Summary
Understanding how neural information is processed in physiological and pathological states would benefit from precise detection, localization and quantification of the activity of all neurons across the entire brain, which has not to date been achieved in the mammalian brain. We introduce a pipeline for high speed acquisition of brain activity at cellular resolution through profiling immediate early gene expression using immunostaining and light-sheet fluorescence imaging, followed by automated mapping and analysis of activity by an open-source software program we term ClearMap. We validate the pipeline first by analysis of brain regions activated in response to Haloperidol. Next, we report new cortical regions downstream of whisker-evoked sensory processing during active exploration. Lastly, we combine activity mapping with axon tracing to uncover new brain regions differentially activated during parenting behavior. This pipeline is widely applicable to different experimental paradigms, including animal species for which transgenic activity reporters are not readily available.
Neuronal activity in the brain is an ever-changing landscape. Signals processed from the outside world and internal organs are constantly shifting activity levels across brain regions to produce relevant responses. Significant efforts have been made in the past decade to record large ensembles of neurons to better understand the network dynamics underlying animal behaviors (Keller and Ahrens, 2015), but obtaining a complete snapshot of the activity of all neurons in the mammalian brain has yet to be achieved. In mammals, large-scale neural activity can be recorded through the animal skull using Electro-Encephalogram Recordings or functional MRI, and single cell recording of large neuronal ensembles can be achieved with multi-channel silicon probes, which provide high temporal resolution, enabling precise discrimination of neuron types by firing patterns (Berényi et al., 2014). Optical techniques, coupled with GCaMP calcium probes, provide live recording of neuronal activity over larger volumes, including activity of most neurons in Zebra fish larva brain (Ahrens et al., 2012), and several thousand neurons in the mouse cortex (Peron et al., 2015). However, because of the size and optical properties of the mouse brain, it is not currently possible to image in vivo deeper than the cortex, although deeper structures can be made accessible using fiber optics, endoscopes (Cui et al., 2014) or by surgically removing the cortex (Lovett-Barron et al., 2014), though at the expense of the field of view.
To assess neuronal activity in regions less accessible to live imaging, Immediate Early Genes (IEGs) such as c-fos, whose expression levels reflect recent changes in neuronal activity, have often been used as proxies. The first in vivo example of c-fos induced expression in neurons was reported in the dorsal horn of the spinal cord following a nociceptive stimulus (Hunt et al., 1987). Since then, the up-regulation of expression of IEGs like c-fos, arc, egr-1, fosb and npas4 have been used as a surrogates for neuronal activity in most neuronal systems and in most regions of the brain. Because they rely on translational machinery, IEGs have low temporal resolution. Up-regulation of their expression usually lags neuronal stimulation by about 30 minutes, and their expression can outlast the end of activity by over 4 hours (Barnes et al., 2015), so that IEGs can be considered to encode a short-term memory trace of the activity (Denny et al., 2014). Although highly informative (Mongeau et al., 2003), unbiased analyses of IEGs expression in the brain are painstakingly arduous to perform, and their low throughput limits their implementation. Serial Two-Photon Tomography (STP) represents a current standard for automated whole brain histological analysis using fluorescent reporters (Ragan et al., 2012), and was more recently used to map c-Fos expression in the mouse brain in an automated way by imaging endogenous GFP fluorescence in a c-Fos-GFP reporter line to comprehensively analyze complex social behaviors (Kim et al., 2015). However, STP tomography-based IEG mapping also has disadvantages. First, it requires advanced instrumentation that is not readily available to most laboratories. Second, there are only a few fluorescent IEG reporter mice, and these have some limitations compared to the detection of native IEGs. For example the Arc-GFP mouse is a genetic knock-in with only one Arc allele remaining active (Wang et al., 2006), while the Arc-dVenus and c-Fos-GFP mice possess transgenes with short promoters that may not exactly replicate the intricate regulation of activity-driven induction in all brain areas. Lastly, a complete recording of all c-Fos+ cells is not possible with STP, as single 2D planes are sampled only every 50 to 100μm from the block, leaving undocumented gaps between each section.
Whole-brain detection of native IEGs would allow the analysis of IEGs for which there are no transgenic reporters and likely improve the reliability of the detection of the arc and c-fos gene products. In fact, whole brain activity mapping with IEGs without sectioning has been shown to be a powerful approach to investigate brain regions active in freely moving zebrafish larvae using image registration (Wee et al., 2015), but the extension of this approach to the much larger mammalian brain has not to date been achieved.
Recently, renewed interest in tissue clearing techniques and volume imaging has driven the development of several families of protocols to perform histological analysis in intact samples (Richardson and Lichtman, 2015). Those techniques are now starting to enable the streamlined analysis of axonal projections or cell positions in intact mouse brains in a manual or semi-automated way (Lerner et al., 2015; Menegas et al., 2015; Schwarz et al., 2015). We reasoned that volume imaging using light sheet fluorescence microscopy (LSFM) coupled with tissue clearing could be a powerful alternative to STP for imaging IEG expression in the intact brain. Imaging IEG expression in the intact brain using the Arc-dVenus fluorescent reporter has been reported along the publication of the CUBIC method and its expanded protocol (Susaki et al., 2014; 2015).
We recently developed iDISCO, a method to immunolabel and image large intact samples, including adult brain (Renier et al., 2014). The ability to immunolabel many samples in parallel and the extremely high transparency achieved makes iDISCO well-suited to map endogenous IEG expression in all brain regions at a higher throughput compared to STP and with greater versatility, thereby complementing the use of transgenic reporters in other whole brain imaging methods.
To enable automated brain-wide analysis of IEGs, it was, however, necessary to develop a new iDISCO-based pipeline on three fronts:
First, we improved the iDISCO protocol to reduce sample shrinkage and better preserve brain morphology, thereby enabling automated registration of the LSFM-imaged sample onto a reference brain atlas for automated comparisons; the resulting protocol is termed iDISCO+.
Second, we optimized parameters to obtain a near isotropic resolution of about 5μm (4.065 × 4.065 × 3μm /pixel) by horizontally scanning the light sheet over a 7 × 7 × 6 mm region of the mouse brain in 1hr at a cellular resolution, using an average of 20GB per sample.
Third, we developed “ClearMap”, a computer program based solely on open-source components and compatible with a desktop workstation, that is applied to the imaging data to count cells in 3D, register them onto a reference atlas, and generate distribution maps and statistical analysis of intact mouse brains in less than 1 hr per sample.
The power and utility of the resulting pipeline is highlighted here in three applications: (i) to document brain regions activated in response to an anti-psychotic drug Haloperidol; (ii) to search for brain regions differentially activated when mice explore a new environment with or without using their whiskers; (iii) and to correlate neuronal projections with activity, illustrated by tracing projections from the galanin-expressing neurons of the medial pre-optic area to map activity in its target regions during parenting behavior in the mouse.
Results
Imaging and registration of IEGs in the mouse brain
The original iDISCO protocol for immunolabeling and brain clearing permits detection of immunolabeled structures throughout the intact adult mouse brain and is compatible with a wide variety of antibodies to diverse epitopes (Renier et al., 2014). In particular, it is compatible with antibodies that detect a variety of IEGs (see below). Specific signals are detected using secondary antibodies coupled to Alexa fluorophores that emit in the far-red spectrum (647 nm laser excitation), to which the cleared brain is essentially transparent (Renier et al., 2014). In the blue-green spectrum (480 nm laser excitation), intrinsic tissue fluorescence highlights the major structures in the brain, such as myelinated tracts, cortical regions and various deep nuclei (Figure 1B and S1A), thus providing a counterstain without the need for counterstaining (Movie S1).
