Summary
Transcription controls splicing and other gene regulatory processes, yet mechanisms remain obscure due to our fragmented knowledge of the molecular connections between the dynamically phosphorylated RNA polymerase II (Pol II) C-terminal domain (CTD) and regulatory factors. By systematically isolating phosphorylation states of the CTD heptapeptide repeat (Y1S2P3T4S5P6S7), we identify hundreds of protein factors that are differentially enriched, revealing unappreciated connections between the Pol II CTD and co-transcriptional processes. These data uncover a role for threonine-4 in 3′ end processing through controlling the transition between cleavage and termination. Furthermore, serine-5 phosphorylation seeds spliceosomal assembly immediately downstream of 3′ splice sites through a direct interaction with spliceosomal subcomplex, U1. Strikingly, threonine-4 phosphorylation also impacts splicing through serving as a mark of co-transcriptional spliceosome release and ensuring efficient post-transcriptional splicing genome-wide. Thus, comprehensive Pol II interactomes identify the complex and functional connections between transcription machinery and other gene regulatory complexes.
Introduction
Transcription regulation controls both RNA transcript levels and the final sequence of the RNA transcript through regulated coordination with co-transcriptional RNA processing. Capping, splicing and termination are coupled to transcription through phosphorylation of the C-terminal domain (CTD) of the largest RNA polymerase II (Pol II) subunit. The CTD extends from the Pol II body and consists of a conserved heptapeptide, Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, repeated 26 times in yeast and 52 times in humans. Five of the seven residues are dynamically phosphorylated during the transcription cycle in a characteristic pattern across gene bodies. (Fuchs et al., 2009; Hintermair et al., 2012; Jeronimo et al., 2013; Komarnitsky et al., 2000; Mayer et al., 2010, 2012;). Phosphorylation of particular CTD residues contributes to the recruitment of key regulatory factors at the right place and time, facilitating connections to co-transcriptional processes. Yet, the extent of connections between CTD phosphorylation marks and RNA processing machinery are not clear. A focus on identifying direct phospho-CTD binding proteins has slowed experimental throughput and overlooked mechanisms that occur through phosphorylation marks blocking regulatory interactions or indirectly connecting transcription to other processes.
The best-characterized CTD residues are Ser2 and Ser5. Pol II complexes containing high levels of phospho-Ser2 (Ser2P) are thought to be involved in the latter stages of transcription as Ser2P rises slowly across gene bodies and peaks at the 3′ ends of genes (Barillà et al., 2001; Kim et al., 2004). A termination factor, Rtt103, directly binds Ser2P, yet is only recruited after the polyadenylation (polyA) site. This specificity is controlled in part by phospho-Tyr1 (Tyr1P), presumably through blocking Rtt103 binding before the polyA site (Mayer et al., 2012). It is unclear whether additional residues contribute to the precise recruitment of Rtt103 and, more generally, how multiple residues might work together in the coupling of other RNA processing events to transcription.
Pol II complexes containing high levels of phospho-Ser5 (Ser5P) are enriched near the transcription start site (TSS) of genes, consistent with the essential role of Ser5P in the earliest mRNA processing event, capping of the nascent RNA (Cho et al., 1997; Komarnitsky et al., 2000; McCracken et al., 1997; Schroeder et al., 2000; Schwer and Shuman, 2011). Recently, a connection between Ser5P and splicing has been suggested. In yeast, Ser5P increases transiently during a splicing-related transcription checkpoint at the 3′ splice site, where Pol II pauses and does not resume transcription until splicing-related events occur (Alexander et al., 2010; Chathoth et al., 2014). However it remains unclear whether there are direct connections between splicing and Ser5P or the CTD in general (Licatalosi et al., 2001). Initial studies indicated a component of the spliceosome (Prp40) could directly bind the CTD (Morris and Greenleaf, 2000). However, other work showed no defect on the co-transcriptional recruitment of the spliceosome in strains where the CTD-binding domain of Prp40 was removed (Görnemann et al., 2011). Thus despite the observation of co-transcriptional splicing and spliceosomal assembly, (Kotovic et al., 2003; Lacadie and Rosbash, 2005; Lacadie et al., 2006; Moore et al., 2006; Tardiff et al., 2006) the degree of coupling between the transcription and splicing machineries, and what role Ser5P may play, remains unclear.
Not all CTD residues are as well characterized as Ser2 and Ser5. For example, Thr4 regulates elongation in human cells but phospho-Thr4 (Thr4P) levels peak after polyA sites (Hintermair et al., 2012). Substitution of valine for each CTD threonine yielded a defect only in 3′ end processing of histone mRNAs in chicken DT40 cells (Hsin et al., 2011) and had minimal impact on the transcriptome in yeast (Rosonina et al., 2014). Yet recent mass spectrometry analysis of the Pol II CTD showed that Thr4P is possibly as abundant as Ser2P, suggesting that key roles for this residue remain to be discovered (Schuller et al., 2016).
To determine unappreciated connections between CTD phosphorylation and RNA processing, we performed an unbiased and comprehensive analysis of phospho-specific Pol II complexes. We developed a sequential purification strategy to purify Pol II complexes enriched for each CTD phosphoisoform and used quantitative label-free mass spectrometry to identify factors that interact with phospho-specifc Pol II complexes compared to all Pol II complexes. Application of this approach to Saccharomyces cerevisiae identified unique phospho-specific Pol II interactomes for the five phosphorylatable CTD residues. The factors enriched in each phospho-CTD interactome allow prediction and discovery of new roles for individual factors and phospho-CTD residues. By combining the interactome data with multiple high-resolution genomic techniques, we describe a previously uncharacterized role for Thr4 in transcription termination in yeast, through the recruitment of termination factors and regulation of Pol II dynamics.
Our proteomic analysis reveals a striking differential enrichment of the spliceosome as a function of CTD phosphorylation. The Ser5P interactome is strongly enriched with U1 spliceosomal components and we demonstrate that Ser5P contributes directly to the co-transcriptional recruitment of the spliceosome. The Thr4P interactome has a complete lack of enriched spliceosome subunits and we observe that this mark peaks in the terminal exon after dissociation of splicing machinery. Thr4 is required in the post-transcriptional completion of splicing, demonstrating that CTD modifications impact both co-transcriptional and post-transcriptional splicing through separate residues. Together, these data comprehensively define the protein factors that associate with Pol II and uncover novel roles for CTD modifications in splicing and termination regulation.
Results
Proteomic Analysis of Phospho-specific Pol II Complexes
To determine the compendium of factors that associate to actively transcribing Pol II, we first analyzed purifications of total Pol II by label-free quantitative mass spectrometry. We adapted the immunoprecipitation approach developed for native elongating transcript sequencing (NET-seq), whereby engaged Pol II complexes are solubilized from chromatin and immunoprecipitated (Churchman and Weissman, 2011). Analysis of enriched factors was consistent with previous Pol II proteomics studies and in identifying many known transcription and RNA processing factors (Figure S1A) (Mosley et al., 2011, 2013; Tardiff et al., 2007).
