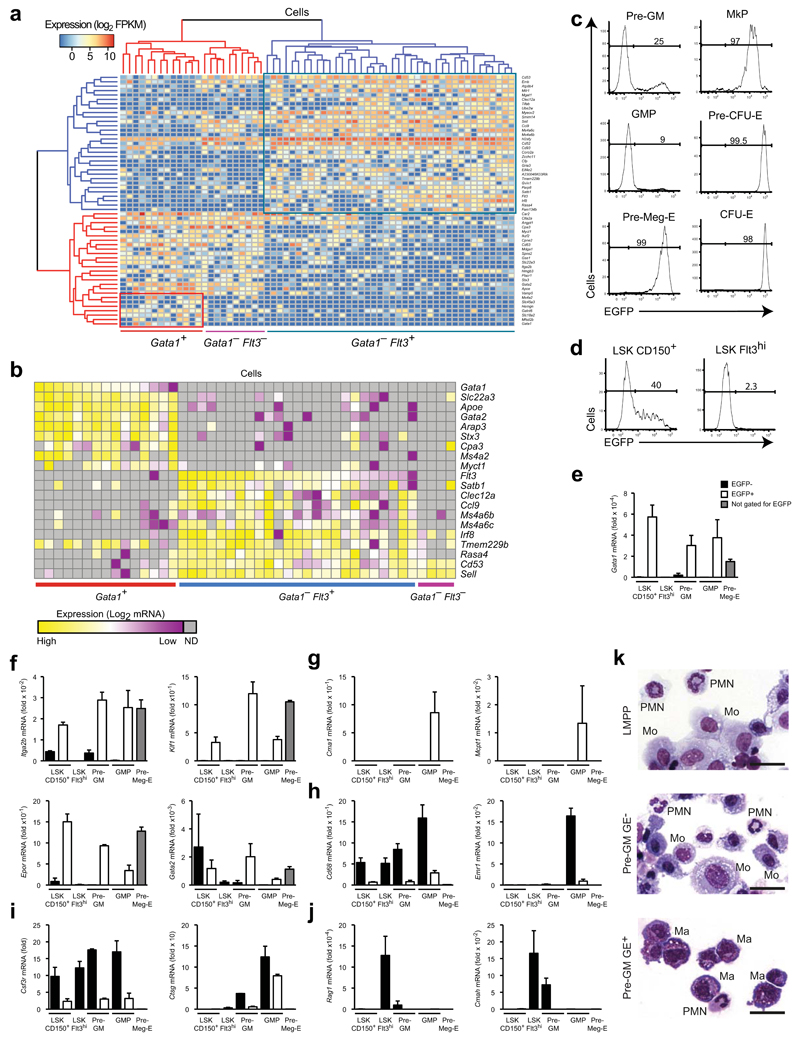
To address if the same myeloid differentiation pathways emerge from CMPs and LMPPs we performed global single cell gene profiling of bone marrow (BM) pre-GMs6, the earliest progenitor predicted to be shared between the two pathways6, 7, 8. RNA sequencing of 63 single pre-GMs was followed by unsupervised clustering using the 100 genes with greatest variance across the entire cell population. This led to the identification of two distinct pre-GM clusters (Supplementary Fig. 1a), and 55 genes showing differential expression between them (P<0.05; FDR<0.05; Supplementary Fig. 1b) as potential sub-population classifiers. Clustering using this gene set identified three distinct pre-GM subpopulations (Gata1+Flt3−, Gata1−Flt3− and Gata1−Flt3+) defined by expression of Gata1 and Flt3 (Fig. 1a), a pattern that was validated by targeted single cell gene expression analysis (Fig. 1b). This analysis also identified Gata1 expression as an optimal classifier, homogeneously and selectively expressed in a distinct subpopulation of Flt3− preGMs, but not detected in Flt3+ preGMs. Notably, Gata1 and Flt3 expression have been used to define CMPs11 and LMPPs12, respectively, within the CD34+LSK population, suggesting they have the potential to identify pre-GM subsets derived from these distinct upstream progenitors.
Figure 1. Gata1 expression identifies distinct myeloid progenitor subsets.
(a) Hierarchical clustering of single pre-GM cells using the gene set from Supplementary Figure 1b. The major molecularly distinct cell clusters are labeled according to Gata1 and Flt3 expression. (b) Heatmap showing expression of genes selectively expressed between the 3 clusters from (a) in single pre-GMs. Expression values were normalized to Hprt expression and shown as deviation from mean expression value of each individual gene. Cells were grouped according to descending Gata1 and Flt3 expression. (c) EGFP expression in pre-GM, GMP, pre-Meg-E, MkP, pre-CFU-E and CFU-E populations from Gata1-EGFPtg/+ adult bone marrow. The percentage of EGFP positive cells is indicated. Gates were set based on wild type cells. Gating strategy for the populations is depicted in Supplementary Figure 1d. (d) EGFP expression in LSKCD150+Flt3− and LSKFlt3hi cells. Gating strategy for populations is depicted in Supplementary Figure 1e. (e-j) Quantitative PCR analysis of mRNA expression relative to Hprt of Gata1 (e), genes associated with megakaryocytes/erythrocytes (f), mast cells (g), monocytes-macrophages (h), neutrophils (i) and lymphoid cells (j) in purified stem- and progenitor populations. (k) Representative morphology of cells from day 8 of cultures of LMPPs, pre-GMs GE- and pre-GMs GE+. Monocytes (Mo), polymorphonuclear granulocytes (PMN), and mast cells (Ma) are indicated. Scale bars: 25μm. Analysis representative of 20 (c) and 5 (d) independent experiments is shown. Gene expression is mean ± s.e.m, n=3 biological replicates, except n=2 for pre-GM EGFP- (e-j).
Gata1 expression defines distinct myeloid progenitors
The Gata1-EGFP transgene used to define CMPs11 lacks key regulatory elements19, 20, 21, and is not expressed in GMPs22. We therefore generated the Gata1-EGFP reporter mouse line using a BAC transgenic construct containing all known Gata1 regulatory sequences, in which an enhanced green fluorescence protein (EGFP) expression cassette replaced the coding part of the second exon of the Gata1 gene (Supplementary Fig. 1c). In these Gata1-EGFP reporter mice distinct GE+ and GE− subsets were observed in pre-GMs and GMPs (Fig. 1c; Supplementary Fig. 1d). Within the BM multi-potent Lin−Sca-1+c-Kit+ (LSK) compartment, HSCs are CD150+LSK23, whereas LMPPs are Flt3hiLSK12. CD150+LSK cells contained a significant fraction of GE+ cells (Fig. 1e; Supplementary Fig. 1e), but only GE− CD150+LSK cells possessed HSC function in vivo (Supplementary Fig. 2a-d). HSCs are therefore defined herein as CD150+GE−LSKs. Similarly, a small fraction (2-3%) of LMPPs had low Gata1-EGFP expression (Fig. 1d). Previous studies showed that approximately 2% of LMPPs possess detectable Mk potential12, and we found this Mk potential to be confined to the small GE+ sub-fraction of Flt3hiLSK cells (Supplementary Fig. 2e), supporting the existence of an LMPP population devoid of platelet-forming capacity12, 24, 25, and leading us to stringently define and to sort LMPPs as Flt3hiGE−LSK in this study. Importantly, Gata1-EGFP expression mirrored endogenous Gata1 mRNA expression both at the population level (Fig. 1e) and in analysis of single pre-GMs (Supplementary Fig. 2f). The Gata1-EGFP reporter therefore identifies transcriptional heterogeneity within the phenotypic HSC, LMPP, preGM and GMP populations.