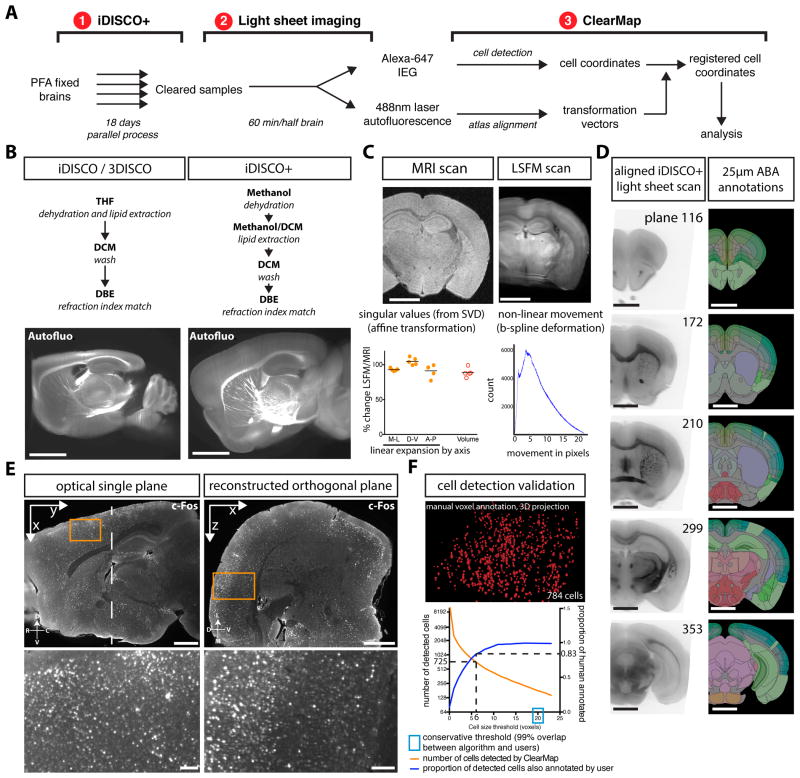
Figure 1. iDISCO+ and ClearMap: a pipeline for cell detection, registration and mapping in intact samples using light sheet microscopy.
A Presentation of the iDISCO+ and ClearMap pipeline. B New clearing strategy enabling morphology preservation. LSFM scans of the autofluorescence of adult mouse brains. Optical plane through brains cleared with either 3DISCO (after iDISCO whole-mount immunolabeling) or iDISCO+. See also Figure S1+ and S2. C Quantification of the shrinkage and deformations induced by the clearing procedure by registering MRI-scanned brains back onto themselves after clearing (n=5). Left graph: the singular values of the coefficient matrix from the affine transformation show a modest linear change in size along the medio-lateral (M-L), dorso-ventral (D-V) and antero-posterior (A-P) axis. The change in volume is given by the determinant of the matrix. Right graph: histogram of the movements of voxels during the non-linear transformations (representative example from 1 brain). See also Figure S2. D Fully automated registration of the LSFM autofluorescence scan to the Allen Brain Institute 25μm reference annotation. E Single optical plane and reconstructed planes showing the isotropic imaging resolution at the center of the imaging lens with a cellular resolution and large field of view, with a blow up of the boxed cortical region. Right panels: orthogonal cross-section at the level of the dotted line with a blow up of the boxed cortical region, showing the minimal degradation of the resolution in the z direction at the center of the lens. See also Figure S1, S4 and S5. F Manual annotation of cells and comparison with the automated detection. All voxels of each cell were manually painted, and the 3D automated cell detection was run on the same dataset. The proportion of detected cells also manually annotated was determined for each value of the threshold for cell volume (in voxels). See also Figure S3. Scale bars are 2mm.
Building on iDISCO, we developed ClearMap, a cell detection and registration pipeline outlined in Figure 1A and Movie S2, which relies on key improvements in iDISCO sample preparation and imaging procedures, as follows.
First, we optimized imaging conditions to achieve the minimal x,y,z pixel size of 4.06 × 4.06 × 3μm/pixel necessary to image a large field of view at cellular resolution while still preserving acquisition speed and a relatively low data footprint (See Methods and Figure S1). We found that imaging at 1.6X with a 2X 0.5NA objective and continuous horizontal scanning of the light sheet provided a resolution sufficient to image single c-Fos positive cells within the sharp center of the lens, which covers the length of the entire mouse forebrain in sagittal orientation (Figure S1A). At this magnification, the optical maximal resolution is almost isotropic at the center of the imaging lens (Figure 1E and S5), so that images acquired in one plane can be combined to give an undistorted representation in the other orthogonal planes. The measured z-resolution (thickness) of the light sheet at the numerical aperture used (0.1) is about 5μm at its thinnest point (manufacturer's information). To maximize the z-resolution throughout the dorso-ventral length of the brain, the thinnest focal point of the light sheet was horizontally scanned and a composite image was created for each plane by using a contrast projection, thus circumventing the Gaussian shape of the beam. While the camera sensor field of view (10.4mm in length) and the tiling of 3 parallel light sheets covers the mouse brain in sagittal orientation from the cerebellum to the olfactory bulb, the sharp center of the imaging lens covers a smaller region (7.2mm in length), which limits the region covered by one tile to the forebrain excluding the olfactory bulbs (Figure S4A). However, the consistency of the sharpness and signal intensity for detected cells throughout this region suggested that it could be possible to build a robust pipeline to automatically analyze the distribution of IEGs within the brain (Figure S1B,C).
Second, several efficient 3D image registration software packages have recently been developed. As mentioned, the intrinsic tissue fluorescence highlighting major structures (Figure 1B and Figure S1A) offers enough detail and contrast to allow in principle for automated registration of scans using signal harvested with the 480nm laser excitation. However, tissue distortion due to shrinkage during 3DISCO clearing (when used after the iDISCO staining) precluded automated image registration (Menegas et al., 2015) (Figure 1B). We traced the major source of shrinkage to TetraHydroFuran used in the 3DISCO protocol, and were able to modify the protocol to dispense with this chemical, replacing it with a combination of Methanol and DiChloroMethane (Figure 1B). With this novel iDISCO protocol, which we term iDISCO+, only minor tissue deformation occurred (see below). The clearing performance of iDISCO+ enables imaging of c-Fos+ cells with a consistent signal throughout the working distance of the objective, well over 6mm deep in the tissue (Figure S1A and B). There was no drop in the number of c-Fos+ cells detected throughout the imaging depth, as the density of c-Fos+ cells was identical on both sides of the midline (figure S1B). We verified that the new clearing protocol remains compatible with all tissues and has an identical clearing performance to that seen with iDISCO/3DISCO (Figure S2G and data not shown).
The ClearMap pipeline includes interfaces to the Elastix open-source software (Klein et al., 2010; Shamonin et al., 2014) to automatically register the iDISCO+ light sheet scans, as previously developed for STP imaged mouse brains (Kim et al., 2015). We quantified the extent of deformation by registering lightsheet scans of cleared half-brains to matched MRI scans of the same samples (Figure 1C and Figure S4A-B). The optimized affine transformation coefficients determined by Elastix indicate a modest linear reduction in size of 7% and 9% in the medio-lateral and rostro-caudal axis respectively and a small linear expansion of 5% along the dorso-ventral axis (n=5). The overall volume shrinkage as determined by the determinant of the transformation matrix was 11%, down from the reported 50% shrinkage observed with the original 3DISCO clearing protocol (Richardson and Lichtman, 2015). For whole brains, which require a little distortion to fit in the MRI tube, we observed similar but smaller coefficients when registering whole brains to the STP reference atlas (5% volume difference, n=6, Figure S4B), suggesting that whole brain morphology may be better preserved during clearing. The average displacement after applying the optimized non-linear (b-spline) transformation from the sample to the reference was 4 voxels (Figure 1C). Most of the non-linear transformations corrected for a small expansion of the ventricular volume and adjustment of the cortical curvature. The new clearing protocol therefore enabled image registration of the cleared samples datasets to the STP reference (STPR) mouse brain aligned to the Allen Brain 25μm reference Atlas, whereas the original clearing protocol would have required custom reference Atlases (Kim et al., 2015) (Figure 1D and Figure S4D-F).