In order to investigate direct and indirect Pol II interactors specific to each CTD phosphoisoform, we developed a native sequential immunoprecipitation (IP) strategy to purify Pol II complexes enriched for different phosphorylated CTD residues (Figure 1A). Our strategy is designed to purify the host of proteins that interact both directly and indirectly with Pol II when a particular residue is phosphorylated. Thus we obtain a comprehensive picture of the co-occurring processes marked by each phosphorylated residue. To this end, all Pol II complexes are first purified via a 3xFLAG epitope tag to isolate the full complement of Pol II phosphoisoforms (5 phosphorylatable residues across 26 repeats). From this pool, highly efficient IPs enrich for sets of phosphoisoforms with well-characterized and specific antisera targeting each phosphorylatable residue (Tyr1P, Ser2P, Ser5P, Thr4P and Ser7P) or with a mock antisera (mCherry) (Figure S1B) (Hintermair et al., 2012; Mayer et al., 2012). Native IP conditions were chosen instead of crosslinking in order to reduce the possibility for false positives. Importantly, the repetitive nature of the CTD permits the purification of factors that directly bind the targeted phospho-CTD residue even in the case that their association is displaced by the antibody interaction. Nevertheless, for direct interactors, our purification strategy has the potential to be less quantitative and may yield false negatives. Each phospho-specific IP purifies a subset of Pol II complexes that are highly enriched for the selected phosphorylated residue, yet are also phosphorylated on other residues across the 26 repeats, thus allowing for the enrichment of indirectly associating factors recruited by frequently co-occurring patterns of Pol II modifications (Figure S1C). Furthermore, our strategy will isolate any proteins that are bound to the nascent RNA. In sum, this approach comprehensively purifies proteins that interact directly and indirectly with Pol II coincident with each phosphorylation mark.
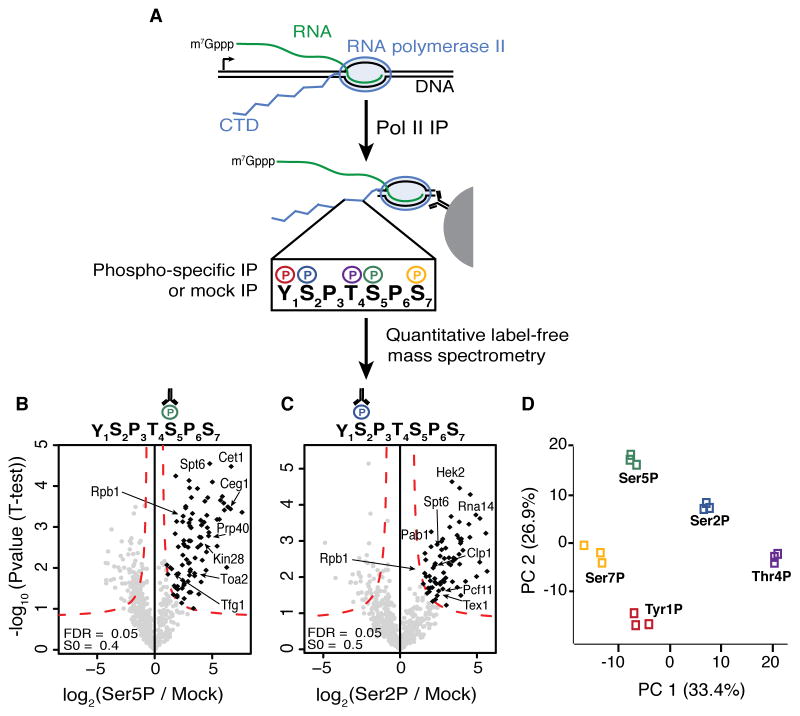
Figure 1. Purification and Analysis of Native Pol II Complexes.
(A) Diagram of native Pol II purification followed by phospho-specific immunoprecipitations (IPs) and quantitative label-free mass spectrometry analysis for each CTD phospho-modification; m7Gppp, RNA cap.
(B, C) Volcano plots comparing Ser5P (B) and Ser2P (C) IPs to mock IPs. Specifically enriched factors determined using an FDR of 0.05 and an S0 value (curve bend, see Supplemental Experimental Procedures for details) of 0.4 for Ser5P and 0.5 for Ser2P are highlighted in black. Factors mentioned in the text are labeled.
(D) Principal component analysis of significantly enriched interactors from each phospho-CTD interactome. PC, principal component.
See also Figure S1.
To identify enriched factors in a quantitative manner, samples from the Pol II IP, each phospho-specific IP, and the mock IP were analyzed by mass spectrometry in biological triplicate. Significantly enriched factors in each phospho-specific IP were determined using quantitative label-free proteomic analysis (Hubner and Mann, 2011; Hubner et al., 2010). Specifically, the phospho-specific:mock IP fold increase of spectral intensities are compared to p-values determined by t-tests, generating a volcano plot (Figures 1B and C; S1A, D–F). Only factors present in all three replicates are analyzed and are identified as significantly enriched using a false discovery rate of 0.05. As the mock IP is done after the first IP, factors enriched with each phosphoisoform represent proteins that are enriched over a background of all Pol II interactors. The stringent nature of this strategy identifies the most highly enriched factors associated with each phosphoisoform. Together, the five Pol II phospho-CTD interactomes contain nearly two hundred enriched factors, including Pol II subunits, RNA processing factors, transcription factors and chromatin-related factors. Our analysis successfully identified factors previously shown to directly associate with different phospho-CTD isoforms, such as enrichment of the mRNA capping complex (Cet1 and Ceg1) and the Ser5 kinase TFIIH in the Ser5P inteactome (Figure 1B) (Komarnitsky et al., 2000; Schroeder et al., 2000) and enrichment of Pcf11 and other cleavage, polyadenylation and export factors (Clp1, Rna14 and Tex1) in the Ser2P interactome (Figure 1C) (Barillà et al., 2001; Buratowski, 2005). Although our focus is not entirely on identifying direct phospho-CTD binders, these results demonstrate that our approach robustly purifies direct Pol II CTD interactors, presumably facilitated by the repetitive nature of the CTD.
Pol II Phospho-specific Interactomes Are Distinct
Utilizing the stringent nature of the interactome data we asked whether the proteins that are highly enriched in each Pol II CTD interactome constitute distinct groups of factors, which would suggest that each Pol II phospho-CTD residue connects to different Pol II regulation modalities and/or co-transcriptional processes. Principal component analysis of all five phospho-CTD interactomes demonstrates that each interactome contains a unique set of factors that are reproducibly identified (Figure 1D) (Tables S1 and S2). Consistently, we observe minimal overlap of factors across all interactomes (Figure 2A). Gene ontology (GO) analysis of all co-purifying factors displays enrichment of many transcription-related GO terms as well as RNA processing and chromatin organization (Figure 2B). Additionally, the phospho-CTD interactomes show enrichment for protein domains involved in chromatin regulation, RNA processing and in promoting protein-protein interactions (Figure 2C). Together these data indicate our approach isolates factors with functions in multiple transcriptional and co-transcriptional processes.
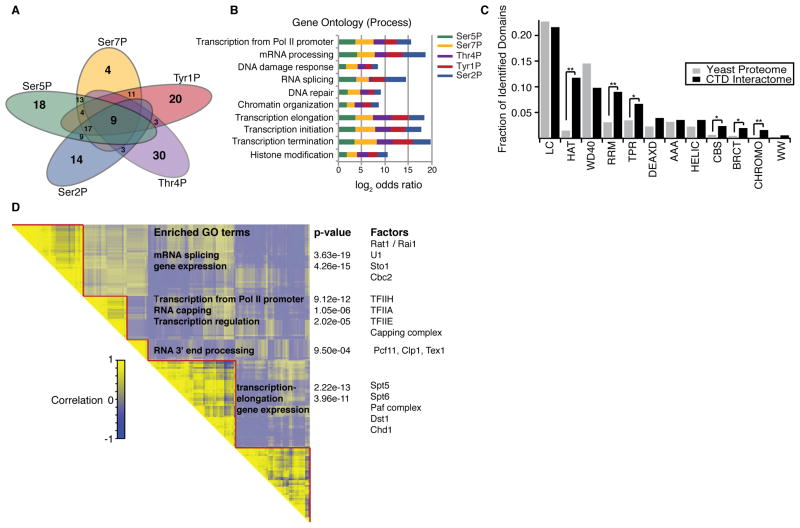
Figure 2. Pol II phospho-CTD interactomes contain unique sets of factors.
