SUMMARY
Human natural killer (NK) cells develop in secondary lymphoid tissues (SLTs) through distinct stages. We identified two SLT lineage (Lin)−CD34−CD117+/−CD94+CD16− “stage 4” subsets according to expression of the C-type lectin-like surface activating receptor, NKp80: NKp80− (stage “4a”) and NKp80+ (stage “4b”). Whereas stage 4b cells expressed more of the transcription factors T-BET and EOMES, produced interferon-gamma, and were cytotoxic, stage 4a cells expressed more of the transcription factors RORγt and AHR and produced interleukin-22, similar to SLT Lin−CD34−CD117+CD94−CD16− “stage 3” cells whose phenotype overlaps with that of Group 3 innate lymphoid cells (ILC3s). Co-culture with dendritic cells or transplantation into immunodeficient mice produced mature NK cells from stage 3 and stage 4a populations. These data identify NKp80 as a marker of NK cell maturity in SLTs and support a model of human NK cell development through a stage 4a intermediate with ILC3-associated features.
eTOC blurb
Human natural killer (NK) cells have potent effector functions against cancer; how NK cells develop in humans is unclear. Freud et al. demonstrate that NKp80 expression marks functionally mature NK cells as they develop in secondary lymphoid tissues. These findings help define the pathway of human NK cell development.
INTRODUCTION
Natural killer (NK) cells are innate lymphoid cells (ILCs) that can kill pathogen-infected and malignant cells as well as modulate other components of the immune system by producing chemokines and cytokines. Numerous recent studies highlight the existence of other ILC populations, and collectively all ILCs are now categorized into three groups according to their differential expression of surface antigens, transcription factors, and cytokines (Spits et al., 2013). NK cells represent a subtype of Group 1 ILCs, and their distinguishing features include 1) expression of the transcription factors, T-BET and EOMES; 2) expression of major histocompatibility complex (MHC) class I molecule-binding receptors, CD94/NKG2 [considered specific for NK cells among ILCs (Spits et al., 2013)] and killer immunoglobulin-like receptors (KIR); 3) production of interferon-gamma (IFN-γ); and 4) the ability to mediate perforin-dependent natural cytotoxicity (Caligiuri, 2008, Cortez et al., 2015). ILC1s represent the other Group 1 ILC subtype (Spits et al., 2013). While ILC1s similarly produce IFN-γ and express T-BET, recent mouse and human studies indicate that they comprise a lineage that is distinct from NK cells in that the former are non-cytolytic and lack expression of numerous NK-associated molecules including EOMES, CD94, CD56, CD16, perforin, granzymes, and KIRs (Bernink et al., 2015, Bernink et al., 2013, Fuchs et al., 2013, Klose et al., 2014).
Studies in both mice and humans indicate that NK cells can develop in multiple tissues including bone marrow (BM), secondary lymphoid tissues (SLTs), the liver, the uterus, and the thymus (Yu et al., 2013). A five-stage model of human NK cell development within SLTs was proposed based on the differential expression of CD34, CD117, CD94, and CD16 among lineage (Lin) negative cells [i.e. cells lacking T, B, dendritic cell (DC), and myelomonocytic associated antigens]: stage 1, CD34+CD117−CD94−CD16−; stage 2, CD34+CD117+CD94−CD16−; stage 3, CD34−CD117+CD94−CD16−; stage 4, CD34−CD117+/−CD94+CD16−; and stage 5, CD34−CD117+/−CD94+/−CD16+ (Freud et al., 2014). The original study characterizing these stages of development (Freud et al., 2006) showed that in bulk cultures CD34+ stage 1 and stage 2 populations are capable of DC, T cell, and NK cell differentiation, whereas the CD34− stage 3 population can give rise to NK cells but not to DCs or T cells. In addition, stage 3 cells lack the two hallmark functions of mature NK cells (i.e. IFN-γ production and perforin-dependent cytotoxicity) that are detected at stage 4 (CD94+). Therefore, it was originally concluded that stage 3 cells are lineage-restricted NK cell precursors and that functional maturity is acquired at stage 4 (Freud and Caligiuri, 2006). This NK cell development model is supported by analysis of in vitro-derived intermediates (Grzywacz et al., 2006).
Subsequent studies showed that cells within the SLT stage 3 population express aryl hydrocarbon receptor (AHR), retinoic acid receptor-related orphan receptor gamma (RORγt), interleukin 22 (IL-22), IL-1R1, and IL-7Rα (CD127) (Cella et al., 2009, Cupedo et al., 2009, Hughes et al., 2010, Hughes et al., 2009, Hughes et al., 2014, Tang et al., 2011). These features have recently also been associated with Group 3 ILCs (ILC3), which in mice comprise a lineage separate from that of conventional Eomes+ NK cells (Hazenberg and Spits, 2014, Satoh-Takayama et al., 2010, Spits et al., 2013, Vonarbourg et al., 2010). To the best of our knowledge it is not yet clear how best to distinguish between these two human SLT populations ex vivo (Freud et al., 2014). Herein we refer to the SLT Lin−CD34−CD117+CD94−CD16− population as “stage 3”.
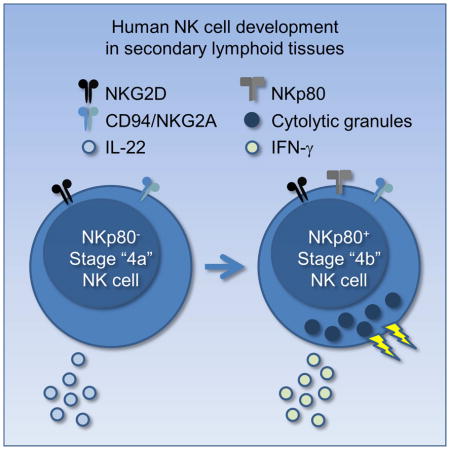
In this study we identified two subsets of SLT stage 4 cells according to the expression of the C-type lectin-like surface activating receptor, NKp80 (Bartel et al., 2013): NKp80− (stage “4a”) and NKp80+ (stage “4b”). Whereas stage 4b cells expressed more of the transcription factors T-BET and EOMES, produced IFN-γ, and were cytotoxic, stage 4a cells on the other hand expressed more RORγt and AHR and produced IL-22. Following in vitro co-culture with DCs or in vivo transplantation into immunodeficient mice, stage 4a cells (as well as stage 3 cells) gave rise to mature NK cells. These data refine the previous model of human NK cell development in SLTs and identify NKp80 as a marker of functional maturity ex vivo. In addition, these data suggest that human NK cells and ILC3s share a close developmental relationship, as we provide compelling evidence that in humans NK cells can develop in vivo through a stage 4a intermediate with ILC3-associated features.
RESULTS
Identification of two SLT stage 4 subsets according to NKp80 expression
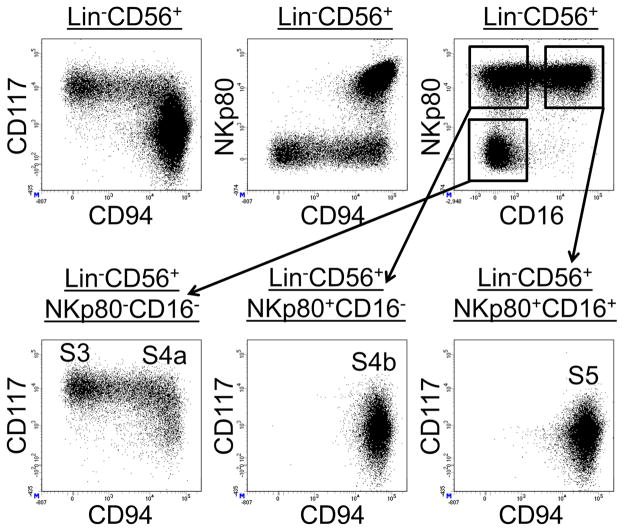
As previously described (Freud and Caligiuri, 2006, Freud et al., 2006) and shown here in Figure 1 (top row, left dot plot), visualizing CD94 versus CD117 expression among total freshly enriched SLT Lin−CD56+ cells revealed a continuum of events between the CD117+CD94− stage 3 and CD117+/−CD94+ stage 4 NK cell populations, suggestive of dynamic NK cell development in situ. We observed that stage 3 cells as well as the intermediate CD94+ cells directly emerging from stage 3 lacked expression of the activating receptor, NKp80 (Figure 1, top row, middle dot plot), which was detectable on the surface of virtually all NK cells in healthy human PB (Figure S1). We also observed that nearly all SLT Lin−CD56+CD16+ cells (i.e. stage 5 cells) co-expressed NKp80 (Figure 1 top row, right dot plot).
Figure 1. Identification of two SLT stage 4 subsets according to NKp80 expression.
Representative flow cytometry analysis of negatively enriched SLT ILCs (n = 10). Dot plots in the top row were gated on SLT Lin−CD56+ lymphocytes. Dot plots in the bottom row were gated on subsets within the SLT Lin−CD56+ population according to NKp80 and CD16 expression as indicated. S3, stage 3; S4a, stage 4a; S4b, stage 4b; S5, stage 5. See also Figure S1.