We next developed a complete suite of open-source image processing tools within our modular ClearMap software to detect cells in 3D and register their coordinates onto the reference atlas, which has a similar workflow to previously used analysis pipelines (Kim et al., 2015; Menegas et al., 2015), and runs under 90 min per brain in fully automated fashion on a desktop workstation, facilitated by our optimized 3D cell detection algorithm and compact dataset. To validate the detected accuracy, the cell detection was run on a dataset for which positive cell voxels were manually annotated by two users. The filters and threshold were tuned until the disagreement ratio between the manual and automated annotation was similar to the ratio between the two users (Figure 1F, S3C and see Extended Methods).
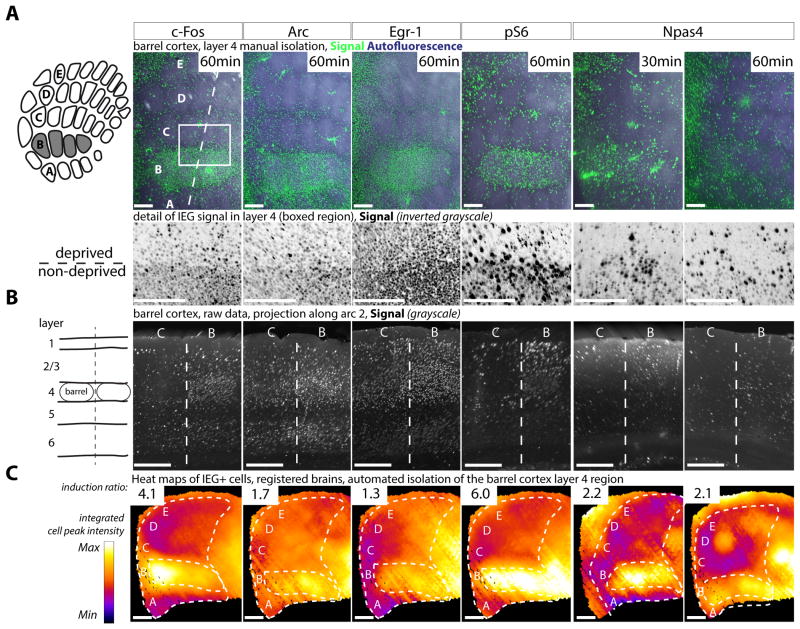
As iDISCO+ relies on immunostaining, it offers flexibility when choosing the IEGs used to map brain activity. To test the ability of the method to detect the induction of known IEGs, we relied on activity evoked in the barrel cortex by whisker stimulation as a tool to validate antibodies for c-Fos, Arc, Egr-1, Phospho-S6 and Npas4. Cortical Layer 4 neurons receiving inputs from the large mystacial whiskers on the snout are topographically organized in the barrel cortex. Their map can be readily visualized using intrinsic tissue fluorescence, which is preserved in the blue-green spectrum in the iDISCO+ protocol. We trimmed all whiskers except select rows or arcs; exposed animals for 1 hr to an enriched environment in which the remaining whiskers were stimulated as the animals explored; processed brains using the iDISCO+ protocol with each of the antibodies, using secondary antibodies conjugated to a fluorophore that emits in the far-red spectrum; and manually segmented cortical layers to visualize the activity map of the barrel cortex in a virtual projection of layer 4, or as a virtual section along the remaining row or arc (Figure 2A and B). The IEGs tested all showed increased staining in the stimulated rows or arcs (B row shown in Figure 2). Of those tested, in this system, c-Fos and Phospho-S6 showed the highest induction in labeled cell density between stimulated and deprived barrels (4.1 and 6 respectively, ratio of the number of cells at the center of B2 over C2). Egr-1 had the highest baseline activity (ratio of 1.3). Arc showed a banded pattern by cortical layers (high expression in layers 6, 4, 3 and 2) and an induction ratio of 1.7 in this paradigm (Figure 2C). Npas4 showed modest up-regulation of expression 30 min and 1 hr after stimulation (2.2 and 2.1 respectively) (Figure 2A and B), well within the induction window reported in vivo for the cortex (Lin et al., 2008; Spiegel et al., 2014).
Figure 2. Compatibility of antibodies to several Immediate Early Genes with whole brain immunolabeling and automated volume analysis.
Whole-mount iDISCO+ immunolabeling for c-Fos, Arc, Egr-1, pS6 and Npas4 of adult mouse brains, with B-row whiskers stimulated for 1h prior to euthanasia. See Table S4 for informations about the antibodies. A Manual projection of the cortical layer 4, caudo-medial region of the barrel cortex (staining and autofluorescence). A zoom of the boxed region is shown below, highlighting the difference of positive cell density between the deprived and non deprived barrels. B 25μm projection along the axis of arc 2 showing the cortical layers at the level of barrels B. C Mapping of each IEG scan to the reference atlas, with automated isolation of the cortical layer 4 of the primary somatosensory cortex. The induction ratio (number of cells detected between the center of B2 and C2 barrels) is indicated. The detected cell centers are voxelized onto spheres of 375μm of diameter to generate a density map. Scale bars are 200μm.
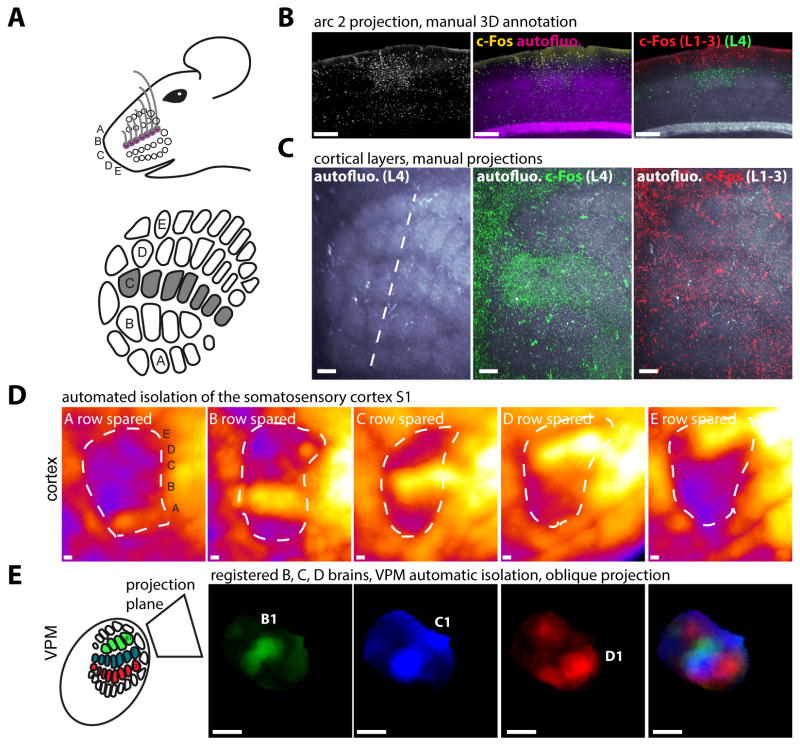
We then used ClearMap to analyze the data obtained with whisker trimming and stimulation for all IEGs and different combinations of rows and arcs, and were able to detect the increased cell density in the corresponding stimulated barrels for various whisker stimulations (Figure 3A-D). To check the accuracy of the registration, we verified that we could use the Atlas annotations to automatically isolate activity maps in the barrel cortex and in its thalamic relay station, the Ventro Postero Medial (VPM) nucleus, from the complete 3D dataset (Figure 3D-E).