(A) Venn diagram of enriched factors from the interactome of each CTD phosphoisoform.
(B) Gene ontology terms enriched in the phospho-CTD interactome data are plotted for each phosphoisoform.
(C) Protein domains enriched in the phospho-CTD interactomes. The abundance of the top 12 protein domains identified in the phospho-CTD interactome compared to their abundance in the yeast proteome. Fisher exact test, * p-value < 0.05, ** p-value < 0.01. LC, low complexity; HAT, half-A-TPR repeat; WD40, repeat ending in tryptophan-aspatic acid repeat; RRM, RNA recognition motif; TPR, tetratricopeptide repeat; DEAXD, DEAD-like helicase; AAA, ATPases; HELIC, helicases; CBS, cystathionine beta synthase domain; BRCT, breast cancer c-terminal; CHROMO, chromatin organization modifier; WW, two conversed Trp resides.
(D) Clustered pairwise correlation matrix comparing phospho-CTD interaction profiles of all enriched factors. GO terms enriched in separate clusters, p-values for the significance of enrichment of each GO term, and representative factors in each cluster are indicated.
We reasoned that proteins with similar phospho-CTD interaction profiles should have related roles in transcription. Pairwise correlation analysis between the phospho-CTD interaction profiles of all enriched factors followed by hierarchical clustering identifies distinct groups of factors (Figure 2D). We find that factors regulating transcription initiation and early transcriptional processes, such as mRNA capping, are highly correlated and are anti-correlated with factors enriched for 3′ end processing of nascent RNA. Also, a majority of transcription elongation factors cluster together, revealing that proteins associated with the three major stages of transcription interact with Pol II in distinct modes with specific phospho-CTD states. These data demonstrate that the interactome of a phospho-CTD residue can be used to predict its function in transcription.
CTD Thr4 and Rtt103 Transition Pol II to Termination
As many functions have been attributed to the well-characterized CTD phosphoisoforms, Ser5P and Ser2P, we utilized the phospho-CTD interactome dataset to explore the lesser-studied Thr4P phosphoisoform. Moreover, mass spectrometry analysis of the Pol II CTD demonstrated that Thr4P is a prevalent modification, possibly as abundant as Ser2P (Schuller et al., 2016; Suh et al., 2016). Purification of Pol II CTD Thr4P complexes was reproducibly efficient (95% average IP efficiency, Figure S1B) and mass spectrometry analysis revealed significant associations between Thr4P and a number of canonical transcription elongation factors (Figure 3A). Interestingly, the transcription termination factor Rtt103 was also enriched with Thr4P. Rtt103 is a CTD binding protein that functions with the exonuclease complex Rat1/Rai1 for termination of polyadenylated transcripts (Buratowski, 2005; Kim et al., 2004) and was recently demonstrated to interact with a CTD containing phosphorylated Thr4 (Suh et al, 2016). To decipher whether interaction of Rtt103 with Pol II complexes is a direct result of Thr4 phosphorylation we tested the ability of the Thr4P antisera to compete with Rtt103 for CTD binding. After purifying Pol II complexes using a FLAG epitope tag on the Rpb3 subunit, antisera directed against Thr4P, Ser2P, Ser5P, no antisera or FLAG peptide (as a positive elution control) was incubated in vast excess (>13 fold higher concentration compared to the IP reactions) with the purified complexes on beads. Supernatant and bead fractions were assayed by Western blot (Figure 3B). As expected, the Ser2P antisera were able to compete with Rtt103 for CTD binding (Kim et al., 2004), and the Ser5P antisera were not effective competition. Consistent with the mass spectrometry data, Thr4P antisera were also able to compete with Rtt103 binding, indicating both residues likely regulate the interaction between Rtt103 and the Pol II CTD.
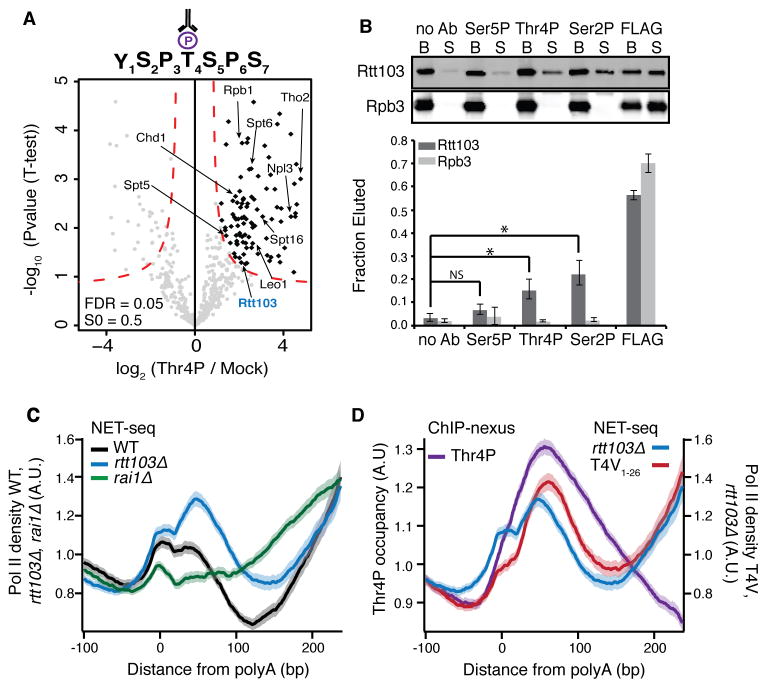
Figure 3. Thr4P regulates Pol II dynamics during transcription termination.
(A) Volcano plot comparing Thr4P IPs to mock IPs. Specifically enriched factors are determined using an FDR of 0.05 and S0 = 0.5 and are highlighted in black. Factors mentioned in the text are labeled, Rtt103 is labeled in blue.
(B) Western blot (top) and quantification (bottom) of the amount of Rtt103 or Rpb3 eluted from purified Pol II complexes by phospho-CTD antisera (no Ab, Ser5P, Ser2P, Thr4P) or FLAG peptide. Competition assays were conducted in biological triplicate. Plotted are mean fraction eluted values, error bars represent standard deviation. NS, not significant; *, p-value < 0.01; Ab, antibody; B, beads; S, supernatant.
(C) Normalized average NET-seq profiles of WT, rtt103Δ, and rai1Δ cells at polyadenylation sites (polyA) of protein coding genes.
(D) Normalized average NET-seq profiles of rtt103Δ and T4V1–26 mutant CTD cells, and normalized average ChIP-nexus profile of Thr4P at the polyA site of protein coding genes. (C) and (D), NET-seq and ChIP-nexus reads for each gene are normalized by total reads for each gene in the analyzed region, shaded areas represent the 95% confidence interval. n=2738. A.U., arbitrary units.
See also Figures S2 and S3.
Surprisingly, the Rat1/Rai1 exonuclease complex was not enriched in the Thr4P interactome, leading us to postulate that Rtt103 and Rat1/Rai1 may regulate separate steps in the termination process. To test this hypothesis we used NET-seq, which maps actively transcribing Pol II complexes at nucleotide resolution genome-wide (Churchman and Weissman, 2011) and analyzed changes in Pol II density in rtt103Δ and rai1Δ deletion mutants after polyA sites. Aggregate analysis of wild-type NET-seq data around polyA sites reveals an increase in Pol II density just after polyA sites where 3′ end cleavage occurs co-transcriptionally (Figure 3C, black line). Subsequent termination leads to a decrease in Pol II density before neighboring genes increase the signal again. Individual deletion of RAI1 and RTT103 caused widespread termination defects, albeit in very different ways (Figure 3C; Figure S2A). Loss of RAI1 (Figure 3C, green line) leads to a steady rise in Pol II density after polyA sites, indicating that transcription proceeds for much longer. In contrast, loss of RTT103 (Figure 3C and D, blue line) results in a pronounced peak of Pol II density directly after polyA sites precisely where Rtt103 occupancy peaks (Figure S2B) (Mayer et al., 2012). Furthermore, termination is not affected as Pol II density decreases normally thereafter, suggesting that instead Rtt103 binding regulates the onset of termination.