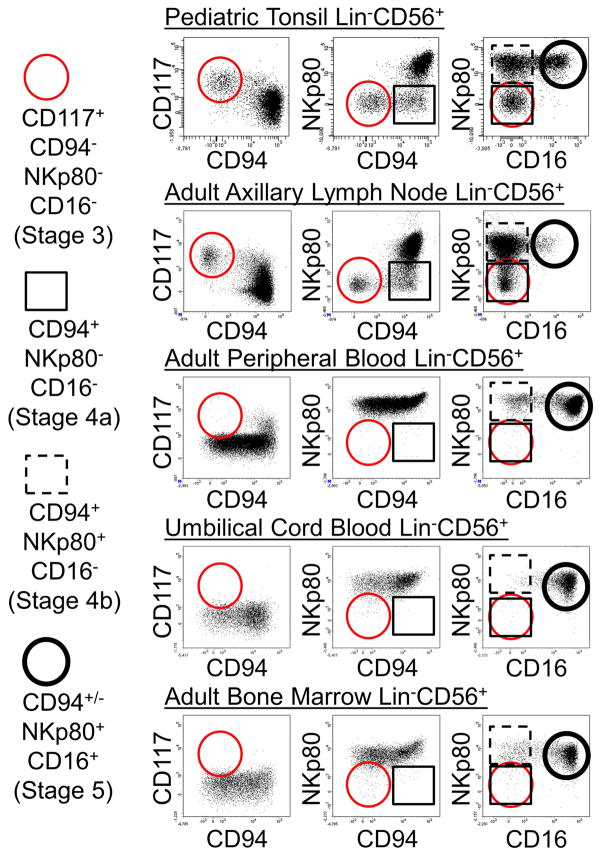
By gating on SLT Lin−CD56+ subsets according to the differential expression of NKp80 and CD16 and then viewing CD94 versus CD117 expression we confirmed that the aforementioned CD94+ intermediate population emerging from stage 3 lacked both NKp80 and CD16 expression (Figure 1, bottom row). These ex vivo flow cytometry data suggested a step-wise acquisition of CD94, NKp80, and CD16 during human NK cell development in SLTs. Moreover, they revealed two distinct SLT Lin−CD94+CD16− stage 4 NK cell subsets based on the differential expression of NKp80. In keeping with the previous model designating stage 4 NK cells as CD94+ and stage 5 NK cells as CD16+ (Freud et al., 2014), we now refer to Lin−CD94+NKp80−CD16− cells as stage “4a”; Lin−CD94+NKp80+CD16− cells as stage “4b”; and Lin−CD94+/−NKp80+CD16+ cells as stage 5 (Figure 1, bottom row). We note that although CD56 was expressed by many stage 3 cells and by most stage 4a, 4b, and 5 cells in SLTs, CD56 was not uniformly expressed by each of these populations (see Figure 4A below) (Freud et al., 2006). Therefore, we did not include CD56 as a defining marker of these populations but rather used it as a relatively specific marker to help visualize patterns of antigen loss and acquisition during NK cell development in SLTs. Using this approach, we also observed that just like stage 3 cells (Lin−CD117+CD94−NKp80−CD16−), stage 4a cells were naturally enriched in SLTs (tonsils and lymph nodes), whereas they were extremely rare if not undetectable in healthy adult PB, umbilical cord blood (UCB), and adult BM samples (Figure 2). In contrast, stage 5 cells represented the major NK cell population in blood and BM.
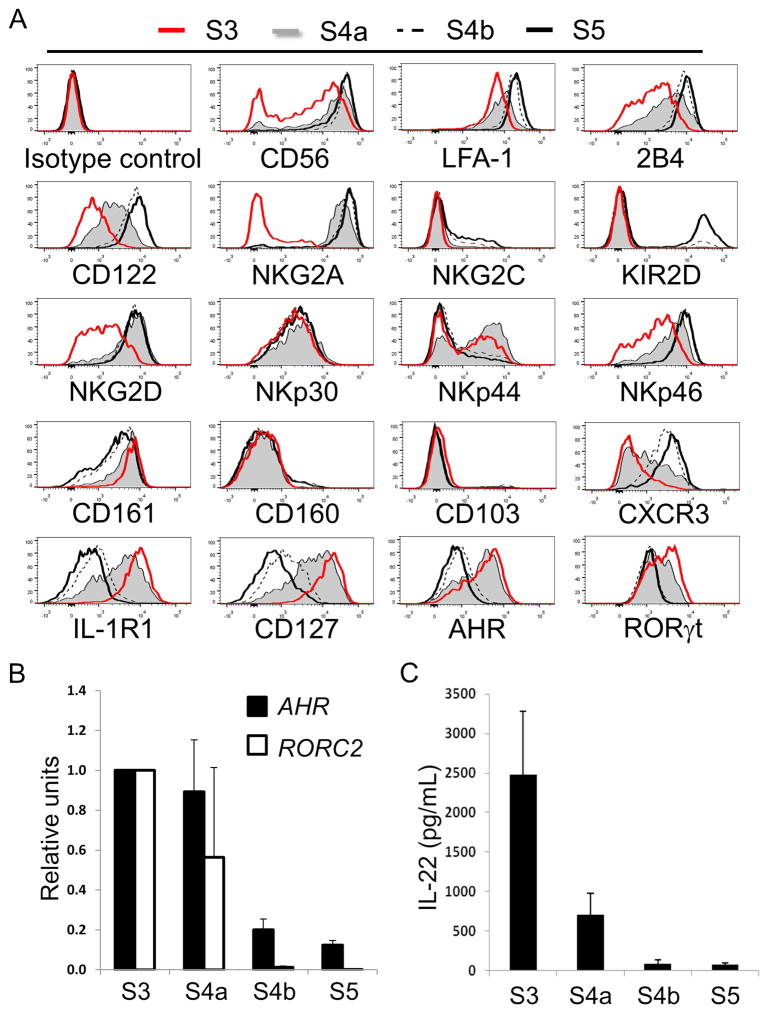
Figure 4. SLT stage 4a cells show both NK- and ILC3-associated features.
(A) Representative flow cytometry analyses of freshly enriched SLT stage 3 - stage 5 cells showing relative surface and intracellular expression of the indicated antigens (n = 4). Histogram data were generated by gating on Lin− ILC populations as defined in Figure 2. (B) Transcriptional expression analysis by qPCR of AHR and RORC2 by freshly purified SLT stage 3 - stage 5 cells. Data are represented as mean ± SEM (n = 6). Decreasing trends of gene expression from stage 3 (values set to 1 in arbitrary units) - stage 5 were statistically significant (p < 0.0001 for each trend) as estimated using linear mixed models based on the ddCT method. (C) Quantitative ELISA data showing IL-22 production by freshly purified SLT stage 3 - stage 5 cells stimulated with IL-1β and IL-23. Data are represented as mean ± SEM (n = 6). A linear mixed effect model was used to evaluate the trend of IL-22 production from stage 3 - stage 5 cells (p < 0.0001). See also Figure S2 and Table S1.
Figure 2. Stage 3 and stage 4a cells are naturally enriched in human SLTs.
Representative flow cytometry analyses of Lin−CD56+ ILCs negatively enriched from pediatric tonsil (n = 10), adult axillary lymph node (n = 4), adult PB (n = 5), UCB (n = 3), and adult BM (n = 3) specimens. Dot plots in each row were gated on Lin−CD56+ lymphocytes. Circles and squares refer to indicated populations in the dot plots.
SLT stage 4a cells lack mature NK cell functions
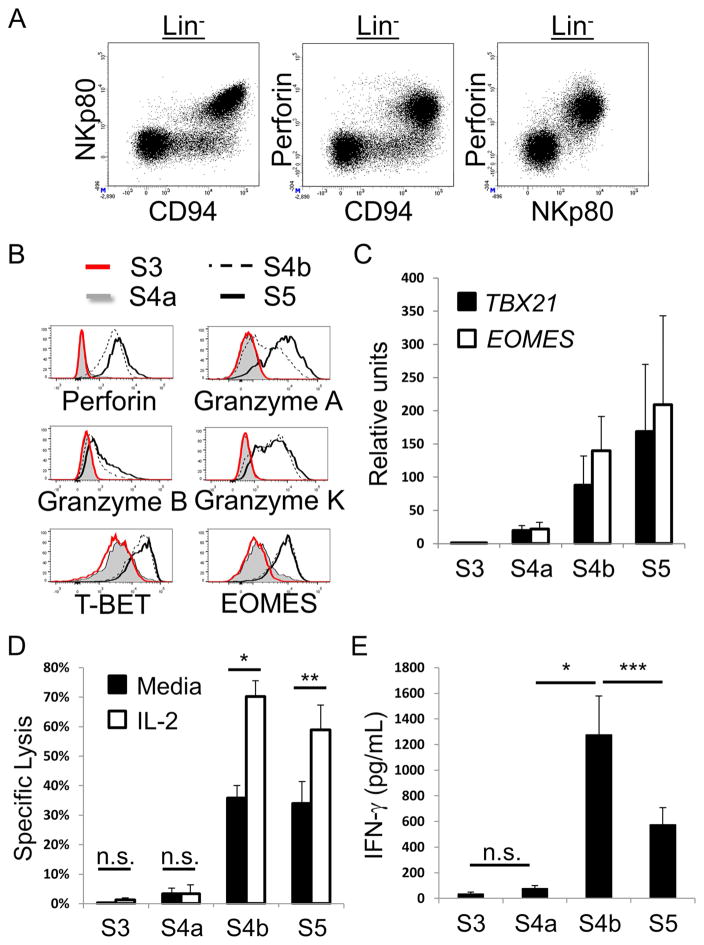
In order to characterize the SLT stage 4a and 4b subsets, we tested these cells for numerous phenotypic and functional parameters ex vivo. Stage 3 and stage 5 cells were tested in parallel for comparison. We first observed a striking correlation between surface NKp80 and intracellular perforin expression among total SLT Lin− cells (Figure 3A), indicating that stage 4a cells, like stage 3 cells, lacked perforin expression. In addition, we observed that stage 3 and stage 4a cells lacked intracellular expression of granzymes A, B, and K, whereas these molecules were at least partially expressed by stage 4b and stage 5 cells (Figure 3B). T-BET and EOMES protein expression were also very low or undetectable within stage 3 and stage 4a cells yet clearly expressed by stage 4b and 5 cells (Figure 3B). These intracellular flow cytometry data were supported by quantitative Real-time reverse transcriptase polymerase chain reaction (qPCR) analyses of TBX21 (encoding T-BET) and EOMES expression by SLT stage 3 – stage 5 cells (Figure 3C) that were freshly purified from SLTs by cell sorting (Figure S2). Consistent with these ex vivo phenotypic and qPCR data, freshly purified SLT stage 3 and stage 4a cells failed to mediate perforin-dependent cytotoxicity against the MHC class I molecule-negative target cell line, K562, whereas stage 4b and stage 5 cells lysed K562 targets (Figure 3D). Moreover, stage 4b and stage 5 cytotoxicity was augmented when IL-2 was present during the 4-hr cytotoxicity assay, whereas IL-2 did not impact the cytotoxic functions of stage 3 and stage 4a cells (Figure 3D). Freshly isolated stage 3 and stage 4a cells also produced negligible amounts of IFN-γ as measured by enzyme-linked immunosorbent assay (ELISA). In contrast, stage 4b and stage 5 cells produced IFN-γ as detected by ELISA (Figure 3E) and by intracellular flow cytometry (Figure S3), and stage 4b cells showed significantly higher production of IFN-γ in comparison to stage 5 cells. Thus stage 4a cells lacked mature NK cell functions ex vivo, and NKp80 expression within the originally described stage 4 population (Freud et al., 2006) closely correlated with mature NK cell functionality in SLTs.