Figure 3. Isolation of neuronal activity by fully automated cell detection and sample registration.
A Whole-mount iDISCO+ immunolabeling of adult mouse brains for c-Fos, with C-row whiskers stimulated for 1h prior to euthanasia (n=3). B Projection along arc 2 of the unprocessed data, with the autofluorescence (in purple) delimiting the barrels. Manual 3D annotation of the upper cortical layers is shown. C Projections of layers 4 (green) and 1,2,3 (red), showing the increased c-Fos+ cell density in the C-row in all upper layers. D Voxelized density maps of the c-Fos+ cells detected in aligned mouse brains. All whisker rows were trimmed except A, B, C, D or E, which were stimulated for 1hr. E Automated isolation of the VPM region, and projection of the aligned density maps on an oblique plane showing the first barrelloid of the stimulated row (B, C or D) and a composite image of the superimposed brains. Abbreviations: VPM: Ventro-Postero Medial Nucleus of the thalamus. Scale bars are 200μm.
Mapping of pharmacologically induced brain activity
Because of their broad spectrum of action, many classical psychoactive drugs affect the function of multiple brain regions. The systematic mapping of brain activity provides the opportunity for an unbiased investigation of regions up or down regulated by such drugs. As an example, we selected the antipsychotic Haloperidol, a well-studied high affinity antagonist of D2 dopamine receptors and low affinity agonist of serotonin receptors.
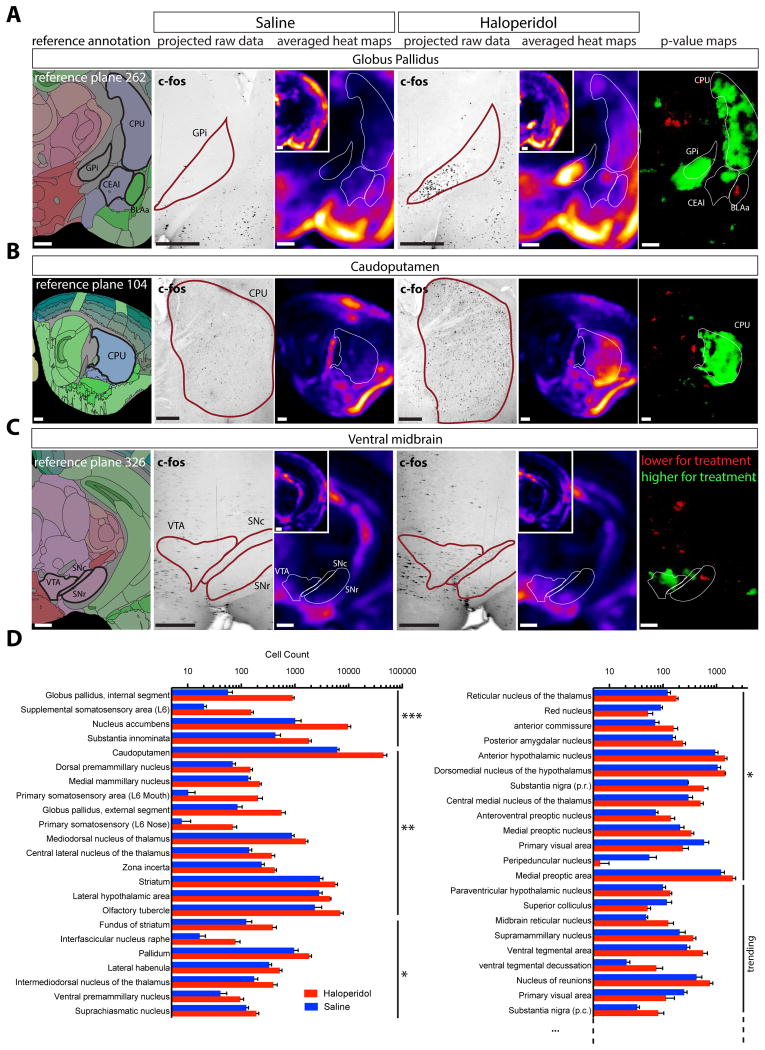
Mice were treated with an acute saline or Haloperidol injection, and their brains were processed through our analysis pipeline (n=4 for each group). ClearMap detected statistically significant increases in activity in hallmark target regions of Haloperidol, including the internal segment of the Globus Pallidus (GPi), the Caudo-Putamen formation (CPu) and the Nucleus Accumbens (NAc), showing that the deepest structures in the samples are effectively labeled by the antibodies to c-Fos (Figure 4 and table S1). To visually parse the data and find activity hot spots, we generated density maps by summing spheres of 375μm (15 pixels) diameter and uniform intensity centered on each cell. Results obtained from automated segmentation of the brain regions were confirmed by inspection of both voxelized density maps and raw data (Figure 4A-C). A useful way to depict differences in cell counts in various regions between experimentally-treated animals versus controls is with the use of P-value maps, in which regions are highlighted only when the P value is below a certain level (typically 0.05 or 0.01, indicated in each case below), thus indicating statistical significance; one color (typically green) is used when the number is higher in treated than controls, and a different color (typically red) when the number is lower (Figure 4). This enables a rapid and unbiased discovery method for identifying responsive regions for further analysis. The density maps, P-value maps and Result Tables can be presented either as total cell counts or by summing the peak intensity of each cell (Figure S6A and B). Using total cell counts, peak intensity counts, or cell size weighted counts yielded very similar results (see Methods), although weighted counts typically yielded greater statistically significant differences of tests versus controls (Figure S6A).
Figure 4. Detection of activity changes in the brain after an acute injection of Haloperidol.
Automated analysis of c-Fos+ cell distribution in mouse brains harvested 3hr after exposure to Haloperidol or saline (n=4). Panels show the reference annotation with details from the averaged density maps (4 brains averaged), p-value maps and examples of the raw data (25μm orthogonal projection) for the following regions: A Globus Pallidus, internal segment with the Central amygdala and striatum (coronal projections), B Striatum (sagittal projection) and C Ventral Tegmental Area and Subtantia Nigra. D Automated segmentation of the cell counts by anatomical regions sorted by p-values. Data are represented as mean ± SEM. The top hits are presented here (***: p < 0.001, **: p < 0.01, *: p < 0.05). See also Figure S6. Abbreviations: BLAa: BasoLateral Amygdala, anterior part, CEAl: Central Amygdala, lateral part, CPU: Caudoputamen nucleus, FS: Fondus of the Striatum, GP: Globus Pallidus internal or external segment, MD: thalamic MedioDorsal Nucleus, NAc: Nucleus Accumbens, SN: Substentia Nigra, pars compacta or reticulata, VTA: Ventral Tegmental Area. Scale bars are 500μm.
These analysis tools again highlighted known areas of c-Fos induction, and also identified many others of interest (Figure 4 and table S1). Interestingly, a few nuclei in table S1, such as the Ventral Tegmental Area (VTA), that trended towards significance but failed to achieve it had in fact a significantly higher activity restricted to a sub-region of the nucleus (here, the lateral part of the VTA) when manually inspected on the density map (Figure 4C and D); the failure to achieve significance across the entire subregion presumably reflects a localized increase in c-Fos activation within the larger, otherwise unchanged subregion. The single cell resolution of our method thus enabled us to detect a change in the detailed activation pattern of a nucleus.