Early separation of macrophage and mast cell potentials
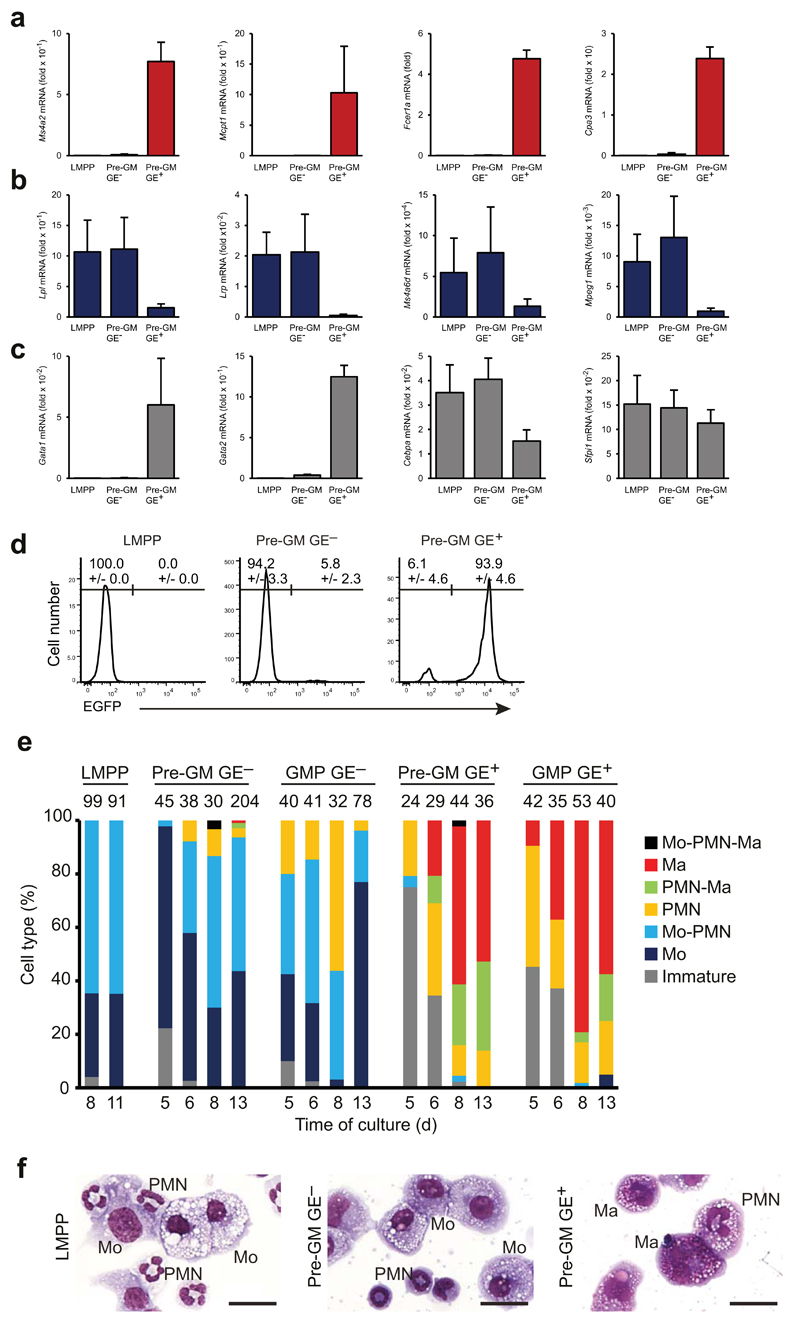
Quantitative PCR of lineage-specific gene expression showed that megakaryocyte-erythroid–affiliated genes (Itga2b, Klf1, Epor, Gata2) were expressed preferentially in GE+ preGM and GMPs, preMegE (Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD150+CD105−), MkP (Lin−Sca-1−c-Kit+CD41+) and preCFU-E (Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD150+CD105+) progenitor cell populations (Fig. 1f), as were mast cell genes (Cma1, Mcpt1; Fig. 1g), whereas macrophage (Cd68, Emr1; Fig. 1h), neutrophil (Csf3r, Ctsg; Fig 1i) and lymphoid genes (Rag1, Cmah; Fig. 1j) were selectively expressed in LMPPs, GE− pre-GMs and GE− GMPs. The divergent expression of macrophage-neutrophil and mast cell genes between GE+ pre-GMs and GMPs and GE− pre-GMs and GMPs, suggested that these progenitors could generate different myeloid cell types. We therefore cultured GE+ pre-GMs and GE− pre-GMs under pan-myeloid conditions (IL-3, IL-5, IL-9, SCF and GM-CSF) for 8 days, and examined the resulting cultures by cytospin morphology and gene expression analysis. Morphological analysis showed that LMPPs and GE− pre-GMs generated monocytes, but no mast cells, whereas GE+ pre-GMs generated mast cells, but no monocytes (Fig. 1k). Polymorphonuclear (PMN) cells were observed in both cultures, with neutrophil morphology (small cell size, highly condensed and segmented nucleus) evident in those derived from GE− preGMs and LMPPs, whereas those derived from GE+ pre-GMs were generally larger with less condensed nuclei (Fig. 1k). Consistent with the observed separation of mast cell and monocyte-macrophage potential, mast cell-specific genes were exclusively expressed in the GE+ pre-GM-derived cultures (Fig 2a), and monocyte-macrophage–specific genes selectively in those derived from LMPPs and GE− pre-GMs (Fig. 2b). Finally, while the genes encoding the myeloid PU.1 and C/EBPα transcription factors were expressed in all cultures, Gata1 and Gata2 expression was confined to cultures derived from GE+ pre-GMs (Fig. 2c,d).
Figure 2. GE- and GE+ progenitor cells have distinct myeloid lineage potentials.
(a-c) Total cell populations produced from LMPPs, pre-GM GE- and pre-GM GE+ progenitors after 8 days of culture were analyzed by quantitative-PCR for expression of genes associated with mast cells (a), monocytes-macrophages (b) and for myeloid-erythroid transcription factors (c). Gene expression is presented relative to Ubc expression. (d) LMPP, pre-GM GE- and pre-GM GE+ populations were cultured for 3 days with mSCF and mIL-3, allowing the cells to reach a GMP stage, as defined by FcγRII/III expression. Histograms show EGFP expression of in vitro derived FcγRII/III+ cells from indicated progenitor populations. Percentages of EGFP negative and positive cells are indicated. (e) Cell type analysis by morphology of cytospins from single cell cultured LMPPs, GE- and GE+ pre-GMs and GMPs, shown as frequency of analyzed clones (numbers on top of bars). Time of culture is indicated in days. Clones were scored as immature only if no differentiated cell type was found. Mo: monocyte, PMN: Polymorphonuclear granulocytes, Ma: mast cell. (f) Morphology of representative mixed lineage clones from cultured single progenitors. Monocytes (Mo), polymorphonuclear granulocytes (PMN), and mast cells (Ma) are indicated. Scale bars: 25μm. Gene expression is mean ±SD, n=6 biological replicates (a,b,c). EGFP expression is mean ±SD, n=2 and n=7 biological replicates for LMPP and pre-GMs respectively (d). Data from (e) is representative for 6 independent experiments.
To determine the frequencies and distribution of granulocyte and monocyte-macrophage lineage potentials within the different progenitor populations at the single cell level we individually cultured sorted progenitor cells and analyzed their myeloid lineage output at several time points. While single LMPPs, GE− pre-GM and GE− GMPs produced large numbers of monocytes, mast cell potential was never detected in cultures of single LMPPs or GE− GMPs, and was very rare (<2% of cultures) in GE− pre-GMs single cell-derived cultures, at all time points investigated. In contrast, monocytes were generated only very rarely (<2% of single cell cultures), whereas mast cell potential was highly abundant, from GE+ pre-GM or GE+ GMPs (Fig. 2e), Combined monocyte and mast cell morphology was exceptionally rare, seen only in 2 of >1000 single cell-derived clones of all progenitor analyzed. Importantly, this was not due to any inability of the culture system to support development of these two cell types simultaneously, as culture of multi-potent HSCs or co-culture of GE+ and GE− pre-GMs generated combined mast cells and monocytes with high frequency (Supplementary Fig. 2g,h). Both monocytes and mast cells were observed in conjunction with other granulocytes with high frequency in single cell-derived clones from GE+ pre-GM cells (22-33%), GE− pre-GM (ca. 50%) and LMPPs (65%), and also in GE+ GMPs and GE− GMPs at lower frequencies (Fig. 2e). The polymorphonuclear cells associated with monocytes showed neutrophil morphology, whereas those associated with mast cells appeared larger with less condensed nuclei (Fig. 2f). These results show that GE+ and GE− myeloid progenitors have distinct lineage potentials.
Eosinophil potential is found in GE+ preGMs and GE+ GMPs
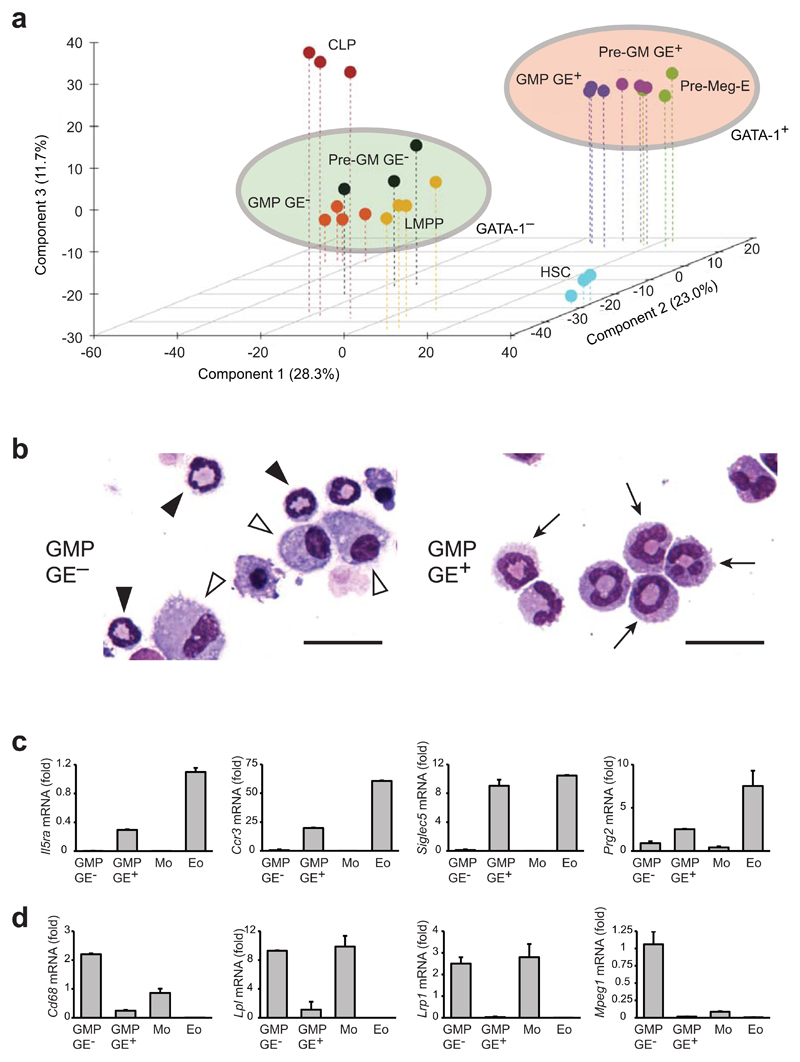
While the above data clearly showed that mast cell and monocyte-macrophage potentials were separated prior to the formation of pre-GMs and GMPs, the nature of the additional granulocyte lineage potentials associated with these progenitors remained unclear. To address this issue we performed Affymetrix-based global gene profiling of pre-GMs, GMPs, HSCs, LMPPs, CLPs (Lin−B220−Sca-1loc-KitloFlt3+IL-7Rα+)26 and preMegEs. Consistent with their similar granulocyte-monocyte lineage readouts, LMPPs, GE− pre-GMs and GE− GMPs clustered together in principal component analysis, in close association with CLPs, whereas GE+ pre-GMs and GE+ GMPs formed a separate cluster, associated with preMegEs (Fig. 3a). Direct comparison of GE+ GMPs and GE− GMP transcriptomes showed that GE+ GMPs were highly enriched for mast cell and eosinophil-specific gene expression, whereas numerous monocyte-macrophage and neutrophil-associated genes were more highly expressed in GE− GMPs (Supplementary Table 1). We therefore examined the possibility that the polymorphonuclear cells derived from GE+ pre-GMs and GE+ GMPs were eosinophils. First, we cultured GE+ GMPs and GE− GMPs in the presence of SCF and IL-5 (Supplementary Fig. 3a), and examined the resulting cultures by morphology and gene expression. Under these conditions GE+ GMPs did not generate mast cells, but only large polymorphonuclear cells with uncondensed nuclear morphology, while GE− GMPs generated cells with monocyte and neutrophil morphology (Fig. 3b). Gene expression analysis indicated the selective expression of eosinophil-specific genes (Il5ra, Ccr3, Siglec5, Prg2) in GE+ GMP cultures (Fig. 3c), and of monocyte-macrophage-specific genes (Cd68, Lpl, Lrp1, Mpeg1) in GE− GMP cultures (Fig. 3d). In both cases the cell-type specificity of gene expression was validated by analysis of macrophages and eosinophils sorted from the peritoneal cavity as controls. Cytospins indicated that the morphology of the in vitro GE+ GMP-derived eosinophil-like cells was similar to that of purified peritoneal eosinophils (Supplementary Fig. 3b). These results indicated the selective development of eosinophils from GE+ GMPs.