Figure 3. NK cell functional maturity correlates with NKp80 expression in SLTs.
(A) Representative flow cytometry analysis of Lin− enriched ILCs from SLT showing the close correlation between surface NKp80 expression and intracellular perforin (n = 4). (B) Representative flow cytometry analyses of freshly enriched SLT stage 3 - stage 5 cells showing intracellular expression of the indicated proteins (n = 4). Histogram data were generated by gating on Lin− ILC populations as defined in Figure 2. (C) Transcriptional expression analysis by qPCR of TBX21 and EOMES by freshly purified SLT stage 3 - stage 5 cells. Data are represented as mean ± SEM (n = 6). Increasing trends of gene expression from stage 3 (values set to 1 in arbitrary units) to stage 5 were statistically significant (p < 0.0001 for each trend) as estimated using linear mixed models based on the ddCT method. (D) Specific lysis of K562 target cells by freshly purified SLT stage 3 - stage 5 cells in media alone or in the presence of IL-2. Data are represented as mean ± SEM (n = 4, n.s. not significant, *p < 0.0001, **p = 0.0003). (E) Quantitative ELISA data showing IFN-γ production by freshly purified SLT stage 3 - stage 5 cells stimulated with IL-12, IL-15, and IL-18. Data are represented as mean ± SEM (n = 7, ***p = 0.0021). See also Figures S2 and S3.
SLT stage 4a cells have both NK- and ILC3-associated features
Additional ex vivo analyses of stage 4a cells from human SLTs revealed that these cells expressed numerous NK cell-associated surface antigens and functional receptors including CD56 as well as lymphocyte function associated antigen (LFA)-1, 2B4, CD122, NKG2D, and NKp46. For most of these surface antigens, with the exception of NKG2D, stage 4a expression levels were intermediate between those of stage 3 cells (partial and/or low) and those of stage 4b and stage 5 cells (high) (Figure 4A). NKp30 was nearly uniformly expressed by SLT stage 3 – stage 5 cells, whereas NKp44 was expressed by subsets of the four populations. Nearly all stage 4a cells expressed NKG2A and lacked NKG2C and KIRs. In contrast, NKG2C+ and KIR+ subsets were detected within the stage 4b and stage 5 populations (Figure 4A). Some CD57 expression, which is associated with terminal differentiation among human PB NK cells (Lopez-Verges et al., 2010), was detected within stage 5 but not within stage 3 – stage 4b populations (n = 4, data not shown).
We also evaluated SLT stage 3 – stage 5 cells for the expression of other ILC-associated surface antigens. CD161 was expressed at high levels on stage 3 and stage 4a cells, whereas stage 4b and stage 5 cells showed variable CD161 expression ranging from high to partially low or negative. CD101, CD103, CD160, and CXCR3 expression have been described on human ILC1 cells (Bernink et al., 2013, Fuchs et al., 2013), and we observed that CD101 and CD103 molecules were detected among minor fractions of stage 3 – stage 5 cells (Figure 4A and data not shown). In contrast, CD160 was primarily restricted to minor subsets of stage 4b and stage 5 cells among total Lin− cells. CXCR3 expression was low or undetectable at stage 3, intermediate at stage 4a, and highest at stages 4b and 5 (Figure 4A).
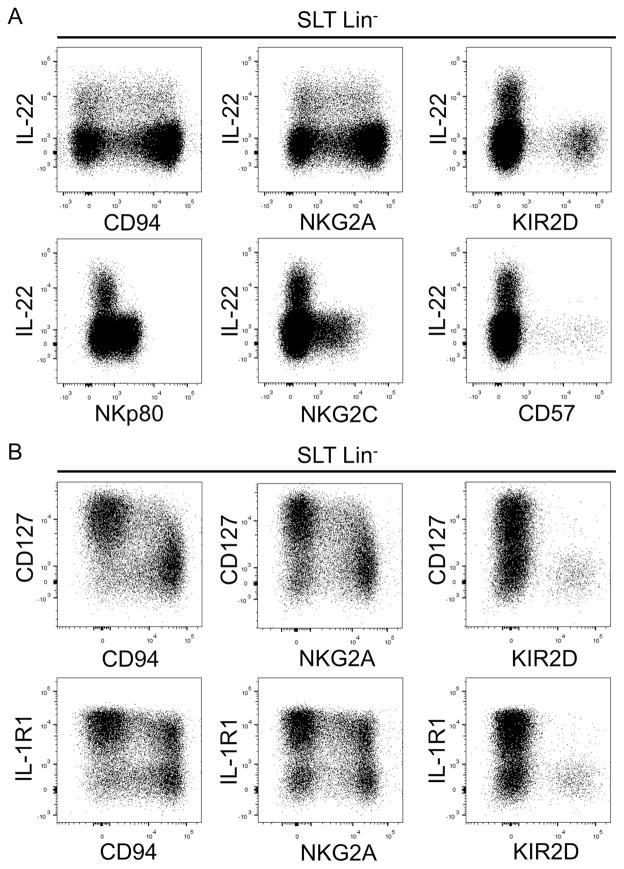
Regarding ILC3-associated features, we observed that similar to stage 3 cells stage 4a cells, which were CD94+, expressed intermediate to high levels of IL-1R1, CD127, AHR, and RORγt (Figure 4A, bottom row). In contrast, stage 4b and stage 5 cells showed low or undetectable expression of these molecules by flow cytometry. We confirmed by qPCR that stage 4a cells expressed mRNA transcripts for AHR and isoform 2 of the RORC locus (RORC2) which encodes for RORγt protein (Figure 4B), and we also observed that stage 4b and stage 5 cells expressed detectable, albeit very low, mRNA from both genes (Figure 4B). As shown in Figure 4C, freshly purified SLT stage 4a cells produced IL-22 following overnight stimulation with IL-1β and IL-23, and the amounts of IL-22 produced were significantly lower compared to donor-matched stage 3 cells. In contrast stage 4b and stage 5 cells produced negligible amounts of IL-22. We further evaluated IL-22 production by intracellular flow cytometry following a 6-hr stimulation of freshly enriched SLT ILCs with phorbol 12-myristate 13-acetate (PMA), ionomycin, and IL-2. As shown in Figure 5A, IL-22 production was detected by Lin−CD94/NKG2A+ cells but not by Lin− cells expressing KIRs, NKp80, NKG2C, CD57, or CD16 (Figure 5A and data not shown). Likewise, many Lin−CD94/NKG2A+ cells coexpressed CD127 and IL-1R1, whereas most Lin−KIR+ cells did not (Figure 5B). Collectively, these immunophenotypic and functional data indicate that SLT stage 4a cells are CD94/NKG2A+NKp80−CD16−KIR−NKG2C−CD57− and have some ILC3-associated features ex vivo.
Figure 5. ILC3-associated features of CD94/NKG2A+ stage 4a cells.
(A) Representative flow cytometry analysis of IL-22 production by SLT enriched ILCs stimulated for 6 hr in PMA, ionomycin, and IL-2 (n = 4). (B) Representative flow cytometry analysis of freshly enriched and unstimulated SLT ILCs (n = 4). Dot plots in A and B were gated on Lin− lymphocytes enriched from pediatric tonsils.