Mapping of cortical activity during an exploration task
Live recording of whole mammalian brain activity is not possible for behaviors involving active exploration, and IEGs can provide a simple solution to map brain activity in freely moving mice. To demonstrate this, we chose to look for brain regions differentially regulated when mice explore a new environment with or without their whiskers. Mice had all whiskers either trimmed or spared, were transferred to an empty cage in the dark for 24 hr, after which they were transferred to a new cage containing an enriched environment for 1 hr in the dark, and then sacrificed.
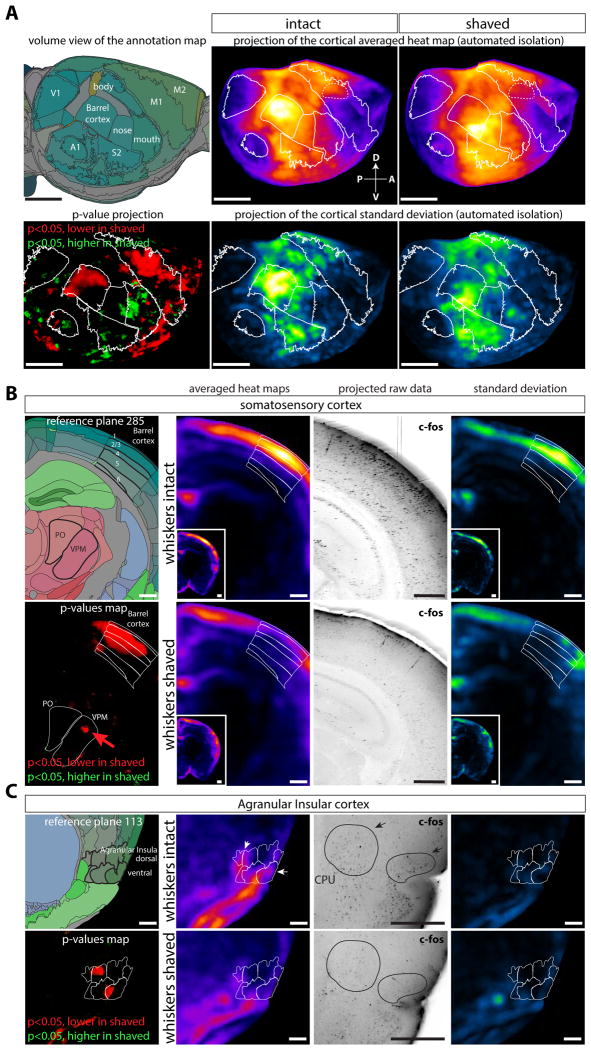
Automated isolation of cortical regions was performed using ClearMap, (Figure 5A) to display averaged density maps as well as their variability shown as standard deviation maps for both groups. While the mapped values for standard deviation are naturally higher in active regions, they provide a visual way to assess the variability and significance of activity differences between groups (see Figure 5C). As expected, in the shaved group, c-Fos positive neuron density was significantly reduced in the caudomedial region of the barrel cortex, the region of the brain that receives input from the whiskers, and also in a patch at the center of the primary motor cortex that controls whisker movements (Figure 5A). Moreover, activity in the dorso-medial part of the VPM of the thalamus (relaying whisker input from the brainstem to the cortex) was also reduced in the shaved group, while other parts of the VPM and PO nucleus (receiving inputs from other parts of the body) had higher densities of active neurons (Figure 5B). There is a strong cross-regulation in the cortex between different sensory modalities (He et al., 2012), and we indeed recorded significant differences in the primary auditory and piriform cortex between shaved and non-shaved groups, but not in the visual cortex (as expected, since the experiment was performed in the dark) (Figure 5A). Interestingly, we noticed a significant depression of the activity in the anterior part of the Agranular Insular Cortex in the shaved animals (Figure 5C), a region not traditionally associated with the sensory integration of whisker-evoked signals. We also observed increased activity in shaved animals in the regions of the somatosensory cortex associated with the nose and mouth (Figure 5A), which suggests that, lacking whiskers, the shaved animals instead actively explored the enriched environment by touching objects with those regions of their faces.
Figure 5. Brain regions differentially regulated by whiskers during an exploration task.
Mice had their whiskers shaved or left intact, and allowed to explore a new cage in the dark for 1hr, then brains where harvested and activity was probed by c-Fos immunolabeling (n=5 (shaved group), n=3 (control group)). A Lateral projections of the reference annotation, averaged density maps, p-value maps and standard deviation maps (automated isolation of the neo-cortex). The caudo-medial part of the barrel shows a decreased activity in the shaved group. B Coronal projection at the level of the barrel cortex and VPM thalamic relay, showing decreased activity in the whisker projection field (arrow in the VPM p-value map). C Decreased activity in the Agranular Insula (anterior part) in the shaved group. Abbreviations: CPU: Caudoputamen nucleus, Po: Posterior nucleus of the Thalamus, VPM: VentroPostero Medial nucleus of the thalamus. Scale bars are 2mm (panel a) and 500μm (panels b and c).
Activity mapping with axonal tracing to investigate parental behavior
While IEG maps obtained from electrical, optogenetic or pharmacological stimulation of specific brain nuclei can provide a good indication that the activity detected arises from connected regions (Bepari et al., 2012), maps derived from natural behaviors cannot provide a definitive indication about the functional relationships between the active regions. Therefore, it is useful to combine axon tracing with IEG mapping, to determine whether regions connected by specific axonal projections show correlation (or anti-correlation) in IEG expression, indicative of potentially functional links.
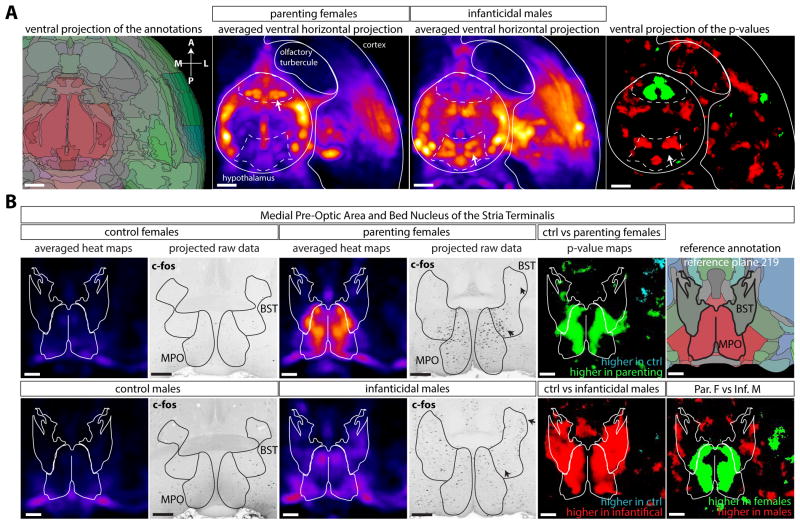
Parental care is a critical behavior hard-wired in the mammalian brain, showing sexual dimorphism in both activity patterns and connectivity (Dulac et al., 2014). Previous work has shown that in laboratory mice, virgin males are aggressive towards pups and switch to parenting behavior after mating (Saal, 1985; Tachikawa et al., 2013). The activity of a group of galanin-expressing neurons located in the Medial Pre-Optic nucleus (MPO) in the hypothalamus has been shown to drive this behavior (Wu et al., 2014). We examined the 3D c-Fos expression patterns in parenting females (n=3) and infanticidal males (n=6), compared to their respective controls (n=6 for each group). The ventral side of the brain showed a clear dichotomy in the distribution of activity between females and males in the hypothalamus (Figure 6A). Females showed a higher density of activated neurons in the anterior region of the hypothalamus, while the posterior hypothalamic region was more active in males. Indeed, as previously described (Wu et al., 2014), the density of active neurons was lower in the MPO in virgin males and control females than in parenting females (Figure 6B), while the opposite was observed in the DMH and PH Nuclei (Figure S7A). The number of active neurons was also higher in the MPO of aggressive males compared to control males, although not as high as in parenting females (Figure 6B). It is possible that the active neurons of the MPO in infanticidal males are a different population than the Gal+ neurons active in parenting females. Indeed the activity patterns in the MPO appear complementary between parenting females and infanticidal males (Figure 6B). The Bed Nucleus of the Stria Terminalis also showed an interesting dichotomy, with more active neurons found ventral to the anterior commissure in parenting females while the dorsal side was more active in infanticidal males.