Figure 3. Identification of lineage potentials associated with the Gata1-expressing and non-expressing myeloid pathways.
(a) Principal component analysis of hematopoietic stem/progenitor cell populations. Gene expression profiles were generated for each cell population using Affymetrix MoGene 1.0 arrays. Genes differentially expressed across the entire experiment (ANOVA P<0.05; CV>0.4; 2.305 genes) were used to calculate the components, and individual expression profiles were plotted against the first 3 components. Clusters containing GATA-1 non-expressing (LMPP, pre-GM GE-, GMP GE-) and GATA-1-expressing progenitors (pre-Meg-E, pre-GM GE+, GMP GE+) are highlighted. (b) Morphology of cultures derived from GMP GE- (left panel) and GMP GE+ (right panel) under eosinophil favoring culture conditions as depicted in Supplementary Figure 3a. Images show typical neutrophil morphology (black arrowheads), monocytes (white arrowheads) and polymorphonuclear cells (black arrows). Scale bars: 25μm. (c, d) Expression of eosinophil genes (c) and monocyte-macrophage genes (d) in GMP cultures, compared to monocytes (Mo) and eosinophils (Eo) purified from peritoneal cavity. Results are presented relative to Hprt expression (mean ±SD) (n=2 biological replicates, each containing 4 technical replicates). For (a) 4 individual gene expression profiles were made for GMP GE- and LMPP, and 3 for other populations.
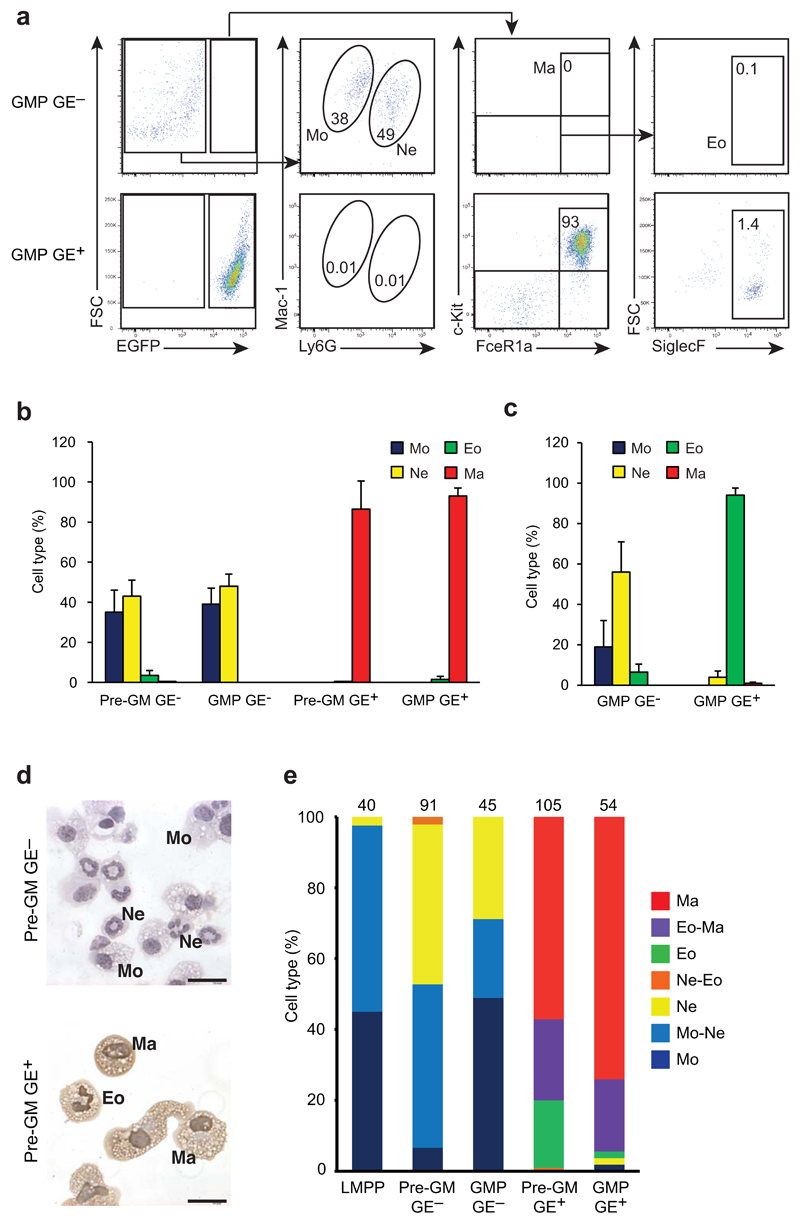
We also performed flow cytometric analysis of the cells differentiated from GE+ pre-GMs and GE− pre-GMs, as well as GE+ GMPs and GE− GMPs following culture under pan-myeloid cytokine conditions. Cells with a FcεRIα+c-Kit+GE+ mast cell surface phenotype were abundant in GE+ pre-GM and GE+ GMP cultures, whereas Mac-1+Ly6GloGE− monocytes and Mac-1+Ly6GhiGE− neutrophils were the principal cell types generated from GE− pre-GMs and GE− GMPs (Fig. 4a,b), indicating that neutrophil potential was selectively associated with GE− pre-GMs and GE− GMPs. Eosinophil, neutrophil and mast cell identities were further confirmed by analysis of the expression of lineage-specific genes (Supplementary Fig. 3c-f). FcεRIα−c-Kit−SiglecF+GE+ eosinophils were inefficiently generated under the pan-myeloid conditions (Fig. 4a,b), likely because of overgrowth by mast cells. To circumvent this, we cultured GMP in the presence of IL-5 and SCF, and observed the selective development of eosinophil-like cells in GE+ GMP cultures, while GE− GMPs gave rise to monocytes and neutrophils, but few or no eosinophils, even under conditions favoring eosinophil development (Fig. 4c). As such, based on analyses of morphology, surface markers and gene expression we concluded that GE+ GMPs gave rise to eosinophils and mast cells, but did not produce detectable neutrophils or monocytes. Finally, to accurately co-localize lineage potentials in single progenitors we combined morphological analysis of cytospins of single cell-derived cultures with immunocytochemistry against EGFP to distinguish GE+ eosinophils from GE− neutrophils (Fig. 4d). GE+ pre-GMs and GE+ GMPs contained a high proportion of bi-potent mast cell-eosinophil progenitors, whereas GE− LMPPs, GE− pre-GMs and GE− GMPs were abundant in bi-potent neutrophil-monocyte progenitors (Fig. 4e). In addition, under pan-myeloid conditions LMPPs, GE+ pre-GMs and GE− pre-GMs we observed the emergence of FcγRII/III+GE+ cells from GE+ pre-GMs and FcγRII/III+GE− cells from GE− pre-GMs and LMPPs in both cases displaying the clonogenic lineage potentials of the corresponding GE+ and GE− GMP population (Supplementary Fig. 4a,b), further supporting the existence of independent and functionally distinct GE+ and GE− progenitor hierarchies. The comparison of gene expression profiles of GE+ GMPs and GE− GMPs identified genes encoding cell surface markers that were preferentially expressed in the GE+ subsets, such as Cd55, Il1rl1, or the GE− subsets, such as Ly6c (Supplementary Table 1). While cell surface expression of Il1rl1 was not detectable on pre-GMs or GMPs with available antibodies, CD55 and Ly6C were expressed selectively on GE+ pre-GMs and GMPs, and GE− pre-GMs and GMPs, respectively (Supplementary Fig. 4c). In agreement with this selective expression, they allowed the separation of progenitors with a restricted monocyte-neutrophil potential (Ly6C+ pre-GMs and GMPs) or restricted mast cell-eosinophil potential (CD55+ pre-GMs and GMPs), without the use of the Gata1-EGFP transgene (Supplementary Fig. 4d). Overall, these results indicate that mast cell-eosinophil potential is found in GE+ preGMs and GE+ GMPs, whereas monocyte-neutrophil potential resides in LMPPs, GE− preGMs and GE− GMPs.
Figure 4. Derivation of eosinophils from GMP GE+ and neutrophils from GMP GE-.