As Rtt103 is enriched in the Thr4P interactome, we postulated that Thr4 aids Rtt103 in regulating Pol II. We mapped Thr4P genome-wide at near nucleotide resolution using exo-nuclease based ChIP-nexus (He et al., 2015), which revealed a peak of threonine-4 phosphorylation downstream of the polyA site, where the peak in Pol II density is observed in the rtt103Δ strain (Figure 3D purple line, and Figure S2B, D, E, F and G). Consistent with our detection of Rtt103 in the Thr4P interactome, the peak in Thr4P density overlaps with Rtt103 density (Figure S2B). To determine whether Thr4 controls 3′ end cleavage and transcription termination, we replaced each Thr4 residue in the endogenous 26 CTD heptapeptide repeats with a valine residue (T4V1–26), representing a non-phosphorylatable state, and with a glutamic acid residue (T4E1–26), mimicking the constitutively phosphorylated state. The T4E1–26 mutant is lethal (Figure S3A), but the T4V1–26 mutant is viable, as observed previously (Rosonina et al., 2014), with a modest growth defect (15% increase in doubling time). NET-seq analysis at the 3′ ends of genes demonstrates that the T4V1–26 strain has a similar post-polyA transcription defect as the rtt103Δ strain with a strong increase in Pol II density after polyA sites, at exactly the location where Thr4P peaks in occupancy (Figure 3D and S2A, red line). A less stringent and inducible T4V mutant was created by generating an allele of RPB1 that encodes a CTD where Thr4 of the first 8 repeats proximal to the Pol II core are mutated to valine (T4V1–8). By placing the endogenous RPB1 under control of a Tet-off promoter, introduction of doxycycline induces incorporation of the mutant CTD into the Pol II complex (Malagon et al., 2006; Morrill et al., 2016.). NET-seq analysis in this T4V1–8 mutant shows defects similar to the T4V1–26 mutant in post-polyA transcription (Figure S3B). RNA-seq analysis of the T4V1–26 strain shows a global downstream shift in the locations of polyA sites, consistent with a possibility of altered cleavage in this strain (Figure S3 C–E). Importantly, no down-regulation of transcription elongation or RNA processing related gene classes was detected in either the T4V1–26 or rtt103Δ mutants (Table S3, Figures S2C, S6E). Together these data lead to a model by which both Ser2P and Thr4P recruit Rtt103 after polyA sites and indicate the presence of a post-polyA checkpoint controlled by Thr4 that ensures robust 3′ end cleavage followed by an efficient transition to transcription termination. Moreover, these data illustrate how analysis of Pol II CTD interactomes reveals distinct roles for CTD modifications and their interacting factors.
CTD Ser5P is Enriched for Spliceosomal Components While Thr4P is Depleted
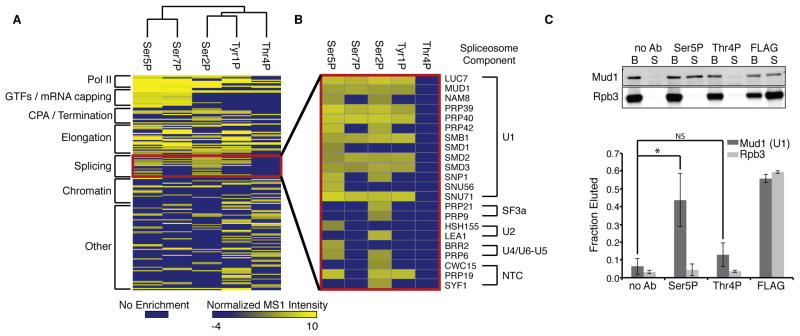
To search for additional connections between Thr4P and RNA processing events, we compared the interactomes by complete-linkage hierarchical clustering. Interestingly, CTD phosphorylation marks with similar ChIP occupancy (Ser5P/Ser7P and Tyr1P/Ser2P) have interactomes that cluster together (Figure 4A), and the Thr4P interactome does not cluster with any other phospho-CTD interactome. A major feature separating the Thr4P interactome is the absence of enriched spliceosomal subunits (Figure 4B) while other phospho-CTD interactomes enrich for 8–18 spliceosomal subunits across five subcomplexes. In contrast, the Ser5P interactome is highly enriched for many components of the U1 spliceosomal complex, purifying nearly 70% of U1 associated proteins. To determine if U1 is associating through a protein-protein interaction with Ser5P we tested the ability of Ser5P antisera to displace U1 from purified Pol II complexes. Ser5P antisera were able to elute nearly half of the associated U1 from the CTD, while Thr4P antisera were unable to significantly compete with U1 binding (Figure 4C). These data suggest that U1 directly associates with Pol II in a Ser5P dependent manner. The difference in enrichment and interaction with the spliceosome between Ser5P and Thr4P suggest each phospho-modification may regulate different stages of splicing.
Figure 4. Phospho-CTD Interactomes Reveal Connection Between Pol II CTD Ser5P, Thr4P and Splicing.
(A) Complete linkage clustering of each Pol II phospho-CTD interactome.
(B) Heatmap of splicing factors specifically enriched in each phospho-CTD Pol II interactome.
(C) Western blot (top) and quantification (bottom) of the amount of Mud1 (U1) or Rpb3 eluted from purified Pol II complexes by phospho-CTD antisera (no Ab, Ser5P, Thr4P) or FLAG peptide. Competition assays were conducted in biological triplicate. Plotted are mean fraction eluted values, error bars represent standard deviation; NS, not significant; *, p-value < 0.05; Ab, antibody; B, beads; S, supernatant.
Co-transcriptional Spliceosome Recruitment is Facilitated by Ser5P
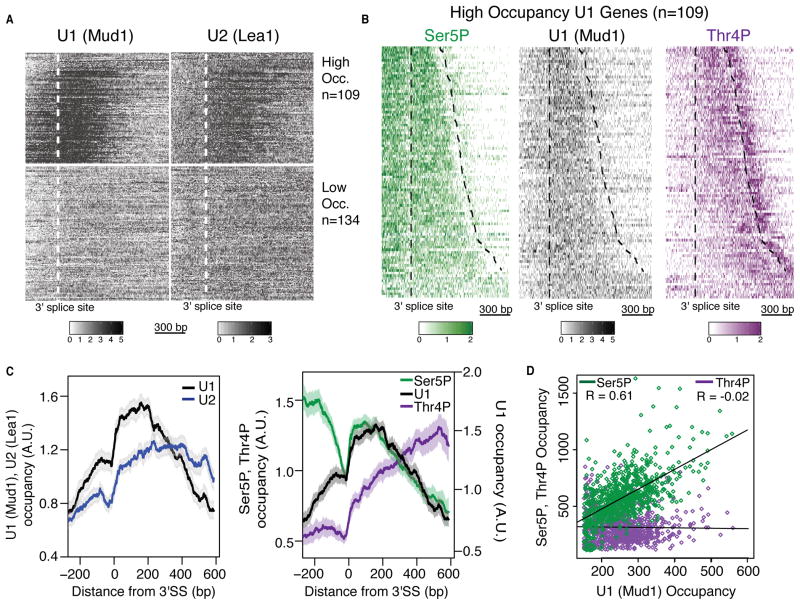
To examine the roles of both Ser5P and Thr4P in coordinating splicing, we first analyzed the co-transcriptional recruitment of the spliceosome through mapping obligatory components of the U1 and U2 spliceosomal subcomplexes, Mud1 and Lea1 respectively. Analysis of U1 and U2 occupancy by ChIP-nexus displays enrichment for spliced genes and matches lower-resolution profiles around the 3′ splice site from U1 and U2 ChIP-chip data (Tardiff et al., 2006) (Figure S4A–E). We observe high levels of U1 and U2 enrichment over spliced genes, and consistent with other studies, we observe low signal over unspliced genes (Figure S4D) (Kotovic et., 2003; Moore et al., 2006; Tardiff et al., 2006). Hierarchical clustering of the data based on U1 occupancy breaks spliceosome occupancy into two clear categories of high and low occupancy genes, which had been observed previously (Figure 5A and Table S4) (Tardiff et al., 2006). The high occupancy genes tend to be highly co-transcriptionally spliced and well expressed, to have longer introns and shorter terminal exons, and to encode ribosomal proteins (Figure S4F) (Carrillo Oesterreich et al., 2010). We focused our analysis on the high occupancy genes. The ChIP-nexus data resolves a rapid and sharp increase in U1 occupancy 10–30 base pairs (bps) downstream of the 3′ splice site that remains high for 200 bps before decreasing (Figure 5B and C). Extending for almost 400 bps, U2 occupancy also increases sharply at the 3′ splice site, albeit more slowly than U1, consistent with ordered spliceosomal assembly (Figure 5C, S4G) (Hoskins et al., 2011).