Dendritic cells promote the differentiation and maturation of stage 3 and stage 4a cells in vitro
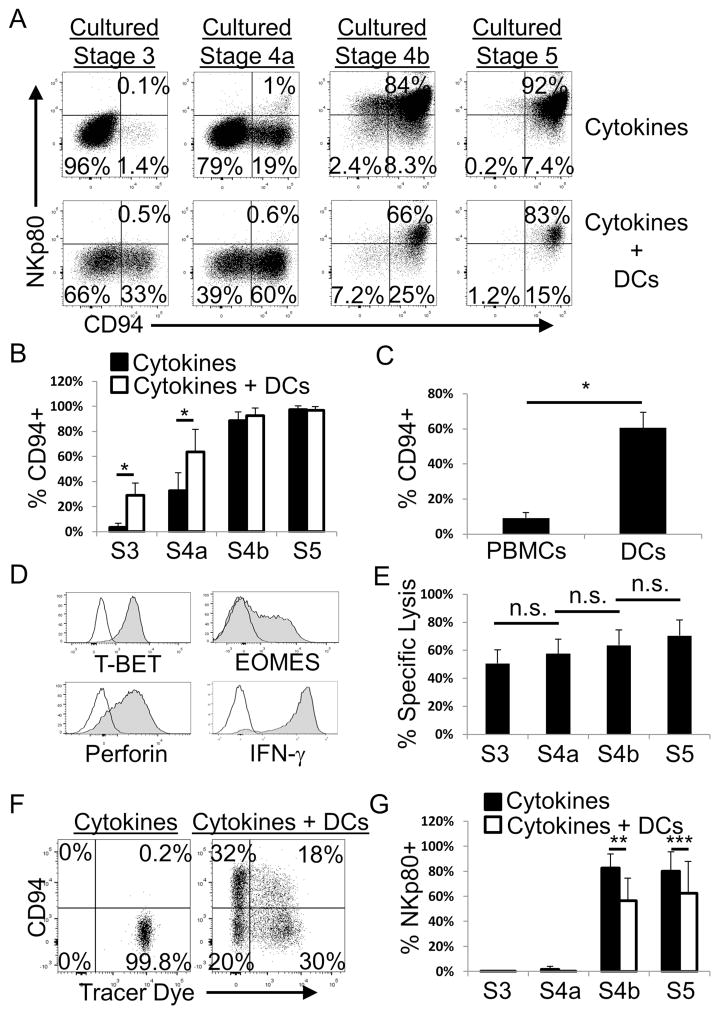
We and others previously demonstrated that human stage 3 cells as well as mature NK cells closely associate with CD11c+ DCs in situ in SLTs (Ferlazzo et al., 2004, Hughes et al., 2010). DCs can also promote mature NK cell homeostasis in vivo in mice (Guimond et al., 2010). We therefore hypothesized that human DCs could promote NK cell differentiation in vitro from human SLT NK cell precursors. Consistent with this hypothesis we observed that co-culture of allogeneic monocyte-derived DCs with either stage 3 or stage 4a cells promoted robust acquisition or maintenance of CD94 expression, respectively, compared to cultures in cytokines [flt3 ligand (FL), c-kit ligand (KL), and IL-15] alone (Figures 6A and 6B). This was observed in bulk cultures (Figure 6A and 6B) as well as in clonal assays comparing individually sorted stage 3 cells cultured with either activated peripheral blood mononuclear cells (PBMCs) or DCs (Figure 6C). Although CD94+ cells were observed in all of the analyzed stage 3-derived clones regardless of culture condition (29 clones total; cloning efficiency 10–15%), the percentages of CD94+ cells per stage 3-derived clone were significantly higher in the presence of DCs compared to PBMCs (Figure 6C).
Figure 6. DCs promote NK cell functional maturation of stage 3 and stage 4a cells in vitro.
(A) Representative flow cytometry analysis following culture of purified SLT stage 3 - stage 5 cells with IL-15, FL, and KL (i.e. “Cytokines”) or with cytokines plus DCs (n = 16). Dot plots were gated on Lin−CD56+ viable lymphocytes. (B) Percentages of CD94+ NK cells among total Lin−CD56+ cells derived in vitro from purified SLT stage 3 - stage 5 cells cultured in cytokines with or without DCs. Data are represented as mean ± SEM (n = 16, *p < 0.0001). (C) Percentages of CD94+ NK cells derived in vitro from freshly purified and individually sorted SLT stage 3 cells expanded in the presence of allogeneic PBMCs and IL-2 or in the presence of IL-15, FL, KL, and DCs. Pooled data from two separate experiments are represented as mean ± SEM (n = 18 total clones for PBMC condition, n = 11 total clones for DC condition, *p < 0.0001). (D) Representative intracellular flow cytometry analysis of Lin−CD94+ NK cells derived in vitro from bulk co-culture of SLT stage 3 cells with cytokines and DCs (n = 4). IFN-γ production was induced by 6 hr stimulation with PMA, ionomycin, and IL-2. Black lines represent isotype control staining, whereas shaded histograms represent staining with antigen-specific antibodies. Data are similar to those generated with Lin−CD94+ cells derived in vitro from stage 4a cultures (n = 4, data not shown). (E) Specific lysis of K562 target cells by purified Lin−CD94+ cells derived in vitro from stage 3 - stage 5 cells cultured with cytokines and DCs. Data are represented as mean ± SEM (n = 5, n.s. not significant). (F) Representative flow cytometry analysis of SLT stage 3 cells that were labeled with a membrane tracer dye at day 0 and then cultured in vitro for two weeks with cytokines alone or cytokines plus DCs (n = 2). (G) Percentages of NKp80+ NK cells among total Lin−CD56+ cells derived in vitro from purified SLT stage 3 - stage 5 cells cultured in cytokines with or without DCs. Data are represented as mean ± SEM (n = 10, **p = 0.0067, ***p = 0.045). For all culture experiments, stage 3 – stage 5 cells were sorted to >99% purity using the strategy depicted in Figure S2. See also Figures S2, S4, and S5.
In contrast to freshly isolated SLT stage 3 and stage 4a cells (Figure 3), the in vitro derived CD94+ cells isolated from stage 3- and stage 4a-initiated bulk co-cultures with DCs expressed T-BET, EOMES, perforin, and IFN-γ (Figure 6D and data not shown). In addition, stage 3- and stage 4a-derived CD94+ cells were cytotoxic against K562 at levels comparable to CD94+ cells generated from stage 4b-initiated and stage 5-initiated cultures (Figure 6E), thus supporting their designation as functionally mature NK cells. The CD94− cells derived in the co-cultures with stage 3 cells and DCs either retained ILC3-associated features or adopted an ILC1-like phenotype, expressing CD161, T-BET and IFN-γ but not EOMES (Figure S4 and data not shown). Human ILC3 conversion into ILC1s was recently reportedly induced by IL-12 (Bernink et al., 2013); however, this endogenous cytokine was not required for the augmentation of CD94+ NK cell differentiation from stage 3 cells in the DC co-culture system we used (Figure S5).
Cultures initiated with DCs alone without SLT-derived cells did not contain any distinct Lin−CD161+CD94+/−CD56+/− ILCs. In addition, we observed similar inductions of CD94 expression on stage 3 cells if they were co-cultured with DCs that had been irradiated (data not shown) validating that these results were not due to the outgrowth of PB NK cells contaminating the DCs. We also performed membrane tracer labeling experiments to confirm that the data were not the result of the selective outgrowth of rare functionally mature CD94+ NK cells contaminating the sorted SLT stage 3 or stage 4a populations. For these experiments, tonsil-derived ILCs were first labeled with a membrane tracer dye, and then tracer dye+ stage 3 cells were sorted to >99% purity (data not shown). The purified tracer dye+ stage 3 cells were then cultured in FL, KL, and a very low dose of IL-15 (10 pg/mL; i.e. a 1,000-fold lower concentration than that used throughout the rest of this study) in order to minimize proliferation in vitro and hence promote retention of the tracer dye. As shown in Figure 6F, after two weeks in cultures containing only cytokines, essentially all cells retained the membrane tracer dye and did not induce CD94 expression. In contrast, in the cultures containing cytokines plus DCs, approximately half of the cultured stage 3 cells induced CD94 (Figure 6F). Despite the fact that at least half of all the cells proliferated, as indicated by the loss of the tracer dye in the presence of the DCs, a substantial proportion of the CD94+ cells were tracer dye+ and thus had not significantly proliferated (Figure 6F). These data confirm that NK cell differentiation, represented by de novo CD94 induction, had occurred from stage 3 cells and that the results were not due to the marked proliferation of rare contaminating CD94+ NK cells.
Despite the acquisition of NK cell maturity-associated phenotypic and functional features following co-culture of DCs with either stage 3 or stage 4a cells, NKp80 surface expression was not induced (Figures 6A and 6G). Moreover, NKp80 expression was significantly downregulated on stage 4b and stage 5 cells following in vitro co-culture with DCs (Figure 6G), similar to what occurs when PB NK cells are stimulated ex vivo (Klimosch et al., 2013). Therefore, while NKp80 expression correlated with NK cell functional maturity among ex vivo tested SLT ILCs, NKp80 expression did not correlate with NK cell differentiation following culture of stage 3 or stage 4a cells with DCs under the in vitro conditions tested here.
