Significance
Promyelocytic leukemia protein (PML) is a component of nuclear domain 10 (ND10) bodies and an antiviral effector of IFN-β. A herpes simplex virus (HSV) protein, ICP0, interacts with PML, merges with ND10 bodies, degrades PML, and ultimately takes over the domain of ND10 bodies. Here we show that viral gene expression and growth are reduced in PML−/− cells infected at low ratios of virus to cells. In essence, the results indicate that HSV-1 evolved the means to take advantage of an inimical cellular defense mechanism to degrade it and at the same time use it to gain access to a nuclear domain essential for efficient replication.
Keywords: ND10, ICP0, Sp100, Daxx, interferon
Abstract
After entry into the nucleus, herpes simplex virus (HSV) DNA is coated with repressive proteins and becomes the site of assembly of nuclear domain 10 (ND10) bodies. These small (0.1–1 μM) nuclear structures contain both constant [e.g., promyelocytic leukemia protein (PML), Sp100, death-domain associated protein (Daxx), and so forth] and variable proteins, depending on the function of the cells or the stress to which they are exposed. The amounts of PML and the number of ND10 structures increase in cells exposed to IFN-β. On initiation of HSV-1 gene expression, ICP0, a viral E3 ligase, degrades both PML and Sp100. The earlier report that IFN-β is significantly more effective in blocking viral replication in murine PML+/+ cells than in sibling PML−/− cells, reproduced here with human cells, suggests that PML acts as an effector of antiviral effects of IFN-β. To define more precisely the function of PML in HSV-1 replication, we constructed a PML−/− human cell line. We report that in PML−/− cells, Sp100 degradation is delayed, possibly because colocalization and merger of ICP0 with nuclear bodies containing Sp100 and Daxx is ineffective, and that HSV-1 replicates equally well in parental HEp-2 and PML−/− cells infected at 5 pfu wild-type virus per cell, but poorly in PML−/− cells exposed to 0.1 pfu per cell. Finally, ICP0 accumulation is reduced in PML−/− infected at low, but not high, multiplicities of infection. In essence, the very mechanism that serves to degrade an antiviral IFN-β effector is exploited by HSV-1 to establish an efficient replication domain in the nucleus.
Several prominent events take place after the entry of herpes simplex virus (HSV) DNA into the nucleus of newly infected cells. Thus, viral DNA becomes coated by repressive proteins, the function of which is to block viral gene expression (1–6); nuclear domain 10 (ND10) bodies colocalize with the viral DNA (7, 8); α or immediate early viral genes are expressed; and one viral protein, ICP0, degrades promyelocytic leukemia protein (PML) and Sp100, two key constituents of ND10 bodies in conjunction with the UbcH5A ubiquitin-conjugating enzyme (9–11). What is left of the ND10 bodies is infiltrated by viral proteins and becomes the viral replication compartment (12–15).
ND10 bodies range between 0.1 and 1 μM in diameter. The composition of ND10 bodies varies depending on the cellular function or in response to stress, such as that resulting from virus infection (16–19). Among the constant components of ND10 are PML, Sp100, and death-domain associated protein (Daxx). PML has been reported to be critical for the recruitment of components and for the organization of the ND10 bodies (18–23). The function of ND10 bodies may vary under different cellular conditions and may also depend on their composition.
A key question that remains unanswered is the function of ND10 bodies in infection, and in particular, why HSV has evolved a strategy that specifically targets PML and Sp100 for degradation. Two clues that may ultimately shed light on the function of ND10 is that exposure of cells to IFN leads to an increase in the number of ND10 bodies and an increase in PML (16, 24–26). The second clue emerged from the observation reported earlier by this laboratory is that pretreatment of murine PML+/+ cells with IFN-β led to a drastic reduction in virus yields. In contrast, exposure of PML−/− cells to IFN-β led to a significantly smaller decrease in virus yields (27). The results suggest PML is an antiviral effector of IFN-β, but many questions regarding the function of PML remain unanswered (28).
In this study, we constructed a PML−/− cell line (1D2) derived from HEp-2 cells. The first part of this report centers on the structure of ND10 bodies bereft of PML and the interaction of these bodies with ICP0. In the second part, we report on the replication of HSV-1 in PML−/− cells. Here we show that HSV-1 replication and the accumulation of ICP0 are significantly reduced in PML−/− cells exposed to low ratios of virus per cell. HSV has evolved a strategy to take advantage of PML before its degradation.
Results
Generation and Properties of PML−/− 1D2 Clone Derived from HEp-2 Cells.
PML is a family of seven isoforms. The largest, PML I, consists of nine exons (29–31). PML−/− cell clones were generated from HEp-2 cells by transfection of clustered regularly interspaced short palindromic repeats [clustered regularly interspaced short palindromic repeats (CRISPR)/cas9 cassette] targeting exon 1 of PML (32–34). The procedure for drug selection and flow cytometry were both performed according to the manufacturer’s instructions and are briefly outlined in Materials and Methods. Of the numerous clones derived from single cells, we selected clone ID2 for further study.
