Abstract
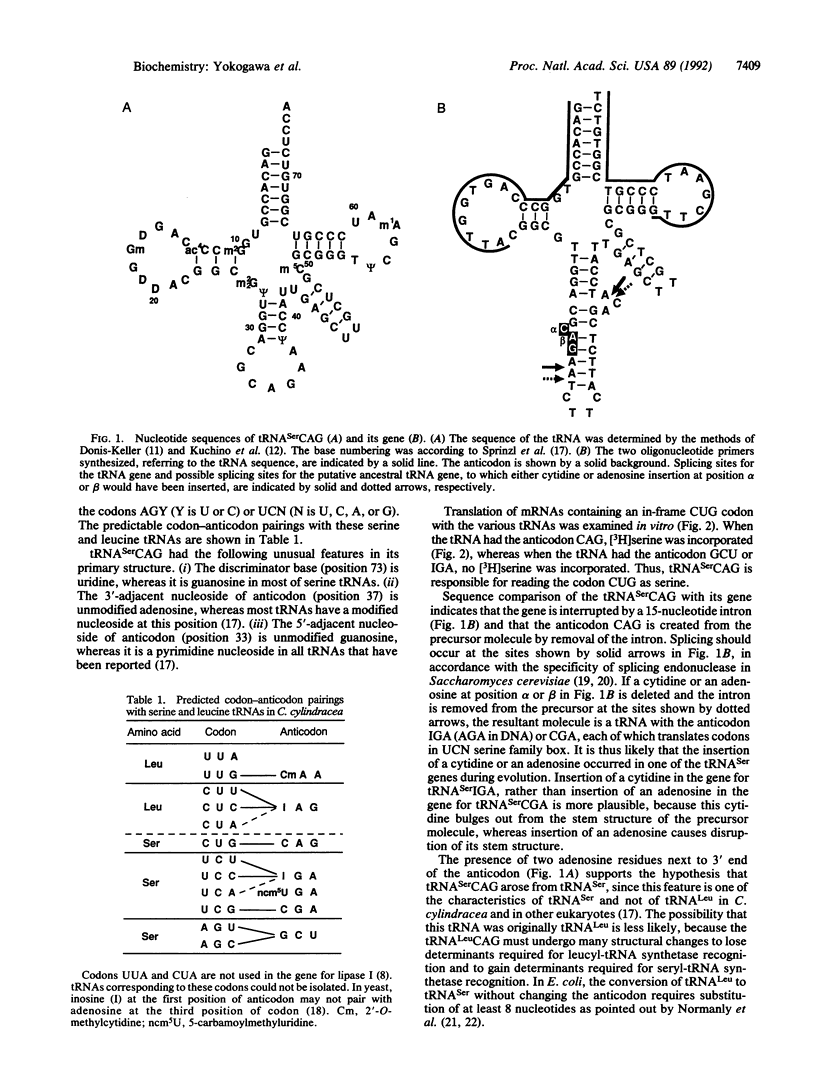
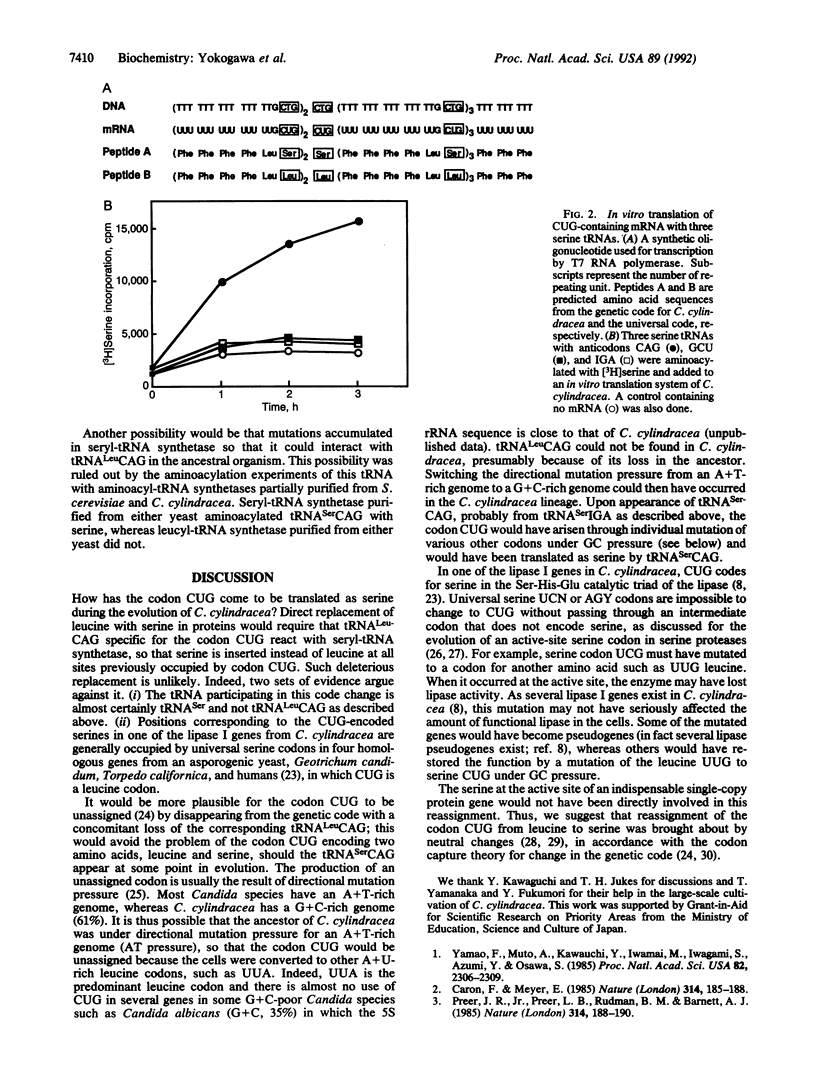
In the asporogenic yeast Candida cylindracea, the codon CUG is read as serine instead of leucine. This is an unusual instance in which the amino acid assignment of a codon deviates from the universal code. To infer the evolutionary process of this change, the tRNA with the anticodon sequence CAG, which is complementary to and thus responsible for translation of the codon CUG, has been identified. Indeed, this tRNA translates an in-frame CUG codon in a synthetic mRNA as serine in an in vitro translation system. The gene for the tRNA is interrupted by an intron in the anticodon loop. Sequence comparisons of the tRNA and its gene suggest that a single cytidine was inserted into the anticodon loop of the gene for tRNA(Ser)IGA during evolution to produce tRNA(Ser)CAG. The tRNA(Ser)CAG may be produced from its precursor molecule containing the cytidine insertion by splicing.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989 Sep 5;209(1):37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988 Aug 11;334(6182):528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- Caron F., Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985 Mar 14;314(6007):185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Pearlman R. E. Isolation and characterization of the actin gene from Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5160–5164. doi: 10.1073/pnas.83.14.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior E., Herrera F., Sadnik I., McLaughlin C. S., Moldave K. The preparation and characterization of a cell-free system from Saccharomyces cerevisiae that translates natural messenger ribonucleic acid. J Biol Chem. 1979 May 25;254(10):3965–3969. [PubMed] [Google Scholar]