Figure 5. Pol II CTD Ser5P and Thr4P regulate spliceosome occupancy and splicing dynamics.
(A) Heatmap of normalized U1 (Mud1) and U2 (Lea1) ChIP occupancies around the 3′ splice site (300 bps upstream and 1000 bps downstream) of non-overlapping intron-containing genes (See Supplemental Experimental Procedures). Genes are classified as high (High Occ.) or low occupancy (Low Occ.) based on U1 recruitment.
(B) Heatmap of normalized Ser5P, U1 and Thr4P ChIP occupancies around the 3′ splice site (300 bps upstream and 1000 bps downstream) of high U1 occupancy genes. Dashed lines represent the 3′ splice site and polyadenylation site respectively.
(C) Left: Normalized average ChIP-nexus profiles of U1 (Mud1) and U2 (Lea1) around the 3′ splice site (3′SS) of high U1 occupancy spliced genes. Normalization of ChIP data to internal spike-ins allows direct comparison of U1 and U2 profiles. Right: Normalized average ChIP-nexus profiles of Ser5P and Thr4P and U1 around the 3′ splice site of high U1 occupancy spliced genes. n=109, Shaded areas represent 95% confidence intervals.
(D) Comparison of U1 (Mud1) and Ser5P (green) or U1 and Thr4P (purple) ChIP-nexus reads from the 3′ splice site to 100 bp past the polyA site of high U1 occupancy spliced genes (n=109). R, Pearson correlation coefficient. (A–C) ChIP-nexus reads for each gene are normalized by total reads for each gene in the analyzed region.
See also Figures S4 and S5
From our Pol II CTD interactomes, we postulated that Ser5P occupancy would mirror U1. ChIP-nexus analysis of Ser5P occupancy revealed a characteristic peak of enrichment near transcription start sites of coding genes (Figure S5A–C). Interestingly, analysis around 3′ splice sites of high U1 occupancy genes displays a sharp increase in Ser5P 10–30 bps downstream of the 3′ splice site that persists for 200 bps, overlaying precisely with the U1 peak (Figure 5B and C). In contrast, Thr4P peaks late into the terminal exon, concomitant with low levels of Ser5P, U1 and U2 (Figure 5B and C). Interestingly, the peak of Thr4P in the terminal exon occurs just prior to polyA sites and is distinct from the peak of Thr4P after polyA sites (Figure 5B, C and S5D). The bimodal Thr4P profile around the polyA sites is not observed at unspliced genes where only a single peak occurs after polyA sites (Figure S2G). These trends can be observed through comparison of raw ChIP-nexus signal between U1 and Ser5P (Pearson correlation, r = 0.61) or Thr4P (Pearson correlation, r = −0.02) across terminal exons (Figure 5D, S5F). These data suggest that Ser5P recruits the spliceosome at the 3′ splice site and that when the spliceosome dissociates, high levels of Thr4P are present. Importantly, U1 and U2 occupancy profiles are not altered in the T4V1–26 mutant (Figure S5E) suggesting that Thr4 does not cause the dissociation of the spliceosome, and that instead high levels of Thr4P act as a mark of spliceosome release.
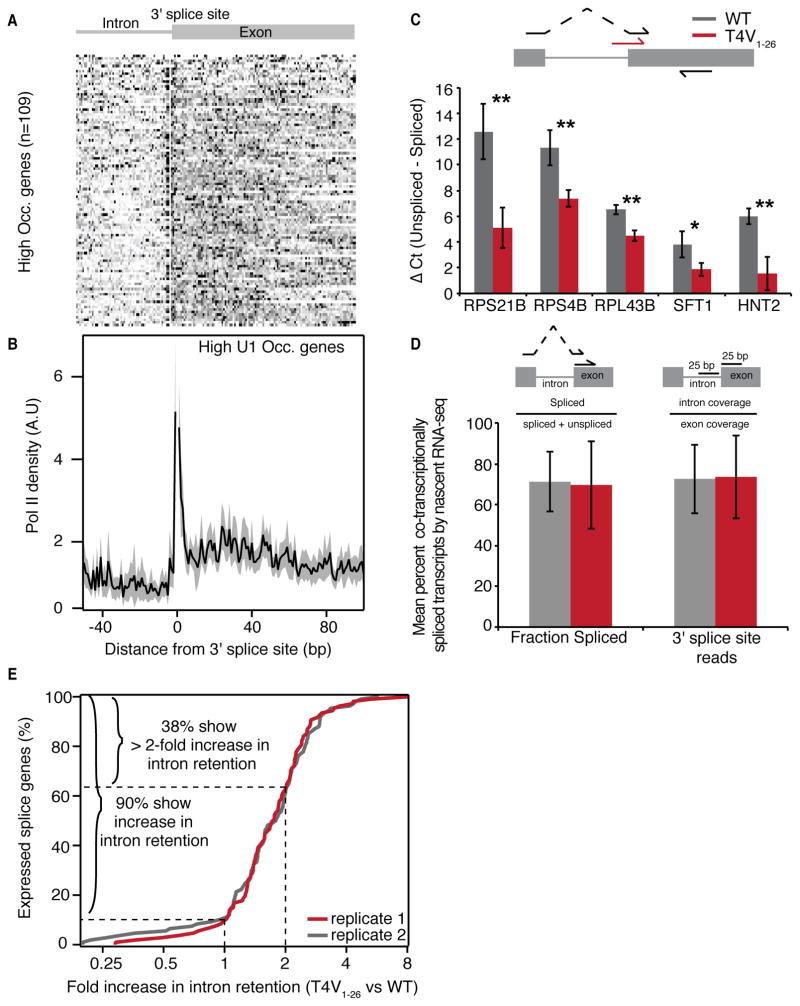
The increase in Ser5P and rapid recruitment of U1 at the 3′ splice site are coincident with a previously proposed splicing checkpoint involving Ser5P and Pol II pausing around the 3′ splice site (Alexander et al., 2010; Chathoth et al., 2014). Furthermore, pausing has been observed elsewhere at spliced genes in yeast (Carrillo Oesterreich et al., 2010). However, these studies have been limited by low resolution or indirect methods to detect transcriptional pausing. We analyzed NET-seq profiles of high-occupancy genes around the 3′ splice site (3′ SS) in an effort to resolve putative splicing checkpoints at nucleotide resolution. Analysis by NET-seq revealed highly reproducible patterns of Pol II pausing at intron-exon junctions as well as increased Pol II density over exons similar to earlier observations in yeast and in human cells (Figures 6A, B and S5G) (Alexander et al., 2010; Jonkers et al., 2014; Mayer et al., 2015; Veloso et al., 2014). Splicing intermediates, such as lariats, are known contaminants in NET-seq data, so the precise nucleotide position where their 3′ ends align are removed from analysis, resulting in a single nucleotide gap. In order to determine whether the remaining signal upstream and downstream of the 3′ SS are due to lariats from missplicing events, we sequenced total nascent RNA in order to analyze the abundance of alternative splicing junctions. Greater than 99% of transcripts are correctly spliced (Figure S5H), strongly indicating that the NET-seq signal around the 3′ SS is due to Pol II pausing. Moreover, pausing at the 3′ SS is observed uniformly across spliced genes with high U1 recruitment (Figure 6A). Together these data suggest coordination between Pol II dynamics, Ser5 phosphorylation and recruitment of the spliceosome during a splicing checkpoint at the 3′ splice site.