(a) FACS analysis of cells produced from GMP GE- and GMP GE+, cultured for 8 days under conditions sustaining all myeloid cell types, identifying monocytes (Mo), neutrophils (Ne), mast cells (Ma) and eosinophils (Eo) with indicated markers. Numbers indicate cells in the gate as percentage of analyzed live single cells. (b) Quantification of identified cell types after 8 day cultures of GMPs and pre-GMs as in (a) shown as percentage of live cells. (c) Quantification of identified cell population as in (a) after culturing GMPs under eosinophil favoring conditions. (d) Immunocytochemistry detecting EGFP expression (brown stain) on cytospins of cultured GE+ pre-GMs. Monocytes (Mo), neutrophils (Ne), mast cells (Ma) and eosinophils (Eo) are shown. Scale bars: 20μm (e) Morphological analysis after staining for EGFP as in (d) of cytospins from single cell cultured LMPPs, and GE- and GE+ pre-GMs and GMPs, cultured for 8 days under pan-myeloid conditions. Cell types are shown as frequency of analyzed clones (numbers on top of bars). FACS quantification show data from n=8 biological replicates and error bars indicate SD (b, c). Data from (e) was accumulated from 3 independent experiments.
Gata1 expression defines two progenitor domains
We further examined the extent to which the identified progenitors possessed other lineage potentials. Gene Set Enrichment Analysis-based27 comparison of lineage programming in − pre-GMs and GE+ pre-GMs showed strong enrichment of Mk-E-associated genes in GE+ pre-GMs, and of lymphoid genes in GE− pre-GMs (Fig. 5a). All the identified pre-GM and GMP populations generated colonies when cultured under myeloid conditions (Fig. 5b). In contrast, megakaryocytic and erythroid colony forming ability was primarily associated with GE+ pre-GMs, which showed a Mk-E potential comparable to that seen in preMegEs (Fig. 5c,d), and contained multi-potent cells with combined granulocyte and Mk-E potential, but no monocyte-macrophage potential (Fig. 5e,f). Conversely, B- and T-lymphoid potential was high in LMPPs, but also significant in GE− pre-GMs, but not GE+ pre-GMs (Fig. 5g; Supplementary Fig. 4e).
Figure 5. Granulocyte-monocyte potentials segregate prior to dissociation from lymphoid and erythro-megakaryocytic lineages.
(a) GSEA-based comparison of GE- and GE+ pre-GM gene expression using CLP (top) and pre-Meg-E (bottom) gene sets. Normalized enrichment score (NES) and P-value are indicated. (b-e) Colony formation evaluated after 8 days of culture of indicated progenitor cells. Type of colonies assayed are (b) myeloid (CFU-GM), (c) erythroid (BFU-E), (d) megakaryocyte (CFU-Mk) and (e) combined myeloid and erythroid (CFU-Mix). (f) Cytospins from single CFU-Mix colonies derived from pre-GM GE+, showing combined erythroid cells (Ery), polymorphonuclear cells (PMN), megakaryocytes (Meg) and mast cells (Ma) but no monocytes. Scale bars: 25μm. (g) Assessment of lymphoid potential of the indicated progenitors. For each population the indicated number of cells/well were plated under B-cell (OP9 stroma) and T-cell (OP9-DL1 stroma) conditions. Bars indicate the percentage of wells producing B-cells (defined as CD19+) (top) or T-cells (defined as Thy1.2+CD25+ or CD4+CD8+) (bottom), respectively. Mean (±s.e.m.) number of positive wells plated with 1, 10 or 50 cells. Data are from 2 experiments each with 24 wells/population and cell density. Number of CFU-GM, BFU-E and CFU-Mk colonies are shown as mean ±s.e.m of 6,7,6,8,4 (b), 8,7,6,6,4 (c) and 7,7,7,6,4 (d) biological replicates for pre-GM GE-, pre-GM GE+, GMP GE-, GMP GE+ and pre-Meg-E resp. from 4 experiments. Numbers of CFU-mix colonies are mean ±SD from 3 biological replicates, each assayed in technical duplicate.
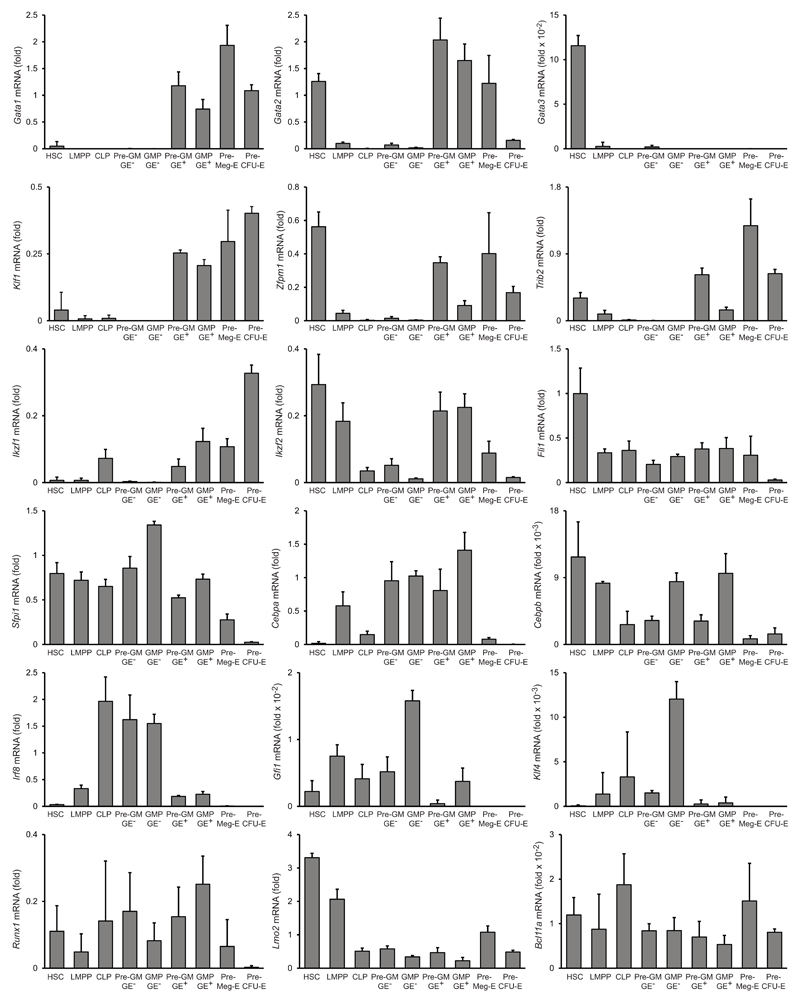
Antagonistic transcription factor pairs, including PU.1–GATA-1 and C/EBP–FOG-1, have been proposed to be co-expressed in multi- or bi-potent progenitors, and resolution of the antagonism to underlie lineage bifurcations28. We therefore measured the expression of these and other key transcription factors in GE+ pre-GMs and GMPs, as well as GE− pre-GMs and GMPs, as well as in HSCs, LMPPs, pre-CFU-Es, pre-MegEs and CLPs, to determine if their expression was consistent with the associated lineage potentials. Most notably, Cebpa, Cebpb and Zfpm1 (encoding FOG-1) were all expressed in GE+ pre-GMs, with Zfpm1 down-regulated and Cebpa and Cebpb up-regulated in GE+ GMPs, The converse change, Zfpm1 upregulation and Cebpa and Cebpb downregulation, was seen in pre-MegEs. The FOG-1 target gene Trib229, which mediates C/EBPα and C/EBPβ degradation30, 31, was up-regulated in preMegEs, consistent with their proposed role in resolving the multi-potency of GE+ pre-GMs29, and with the observed ability of FOG-1 to suppress eosinophil and mast cell lineage differentiation32, 33. Sfpi1 (encoding PU.1) and Gata1 were expressed in GE+ pre-GMs, with Sfpi1 down-regulated in Mk-E-committed pre-MegEs. Gata1 and Sfpi1 were co-expressed in GE+ GMPs, indicating that down-regulation of GATA-1 is not decisive for commitment of GE+ pre-GMs to granulocyte lineages, and consistent with PU.1 being required for mast cell differentiation34. Finally, key transcriptional regulators of monocyte-macrophage (Irf8, Klf4)35 and neutrophil (Gfi1)36 differentiation were highly expressed in LMPP and GE− pre-GM and GMP populations compared to GE+ pre-GMs and GMPs (Fig. 6). These data identify LMPPs and GE+ preGMs as distinct multi-potent progenitors with non-overlapping lineage potentials, the former producing lymphocytes, neutrophils and monocuytes-macrophages, and the latter erythrocytes, platelets, eosinophils and mast cells.
Figure 6. Gene expression in hematopoietic stem/progenitor populations.
Quantitative PCR analysis of mRNAs of indicated genes in hematopoietic stem and progenitor populations as indicated. Data show mRNA expression relative to Hprt. (mean ±SD, n=3 biological replicates for each population, except n=2 for preGM GE-).