Figure 6. Volume survey of brain activity applied to parental behavior in the mouse.
Brains obtained from parenting (nesting) female mice, aggressive infanticidal males or their respective controls (no pups) were whole-mount stained for c-Fos, imaged and subjected to automated analysis. A Detail of a ventral horizontal projection (2mm) of the annotation map, averaged female and males density maps showing the hypothalamus and ventral cortical regions. The c-Fos averaged density maps (n=6 per group, except parenting females n=3) show the increased activity in the rostral part of the hypothalamus in females and in the caudal part of the hypothalamus in the males (arrows). B Coronal projections of the reference annotation map, c-Fos density maps, p-value maps and unprocessed data at the level of the MPO (top panels) or Posterior Hypothalamic nucleus (bottom panels). See also Figure S7. Abbreviations: BST: Bed Nucleus of Stria Terminalis, MPO: Medial Pre-optic nucleus. Scale bars are 500μm.
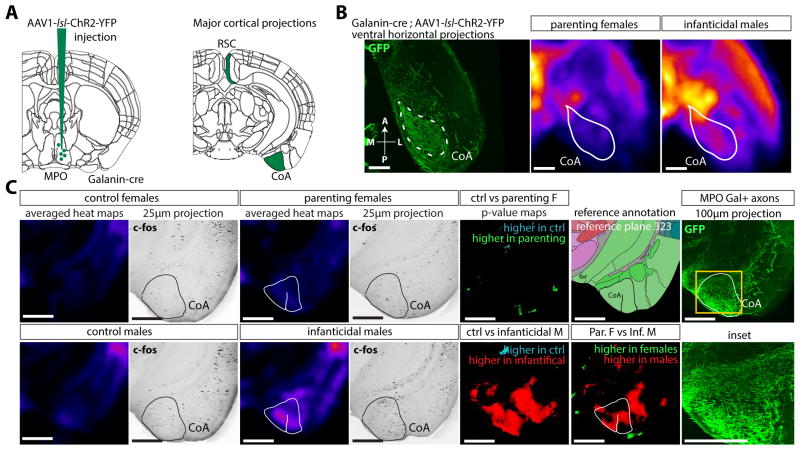
To test whether we could correlate the activity of MPO neurons during parenting tasks to a modulation of activity in other regions of the brain, we injected a floxed eYFP expressing virus into the MPO of Gal::Cre virgin females and virgin males. Using iDISCO+, we double-immunolabeled the samples for c-Fos and YFP expression. Volume imaging showed that the Galanin neurons in the rostral part of the hypothalamus had extensive projections to the thalamus, ipsilateral olfactory bulb, retro-splenial cortex, amygdalar cortex, pons and peri-acqueductal grey (Movie S3). Very extensive projections from the MPOGal neurons were seen to the Cortical Amygdalar Area (CoA) in both males and females (Figure 7B). The c-Fos density in the CoA was found to be higher in infanticidal males than in control males and both parenting and control females, specifically in the region receiving the bulk of the projections from the hypothalamus (Figure 7C). MPO neurons are inhibitory as evidenced by Gad1 expression (Wu et al., 2014), so it is possible that the increased activity in the MPOA neurons could drive a tonic inhibition of the CoA, which could act as a brake when the global activity in the MPO is high in parenting females. These results show that our method can be used to automatically map the position of c-Fos positive neurons in the brain, and correlate active and inactive regions to virally traced axonal projections, thereby generating hypotheses for further testing.
Figure 7. Correlated tracing of axons from the MPO with activity mapping during parental tasks.
A Viral tracing of the Galanin+ projections from the MPO with a AAV1-FLEx-ChR2-eYFP virus injected in the MPO region of Galanin-cre mice, imaged with iDISCO+. B-C Extensive cortical projections are seen in the Retro-Splenial Cortex and Cortical Amygdala. Overlay of the volume tracing data with the averaged activity density maps of the females and males groups in horizontal projections (B) and coronal projections (C). Increased activity in the infanticidal males, significantly higher than in any other group, is seen in the CoA region receiving the bulk of the Galanin+ projections from the hypothalamus. Abbreviations: BST: Bed Nucleus of Stria Terminalis, CoA: Cortical Amygdala, RSC: RetroSplenial Cortex, MPO: Medial Pre-optic nucleus. Scale bars are 500μm.
Discussion
Unbiased mapping of neuronal activity can be a valuable exploratory tool to guide the discovery of new functions for brain regions (refs (Kim et al., 2015; Mongeau et al., 2003), and this study), but time and low throughput have been major hurdles to performing unbiased screens based on IEG expression, thereby limiting their application. Here, we present a method combining whole-mount immunolabeling, brain clearing, volume imaging and automated 3D image analysis and atlas registration to map active neurons in an intact mouse brain using IEGs as a proxy for neuronal activity with very high throughput and straightforward implementation even for small laboratories. The pipeline is enabled by an improved optical clearing protocol (iDISCO+) that preserves the morphology of the samples, thereby permiting automated registration to a reference atlas. While used for the brain here, this protocol and the ClearMap software could easily be used to map objects in other adult tissues or embryos in the mouse or other species. As ClearMap can accommodate any atlas, users can also create their own reference files using open-source softwares such as ITK-snap (Yushkevich et al., 2006).
LSFM is the method of choice for fast imaging of large samples. Choosing an imaging resolution adequate for the desired biological question is paramount in volume imaging because the amount of data can quickly become unwieldy, and also to ensure fast imaging and data handling. As the signal for c-Fos is restricted to the cell nucleus, the necessary resolution is decreased. Therefore, the ultramicroscope's optical design, which enables very large fields of view with low-magnification optics is well suited for this type of imaging. A newly released 4X lens corrected for high refraction indices further improves the imaging quality, especially at the edge of the field of view. We tested with success higher resolution acquisitions by covering the whole brain with 12 tiles at 4X (1.6 × 1.6μm/pixels) using the stitching algorithm provided by Vision4D (Arivis). This strategy provides a better imaging quality, especially at the corners of the sample, but it increases by 10 fold the data footprint (200GB/brain instead of 20GB/brain) and processing time. Therefore, we deemed tiling 4X scans unnecessarily complex for our particular application.
ClearMap is readily applicable to the detection and mapping of cells detected using nuclear markers, such as transcription factors. The modular nature of the software makes it easily expandable to the detection of more complex objects such as neurons labeled with a cytosolic reporter through the implementation of machine learning filters (Ilastik (Sommer et al.), which enables this, is already included in ClearMap).
We have shown here that our pipeline is ideally suited for the mapping of neuronal activity in the intact mouse brain for the discovery of novel brain regions activated by drugs, sensory deprivation, and natural behaviors.
Mapping brain regions activated by chemical compounds will greatly benefit from this streamlined approach. Haloperidol is a broad spectrum neuroleptic; for instance, in addition to being a strong D2 antagonist, it is also a low affinity agonist of serotonin receptors (Roth et al., 1992). It is therefore not surprising that strong effects on c-Fos expression are seen outside of regions known to express high levels of D2 receptors (Sumner et al., 2004), e.g. in the somatosensory cortex (in particular the mouth area of S1) and the Red Nucleus (Figure 4D and Figure S6B). Interestingly, the latter regions correlate with two known secondary effects of Haloperidol, dry mouth and dystonia. Mapping neuronal activity evoked by drugs might thus help screen for unwanted secondary effects in pre-clinical trials.