Figure 6. Pol II CTD Thr4 regulates post-transcriptional splicing.
(A) Heatmap of normalized NET-seq reads around the 3′ splice site of high U1 occupancy genes.
(B) Normalized average NET-seq reads of around the 3′ splice site of high U1 occupancy genes. NET-seq reads for each gene are normalized by total reads for each gene in the analyzed region. Shaded areas represent 95% confidence intervals. Splicing intermediates are contaminants in NET-seq data, so the precise 1 bp positions where they align are removed from analysis and lead to a gap in the Pol II density profiles. A.U., arbitrarty units, n=109.
(C) Quantification of the mean difference in unspliced versus spliced transcripts in WT and T4V1–26 CTD mutant cells as determined by qPCR of splice junctions at five splice genes. Error bars represent standard deviation of three biological replicates. * p-value < 0.05, ** p-value < 0.01. Top: diagram depicting primer pairs used to determine spliced (black forward arrow) and unspliced (red forward arrow) transcript abundance.
(D) Mean fraction of co-transcriptionally spliced transcripts as determined by nascent RNA-seq in WT (black) or the T4V1–26 CTD mutant (red) in high U1 occupancy genes. Two metrics were used to determine co-transcriptionally spliced reads: Fraction spliced, which calculates the fraction of spliced reads by dividing the number of spliced reads by the total number of reads (spliced + unspliced) spanning the 3′ splice site and 3′ splice site coverage which uses the ratio of intron to exon coverage in a region spanning 25 bp up or downstream of the 3′ splice site and subtracting this value from 1. Top: diagrams depicting the fraction spliced and 3′ splice site metrics. Error bars represent standard deviation.
(E) Distribution of the increase in intron retention in the T4V1–26 CTD mutant vs WT cells for two biological replicates (n= 107 and 108 genes). Genes included in the analysis contained reliably quantifiable intron coverage as determined by RNA standards in both samples (Figure S6B and Supplemental Experimental Procedures).
See also Figures S5 and S6.
We reasoned that if Ser5P is responsible for the rapid and precise recruitment of U1 directly following 3′ splice sites, then a loss of Ser5 would disrupt splicing. As complete loss of Ser5 is lethal, we created an allele of the RPB1 gene that encodes the CTD where Ser5 of the first 8 repeats proximal to the Pol II core are mutated to alanine (S5A1–8), similar to the T4V1–8 strain. We also constructed similar alleles for Ser2 (S2A1–8) and for Ser7 (S7A1–8). Here the endogenous RPB1 is again controlled by a Tet-off promoter, and we investigated the splicing phenotype in total RNA of these strains as well as the T4V1–8 strain after exposing the cells to doxycycline. RT-qPCR analysis of splice junctions revealed that in contrast to S2A1–8 and S7A1–8, both S5A1–8 and T4V1–8 lead to intron retention at the RPL34B gene (Figure S6A), demonstrating the functional importance of Ser5P for efficient splicing, presumably through the recruitment of U1.
Thr4 is Responsible for Efficient Post-transcriptional Splicing Genome-wide
The pre-polyA peak of Thr4P specifically at spliced genes, a lack of enriched spliceosomal subunits in the Thr4P interactome and the splicing defect observed in the T4V1–8 strain lead us to postulate that Thr4 has a role in splicing. The splicing defect caused by the T4V1–8 mutation is likely to be a modest phenotype as there are still 18 wild type repeats. Indeed, RT-qPCR analysis of the T4V1–26 strain revealed a much stronger splicing defect at 5 genes (Figure 6C). To ask whether the splicing defect arises co-transcriptionally, we generated nascent RNA-seq data for WT and T4V1–26 cells. Using two different metrics, we show that 73% of transcripts are co-transcriptionally spliced, consistent with previous reports, with no difference between WT and T4V1–26 cells (Carrillo Oesterreich et al., 2010) (Figure 6D). Consistently, we observe no defect in the co-transcriptional recruitment of U1 or U2 in the T4V1–26 strain (Figure S5E). We next considered whether post-transcriptional splicing is globally impacted by a replacement of Thr4 with valine by performing total RNA-seq. The inclusion of RNA standards allowed for an accurate and quantitative measure of intron retention genome-wide (Figure S6B–E). Comparison of T4V1–26 and wild type strains reveal 90% of spliced genes have increased intron retention in the T4V1–26 strain and that 38% display a greater than two-fold increase (Figure 6E, and S6F–H). Importantly, no down-regulation of transcription elongation, splicing, RNA processing or RNA turnover related gene classes was detected (Table S3). Furthermore, a re-analysis of RNA-seq data from a T4V mutant reported in Rosonina et al also reveals a splicing defect (Rosonina et al., 2014) (Figure S6I). Thus Thr4 does not impact co-transcriptional splicing, and is instead a critical residue for ensuring efficient splicing post-transcriptionally.
Discussion
Transcription elongation proceeds through multiple stages, marked by distinct CTD phosphorylation patterns and by the set of factors regulating Pol II to coordinate co-transcriptional processes. Here we combined phospho-specific CTD Pol II purification with quantitative mass spectrometry to identify the interactomes of each phospho-CTD residue. Importantly, we observe minimal overlap of factors across phospho-CTD interactomes, emphasizing the unique roles of each phosphorylation mark. The interactomes reveal the complement of factors interacting with Pol II throughout transcription elongation, and will serve as a resource for future studies on the regulation of transcription and co-transcriptional processes.
The Ser5P and Ser2P modifications have been studied extensively, but recent studies have demonstrated that all the phospho-modifications of the CTD can regulate the transcription process (Descostes et al., 2014; Egloff et al., 2007; Hintermair et al., 2012; Hsin et al., 2011, 2014a, 2014b; Mayer et al., 2012; Rosonina et al., 2014). Furthermore, our work and that of others suggest that factors interact with Pol II in a highly regulated manner requiring multiple inputs (presence or absence of particular CTD phosphorylation marks, cis-acting elements in the nascent RNA, etc.) (Kim et al., 2004; Komarnitsky et al., 2000; Lacadie et al., 2005, 2006; Licatalosi et al., 2001; McCracken et al., 1997; Mayer et al., 2010, 2012; Moore et al., 2006; Schwer and Shuman 2011; Suh et al., 2016). For example, Ser2P and the lack of Tyr1P are proposed to facilitate the localization of Rtt103 to Pol II (Mayer et al., 2012). Here we describe Thr4P as an additional input to this process. Our data are consistent with a model whereby Thr4 phosphorylation aids the recruitment of Rtt103 to the transcription elongation complex, followed by a Rtt103/Thr4 regulated release of Pol II from a region of pausing after polyA sites (Figure 7). These results raise the possibility that Pol II slows transcription until the arrival of Rtt103 and the onset of termination. Thus Thr4 joins Tyr1 and Ser2 in facilitating the precise and timely association of Rtt103 with the transcription elongation complex, indicating that the “CTD code” includes intricate CTD phosphorylation combinations to orchestrate the connections between transcription and co-transcriptional processes. Interestingly, the recruitment of Rtt103 may be even more complicated as we also observe a strong enrichment for Rtt103 with Pol II CTD Ser7P (Table S1).