Functional heterogeneity of GE− pre-GMs
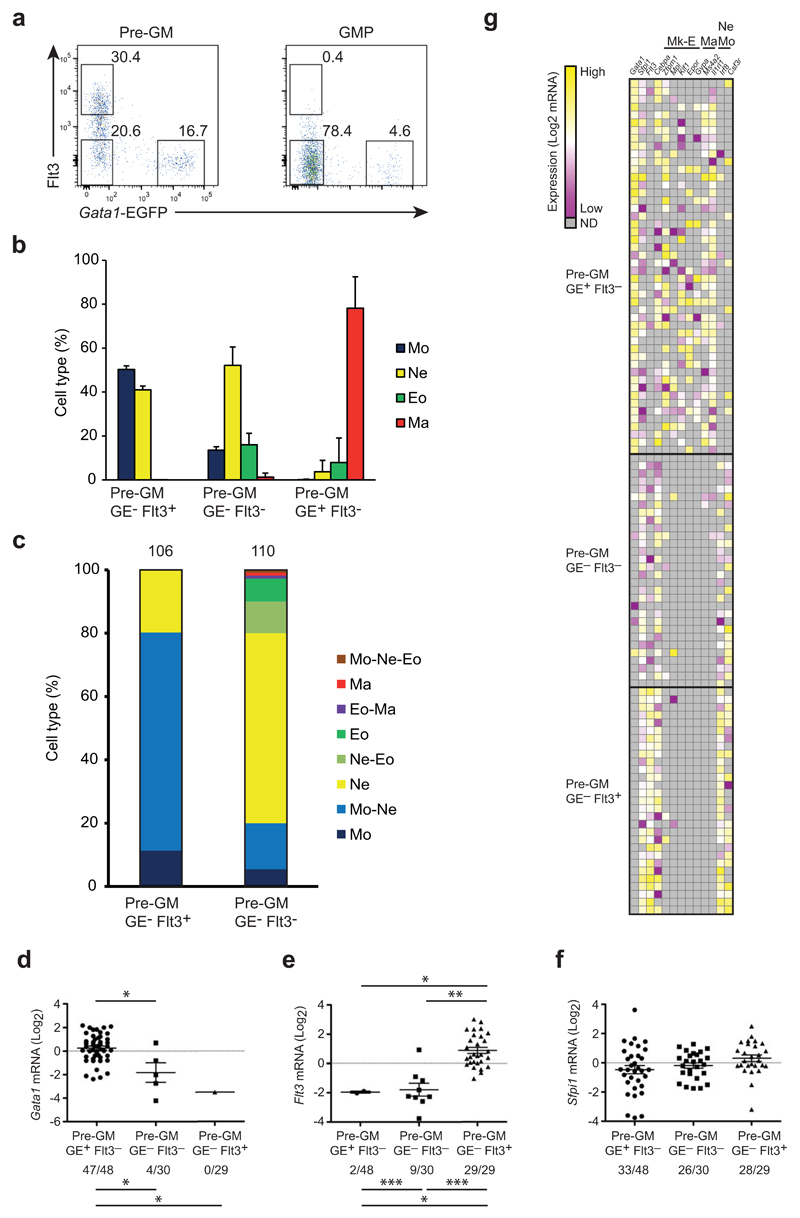
Lymphoid lineage potential has been associated with Flt3+ pre-GM37. By flow cytometry we observed that Flt3+ pre-GMs were exclusively GE− (Fig. 7a). The selective presence of lymphoid potential in Flt3+GE− pre-GMs, but not Flt3−GE− pre-GMs raised the possibility that other lineage potentials also differed between these two populations. Bulk and single cell cultures showed that both Flt3+ and Flt3−GE− pre-GMs generated monocytes and neutrophils (Fig. 7b,c), with the Flt3−GE− pre-GMs perhaps more biased towards neutrophil production. While mast cells did not develop from either progenitor population, a small number of eosinophils developed in the bulk GE−Flt3− pre-GM cultures (Fig. 7b), and 7% of clones derived from single GE−Flt3− pre-GMs showed combined neutrophil and eosinophil morphology (Fig. 7c). These results indicate that a minor, but distinct fraction of GE−Flt3− pre-GM progenitor are restricted to neutrophil and eosinophil differentiation. qRT-PCR profiling of single pre-GMs confirmed expression of Gata1 in GE+ preGMs and of Flt3, with Sfpi1 mRNA expression being similar across the 3 pre-GM subsets (Fig. 7d-f), consistent with their myeloid progenitor identity. Mk-E and mast lineage-specific genes were co-expressed in >90% of single GE+ pre-GMs and antagonistic transcription factor pairs, such as Sfpi1-Gata1 and Cebpa-Zfpm1 were co-expressed with high frequency (32/48 cells and 17/48 cells, respectively) in single GE+ pre-GMs, suggesting the molecular properties of multi-potent progenitors. Co-expression of Sfpi1-Gata1 and Cebpa-Zfpm1 was not observed in GE−Flt3+ or GE−Flt3− pre-GMs, which instead had high expression of monocyte-macrophage and neutrophil genes such as Irf8, Csf3r, consistent with the presence of these lineage potentials in both GE− pre-GM sub-populations (Fig. 7g). In addition, the proportion of Flt3+ pre-GMs was decreased in Flt3 ligand null mice (Supplementary Fig. 5a,b), suggesting that Flt3 signaling regulates the Flt3+ pre-GM subset. In contrast, GE+ pre-GMs and GE+ GMPs were maintained in Gata1fl/Y;Mx1-Cre conditional knockout mice after poly(I-C)–induced deletion (Supplementary Fig. 5c,d). Consistent with previous findings38, Gata1 null GE+ progenitors differentiated into mast cells in vitro (Supplementary Fig. 5e,f), possibly due to redundancy with Gata2 in these progenitors. The gene expression of the identified myeloid progenitors is therefore consistent with their observed lineage potentials.
Figure 7. Flt3 expression subdivides the pre-GM GE- population.
(a) FACS profiles showing Flt3 and EGFP expression in pre-GMs and GMPs from Gata1-EGFPtg/+ BM. Numbers indicate percentage of cells within each gate. (b) 100 cells of indicated pre-GMs were grown for 8 days and cell type was analyzed as in Fig. 4a. and shown as percentage of analyzed cells. (c) Single pre-GMs were grown for 8 days under pan-myeloid conditions and cytospins stained as in Fig. 4d. Cell types are shown as percentage of total analyzed clones, indicated on top of bars. (d-f) Gata1 (d), Flt3 (e) and Spfi1 (f) expression in single cells of indicated progenitors, shown as mRNA (Log2), normalized to Hprt and deviated from the mean. Numbers below graphs indicate cells positive for respective gene expression out of total number of cells. (g) Heatmap showing expression of genes including megakaryocyte/erythrocyte (Mk-E), mast cell (Ma) and neutrophil-monocyte (Ne Mo) related genes in single pre-GMs. Values were normalized to Hprt and shown as deviation from mean of each individual gene. FACS plots are representative for 10 independent experiments (a). Percentage of cell type (b) is mean ±SD from 12 (Pre-GM GE-Flt3+ and Pre-GM GE-Flt3-) and 7 (Pre-GM GE+Flt3-) biological replicates from 2 experiments. Analyzed clones (c) are accumulated from 2 independent experiments. Mann-Whitney U-test was used for expression levels (above graphs), Fisher’s exact test was used to compare expression frequencies between population (below graphs) (*p<0.0001, **p=0.001, ***p=0.002) (d).
In vivo myeloid potentials of GE+ and GE− progenitors
Finally, we investigated in vivo lineage potentials by co-transplanting Gata1+ and Gata1− progenitor populations (CD45.2 and CD45.1/2) intrafemorally into non-irradiated, c-Kit hypomorphic W41/W41 recipients (CD45.1) (Supplementary Fig. 6a). Flow cytometry analysis (after 7 and 10-11 days) indicated that Lin−CD11bloLy6Ghi neutrophils and Lin−CD11bhiLy6G−c-Kit−FcεR1α−SiglecF− monocytes were generated by Gata1− GMPs and GE−Flt3+ pre-GMs, and Lin−CD11bloLy6G−c-Kit−FcεR1α−SiglecF+ eosinophils by GE+ GMPs and GE+ pre-GMs, in both the spleen and bone marrow of reconstituted mice (Fig. 8a and Supplementary Fig. 6b,c). However, overall engraftment levels, in particular of eosinophils, were low, and no Lin−Ly6G−c-Kit+FcεR1α+ mast cells were detected. To overcome this obstacle, we tested the ability of GMP and pre-GM progenitors to engraft after intraperitoneal injection, and found that total engraftment of the peritoneum by both pre-GMs and GMPs was 1-2 orders of magnitude higher compared to that observed in bone marrow and spleen after intrafemoral transplant (Supplementary Fig. 6d), and also mast cell development was readily detected (Fig. 8b). Following intraperitoneal transplantation, GE+ GMPs generated exclusively peritoneal mast cells and eosinophils, whereas only monocytes and neutrophils were derived from GE− GMPs (Fig. 8c). Similarly, GE+ pre-GMs produced predominantly mast cells and eosinophils, whereas GE−Flt3+ pre-GMs and LMPPs generated almost exclusively neutrophils and monocytes (Fig. 8d,e). The cellular phenotypes defined by flow cytometry were further validated by morphology (Supplementary Fig. 6e-h) and selective GE+ expression in eosinophils and mast cells (Supplementary Fig. 6i). Low numbers of neutrophils (but no monocytes) were produced by GE+ pre-GMs, and LMPPs and GE−Flt3+ pre-GMs generated limited numbers of eosinophils (but no mast cells), indicating that the more upstream myeloid progenitors (LMPPs and pre-GMs) retain some lineage plasticity, restricted to the eosinophil and neutrophil lineages in vivo, which is lost in both GE+ GMP and GE− GMPs (Fig. 8c-e). These results confirmed the same lineage potentials in vivo as observed in vitro for both GE+ myeloid progenitors (GE+ preGMs, GE+ GMPs) and GE− progenitors (LMPPs, GE− preGMs, GE− GMPs). We therefore propose to designate GE+ preGMs as candidate erythroid-megakaryocyte-primed multi-potent progenitors (EMkMPPs), and GE+ GMPs as eosinophil-mast cell progenitors (EoMPs). In contrast, since GE− preGMs and GE− GMPs generate predominantly neutrophils and monocytes, we propose to designate them pre-neutrophil-monocyte (preNM) and neutrophil-monocyte progenitors (NMPs), respectively. In different current hematopoietic models all monocyte-macrophage and granuolcyte lineage potentials are proposed to reside in one common GMP2, 8, 9, 10, 12, 39, 40 (Supplementary Fig. 7a,b). Instead our findings implicate the organization of GE+ progenitors (EMkMPP, EoMP) and GE− progenitors (LMPP, preNM and NMP) into largely separate hierarchies that generate lymphoid cells, neutrophils and monocytes-macrophages, and eosinophils, mast cells, platelets and erythroid cells, respectively (Supplementary Fig. 7c,d).
Figure 8. in vivo myeloid reconstitution of GE- and GE+ myeloid progenitor cells.