- Hanyu N., Kuchino Y., Nishimura S., Beier H. Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986 Jun;5(6):1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin D. M. Evolution of an active-site codon in serine proteases. Nature. 1988 Dec 1;336(6198):429–430. doi: 10.1038/336429b0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Honda H., Taniguchi-Morimura J., Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989 Sep 14;341(6238):164–166. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Recent development of the neutral theory viewed from the Wrightian tradition of theoretical population genetics. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):5969–5973. doi: 10.1073/pnas.88.14.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu N., Nishimura S. Analysis of modified nucleosides and nucleotide sequence of tRNA. Methods Enzymol. 1987;155:379–396. doi: 10.1016/0076-6879(87)55026-1. [DOI] [PubMed] [Google Scholar]
- Meyer F., Schmidt H. J., Plümper E., Hasilik A., Mersmann G., Meyer H. E., Engström A., Heckmann K. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., Abelson J. Changing the identity of a transfer RNA. Nature. 1986 May 15;321(6067):213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- Oba T., Andachi Y., Muto A., Osawa S. CGG: an unassigned or nonsense codon in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):921–925. doi: 10.1073/pnas.88.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Muto A., Jukes T. H., Ohama T. Evolutionary changes in the genetic code. Proc Biol Sci. 1990 Jul 23;241(1300):19–28. doi: 10.1098/rspb.1990.0060. [DOI] [PubMed] [Google Scholar]
- Preer J. R., Jr, Preer L. B., Rudman B. M., Barnett A. J. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985 Mar 14;314(6007):188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988 Nov 18;55(4):719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- Schrag J. D., Li Y. G., Wu S., Cygler M. Ser-His-Glu triad forms the catalytic site of the lipase from Geotrichum candidum. Nature. 1991 Jun 27;351(6329):761–764. doi: 10.1038/351761a0. [DOI] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]