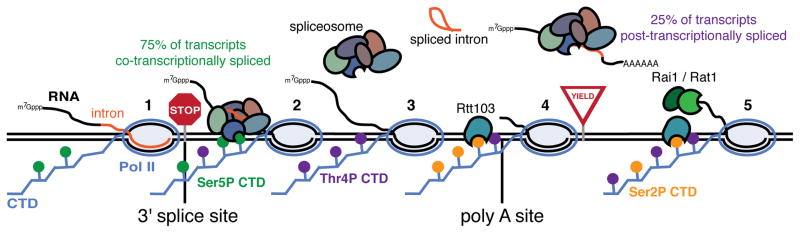
Figure 7. Coupling of transcription to RNA processing by Ser5P and Thr4P.
Model displaying how Ser5 and Thr4 phosphorylation coordinates the pausing of Pol II the at the 3′ splice site (1), recruitment of the splicing machinery (2), completion of splicing and release of the splicoesome (3), pausing for 3′ end processing and recruitment of Rtt103 (4) and the transition from 3′ end processing to termination (5).
Our data reveal that the coupling of transcription to splicing is also controlled by multiple inputs, revealing CTD Ser5 and Thr4 as being critical links between transcription and splicing. Our analyses suggest a model in which after Pol II releases from a 3′ splice site pause, the U1 spliceosomal subcomplex is rapidly recruited to Pol II through physical interaction with Ser5P. U2 subsequently associates, which presumably leads to co-transcriptional splicing (Figure 7). Interestingly, Mud1 (U1) does not co-localize with the canonical Ser5P peak at the 5′ end of all genes, indicating that Ser5P is not sufficient for U1 recruitment. Certainly known interactions between U1 and the nascent RNA likely serve as additional inputs to ensure proper localization (Lacadie et al., 2005). After 3′ splice sites, Thr4P levels increase steadily and peak after U1 and U2. These occupancy data and the lack of spliceosomal subunits in the Thr4P interactome suggest that high levels of Thr4P is a mark of spliceosomal release. Interestingly, we observe that Thr4 is strongly connected to post-transcriptional splicing, but how is this mediated? A modest peak of spliceosome recruitment occurs after polyA sites where both U1 and U2 are simultaneously re-recruited (Figure S5D). As links between splicing, 3′ end processing and termination have been described (Albulescu et al., 2012; Martins et al., 2011) an intriguing possibility is that Thr4 coordinates the commitment to post-transcriptional splicing during 3′ end processing and termination. In sum, the phospho-specific Pol II interactomes transform our understanding of critical co-transcriptional gene regulatory mechanisms through disentangling the complex interactions of CTD phosphorylation and regulatory factors.
Experimental Procedures
CTD Immunoprecipitation
Yeast cultures grown to mid-log phase in YPD were flash frozen in liquid nitrogen and pulverized by a ball-bearing mixer mill (Retsch MM400). 1 gm of grindate was resuspended in 5 mL of 1X lysis buffer (20 mM HEPES pH 7.4, 110 mM KOAc, 0.5 Triton X-100, 0.1% Tween 20, 10 mM MnCl2, 8 U mL−1 RNasin (Promega), 1X cOmplete EDTA free protease inhibitor cocktail (Roche), 1X PhosSTOP (Roche)). 660 U of RQ1 DNase (Promega) was added and lysate was incubated for 20 min on ice and centrifuged at 20,000 xg for 10 min at 4°C. Supernatant was added to 500 μL of ANTI-FLAG M2 affnity gel (Sigma-Aldrich) and rotated at 4°C for 1 hr. IPs were washed 3X for 5 min at 4°C with 10 mL of 1X wash buffer (20 mM HEPES pH 7.4, 110 mM KOAc, 0.5 Triton X-100, 0.1% Tween 20, 8 U mL−1 RNasin, 1 mM EDTA), and eluted (2x) in 300 μL of 2 mg/mL 3X FLAG peptide (Sigma-Aldrich). Elutions were combined, 100 μL of eluet was used for mass spectrometry analysis. For phospho-CTD IPs, 250 μL of the combined elution was added to magnetic beads covalently coupled to 33 μg of phospho-specific CTD antibodies (Active Motif; Ser5P, 3E8 (61085); Ser7P, 4E12 (61087); Ser2P, 3E10 (61083); Tyr1P, 3D12 (61383); Thr4P, 6D7 (61361)), or 33 μg of mCherry rat monoclonal antibody (Life Technologies, M11217) for mock IPs and incubated at 4°C for 30 min. Samples were then washed 3X by pipetting and one time by rotating for 5 min in 1 mL of wash buffer at 4°C. and eluted using 0.1M Gly pH 2.
NET-seq and nascent RNA-seq
NET-seq growth conditions, IPs, and isolation of nascent RNA and library construction were carried out as described in (Churchman and Weissman, 2012). NET-seq linker ligation was done directly to the 3′ end of isolated nascent RNA. For nascent RNA-seq the RNA was fragmented followed by dephosphorylation and linker ligation. Libraries were then subjected to 3′ end sequencing.
RNA-seq
Total RNA from mid-log yeast cultures was harvested using standard hot phenol-chloroform extraction technique. ERCC RNA standard mix 1 (Life Technologies) was added and rRNA was depleted using the Ribo-Zero Gold rRNA removal kit for yeast (Illumina). Library generation was carried out according to the procedure in (Churchman and Weissman, 2012) followed by 3′ end sequencing.
ChIP-nexus
Methods to adapt ChIP-nexus to yeast were obtained through personal communication from Stephen Doris and Fred Winston and will be published elsewhere. Library generation was carried out as described in He et al., 2015.
Mass Spectrometry Analysis
TCA precipitated samples were separated on 12% acrylamide gels, extracted and submitted to the Taplin Mass Spectrometry Facility at Harvard Medical School for analysis. All mass spectrometry data analysis was done using the Perseus software (Hubner and Mann, 2011; Hubner et al., 2010). Summed MS1 intensities for the triplicate IPs was loaded into Perseus along with the respective mock control dataset. Datasets were log2 transformed and filtered for proteins present in all three specific or mock IPs. Missing values were imputed and data were normalized by median subtraction. Enriched proteins were defined using a false discovery rate (FDR) of 0.05.
Processing and Alignment of Sequencing Data
NET-seq and RNA-seq Reads were trimmed and aligned using TopHat2 and library generation artifacts removed. For NET-seq the 5′ end of the sequencing, which corresponds to the 3′ end of the nascent RNA fragment, is recorded with a custom python script using HTSeq package. For RNA-seq and nascent RNA-seq the same script was used, this time recording the entire read. NET-seq and nascent RNA-seq data are normalized by million mapped reads. Positions of splicing intermediates in NET-seq data are removed from analysis. ChIP-nexus reads are selected for the fixed barcode, trimmed and aligned using Bowtie. Plus and minus strand coverage are combined and the 5′ end of the sequencing read is recorded and normalized by the spike-in, corresponding to the number of mapped S. pombe reads.