(a) Representative gating strategy for identification of peritoneal neutrophils (Ne), mast cells (Ma), monocytes (Mo) and eosinophils (Eo). (b) Representative gating strategy for identification of donor derived CD45.1/2 or CD45.2 mast cells (Ma), eosinophils (Eo), neutrophils (Ne) and monocytes (Mo) 7 (GMP) and 11 (pre-GM) days following IP transplantation. Percentages shown are relative to total cells generated from CD45.1/2 and CD45.2 cells, respectively. (c-e) Quantification of in vivo contribution of GE+ and GE- GMPs (c), GE+Flt3- and GE-Flt3+ pre-GMs (d) and LMPPs (e) to peritoneal mast cells, eosinophils, neutrophils and monocytes at indicated days after intraperitoneal transplantation of 1000-4000 cells/donor. Donor derived cells per million analyzed MNCs per 1000 injected cells is shown. * indicates that reconstitution was below detection limit (see methods and Supplementary Fig. 6).
Discussion
We here prospectively isolate and characterize distinct, myeloid progenitor populations restricted to either neutrophil-monocyte (preNM, NMP) or eosinophil-mast cell fate (EoMP), as well as candidate multi-potent progenitors with combined megakaryocyte-erythroid and eosinophil-mast cell potential (EMkMPPs). In addition, we find that LMPPs produce neutrophils and monocytes, but no mast cells and few or no eosinophils.
While existing models imply that segregation of monocyte-macrophage and granulocyte potentials occurs downstream of a common GMP, our results suggest that the separation of Gata1-expressing lineages (megakaryocytes, erythrocytes, eosinophils and mast cells) and Gata1-non-expressing lineages (lymphocytes, neutrophils, monocytes) represents an early lineage bifurcation of the multi-potent HSCs. These findings substantially change our view of how myeloid cell types are specified, by suggesting that mast cells and eosinophils are generated from an EMkMPP progenitor shared with platelets and erythrocytes, rather than a common GMP. This type of lineage relationship has previously been suggested based on observations of myeloid malignancies41. Platelets have important innate immune functions, acting as sentinels and providing chemotaxis of myeloid effector cells42, and the platelet-ancestral thrombocyte is capable of bacterial phagocytosis43. These are all functions shared with mast cells44, consistent with mast cells and platelets having evolved through specialization from a common ancestral cell with both hemostatic and phagocytic function. Our results suggest that eosinophils may have evolved from the same GATA-1–expressing hematopoietic branch, possibly as an adaptation to parasites. In contrast, we find that monocytes-macrophages are developmentally closely related to lymphoid cells, as has already been suggested by the identification of progenitors restricted to macrophage and lymphoid differentiation in both fetal45, 46, 47 and adult hematopoiesis48.
Recent lineage analysis of myeloid colonies from heterogeneous human CD34+ hematopoietic cells are compatible with a similar separation of human GATA-1 expressing granulocyte lineages along with megakaryocytic and erythroid lineage potentials, with the important distinction that also considerable macrophage potential was observed together with the GATA-1 expressing lineages49 This might be related to the investigated human cell populations typically also possessing extensive lymphoid potential of unknown relationship to the investigated granulocyte lineages.
CMPs have been defined as possessing myeloid and megakaryocyte-erythroid lineage potentials, but lacking lymphoid lineage potentials11, 50, and would potentially generate both preNMs-NMPs and MkEMPPs-EoMPs. However, it has not been established if the CMP population as currently defined (Lin−Sca-1+c-Kit+CD34+CD41hi)50 contains cells that possess monocyte-macrophage, as well as all granulocyte potentials at the clonal level, and further studies will therefore be required to firmly establish its relationship to the myeloid progenitor populations identified here. In addition, our studies do not preclude that an upstream progenitor with the same lineage potentials as the EMkMPP exists, for instance within the proposed CMP compartment.
In summary, our findings provide an improved cellular and molecular template for elucidating the evolutionary and developmental relationships of hematopoietic cell types, as well as the identification of cells-of-origin of hematopoietic malignancies, and underscore the power of single cell transcriptome analysis to simultaneously resolve cellular heterogeneity and to identify the molecular determinants selectively and uniformly expressed in novel cell subsets, allowing for their purification and functional characterization.
Online Methods
Mouse lines
The Gata1-EGFP transgene was generated by bacterial artificial chromosome recombinant engineering in bacteria at the EMBL Genome Engineering Core Facility (http://www.embl.it/services/genome-engineering-service/index.html). In this transgene an EGFP-polyA cassette replaced the coding part of the second exon of the mouse Gata1 gene. Gata1-EGFP mice were generated by intracytoplasmic sperm injection51 at the EMBL Monterotondo Transgenic Facility (http://www.embl.it/services/transgenic_facility/index.html). Mice were backcrossed for more than 6 generations onto a C57Bl/6J background. Mice were bred and maintained at the animal facilities of Edinburgh University and Oxford University, UK. Experimental protocols were approved by the Edinburgh University School of Biological Sciences Ethical Review Committee, and the Oxford University Clinical Medicine Ethical Review Committee. Flt3l knockout52, Kit W41/W41 (https://www.jax.org/strain/000119), Gata1 conditional knockout mice53 and Mx1-Cre transgenic mice54 were previously generated. All experiments were performed under projects licenses from the UK Home Office. Mice were sacrificed for harvesting hematopoietic cell populations at an age of 6-17 weeks. Both male and female mice were used.
FACS and flow cytometry
BD LSRFortessa, LSRII , a FACS Aria II, AriaIII and FACSAria Fusion (BD Biosciences) were used for flow cytometry and cell sorting. Cells were prepared for cell sorting by either CD117 enrichment by autoMACS Pro Separator (Miltenyi Biotec)26 or lineage depletion using immunomagnetic beads12. In stainings where anti-FcγRII/III antibody was not included, cells were incubated with Fc-block. FlowJo analysis software (TreeStar) was used for subsequent data analysis. Antibodies used are listed below. Populations were defined as follows: HSCs: Lin−Sca-1+c-Kit+Flt3−CD150+Gata1-EGFP−, LMPP: Lin−Sca-1+c-Kit+Flt3hiGata1-EGFP−, CLP: Lin−B220−Sca-1loc-KitloFlt3+IL-7Rα+, MkP: Lin−Sca-1−c-Kit+CD150+CD41+, pre-GM: Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD105−CD150−, GMP: Lin−Sca1−c-Kit+CD41−CD150−FcγRII/III+; preMegE: Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD150+CD105−, preCFU-E: Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD150+CD105+, CFU-E: Lin−Sca-1−c-Kit+CD41−FcγRII/III−CD150−CD105+. Eosinophils from the peritoneal cavity: Lin(CD4, CD5, CD8, Ter119, B220)− CD11bloLy6G−SiglecF+FcεR1α−F4/80−Gata1-EGFP+. Macrophages from the peritoneal cavity: Lin(CD4, CD5, CD8, Ter119, B220)−CD11b+Ly6G−SiglecF−FcεR1α−F4/80+Gata1-EGFP−. Neutrophils from peripheral blood: Lin(CD4, CD5, CD8, Ter119, B220)−CD11bloLy6G+SiglecF-FcεR1α−F4/80−. See also Transplantation studies.
Transplantation studies
In order to directly compare the in vivo potential of pre-GM and GMP progenitors subfractionated based on Gata1 expression, equal numbers of GE+ and GE− cells were co-transplanted competitively into W41/W41 recipient mice (5-10 weeks of age) on a C57Bl/6 and CD45.1 background. By utilizing Gata1-EGFPtg/+ mice on a CD45.2 or a mixed CD45.1/2 background, the contribution of the two competitor populations to the myeloid lineages could be separated and distinguished from each other endogenous cells based on CD45 allotype, and directly compared in a single recipient. Briefly, GE+ GMPs or GE+Flt3− pre-GMs were sorted from a CD45.2 donor and mixed with equal cell numbers of GE− GMPs or GE−Flt3+ pre-GMs, or vice versa. LMPP cells were isolated from either CD45.1/2 or CD45.2 Gata1-EGFP mice. Purified cells were either injected intraperitoneally (I.P.; 1000-4000 cells from each donor) or intra-femorally (I.F.; 2000-11000 cells from each donor) into W41/W41 CD45.1 mice. For I.P. transplantation, recipients were I.P. injected 12-24 hours before transplantation with 1.5ml of distilled water to deplete endogenous mast cells55.
Contribution to the myeloid lineages was assessed by peritoneal lavage 4-20 days post-injections for I.P. injected mice and by analysis of injected femur and spleen 7 and 10 days post-injection for I.F. injected mice. The contribution of CD45.2 and CD45.1/2 donor cells to mast cells, eosinophils, monocytes and neutrophils was assessed by flow cytometry using the following markers: mast cells: Lin−Ly6G−c-Kit+Fcer1α+; eosinophils: Lin−CD11bloLy6G−c-Kit−Fcer1α−SiglecF+; neutrophils: Lin−Cd11bloLy6Ghi; monocytes: Lin−CD11bhiLy6G−c-Kit−Fcer1α−SiglecF−. Lin: CD4, CD5, CD8a, CD19, Ter119. The contribution of the individual donors (CD45.2 or CD45.1/2) to each lineage was adjusted to the number of cells injected and number of total mononuclear cells analyzed, and expressed as cells per million mononuclear cells per 1000 transplanted cells analyzed. Average values below 2 events per 1 million cells analyzed per 1000 cells injected were not above the non-specific background, and therefore not used to evaluate lineage-contribution of donor-derived cells.