Activity mapping through IEGs also enables the characterization of active regions in freely moving and behaving animals. We investigated whisker-evoked activity in a free exploration paradigm. Outside the somatosensory cortex, the Agranular Insula was one of the regions whose activity was most significantly depressed in the absence of whiskers. While the adjacent posterior Granular Insula is involved in multi-sensory integration (Gogolla et al., 2014), little is known about the role of the Agranular Insula in rodents (AI), owing to its difficult accessibility to imaging. While the AI is not usually considered to be part of the “somatosensory Insula” (Rodgers et al., 2008), it is connected to the barrel cortex (Zingg et al., 2014). Our data therefore suggest a possible role of this region in sensory integration that may be associated with place working memory.
We also combined activity mapping with axon tracing, to enable correlation between regions differentially active between groups and their connectivity.
Maternal or paternal behavior is hard-wired in the brain of parenting species, in a highly conserved fashion (Dulac et al., 2014). Brain regions responding to parental care have been extensively studied through IEGs (Tachikawa et al., 2013; Wu et al., 2014). Our unbiased screen for regions differentially active in the brains of a parenting female versus an aggressive male yielded a very high number of positive hits, which deserve closer examination as possibly being involved in parenting behavior. The combination of viral tracing and activity mapping further allowed us to discover a previously undocumented projection from the galanin-expressing neurons of the MPO to the posterior part of the Cortical Amygdala. The registered c-Fos+ cell maps showed increased activity in the males matching the projection pattern, which led us to hypothesize a direct role of the MPO on cortical activity. Further experiments are needed to explore in detail the functional interactions between those regions, e.g. by using stimulations with DREAAD receptors or via Optogenetics to map functionally connected regions through their induced activity.
In summary, the pipeline we describe here permits mapping of neuronal activity throughout the brain using IEGs as proxies in a fast and robust manner. This pipeline permits the rapid discovery of similarities and differences in activity patterns between animals, as well as the correlation of activity with connectivity, thus dramatically facilitating the generation and testing of hypotheses regarding the brain-wide basis of perception, behavior, and responses to drugs and other perturbations.
Methods
Animals
Animals were maintained on 12 h:12 h light/dark cycle with food and water available ad libitum. All surgical and behavioral procedures in mice were performed in compliance with NIH guidelines and protocols approved by the IACUC of the Rockefeller University and of the Harvard University. The Gal::Cre BAC transgenic line (STOCK Tg(Galcre)KI87Gsat/Mmucd, 031060-UCD) was imported from the Mutant Mouse Regional Resource Center and is described (Wu et al., 2014).
iDISCO+ sample processing
Modifications and continuous updates to the protocol can be found at http://www.idisco.info.
iDISCO+ protocol
Adult mice were euthanized with a rising gradient of CO2 and fixed with an intracardiac perfusion of 4% PFA in PBS. All harvested samples were post-fixed overnight at 4°C in 4% PFA in PBS. Samples were processed with the iDISCO immunolabeling protocol, as detailed in the supplementary methods and in the bench protocol.
Light sheet imaging
Cleared samples were imaged in sagittal orientation (right lateral side up) on a light-sheet microscope (Ultramicroscope II, LaVision Biotec) equipped with a sCMOS camera (Andor Neo) and a 2X/0.5 objective lens (MVPLAPO 2x) equipped with a 6mm working distance dipping cap. Version v144 of the Imspector Microscope controller software was used. The samples were scanned with a step-size of 3μm using the continuous light sheet scanning method with the included contrast blending algorithm for the 640nm and 595nm channels (20 acquisitions per plane), and without horizontal scanning for the 480nm channel.
Image processing and analysis
For display purposes in the figures, a gamma correction of 1.47 was applied on the raw data obtained from the light-sheet microscope (but not on the density maps, which are linearly scaled). Maximum projections were performed using ImageJ (NIH, http://imagej.nih.gov/ij/). Imaris (Bitplane, http://www.bitplane.com/imaris/imaris) was used for the manual 3D annotations in figure 2, 3 and movies. Imaris was also used to generate the orthogonal projections of the unprocessed data in all figures.
ClearMap analysis
All analysis and quantifications where performed with the newly developed open source ClearMap software as shown in the example scripts (Supplementary data 2), and the associated documentation. The latest version of ClearMap can be downloaded from www.idisco.info.
The cell detection is tailored for cell nuclei and uses a background subtraction via morphological opening, followed by a sequence of filters, morphological operations and a 3D peak detection. We chose a threshold of 20 voxels for the cell size. The background was removed by subtraction of the morphological opened image with a disk shape structure element with main axis of 7 pixels of diameter. Cells were detected from peaks and subsequent watershedding, removing background pixels below an intensity cutoff of 700. Density maps were generated by summing spheres of 375μm diameter and uniform intensity centered on each cell. Cell density maps and statistics are presented in the figures. Samples were registered using the average autofluorescence STPR brain (Kim et al., 2015) registered to the Allen Brain Institute 25μm map, and companion annotation map (http://alleninstitute.org/).
Statistics
Cell counts or intensities of each sample in considered regions or annotated brain areas between different groups were compared using the independent two sample student t-test assuming unequal variances. Statistical tests were performed numerically using the SciPy statistics library (http://www.scipy.org/). Using ClearMap as a discovery tool p-values were corrected for multiple comparison to q-values to control for false-discovery rate (Benjamini and Hochberg, 1995).
MRI scanning
Wild type adult littermates were transcardially perfused with 0.9% saline and 4% PFA. The brains were dissected, post-fixed, cut sagitally in half (to fit in the MRM scanner tube without deformation) and stored in PB until imaging.
All MRM scans were performed on a 9.4T Bruker vertical bore magnet. Fomblin was used to seal the specimen to prevent dehydration and as the same time to prevent susceptibility effects.
Haloperidol injections
Adult (10 weeks old) male littermate mice were individually housed for 5 days and on the 6-th day were injected i.p. either 1 mg/kg Haloperidol or vehicle (0.3%-TWEEN-80 in 0.9% saline). The mice were euthanized 3 hours after injection, transcardially perfused with 0.9% saline and 4% PFA. The brains were dissected out, post-fixed overnight in 4% PFA at 4°C and washed in PB until entering the iDISCO+ pipeline.
New environment exploration
Mice were anesthetized with a mixture of Ketamine / Xylazine (Ketamine at 0.1mg/g, Xylazine at 0.01mg/g) and their whiskers were trimmed or left intact (sham operation for the intact-whiskers group). Mice were then left to recover single-housed in an empty cage with access to food and water for 24h in the dark. Mice where then transferred to a new cage containing an enriched environment crowded with cardboard tubes and wire tube racks for 1h to explore. Mice were then promptly anesthetized, perfused and processed according to the iDISCO+ protocol.
Viral injections
Viral tracing experiments were performed in Gal::Cre mice at ∼8-12 weeks of age. Mice were anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine via intra-peritoneal injection. 500 nl of AAV1-EF1a-DIO-hChR2(H134R)eYFP (UPenn Vector Core) were injected into the right medial preoptic area. Two weeks later, mice underwent behavioral testing.
Parental behavior assay
Parental behaviour assays were performed in sexually naive Gal-cre females at ∼8-12 weeks of age, 2 weeks after viral injection. Animals were individually housed for one week before testing. On testing day, animals were habituated to the testing arena for 30min. Two C57BL/6J pups (age P1-3) were then introduced into the cage and placed at the farthest corners from the resident's nest. A timer was started at the first retrieval of a pup to the nest. 90 min after retrieval, mice were deeply anesthetized with isoflurane and transcardially perfused with ice-cold PBS followed by 4% paraformaldehyde (PFA, in PBS).