SLT stage 3 and stage 4a cells undergo terminal NK cell development in vivo
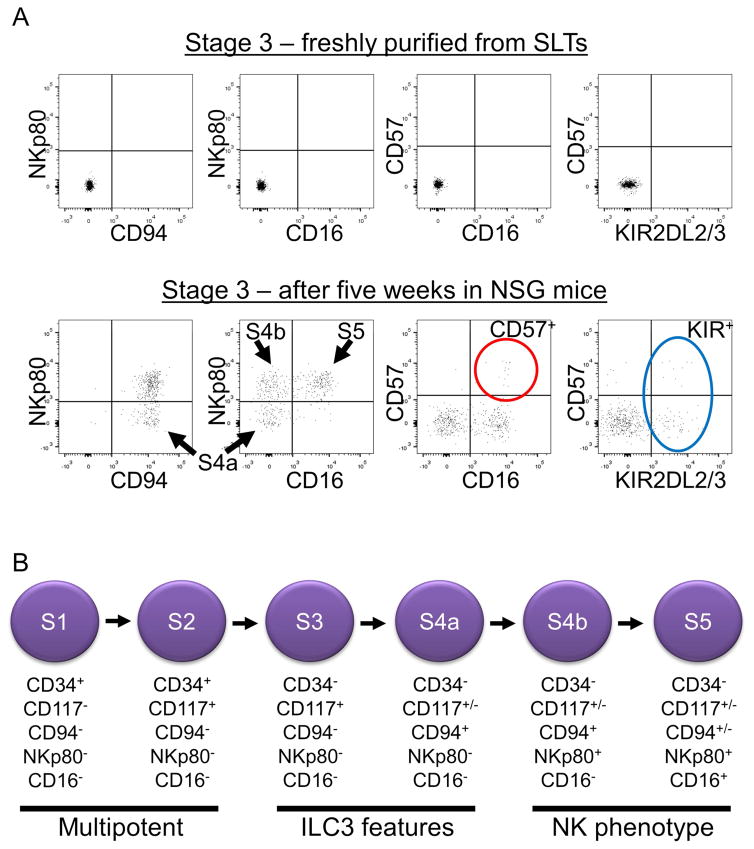
To test the NK cell developmental potential of SLT-derived stage 3 and stage 4a cells in a more complex in vivo environment, these cells were sorted to >99% purity and then each population was transplanted into busulfan-pretreated NOD-scid IL2Rgammanull (NSG) immunodeficient mice that were then injected twice weekly with human IL-15. Analysis of surface CD94, NKp80, and CD16 expression after 5 weeks among human CD45+ cells revealed that transplanted stage 3 and stage 4a cells each showed evidence of downstream NK cell maturation in the spleens (Figures 7A and S6) as well as in the blood and BM (data not shown) of recipient NSG mice. No human CD45+ lymphocytes were detected in control mice that had not been transplanted with human cells (data not shown). Moreover, the human CD45+ cells retrieved from transplanted mice were Lin−CD56+, consistent with NK cells (data not shown). In contrast to the observed NK cell differentiation in vitro (Figure 6), NKp80 was induced on the in vivo differentiated cells derived from transplanted stage 3 or stage 4a cells. In addition, variable proportions of Lin−CD94+NKp80+CD16+ (stage 5) cells were derived in vivo, and some also expressed CD57 and/or KIRs (Figures 7A and S6). Similar findings were observed when purified SLT stage 4b and stage 5 cells (sorted as CD57−KIR−, not shown) were transplanted into NSG mice (Figure S6). Thus, under these in vivo conditions, SLT stage 3 cells and stage 4a cells generated Lin−CD56+CD94+NKp80+CD16+CD57+/−KIR+/− cells bearing the immunophenotype associated with terminally differentiated human NK cells (Bjorkstrom et al., 2010, Luetke-Eversloh et al., 2013). Collectively, these data support a model of human NK cell development in SLTs that incorporates NKp80 expression into the existing framework of CD34, CD117, CD94, and CD16 expression (Freud and Caligiuri, 2006, Freud et al., 2014, Luetke-Eversloh et al., 2013, Yu et al., 2013) (Figure 7B).
Figure 7. Stage 3 cells develop into NKp80+CD16+/− NK cells in vivo supporting a model of human NK cell development.
(A) Representative flow cytometry analysis of SLT stage 3 cells freshly sorted to >99% purity using the strategy depicted in Figure S2 (top row) and after five weeks in vivo following transplantation into an NSG mouse treated twice weekly with human IL-15 (n = 4). Dot plots in the top row were gated on viable lymphocytes. Dot plots in the bottom row were gated on viable human CD45+ lymphocytes isolated from the spleen of the engrafted NSG mouse. Quadrants were set with the aid of isotype control staining. As shown in the bottom row, transplanted stage 3 cells gave rise to stage 4a, 4b, and 5 cells (black arrows). Cells expressing the NK cell terminal maturation-associated markers, CD57 (red circle) and KIR (blue oval) were also detected. (B) Proposed model of NK cell development in SLTs according to the expression of CD34, CD117, CD94, NKp80, and CD16. See also Figure S6.
DISCUSSION
In this study we identified a Lin−CD117+/−CD94+NKp80−CD16− stage “4a” population that was naturally restricted to human SLTs and that appeared to emerge directly from the Lin−CD117+CD94−NKp80−CD16− stage 3 population. While stage 4a cells expressed many NK cell markers, including CD94/NKG2A and NKG2D, they lacked hallmark features of mature NK cells and rather showed an ILC3-like profile, similar to stage 3 cells, including expression of RORγt, AHR, IL-1R1, CD127, and IL-22. In contrast, Lin−CD117+/−CD94+NKp80+CD16− stage “4b” cells showed negligible ILC3-associated features ex vivo and instead expressed T-BET, EOMES, and IFN-γ, and they could also mediate perforin-dependent cytotoxicity. We also demonstrated that SLT stage 3 cells and stage 4a cells acquired phenotypic and functional properties of mature NK cells following in vitro co-culture with DCs as well as following in vivo transplantation into NSG mice treated with IL-15. Thus, the stage 4a population appears to represent a naturally occurring phenotypic and functional developmental intermediate between the stage 3 cell and the stage 4b NK cell in human SLTs.
Several points can be taken from this study. First, the demonstration that NKp80 expression closely correlates with NK cell functional maturity in SLTs suggests that NKp80 may fulfill an important regulatory role during the maturation process in SLTs, potentially related to the acquisition of cytotoxic and/or cytokine-producing functions. The fact that NKp80− stage 4a cells expressed CD94/NKG2A yet were not mature in terms of NK-related functions is also intriguing in light of compelling mouse and human data indicating that MHC class I molecule binding inhibitory receptors influence NK cell maturation, a process commonly referred to as “licensing” or “education” (Anfossi et al., 2006, Kim et al., 2005). Perhaps the transition from stage 4a to stage 4b represents the initial education event during human NK cell development in SLTs. Further studies are warranted to investigate the roles of these surface receptors during human NK cell development.
The identification of the stage 4a population that is naturally enriched in SLTs provides additional evidence that human NK cell development occurs in these tissues. The notion of extramedullary human NK cell development is supported by the prior discovery of NK cell progenitors in SLTs (Freud et al., 2005) as well as by the detection of unique tissue-resident NK cell populations in other anatomical locations such as the liver and uterus (Moffett and Colucci, 2014, Sojka et al., 2014, Yu et al., 2013). Future studies are warranted to determine if similar Lin−CD94+NKp80−CD16− stage 4a cells are present in these other extramedullary tissues, if such cells show ILC3-associated features, and/or if NK cells derive in situ in these tissues potentially through a stage 4a intermediate. The fact that stage 4a cells were not enriched within the total Lin−CD56+ population in adult BM suggests that human NK cell development, if present in BM, proceeds through a different pathway and/or that the pathway we describe is not prominent there.
Our study also provides evidence supporting a role for DCs in augmenting human NK cell maturation. Previous studies indicated that mature NK cells interact with DCs in SLTs and that reciprocal interactions between these two cell types are important in both homeostatic and inflammatory settings (Cooper et al., 2004, Ferlazzo et al., 2004, Guimond et al., 2010, Moretta, 2005). We have also previously reported that stage 3 cells are in close proximity to CD11c+ DCs in human SLTs (Hughes et al., 2010). In the current study, we observed a significant induction of CD94 that was accompanied by T-BET, EOMES, perforin, and IFN-γ expression when purified stage 3 cells were cultured in the presence of FL, KL, IL-15, and DCs but not when they were cultured with cytokines alone. This DC-mediated effect was not reliant on endogenous or exogenous IL-12. We also observed the conversion of stage 3 cells into Lin−CD161+CD94−T-BET+EOMES−IFN-γ+ ILC1s in the DC co-cultures, similar to the recently described conversion of human ILC3s into ILC1s (Bernink et al., 2015). We therefore hypothesize that DCs can support the differentiation and maturation of ILC1s as well as NK cells in situ in SLTs, and future studies are warranted to test this hypothesis and to elucidate the potential mechanism(s) involved.
Lastly, these data provide evidence that in contrast to mouse NK cells at least some human NK cells likely share a close developmental relationship with ILC3s. Previous murine Rorc2 fate-mapping studies indicate murine ILC3s (Rorc2 fate-map+) comprise a lineage that is separate from that of conventional NK cells (Rorc2 fate-map−) (Satoh-Takayama et al., 2010, Vonarbourg et al., 2010). These fate-mapping data coupled with the recent identification in mice of common helper ILC progenitor cells capable of giving rise to ILC1s, ILC2s, and ILC3s but not to NK cells (Constantinides et al., 2014, Klose et al., 2014) provided a clear rationale for empirically excluding any human RORγt+ cell as representing a physiologic NK cell precursor. Nonetheless, we show here that at the population level stage 3 – stage 5 cells in SLTs each expressed at least low levels of detectable RORC2 transcript if not RORγt protein, and others have shown that SLT Lin−CD34+CD45RA+CD117+ stage 2 cells express RORγt (Cupedo et al., 2009, Montaldo et al., 2014). We also recently demonstrated that human PB CD56bright NK cells, which express CD94, CD117, CD127, and NKp80 (Cooper et al., 2001, Matos et al., 1993, Romagnani et al., 2007, Vitale et al., 2001, Vosshenrich et al., 2006), constitutively express RORC2 and can derive from an SLT RORγt+ common ILC progenitor population (Scoville et al., 2016). These data indicate that in humans at least some PB NK cells would be fate-map+ for RORC2. Thus, there appears to be a distinct difference in the lineage fidelities of RORC2 and Rorc2 expression in humans and in mice, respectively, and at least some human NK cells can develop through a RORγt+ pathway. However, this is not to say that all human NK cells develop in this manner, and other pathways of human NK cell development may exist in vivo. Of note, a recent study demonstrated that human patients with RORγt deficiency had reduced PB ILC3s but normal numbers of PB NK cells (Okada et al., 2015). The NK cells in those patients were not evaluated functionally; however, those data indicate that at least some human NK cells do not require RORγt for their development in vivo.