Gata1 knock out studies
Chimeric conditional Gata1cKO mice were established by co-transplanting 0.5x106 Gata1cKO (Gata1fl/yMx1-Cretg/+Gata1-EGFPtg/+, CD45.2) and 0.5x106 wild type bone marrow cells (CD45.1) into lethally irradiated CD45.1/2 recipients. For the non-deletion control, Gata1fl/yMx1-Cre+/+Gata1-EGFPtg/+, CD45.2 were used. After 6 weeks, engraftment was established by analysis of CD45 allotypes in peripheral blood, and the CD45.2 cells contributed between 30-60%. To induce deletion of the Gata1 gene by CRE, mice were treated with 225µg poly(I-C) 8-10 weeks after transplantation by 3 intra-peritoneal injections every other day. Since Gata1 is essential for differentiation to erythroid progenitors and erythrocytes29, Gata1 deletion was confirmed 8-9 weeks after poly(I-C) treatment by absence of CD45.2 CFU-E cells or EGFP signal in erythroid cells.
In vitro cultures
To test myeloid potential, cells were cultured in IMDM with L-glutamine (Invitrogen), 20% heat inactivated fetal calf serum (Gibco), penicillin/streptomycin (Invitrogen) and 10-4M β-mercaptoethanol (Sigma), supplemented with 20ng/ml mSCF (Peprotech), 20ng/ml mIL-3 (Peprotech), 10ng/ml mGM-CSF (Peprotech), 50ng/ml mIL-5 (R&D Systems) and 50ng/ml mIL-9 (R&D Systems) at 37°C, 5% CO2. Bulk cultures of LMPP and pre-GM were started with 100-20.000 cells per culture. Myeloid potential of single cells was tested by sorting single cells directly into Terasaki plates, unless stated differently in the text, each well containing 20μl medium. GMP cells were derived in vitro by culturing progenitors (pre-GM or LMPP) for 3 days with 100ng/ml mSCF and 10ng/ml mIl-3 in round bottom 96 well plates and FcγRII/III+ cells were in this experiment considered as GMPs. In vitro derived GMPs were manually plated at an average of 5 cells/well into 60 well Terasaki plates. For eosinophil favouring culture conditions, GMP cells (200 cells/culture) were grown with 20ng/ml mSCF, 10ng/ml mGM-CSF and 50 ng/ml Il-5 in round bottom 96 well plates. After 5 days, the cells were washed and put back in culture for another 2 days without mSCF and mGM-CSF. All cytospins were prepared with a Shandon cytospin at 1000 RPM with low acceleration, followed by May-Grünwald-Giemsa stain (VWR).
Immunocytochemistry
Air dried cytospins were fixed with 4% PFA in PBS for 5 minutes. Slides were first blocked with PBS/5%BSA containing Fc-block for 30 min at RT and then with 3% H2O2 in water for 5 minutes. Slides were incubated with chicken polyclonal anti-GFP (Abcam, ab13970) in PBS/5% BSA for 4 hours at room temperature and next with peroxidase donkey anti-chicken antibody (Stratech, 703-035-155-JIR) for 1 hour at room temperature. Peroxidase was detected with DAB substrate kit (Vector Laboratories, SK-4100) according to manufacturer’s instructions. Cells were counterstained with Harris Modified Hematoxylin Solution (Sigma-Aldrich, HHS32)) for 10 minutes.
Colony forming assays
For evaluation of erythroid potential from progenitor populations, cells were seeded in Methocult M3436 and myeloid potential was evaluated with Methocult M3534. Colonies were scored after 8 days of culture. CFU-Mix colonies were scored in Methocult M3434 based on colonies containing red cells after 8 days of culture. Megakaryocytic potential was evaluated using the Megacult collagen-based assay. All these media were from StemCell Technologies, Vancouver, Canada. Megakaryocyte cultures were supplemented with 10ng/ml mIL-3, 20ng/ml hIL-6 (Peprotech), 50ng/ml hIL-11 (Peprotech), 50ng/ml hThpo (Peprotech), and megakaryocytes detected using acetylthiocholiniodide staining (Sigma) according to manufacturer’s instructions after 7-8 days culture. For evaluation of Mk potential from single GE+ and GE− Lin−Sca-1+c-Kit+Flt3hi cells, these cells were sorted into X-vivo15 medium containing glutamax and gentamycin (BioWhittaker) supplemented with 10% fetal calf serum (Hyclone), 10-4M β-mercaptoethanol, 50ng/mL mSCF, 50ng/mL hFLT3 ligand (hFL; Immunex), 50ng/mL hThpo, 20ng/mL mIL-3 and 5U/ml hEPO (Roche). For each experiment, 240 cells were manually plated at 1 cell per well into 60 well Terasaki plates. Mk potential was evaluated 8 or 12 days after culture using an inverted microscope.
Measurement of lymphoid lineage potentials
B and T lymphocyte potentials of purified progenitor populations were evaluated following co-culture on OP9 and OP9-DL1 stroma cells, respectively (kindly provided by Dr. A. Cumano, Pasteur Institute, Paris, France). LMPPs, GE+ and GE− pre-GMs, and GE+ and GE− GMPs were seeded onto stromal monolayers. OP9 cultures were supplemented with 25ng/ml mSCF, 25ng/ml hFL (Peprotech) and 10ng/ml hIL-7 (Peprotech), while OP9-DL1 cultures were supplemented with 25ng/ml mSCF and 25ng/ml hFlt3L. SCF was removed after 7 days of culture. Readout was by FACS at 14-21 days of culture. B cells were defined as CD19+ and T cells were defined as CD4+CD8+ and/or Thy1.2+CD25+ (Supplementary Fig. 4e,f).
Quantitative PCR
For gene expression analysis of bulk cultures and cultured cell populations, RNA was purified from 2000-6000 cells per replicate with Trizol (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed with SuperScript VILO cDNA synthesis kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). For single cells and progenitor populations (200 cells per replicate), CellsDirect™ One-Step qRT-PCR kit (Life technologies, 11753-100) was used according to manufacturer’s protocol for preparation and amplification of cDNA. The BioMark 96.96, 48.48 or 192.24 Dynamic Array platform (Fluidigm) and either Taqman® assays (for progenitor populations and single cells) or the Universal probe library system (Roche) (for bulk cultures and cultured cell populations) were used according to the manufacturers’ instructions. UPL primers and probes were chosen by the Assay Design Center web service for mouse genes. Default parameters were used and highest-ranking primers-probe combination was chosen for each gene.
Analysis of quantitative PCR of single cells
ΔCt values relative to Hprt (Ct(Hprt) - Ct(Gene)) were zero centered for each gene by subtraction of the mean value of all positive cells for the gene. These normalized values were used to generate heat maps using GENE-E software.
Microarray gene profiling and bioinformatics analysis
Global gene expression was analyzed in FACS purified HSCs, LMPPs, CLPs, EGFP+ and EGFP− pre-GM, GE+ and GE− GMPs, and pre-MegEs. For all cell populations, 3-4 individually sorted samples from different pools of mice were prepared. For each sample, 5000 cells were sorted directly into RLT buffer (Qiagen) and RNA was extracted according to the manufacturer's instructions (Qiagen). Microarray analysis was performed by the Paterson Institute Microarray Service, Manchester UK, using MoGene 1.0ST arrays. Data were RMA normalized56. Custom R scripts were used to perform principal component analysis (PCA) on 2,305 genes that showed high variation of gene expression across populations (ANOVA with adjusted P values<0.05 and coefficient of variation (CV) equal to or larger than 0.4). GSEA27 was performed using CLP and preMegE gene sets previously described6.
Single cell RNA sequencing; cell capture and preparation of cDNA
5000 pre-GM cells from wild type C57BL/6 male mice (CD45.1) were sorted in 2 μl PBS/50%FCS. Single cells were captured on a small-sized (5-10μm cell diameter) C1™ Single-Cell Auto Prep IFC for mRNA Sequencing (Fluidigm) using the Fluidigm C1 system. Cells were loaded onto the chip at a concentration of ~300 cells/μl and imaged by phase-contrast microscopy to check single cell per capture site. 64 cells were captured and used for RNA-seq. Cells were lysed and cDNA prepared on the C1 fluidigm chip according to manufacturer’s protocol, using SMARTer Ultra Low RNA kit for Illumina (Clontech).
Single cell RNA sequencing; library construction
Single-cell Library size distribution was checked on a High-Sensitivity DNA chip (Agilent Bioanalyzer) (average concentration 239pg/μl, SD 149pg/μl). 1ng of cDNA was used for tagmentation with Nextera XT DNA Sample Preparation kit (Illumina). Purification was done with a 1:1 ratio of AMPure XP beads with a final elution in 25μl of Resuspension Buffer (Nextera). Samples were loaded on a High-Sensitivity DNA chip to check the size and quality of the indexed library (average size 612bp, SD 104), while quantification was done with Qubit High-Sensitivity DNA kit (Invitrogen) (average concentration 347ng/ml SD 95). Libraries were pooled to a final concentration of 10nM and were sequenced with Illumina HiSeq 2000 (single end 50 basepair reads) at The Wellcome Trust Centre for Human Genetics at the University of Oxford.
Single cell RNA sequencing; data analysis and visualization
51 bp reads were aligned to the murine genome (mm9) using STAR57 and default parameters. See (Supplementary Fig. 8) for the information about the number of uniquely mapped reads, percentage of mapping, and the number of detected genes per each single cell). Non-uniquely mapped reads were discarded. Transcript expression values were quantified using Cufflinks58 as fragments per kilobase of transcript per million mapped reads (FPKM). Overlapping transcripts were collapsed giving the highest one expression value per gene loci. One cell was removed from the analysis due to the low detection number of genes (sample81, Supplementary Fig. 8). Custom R scripts were used to perform PCA, hierarchical clustering, and visualization on the log2 of FPKM scale. In brief, PCA analysis was performed on all cells using all genes, which have the average expression values >=1. We selected top 100 genes with the highest absolute correlation coefficient (PCA component loadings) in one of the first three components for the hierarchical clustering analysis. Genes differentially expressed between the two main cellular were identified using ANOVA (P<0.001; FDR<0.05) and used for the final clustering of single cells.