Gene Expression and Intron Retention Analysis
All RNA-seq reads were normalized using ERCC RNA standards mix 1 (Life Technologies) by the number of 104 uniquely mapping ERCC reads. For gene expression analysis the reads per gene were calculated for genes that were expressed in at least one replicate in either the WT or T4V strains. For intron retention analysis the normalized reads per length, in Kb (RPK), was calculated for each intron and exon in non-overlapping spliced genes as well as spliced genes where overlap did not occur on the same strand. To determine which RPK values could be reliably quantified the concentration of ERCC standards was plotted against the RPK scores for the ERCC standards in each library (Figure S5A). For intron retention analysis only those genes with intron and exon RPK values greater than 75 in both WT and T4V samples were analyzed.
Metagene Analysis
NET-seq and ChIP-nexus reads from wild type or mutant strains were scored around the TSS, polyA and the 3′ splice sites of non-overlapping genes in 1bp bins using the deepTools program. The TSS and polyA average profiles were calculated using a sliding window of 25 bp for non-overlapping protein coding genes with an RPKM > 10 in the WT NET-seq data and are at least 500 bp long. For average profiles around the 3′ splice site high occupancy non-overlapping spliced genes as well as spliced genes where overlap did not occur on the same strand were analyzed.
RT-qPCR
Yeast was cultured and RNA extracted as described in RNA-seq methods. For CTD1–8 mutants cells were grown in SC-leu media containing 2% glucose and 50 μg/mL doxycycline (Clontech). Total RNA was harvested using standard hot phenol-chloroform extraction methods, ethanol precipitated, and DNase treated using RQ1 DNase (Promega). cDNA was then generated from gene specific RT primers using SuperScript III (Life Sciences). qPCR was done using SSoFast Supermix from Biorad and carried out on a Biorad CFX384 Real-Time System with a C1000 Theromycylcer.
Antibody Competition Assay
Pol II was immunopurified as described above. Samples were split into 5 aliquots of 200 μL each. To each sample 100 μL of wash buffer containing 0 or 20ug of phospho-CTD antibody or 100 μL of 2 mg/mL FLAG peptide was added. Samples were incubated for 30 minutes at 4°C followed by centrifugation to collect the beads. The beads (B) and supernatant (S) fractions were then analyzed by Western blot.
Statistical Methods
P-values for mass spectrometry analysis, antibody competition assays and qPCR data were calculated using two-sample, two-tailed t-tests. FDRs in volcano plot analysis were calculated using a permutation based FDR for each dataset. P-value in Figure S3E was calculated using a paired two-sample t-test. P-values for GO term enrichment were obtained from GO term finder at the Saccharomyces Genome Database website. P-values comparing protein domain enrichment were calculated using Fisher’s exact test.
Supplementary Material
Acknowledgments
We thank S. Buratowski, C. Guthrie, S. Gygi, R. Kingston, F. Winston and the Churchman laboratory for advice and discussions; S. Buratowski, M. Couvillion, A. Mayer and F. Winston for their critical reading of the manuscript; U. Eser and J. di Iulio for help with data analysis. We thank the Taplin Mass Spectrometry Facility and The Bauer Core Facility at Harvard University for mass spectrometry assistance and next generation sequencing.
This work was supported by US National Institutes of Health NHGRI grants R01HG007173 to L.S.C., a Damon Runyon Dale F. Frey Award for Breakthrough Scientists (to L.S.C.), and a Burroughs Wellcome Fund Career Award at the Scientific Interface (to L.S.C). K.M.H. was supported by US National Science Foundation Graduate Research Fellowship DGE1144152.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures and four tables.
Accession Numbers:
All NET-seq, RNA-seq and ChIP-nexus data sets are available at GEO under the accession number GSE68484
Author Contributions:
K.M.H. and L.S.C. designed experiments and wrote the manuscript. K.M.H. performed IP experiments, analyzed mass spectrometry data, generated mutant strains, performed and analyzed ChIP nexus and qPCR experiments. M.M.M and S.M.F and K.M.H designed and synthesized the CTD mutant plasmids. K.M.H, K.L.T. and E.E.S. generated NET-seq libraries; K.L.T. generated RNA-seq libraries and qPCR data. K.M.H. analyzed sequencing data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albulescu LOO, Sabet N, Gudipati M, Stepankiw N, Bergman ZJ, Huffaker TC, Pleiss JA. A quantitative, high-throughput reverse genetic screen reveals novel connections between Pre-mRNA splicing and 5′ and 3′ end transcript determinants. PLoS Genet. 2012;8:e1002530. doi: 10.1371/journal.pgen.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillà D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell. 2014;53:779–790. doi: 10.1016/j.molcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Native elongating transcript sequencing (NET-seq) Curr Protoc Mol Biol. 2012;Chapter 4(Unit 4.14.1–17) doi: 10.1002/0471142727.mb0414s98. [DOI] [PubMed] [Google Scholar]
- Descostes N, Heidemann M, Spinelli L, Schüller R, Maqbool MA, Fenouil R, Koch F, Innocenti C, Gut M, Gut I, Eick D, Andra J. Tyrosine phosphorylation of RNA Polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife. 2014:e02105. doi: 10.7554/eLife.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochim Biophys Acta. 2009;1789:26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J, Barrandon C, Hujer K, Rutz B, Rigaut G, Kotovic KM, Faux C, Neugebauer KM, Séraphin B. Cotranscriptional spliceosome assembly and splicing are independent of the Prp40p WW domain. RNA. 2011;17:2119–2129. doi: 10.1261/rna.02646811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, Chapman R, Eick D. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JPP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JPP, Li W, Hoque M, Tian B, Manley JL. RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife. 2014a;3:e02112. doi: 10.7554/eLife.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JPP, Xiang K, Manley JL. Function and control of RNA polymerase II C-terminal domain phosphorylation in vertebrate transcription and RNA processing. Mol Cell Biol. 2014b;34:2488–2498. doi: 10.1128/MCB.00181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. The Journal of Cell Biology. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Bataille AR, Robert F. The Writers, Readers, and Functions of the RNA Polymerase II C-Terminal Domain Code. Chem Rev. 2013;113:8491–8522. doi: 10.1021/cr4001397. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Molecular and Cellular Biology. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA: 5′ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley D. Functional Interaction of Yeast Pre-mRNA 3′ End Processing Factors with RNA Polymerase II. Mol Cell. 2001;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Morrill SA, Exner AE, Babokhov M, Reinfeld BI, Fuchs SM. DNA Instability Maintains the Repeat Length of the Yeast RNA Polymerase II C-terminal Domain. J Biol Chem. 2016 doi: 10.1074/jbc.M115.696252. in press (3/29/16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- Mosley AL, Sardiu ME, Pattenden SG, Workman JL, Florens L, Washburn MP. Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000687. M110 000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Hunter GO, Sardiu ME, Smolle M, Workman JL, Florens L, Washburn MP. Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol Cell Proteomics. 2013;12:1530–1538. doi: 10.1074/mcp.M112.024034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Yurko N, Li W, Hoque M, Tian B, Manley JL. Threonine-4 of the budding yeast RNAP II CTD couples transcription with Htz1-mediated chromatin remodeling. Proc Natl Acad Sci USA. 2014;111:11924–11931. doi: 10.1073/pnas.1412802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller R, Forne I, Straub T, Schreieck A, Texier Y, Shah N, Decker T, Cramer P, Imhof A, Eick D. Heptad-Specific Phosphorylation of RNA Polymerase II CTD. Mol Cell. 2016;61:305–314. doi: 10.1016/j.molcel.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43:311–318. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Ficarro S, Kan U, Chun Y, Marto J, Buratowski S. Direct Analysis of Phosphorylation Sites on the Rpb1 C-Terminal Domain of RNA Polymerase II. Mol Cell. 2016;61:297–304. doi: 10.1016/j.molcel.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci USA. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.