In light of the aforementioned mouse and human Rorc2 and RORC2 expression data, respectively, coupled with the demonstration here that SLT stage 4a cells express CD94/NKG2A, have an ILC3-like profile, and in many ways have features intermediate between those of stage 3 cells and stage 4b cells, the distinction between stage 3 NK cell precursors (presumed RORγt−) and ILC3s (RORγt+) in humans is unclear. We performed qPCR on 30 individually sorted stage 3 cells from each of three separate donor tonsil samples, and we detected RORC2 expression in 78% of cells (Table S1). While the positive control gene, CD45, was also detected in only 78% of sorted cells, these single cell qPCR data suggest that not all stage 3 cells necessarily express RORC2 and that there may be heterogeneity within this population. Indeed other groups have investigated the latter possibility. For example, Ahn et al. showed that CD7 and LFA-1 could distinguish stage 3 NK cell precursors from ILC3s within the Lin−CD117+CD94− population derived in vitro following culture of human UCB CD34+ cells (Ahn et al., 2013). However, here we demonstrated that all freshly purified SLT stage 3 cells (as well as all stage 4a, 4b, and 5 cells) expressed LFA-1. In addition, we have observed that some cells within stages 3 – 5 in SLTs are CD7− (A.G.F. and K.A.K., unpublished data). Therefore, LFA-1 and/or CD7 are not sufficient ex vivo to distinguish SLT NK cell precursors from ILC3s.
In a separate study working with freshly purified human tonsil Lin−CD161+CD117+ cells, Crellin et al. showed that a minor CD127− fraction preferentially generated NK cells following in vitro culture with cytokines and stimulated PBMCs, whereas the predominant CD127+ fraction primarily retained ILC3 features under these culture conditions (Crellin et al., 2010). The authors concluded that CD127 expression distinguishes stage 3 NK cell precursors from ILC3s. However, in that study, CD94 was not used to exclude mature NK cells during the purification procedure, and as shown here and also as reported by others mature SLT NK cells and PB CD56bright NK cells can express CD127 (Romagnani et al., 2007, Vosshenrich et al., 2006). In addition, we have previously demonstrated (Freud et al., 2006, Hughes et al., 2014) and also show here with our DC co-culture and in vivo transplantation experiments that NK cell differentiation is significantly influenced by the microenvironment. Thus, it is possible that the results and conclusions presented in the Crellin et al. study were impacted by the culture conditions employed and/or by the preferential expansion of CD94+ NK cells that may have been present in the starting Lin−CD161+CD117+CD127− populations. As such and in light of the data presented here, to the best of our knowledge there is still no reliable method to distinguish stage 3 NK cell precursors from ILC3s ex vivo, and it remains possible that these populations comprise the same cell type. Further investigation is needed.
In summary, we have identified two distinct subsets of SLT stage 4 cells: an NKp80− stage 4a population with both NK- and ILC3-associated features as well as an NKp80+ stage 4b population with phenotypic and functional attributes most similar to PB CD56bright NK cells. Our combined ex vivo, in vitro, and in vivo data indicate that NKp80 marks a critical step in human NK cell development and that a physiologic pathway of human NK cell development likely exists in SLTs through a stage 4a intermediate with ILC3-associated features.
EXPERIMENTAL PROCEDURES
Tissue Collection
All tissues were collected under protocols approved by the OSU Institutional Review Board and performed in accordance with approved guidelines. Fresh human pediatric tonsils were obtained through the NCI approved Cooperative Human Tissue Network (CHTN) from Nationwide Children’s Hospital, Columbus, Ohio. Adult PB leukopaks were obtained from the American Red Cross, Columbus, Ohio. Human UCB and BM specimens were obtained through the Translational Cell Processing Core at Cincinnati Children’s Hospital Medical Center (CCHMC). Clinically benign adult axillary lymph nodes were procured from breast cancer patients undergoing elective prophylactic or therapeutic mastectomies.
Cell Isolation
ILCs were enriched from SLTs as described (Scoville et al., 2015). Briefly, single cell suspensions were generated from SLTs by automated dissociation using the GentleMACs benchtop instrument (Miltenyi Biotec) on the “Spleen 1” setting, according to manufacturer’s instructions. The cells were then resuspended in fetal bovine serum (FBS) (Sigma Aldrich), leukocyte-depleted allogeneic AB Rh+ red blood cells (RBC) (The American Red Cross, Columbus, Ohio), and a custom Human NK cell enrichment RosetteSep reagent (StemCell Technologies) containing a cocktail of bivalent antibodies against glycophorin A and CD3, CD4, CD19, CD20, CD36, CD66b, and CD123. ILCs were enriched from adult human leukopak (i.e. PB), BM, and UCB samples using the standard Human NK cell enrichment RosetteSep reagent. PB monocytes were enriched from adult leukopaks using the Human Monocyte enrichment RosetteSep reagent (StemCell Technologies). Samples mixed with RosetteSep reagents were incubated for 30 minutes at room temperature on a nutator, diluted in phosphate-buffered saline (GE Healthcare), layered over Ficoll-Paque PLUS (GE Healthcare), and then centrifuged at 754 × g for 30 minutes at room temperature with the brake off. The monolayers were harvested, and remaining RBCs were removed using an ammonium chloride RBC lysis buffer (StemCell Technologies). Monocytes were further enriched using the human CD14+ cell isolation kit (Miltenyi Biotec) followed by fluorescence-activated cell sorting (FACS) purification of CD14+ cells to >99% purity using a FACSAriaII sorter (BD Biosciences). For all experiments NK cell developmental stages were sorted to >99% pure from tonsil ILC-enriched mononuclear fractions using a FACSAriaII sorter. Stage 3 cells were sorted as Lin−CD117+CD94−NKp80−CD16−; stage 4a cells were sorted as Lin−CD94+NKp80−CD16−; stage 4b cells were sorted as Lin−CD94+NKp80+CD16−; and stage 5 cells were sorted as Lin−CD94+NKp80+CD16+ (Figure S2). Throughout the manuscript, “Lin” refers to the following antigens: CD3, CD14, CD19, CD20, and CD34.
Flow cytometry
Flow cytometry immunophenotyping was performed using an LSRII cytometer (BD Biosciences) as previously described (Scoville et al., 2015). Isotype-matched or unstimulated controls were used when appropriate to determine the level of nonspecific staining. Sytox V500 (Life Technologies) and AquaBlue (Life Technologies) viability dyes were utilized to exclude non-viable cells in the analyses. The Supplemental Experimental Procedures contains a list of the antibodies used. Data were analyzed using either FACSDiva (BD Biosciences) or FlowJo (TreeStar) software.
Real-time RT-PCR
mRNA was isolated and purified using the Total RNA Purification Kit according to the manufacturer’s instructions (Norgen Biotek), and cDNA was synthesized using the Superscript VILO Master Mix (Life Technologies). A Viia7 Real-Time PCR System (Life Technologies) was used to run standard qPCR reactions using custom primers (Sigma Aldrich) specifically targeting the following genes: TBX21, EOMES, RORC2, and AHR. The Supplemental Experimental Procedures contains the primer sequences for each gene tested. Gene expression was normalized to the GAPDH mRNA internal control (ΔCt = Ct gene of interest - Ct GAPDH). Relative mRNA expression for each target was calculated as 2−(ΔCt).
Cytokine production and cytotoxicity assays
Purified SLT cells were resuspended at 5E4 cells/mL in RPMI-1640 culture media containing 10% fetal bovine serum (FBS) (Sigma Aldrich), 1% antibiotic/antimycotic (Life Technologies), and 1% Hepes Buffer (Life Technologies). 200 μL was added to each well of a 96-well round-bottom plate (Corning), and the cells were stimulated for 16 hr at 37 ºC in media only; in 10 ng/mL recombinant human IL-1β (Miltenyi Biotec) plus 10 ng/mL recombinant human IL-23 (Miltenyi Biotec); or in 10 ng/mL recombinant human IL-12 (Amgen), 10 ng/mL recombinant human IL-15 (Miltenyi Biotec), and 10 ng/mL recombinant human IL-18 (Amgen). Following the stimulations, supernatants were frozen at −80 ºC until they were thawed and analyzed by ELISA as previously described (Freud et al., 2006). For intracellular cytokine detection by flow cytometry, tonsil ILC-enriched mononuclear cells were incubated with either cytokines or with a combination of PMA/Ionomycin (eBiosciences), and 1 nM recombinant human IL-2 (PeproTech) plus Brefeldin A (BD Biosciences) at 37 ºC. After 6 hr, the cells were harvested, stained with antibodies against cell surface antigens, and then permeabilized using Cytofix/Cytoperm (BD Biosciences). The cells were then stained with antibodies against IL-22 and/or IFN-γ and finally fixed with 1% formalin prior to performing flow cytometry. For cytotoxicity assays, purified cells were tested against K562 targets at a 40:1 effector:target cell ratio in 4-hr 51Cr release assays in RPMI-1640 medium with or without 1 nM IL-2 as previously described (Freud et al., 2005).