Statistical analysis
Student’s t-test was used to determine statistical significance for normally distributed data. For single cell expression levels the Mann-Whitney U-test was used, and Fisher’s exact test was used to compare expression frequencies at the single cell level between populations. No statistical method was used to predetermine sample size, and experiments were not randomized. No data were excluded from analysis. The Investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
Acknowledgements
We thank the EMBL Monterotondo Gene Expression Service and Transgenic Core Facility for generating the Gata1-EGFP BAC and the corresponding transgenic mouse line, and FACS facilities at the ISCR (supported by Wellcome Trust Equipment Grant WT087371MA (C.N.) and WIMM (supported by MRC Grants MC_UU_12009, MC_UU_12025 and G0902418) for flow cytometry and cell sorting. This work was supported by the Association for International Cancer Research grant 11-0724 (C.N.), a Medical Research Council, UK Program Grant (H4RPLK0) and Unit Grant (MC_UU_12009/5) (S.E.W.J.), an MRC Strategic Grant (G0701761), Program Grant (G0900892) and Unit Grant (MC_UU_12009/7) (C.N.) and by the European Commission FP7 projects CardioCell (to C.N.) and EuroSyStem (S.E.W.J) projects.
Footnotes
Accession codes.
The microarray data used in this paper have been deposited in the NCBI GEO database (http://www.ncbi.nih.gov/geo) with the accession number GSE49241.
Author contributions.
C.N., S.E.W.J., R.D., N.B.V., P.W., E.S. and A.M. designed the experiments, R.D., N.B.V., P.W. A.Ga., A.Z., M.L., A.Gr. and A.Gi. performed the experiments, S.T. performed gene expression analysis, E.M. generated the Gata1-EGFP BAC transgenic mouse line, C.N., S.E.W.J. and R.D. wrote the paper.
Competing financial interest statement.
The authors have no competing financial interests.
Antibodies
Information on antibodies (including clone, concentration and vendor) is provided in Supplementary Table 2
Taqman assays
Taqman assays: Information on these assays is provided in Supplementary Table 3.
References
- 1.Koenderman L, Buurman W, Daha MR. The innate immune response. Immunol Lett. 2014;162(2PB):95–102. doi: 10.1016/j.imlet.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metcalf D. Lineage commitment and maturation in hematopoietic cells: the case for extrinsic regulation. Blood. 1998;92(2):345–347. discussion 352. [PubMed] [Google Scholar]
- 4.Sadrzadeh H, Abdel-Wahab O, Fathi AT. Molecular alterations underlying eosinophilic and mast cell malignancies. Discov Med. 2011;12(67):481–493. [PubMed] [Google Scholar]
- 5.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14(5):302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 6.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1(4):428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Luc S, Buza-Vidas N, Jacobsen SE. Delineating the cellular pathways of hematopoietic lineage commitment. Semin Immunol. 2008;20(4):213–220. doi: 10.1016/j.smim.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Orkin SH, Zon LI. SnapShot: hematopoiesis. Cell. 2008;132(4):712. doi: 10.1016/j.cell.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 11.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1(4):416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20(21):3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93(22):12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. Embo J. 1997;16(13):3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195(11):1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. 2003;197(3):281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nei Y, Obata-Ninomiya K, Tsutsui H, Ishiwata K, Miyasaka M, Matsumoto K, et al. GATA-1 regulates the generation and function of basophils. Proc Natl Acad Sci U S A. 2013;110(46):18620–18625. doi: 10.1073/pnas.1311668110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasinski M, Keller P, Fujiwara Y, Orkin SH, Bessler M. GATA1-Cre mediates Piga gene inactivation in the erythroid/megakaryocytic lineage and leads to circulating red cells with a partial deficiency in glycosyl phosphatidylinositol-linked proteins (paroxysmal nocturnal hemoglobinuria type II cells) Blood. 2001;98(7):2248–2255. doi: 10.1182/blood.v98.7.2248. [DOI] [PubMed] [Google Scholar]
- 20.Drissen R, Guyot B, Zhang L, Atzberger A, Sloane-Stanley J, Wood B, et al. Lineage-specific combinatorial action of enhancers regulates mouse erythroid Gata1 expression. Blood. 2010;115(17):3463–3471. doi: 10.1182/blood-2009-07-232876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki M, Moriguchi T, Ohneda K, Yamamoto M. Differential contribution of the Gata1 gene hematopoietic enhancer to erythroid differentiation. Mol Cell Biol. 2009;29(5):1163–1175. doi: 10.1128/MCB.01572-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, et al. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005;201(12):1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126(2):415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Luc S, Anderson K, Kharazi S, Buza-Vidas N, Boiers C, Jensen CT, et al. Down-regulation of Mpl marks the transition to lymphoid-primed multipotent progenitors with gradual loss of granulocyte-monocyte potential. Blood. 2008;111(7):3424–3434. doi: 10.1182/blood-2007-08-108324. [DOI] [PubMed] [Google Scholar]
- 26.Luc S, Luis TC, Boukarabila H, Macaulay IC, Buza-Vidas N, Bouriez-Jones T, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13(4):412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1(1):57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 29.Mancini E, Sanjuan-Pla A, Luciani L, Moore S, Grover A, Zay A, et al. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. Embo J. 2012;31(2):351–365. doi: 10.1038/emboj.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10(5):401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J Biol Chem. 2007;282(33):24075–24082. doi: 10.1074/jbc.M701409200. [DOI] [PubMed] [Google Scholar]
- 32.Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, et al. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000;14(19):2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205(3):611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17(5):665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 35.Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121(10):1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18(1):109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 37.Chi AW, Chavez A, Xu L, Weber BN, Shestova O, Schaffer A, et al. Identification of Flt3CD150 myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential. Blood. 2011;118(10):2723–2732. doi: 10.1182/blood-2010-09-309989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohneda K, Moriguchi T, Ohmori S, Ishijima Y, Satoh H, Philipsen S, et al. Transcription factor GATA1 is dispensable for mast cell differentiation in adult mice. Mol Cell Biol. 2014;34(10):1812–1826. doi: 10.1128/MCB.01524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 40.Luc S, Buza-Vidas N, Jacobsen SE. Biological and molecular evidence for existence of lymphoid-primed multipotent progenitors. Ann N Y Acad Sci. 2007;1106:89–94. doi: 10.1196/annals.1392.023. [DOI] [PubMed] [Google Scholar]
- 41.Arock M, Schneider E, Boissan M, Tricottet V, Dy M. Differentiation of human basophils: an overview of recent advances and pending questions. J Leukoc Biol. 2002;71(4):557–564. [PubMed] [Google Scholar]
- 42.Vieira-de-Abreu A, Campbell RA, Weyrich AS, Zimmerman GA. Platelets: versatile effector cells in hemostasis, inflammation, and the immune continuum. Semin Immunopathol. 2012;34(1):5–30. doi: 10.1007/s00281-011-0286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagasawa T, Nakayasu C, Rieger AM, Barreda DR, Somamoto T, Nakao M. Phagocytosis by Thrombocytes is a Conserved Innate Immune Mechanism in Lower Vertebrates. Front Immunol. 2014;5:445. doi: 10.3389/fimmu.2014.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abraham SN, John AL., St Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10(6):440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cumano A, Paige CJ, Iscove NN, Brady G. Bipotential precursors of B cells and macrophages in murine fetal liver. Nature. 1992;356(6370):612–615. doi: 10.1038/356612a0. [DOI] [PubMed] [Google Scholar]
- 46.Lacaud G, Carlsson L, Keller G. Identification of a fetal hematopoietic precursor with B cell, T cell, and macrophage potential. Immunity. 1998;9(6):827–838. doi: 10.1016/s1074-7613(00)80648-2. [DOI] [PubMed] [Google Scholar]
- 47.Kawamoto H, Ohmura K, Katsura Y. Direct evidence for the commitment of hematopoietic stem cells to T, B and myeloid lineages in murine fetal liver. Int Immunol. 1997;9(7):1011–1019. doi: 10.1093/intimm/9.7.1011. [DOI] [PubMed] [Google Scholar]
- 48.Montecino-Rodriguez E, Leathers H, Dorshkind K. Bipotential B-macrophage progenitors are present in adult bone marrow. Nat Immunol. 2001;2(1):83–88. doi: 10.1038/83210. [DOI] [PubMed] [Google Scholar]
- 49.Gorgens A, Radtke S, Mollmann M, Cross M, Durig J, Horn PA, et al. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell Rep. 2013;3(5):1539–1552. doi: 10.1016/j.celrep.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Miyawaki K, Arinobu Y, Iwasaki H, Kohno K, Tsuzuki H, Iino T, et al. CD41 marks the initial myelo-erythroid lineage specification in adult mouse hematopoiesis: redefinition of murine common myeloid progenitor. Stem Cells. 2015;33(3):976–987. doi: 10.1002/stem.1906. [DOI] [PubMed] [Google Scholar]
- 51.Moreira PN, Pozueta J, Giraldo P, Gutierrez-Adan A, Montoliu L. Generation of yeast artificial chromosome transgenic mice by intracytoplasmic sperm injection. Methods Mol Biol. 2006;349:151–161. doi: 10.1385/1-59745-158-4:151. [DOI] [PubMed] [Google Scholar]
- 52.McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95(11):3489–3497. [PubMed] [Google Scholar]
- 53.Lindeboom F, Gillemans N, Karis A, Jaegle M, Meijer D, Grosveld F, et al. A tissue-specific knockout reveals that Gata1 is not essential for Sertoli cell function in the mouse. Nucleic Acids Res. 2003;31(18):5405–5412. doi: 10.1093/nar/gkg723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science (New York, NY. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 55.Kanakura Y, Kuriu A, Waki N, Nakano T, Asai H, Yonezawa T, et al. Changes in numbers and types of mast cell colony-forming cells in the peritoneal cavity of mice after injection of distilled water: evidence that mast cells suppress differentiation of bone marrow-derived precursors. Blood. 1988;71(3):573–580. [PubMed] [Google Scholar]
- 56.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.