Infanticide behavior assay
Infanticide behavior assays were performed in sexually naive C57BL/6J males at ∼8-12 weeks of age that had not been exposed to pups. Animals were individually housed for one week before testing. On testing day, animals were habituated to the testing arena for 2h with hydrogel and food in the cage. Two C57BL/6J pups (age P1-3) were then introduced into the cage and placed at the farthest corners from the resident's nest. The first olfactory investigation marked the beginning of the assay, which was continued if the resident attacked at least one pup. When a pup was attacked, the assay was ended immediately and the wounded pup was euthanized. 90 min after first olfactory investigation, infanticidal males were euthanized, perfused and their brains collected and post-fixed for iDISCO treatment.
Supplementary Material
Acknowledgments
We would like to thank Nils Brose for helpful discussions. We are also grateful to Yingxi Lin for sharing reagents and insights. We would also like to thank members of the Tessier-Lavigne lab for helpful discussions. Our gratitude goes to the Rockefeller University Bio-Imaging Resource Center, and in particular Pablo Ariel, Kaye Thomas, Tao Tong and Alison North. N.R. is supported by a Shelby White and Leon Levy fellowship. E.L.A. is supported by a fellowship from the Rockefeller University Women & Science Initiative. J.K. is supported by a Human Frontier Long-Term Fellowship, an EMBO Long-Term Fellowship and a Sir Henry Wellcome Fellowship. A.E.A. is supported by the Eunice Kennedy Shriver National Institute of Child Health and Development of the National Institutes of Health under award number F32HD078040 and by a NARSAD Young Investigator award. This work was supported by NIH grant 1R01HD082131-01A1 (to C.D.) and by the Rockefeller University (to M.T.-L.). C.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author's contributions: NR, ELA, OO, CD, PO and MTL designed the study. CK developed ClearMap, with help from KUV. ELA and NR performed the sensory deprivation experiments. JK and AEA performed the social behavior experiments and viral tracing. LK performed the drug injection experiment and prepared brains for MRI imaging. YZ, VXW and CYT performed the MRI scans. ZW developed the iDISCO+ protocol, RA and NR tested and characterized its applications. NR analyzed the data with help from CK. NR and MTL wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahrens MB, Li JM, Orger MB, Robson DN, Schier AF, Engert F, Portugues R. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SJ, Sammons RP, Jacobsen RI, Mackie J, Keller GB, Keck T. Subnetwork-Specific Homeostatic Plasticity in Mouse Visual Cortex In Vivo. Neuron. 2015;86:1290–1303. doi: 10.1016/j.neuron.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B … 1995 [Google Scholar]
- Bepari AK, Sano H, Tamamaki N, Nambu A, Tanaka KF, Takebayashi H. Identification of optogenetically activated striatal medium spiny neurons by Npas4 expression. PLoS ONE. 2012;7:e52783. doi: 10.1371/journal.pone.0052783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berényi A, Somogyvári Z, Nagy AJ, Roux L, Long JD, Fujisawa S, Stark E, Leonardo A, Harris TD, Buzsáki G. Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals. Journal of Neurophysiology. 2014;111:1132–1149. doi: 10.1152/jn.00785.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Luo G, Pham MD, Lovinger DM, Vogel SS, Costa RM. Deep brain optical measurements of cell type-specific neural activity in behaving mice. Nat Protoc. 2014;9:1213–1228. doi: 10.1038/nprot.2014.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, Hen R. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, O'Connell LA, Wu Z. Neural control of maternal and paternal behaviors. Science. 2014;345:765–770. doi: 10.1126/science.1253291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Takesian AE, Feng G, Fagiolini M, Hensch TK. Sensory integration in mouse insular cortex reflects GABA circuit maturation. Neuron. 2014;83:894–905. doi: 10.1016/j.neuron.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Petrus E, Gammon N, Lee HK. Distinct sensory requirements for unimodal and cross-modal homeostatic synaptic plasticity. J Neurosci. 2012;32:8469–8474. doi: 10.1523/JNEUROSCI.1424-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328:632–634. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Ahrens MB. Visualizing Whole-Brain Activity and Development at the Single-Cell Level Using Light-Sheet Microscopy. Neuron. 2015;85:462–483. doi: 10.1016/j.neuron.2014.12.039. [DOI] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015;10:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, et al. Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell. 2015;162:635–647. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV, Losonczy A. Dendritic inhibition in the hippocampus supports fear learning. Science. 2014;343:857–863. doi: 10.1126/science.1247485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, Osten P, Uchida N, Watabe-Uchida M. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. Elife. 2015;4 doi: 10.7554/eLife.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongeau R, Miller GA, Chiang E, Anderson DJ. Neural correlates of competing fear behaviors evoked by an innately aversive stimulus. J Neurosci. 2003;23:3855–3868. doi: 10.1523/JNEUROSCI.23-09-03855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peron SP, Freeman J, Iyer V, Guo C, Svoboda K. A Cellular Resolution Map of Barrel Cortex Activity during Tactile Behavior. Neuron. 2015;86:783–799. doi: 10.1016/j.neuron.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers KM, Benison AM, Klein A, Barth DS. Auditory, somatosensory, and multisensory insular cortex in the rat. Cereb Cortex. 2008;18:2941–2951. doi: 10.1093/cercor/bhn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Ciaranello RD, Meltzer HY. Binding of typical and atypical antipsychotic agents to transiently expressed 5-HT1C receptors. J Pharmacol Exp Ther. 1992;260:1361–1365. [PubMed] [Google Scholar]
- Saal Vom FS. Time-contingent change in infanticide and parental behavior induced by ejaculation in male mice. Physiology & Behavior. 1985;34:7–15. doi: 10.1016/0031-9384(85)90069-1. [DOI] [PubMed] [Google Scholar]
- Schwarz MK, Scherbarth A, Sprengel R, Engelhardt J, Theer P, Giese G. Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS ONE. 2015;10:e0124650. doi: 10.1371/journal.pone.0124650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamonin DP, Bron EE, Lelieveldt BPF, Smits M, Klein S, Staring M. Fast Parallel Image Registration on CPU and GPU for Diagnostic Classification of Alzheimer'. s Disease Front Neuroinform. 2014;7 doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C, Straehle C, Köthe U, Hamprecht FA. Ilastik: Interactive learning and segmentation toolkit. 2011 8th IEEE International Symposium on Biomedical Imaging. 2011:230–233. ISBI. [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner BEH, Cruise LA, Slattery DA, Hill DR, Shahid M, Henry B. Testing the validity of c-fos expression profiling to aid the therapeutic classification of psychoactive drugs. Psychopharmacology (Berl) 2004;171:306–321. doi: 10.1007/s00213-003-1579-7. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, Ueda HR. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- Tachikawa KS, Yoshihara Y, Kuroda KO. Behavioral transition from attack to parenting in male mice: a crucial role of the vomeronasal system. J Neurosci. 2013;33:5120–5126. doi: 10.1523/JNEUROSCI.2364-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. Vivo Two-Photon Imaging Reveals a Role of Arc in Enhancing Orientation Specificity in Visual Cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Wee CL, Naumann EA, Nnaemeka O, Schoppik D, Fitzgerald JE, Portugues R, Lacoste AMB, Riegler C, Schier AF, Randlett O, et al. Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods. 2015:1–12. doi: 10.1038/nmeth.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Autry AE, Bergan JF, Watabe-Uchida M, Dulac CG. Galanin neurons in the medial preoptic area govern parental behaviour. Nature. 2014;509:325–330. doi: 10.1038/nature13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.