Cell culture
Purified tonsil-derived populations were cultured for two weeks at a concentration of 5E3 cells/mL in 200 μL culture medium per well in 96-well flat-bottom plates (Corning) in the presence or absence of 1.5E5 allogeneic monocyte-derived DCs (see below). For some experiments, the DCs were irradiated (3E3 rad) prior to co-culture. The culture medium for these experiments consisted of MEM-α (Life Technologies) plus 10% FBS, 1% antibiotic/antimycotic, 1% Hepes Buffer, recombinant human KL (100 ng/mL) (Amgen), recombinant human FL (100 ng/mL) (Miltenyi Biotec), and recombinant human IL-15 (10 ng/mL) (Miltenyi Biotec). For cloning experiments with DCs, single stage 3 cells were individually sorted from bulk sorted stage 3 preparations (i.e. two-rounds of sorting) into wells of 96-well round-bottom plates containing 200 μL MEM-α media with 5E4 DCs. For cloning experiments with PBMCs, individual stage 3 cells were directly sorted into wells of 96-well round-bottom plates containing 5E4 irradiated (3E3 rad) PBMCs pooled from three allogeneic PB samples in RPMI-1640 media plus 10% FBS, 1% antibiotic/antimycotic, 1% Hepes Buffer, 1 nM recombinant human IL-2, and 5E4 Human T-Activator CD3/CD28 Dynabeads® (Life Technologies). Fresh irradiated PBMCs and activator beads were replenished at day 10. Clones were grown for four weeks, and wells with visible growth after this time period were analyzed by flow cytometry. For all cultures, half the media was replaced with 2X cytokines every 3–5 days. In order to derive DCs in vitro, purified monocytes were cultured for six days in MEM-α medium plus 10% FBS, 1% antibiotic/antimycotic, 1% Hepes Buffer, 10 ng/mL recombinant human IL-4 (Miltenyi Biotec), and 10 ng/mL recombinant human GM-CSF (Miltenyi Biotec). Half the medium was then replaced with 20 ng/mL IL-4 and GM-CSF plus 20 ng/mL recombinant human TNF-α (Miltenyi Biotec) and 20 ng/mL purified lipopolysaccharide (Sigma Aldrich) for two more days of culture to mature the DCs. For the cell membrane tracer experiments, enriched tonsil-derived ILCs were stained with antibodies for cell sorting as well as with 5 μM of a membrane tracer dye (Cell Proliferation Dye eFluor® 670, eBioscience), and then tracer dye+ stage 3 cells were sorted to >99% purity. The sorted cells were then co-cultured in bulk for two weeks with or without DCs as described above albeit with IL-15 used at a concentration of 10 pg/mL.
Transplantation experiments in NSG mice
The in vivo experiments with NSG mice (The Jackson Laboratory) were performed in accordance with an approved OSU Institutional Animal Care and Use Committee protocol. Six to ten week old female NSG mice were treated with 25 mg/kg busulfan one day prior to injection of purified human tonsil-derived cells in order to promote engraftment. The next day 5E3 – 1E5 purified NK cell developmental intermediates were injected intravenously into the tail veins of the NSG mice immediately after sorting. At day 0 and twice weekly thereafter for five weeks the mice were given intraperitoneal injections of recombinant human IL-15 (0.5 μg). Five weeks following transplantation, the mice were sacrificed, and their spleens, blood, and BM cells were harvested and then analyzed by flow cytometry. During the latter, murine leukocytes were excluded using an anti-mouse CD45.1 antibody (BD Biosciences). Human leukocytes were identified with an anti-human CD45 antibody (Miltenyi Biotec).
Statistical analysis
Linear mixed effects models were used for analysis to take account of the correlation among observations from the same donor. The differences between conditions were estimated from those models. Holm’s procedure was used to adjust for multiple comparisons. After adjustment for multiple comparisons, p-values <0.05 were considered significant. Summary statistics, such as mean and standard deviation, were provided for all variables. SAS 9.4 was used for analysis (SAS Institute Inc., NC).
Supplementary Material
Highlights.
NKp80 is expressed on a subset of CD94+ NK cells in secondary lymphoid tissues.
CD94+NKp80− NK cells have ILC3-associated features.
CD94+NKp80+ NK cells produce IFN-γ and kill MHC class I− target cells.
CD94+NKp80− NK cells are precursors to CD94+NKp80+ NK cells.
Acknowledgments
This work was supported by NCI grants (MAC) CA095426, CA163205, CA16058, CA068458 MAC, and funds from Pelotonia Fellowship. We thank the Mt. Auburn Ob-Gyn associates and delivery nursing staff at Christ Hospital, the Cell processing and Manipulation Core in the Translational Cores, and the Physicians and Nurses at CCHMC for collecting and processing healthy donor UCB and adult BM samples. We also thank the staff at the CHTN from Nationwide Children’s Hospital, Columbus, Ohio as well as at the flow cytometry shared resource from the OSU Comprehensive Cancer Center. We thank Tatiana Oberyszyn for her insightful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
A.G.F., K.A.K, S.D.S., B.L.M., S. C., and Y.Y. designed, performed, and interpreted experiments. X.M. and X.Z. performed statistical analyses. A.G.F., T.H., R.A.B., P.P., J.Y., W.E.C., and M.A.C. interpreted results and wrote the manuscript. W.E.C. also procured lymph node specimens.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AHN YO, BLAZAR BR, MILLER JS, VERNERIS MR. Lineage relationships of human interleukin-22-producing CD56+ RORgammat+ innate lymphoid cells and conventional natural killer cells. Blood. 2013;121:2234–43. doi: 10.1182/blood-2012-07-440099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANFOSSI N, ANDRE P, GUIA S, FALK CS, ROETYNCK S, STEWART CA, BRESO V, FRASSATI C, REVIRON D, MIDDLETON D, ROMAGNE F, UGOLINI S, VIVIER E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- BARTEL Y, BAUER B, STEINLE A. Modulation of NK cell function by genetically coupled C-type lectin-like receptor/ligand pairs encoded in the human natural killer gene complex. Front Immunol. 2013;4:362. doi: 10.3389/fimmu.2013.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNINK JH, KRABBENDAM L, GERMAR K, DE JONG E, GRONKE K, KOFOED-NIELSEN M, MUNNEKE JM, HAZENBERG MD, VILLAUDY J, BUSKENS CJ, BEMELMAN WA, DIEFENBACH A, BLOM B, SPITS H. Interleukin-12 and −23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 2015;43:146–60. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- BERNINK JH, PETERS CP, MUNNEKE M, TE VELDE AA, MEIJER SL, WEIJER K, HREGGVIDSDOTTIR HS, HEINSBROEK SE, LEGRAND N, BUSKENS CJ, BEMELMAN WA, MJÖSBERG JM, SPITS H. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–9. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- BJORKSTROM NK, RIESE P, HEUTS F, ANDERSSON S, FAURIAT C, IV, ARSSON MA, BJORKLUND AT, FLODSTROM-TULLBERG M, MICHAELSSON J, ROTTENBERG ME, GUZMAN CA, LJUNGGREN HG, MALMBERG KJ. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–64. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- CALIGIURI MA. Human natural killer cells. Blood. 2008;112:461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CELLA M, FUCHS A, VERMI W, FACCHETTI F, OTERO K, LENNERZ JK, DOHERTY JM, MILLS JC, COLONNA M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–5. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONSTANTINIDES MG, MCDONALD BD, VERHOEF PA, BENDELAC A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COOPER MA, FEHNIGER TA, CALIGIURI MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–40. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- COOPER MA, FEHNIGER TA, FUCHS A, COLONNA M, CALIGIURI MA. NK cell and DC interactions. Trends Immunol. 2004;25:47–52. doi: 10.1016/j.it.2003.10.012. [DOI] [PubMed] [Google Scholar]
- CORTEZ VS, ROBINETTE ML, COLONNA M. Innate lymphoid cells: new insights into function and development. Curr Opin Immunol. 2015;32:71–7. doi: 10.1016/j.coi.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRELLIN NK, TRIFARI S, KAPLAN CD, CUPEDO T, SPITS H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med. 2010;207:281–90. doi: 10.1084/jem.20091509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUPEDO T, CRELLIN NK, PAPAZIAN N, ROMBOUTS EJ, WEIJER K, GROGAN JL, FIBBE WE, CORNELISSEN JJ, SPITS H. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- FERLAZZO G, PACK M, THOMAS D, PALUDAN C, SCHMID D, STROWIG T, BOUGRAS G, MULLER WA, MORETTA L, MUNZ C. Distinct roles of IL-12 and IL-15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc Natl Acad Sci U S A. 2004;101:16606–11. doi: 10.1073/pnas.0407522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREUD AG, BECKNELL B, ROYCHOWDHURY S, MAO HC, FERKETICH AK, NUOVO GJ, HUGHES TL, MARBURGER TB, SUNG J, BAIOCCHI RA, GUIMOND M, CALIGIURI MA. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- FREUD AG, CALIGIURI MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- FREUD AG, YOKOHAMA A, BECKNELL B, LEE MT, MAO HC, FERKETICH AK, CALIGIURI MA. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203:1033–43. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREUD AG, YU J, CALIGIURI MA. Human natural killer cell development in secondary lymphoid tissues. Semin Immunol. 2014;26:132–7. doi: 10.1016/j.smim.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUCHS A, VERMI W, LEE JS, LONARDI S, GILFILLAN S, NEWBERRY RD, CELLA M, COLONNA M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 2013;38:769–81. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRZYWACZ B, KATARIA N, SIKORA M, OOSTENDORP RA, DZIERZAK EA, BLAZAR BR, MILLER JS, VERNERIS MR. Coordinated acquisition of inhibitory and activating receptors and functional properties by developing human natural killer cells. Blood. 2006;108:3824–33. doi: 10.1182/blood-2006-04-020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUIMOND M, FREUD AG, MAO HC, YU J, BLASER BW, LEONG JW, VANDEUSEN JB, DORRANCE A, ZHANG J, MACKALL CL, CALIGIURI MA. In vivo role of Flt3 ligand and dendritic cells in NK cell homeostasis. J Immunol. 2010;184:2769–75. doi: 10.4049/jimmunol.0900685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAZENBERG MD, SPITS H. Human innate lymphoid cells. Blood. 2014 doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- HUGHES T, BECKNELL B, FREUD AG, MCCLORY S, BRIERCHECK E, YU J, MAO C, GIOVENZANA C, NUOVO G, WEI L, ZHANG X, GAVRILIN MA, WEWERS MD, CALIGIURI MA. Interleukin-1beta selectively expands and sustains interleukin-22+ immature human natural killer cells in secondary lymphoid tissue. Immunity. 2010;32:803–14. doi: 10.1016/j.immuni.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES T, BECKNELL B, MCCLORY S, BRIERCHECK E, FREUD AG, ZHANG XL, MAO HY, NUOVO G, YU JH, CALIGIURI MA. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the T(H)17 cytokine interleukin-22. Blood. 2009;113:4008–4010. doi: 10.1182/blood-2008-12-192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES T, BRIERCHECK EL, FREUD AG, TROTTA R, MCCLORY S, SCOVILLE SD, KELLER K, DENG Y, COLE J, HARRISON N, MAO C, ZHANG J, BENSON DM, YU J, CALIGIURI MA. The Transcription Factor AHR Prevents the Differentiation of a Stage 3 Innate Lymphoid Cell Subset to Natural Killer Cells. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM S, POURSINE-LAURENT J, TRUSCOTT SM, LYBARGER L, SONG YJ, YANG L, FRENCH AR, SUNWOO JB, LEMIEUX S, HANSEN TH, YOKOYAMA WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- KLIMOSCH SN, BARTEL Y, WIEMANN S, STEINLE A. Genetically coupled receptor-ligand pair NKp80-AICL enables autonomous control of human NK cell responses. Blood. 2013;122:2380–9. doi: 10.1182/blood-2013-01-479790. [DOI] [PubMed] [Google Scholar]
- KLOSE CS, FLACH M, MÖHLE L, ROGELL L, HOYLER T, EBERT K, FABIUNKE C, PFEIFER D, SEXL V, FONSECA-PEREIRA D, DOMINGUES RG, VEIGA-FERNANDES H, ARNOLD SJ, BUSSLINGER M, DUNAY IR, TANRIVER Y, DIEFENBACH A. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–56. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- LOPEZ-VERGES S, MILUSH JM, PANDEY S, YORK VA, ARAKAWA-HOYT J, PIRCHER H, NORRIS PJ, NIXON DF, LANIER LL. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116:3865–74. doi: 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUETKE-EVERSLOH M, KILLIG M, ROMAGNANI C. Signatures of human NK cell development and terminal differentiation. Front Immunol. 2013;4:499. doi: 10.3389/fimmu.2013.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATOS ME, SCHNIER GS, BEECHER MS, ASHMAN LK, WILLIAM DE, CALIGIURI MA. Expression of a functional c-kit receptor on a subset of natural killer cells. J Exp Med. 1993;178:1079–84. doi: 10.1084/jem.178.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOFFETT A, COLUCCI F. Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest. 2014;124:1872–9. doi: 10.1172/JCI68107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONTALDO E, TEIXEIRA-ALVES LG, GLATZER T, DUREK P, STERVBO U, HAMANN W, BABIC M, PACLIK D, STÖLZEL K, GRÖNE J, LOZZA L, JUELKE K, MATZMOHR N, LOIACONO F, PETRONELLI F, HUNTINGTON ND, MORETTA L, MINGARI MC, ROMAGNANI C. Human RORγt(+)CD34(+) cells are lineage-specified progenitors of group 3 RORγt(+) innate lymphoid cells. Immunity. 2014;41:988–1000. doi: 10.1016/j.immuni.2014.11.010. [DOI] [PubMed] [Google Scholar]
- MORETTA A. The dialogue between human natural killer cells and dendritic cells. Curr Opin Immunol. 2005;17:306–11. doi: 10.1016/j.coi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- OKADA S, MARKLE JG, DEENICK EK, MELE F, AVERBUCH D, LAGOS M, ALZAHRANI M, AL-MUHSEN S, HALWANI R, MA CS, WONG N, SOUDAIS C, HENDERSON LA, MARZOUQA H, SHAMMA J, GONZALEZ M, MARTINEZ-BARRICARTE R, OKADA C, AVERY DT, LATORRE D, DESWARTE C, JABOT-HANIN F, TORRADO E, FOUNTAIN J, BELKADI A, ITAN Y, BOISSON B, MIGAUD M, ARLEHAMN CS, SETTE A, BRETON S, MCCLUSKEY J, ROSSJOHN J, DE VILLARTAY JP, MOSHOUS D, HAMBLETON S, LATOUR S, ARKWRIGHT PD, PICARD C, LANTZ O, ENGELHARD D, KOBAYASHI M, ABEL L, COOPER AM, NOTARANGELO LD, BOISSON-DUPUIS S, PUEL A, SALLUSTO F, BUSTAMANTE J, TANGYE SG, CASANOVA JL. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–13. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROMAGNANI C, JUELKE K, FALCO M, MORANDI B, D’AGOSTINO A, COSTA R, RATTO G, FORTE G, CARREGA P, LUI G, CONTE R, STROWIG T, MORETTA A, MÜNZ C, THIEL A, MORETTA L, FERLAZZO G. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178:4947–55. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- SATOH-TAKAYAMA N, LESJEAN-POTTIER S, VIEIRA P, SAWA S, EBERL G, VOSSHENRICH CA, DI SANTO JP. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J Exp Med. 2010;207:273–80. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCOVILLE SD, KELLER KA, CHENG S, ZHANG M, ZHANG X, CALIGIURI MA, FREUD AG. Rapid Column-Free Enrichment of Mononuclear Cells from Solid Tissues. Sci Rep. 2015;5:12490. doi: 10.1038/srep12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCOVILLE SD, MUNDY-BOSSE BL, ZHANG MH, CHEN L, ZHANG X, KELLER KA, HUGHES T, CHEN L, CHENG S, BERGIN SM, MAO HC, MCCLORY S, YU J, CARSON WE, 3RD, CALIGIURI MA, FREUD AG. A Progenitor Cell Expressing Transcription Factor RORgammat Generates All Human Innate Lymphoid Cell Subsets. Immunity. 2016;44:1140–1150. doi: 10.1016/j.immuni.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOJKA DK, PLOUGASTEL-DOUGLAS B, YANG L, PAK-WITTEL MA, ARTYOMOV MN, IV, ANOVA Y, ZHONG C, CHASE JM, ROTHMAN PB, YU J, RILEY JK, ZHU J, TIAN Z, YOKOYAMA WM. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPITS H, ARTIS D, COLONNA M, DIEFENBACH A, DI SANTO JP, EBERL G, KOYASU S, LOCKSLEY RM, MCKENZIE AN, MEBIUS RE, POWRIE F, VIVIER E. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–9. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- TANG Q, AHN YO, SOUTHERN P, BLAZAR BR, MILLER JS, VERNERIS MR. Development of IL-22-producing NK lineage cells from umbilical cord blood hematopoietic stem cells in the absence of secondary lymphoid tissue. Blood. 2011;117:4052–5. doi: 10.1182/blood-2010-09-303081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VITALE M, FALCO M, CASTRICONI R, PAROLINI S, ZAMBELLO R, SEMENZATO G, BIASSONI R, BOTTINO C, MORETTA L, MORETTA A. Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur J Immunol. 2001;31:233–42. doi: 10.1002/1521-4141(200101)31:1<233::AID-IMMU233>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- VONARBOURG C, MORTHA A, BUI VL, HERNANDEZ PP, KISS EA, HOYLER T, FLACH M, BENGSCH B, THIMME R, HÖLSCHER C, HÖNIG M, PANNICKE U, SCHWARZ K, WARE CF, FINKE D, DIEFENBACH A. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity. 2010;33:736–51. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOSSHENRICH CA, GARCIA-OJEDA ME, SAMSON-VILLEGER SI, PASQUALETTO V, ENAULT L, RICHARD-LE GOFF O, CORCUFF E, GUY-GRAND D, ROCHA B, CUMANO A, ROGGE L, EZINE S, DI SANTO JP. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–24. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- YU J, FREUD AG, CALIGIURI MA. Location and cellular stages of natural killer cell development. Trends Immunol. 2013 doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.