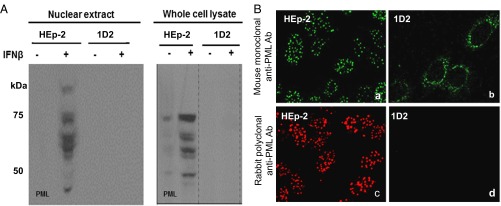
Two series of experiments were done to verify the absence of PML in 1D2 cells. In the first, we took advantage of earlier studies showing that PML is amplified in cells exposed to IFN-β (26). Specifically, after overnight exposure to IFN-β, extracts of isolated nuclei or total cell lysates were solubilized, subjected to electrophoresis in a denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with rabbit polyclonal anti-PML antibody. As shown in Fig. 1A, the antibody to PML reacted with multiple protein bands present in HEp-2 cells. The antibody did not react with proteins present in lysates of 1D2 cells.
Fig. 1.
Confirmation of the absence of PML in 1D2 clone. (A) Parental HEp-2 and 1D2 cell cultures were exposed to IFN-β (1,000 U/mL). After 24 h, the cells were harvested, and lysates of nuclei or of whole cells were subjected to electrophoresis in denaturing gels transferred to a nitrocellulose sheet and reacted with rabbit polyclonal antibody against PML. (B) HEp-2 or 1D2 cells grown on four-well slides were fixed and reacted with mouse monoclonal antibody to PML (a and b) or rabbit polyclonal antibody to PML (c and d). (Magnification: 100×.)
In the second series of experiments, parental HEp-2 or ID2 cells grown on four-well slides and exposed overnight to IFN-β were reacted with mouse monoclonal antibody to PML (Fig. 1 B, a and b) or with rabbit polyclonal antibody to PML (Fig. 1 B, c and d). HEp-2 cells exhibited typical intranuclear clusters of ND10 bodies containing PML. No such structures were observed in 1D2 cells. We did note that ID2 cells contained a ring of green fluorescent puncta observed in cells reacted with the mouse monoclonal antibody. These rings of fluorescent puncta in the perinuclear space were also present in parental HEp-2 cells but were overshadowed by the ND10 bodies. The presence of the fluorescent puncta in both HEp-2 and 1D2 cells suggests they are not derived from truncated PML proteins. Moreover, the observation that these puncta were observed in cells reacted with mouse monoclonal antibody, but not with the rabbit polyclonal antibody against PML, reinforces the conclusion that the antigens reacting with the monoclonal antibody are not derived from PML. We conclude that PML is not expressed in 1D2 cells.
Effect of IFN-β on Accumulation of Sp100 and Daxx mRNAs.
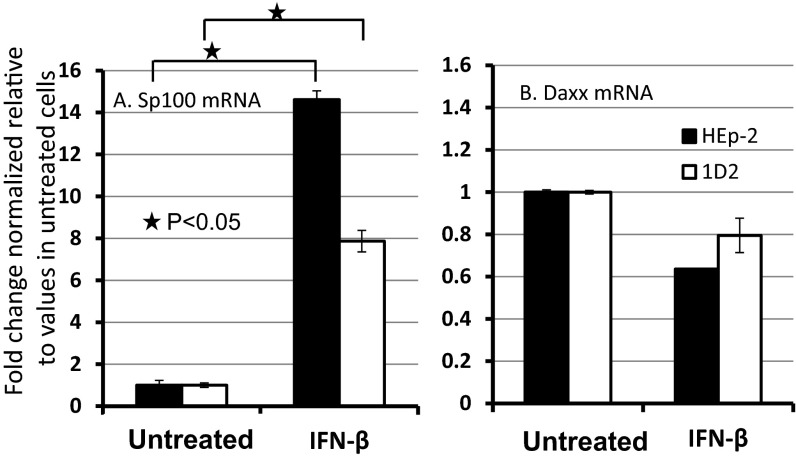
In the experiments described here, we examined the distribution of Sp100 and Daxx in parental HEp-2 cells and in the PML−/− 1D2 clone. Here (Fig. 2), we report that exposure of both parental and 1D2 mutant cells to IFN-β enhanced the accumulation of Sp100 but had no significant effect on the accumulation of Daxx in either the parental HEp-2 or 1D2 cells. The procedures used in this study are described in Materials and Methods.
Fig. 2.
Sp100 and Daxx respond differently to exposure of HEp-2 or 1D2 cells to IFN-β. Parental HEp-2 PML+/+ cells or PML−/− 1D2 cell cultures were mock treated or exposed to IFN-β (1,000 U/mL). After 24 h, the cells were harvested and the relative amounts of Sp100 and Daxx were quantified, as described in Materials and Methods. mRNA levels of Sp100 and Daxx were normalized to 18S RNA, and their relative value in untreated group were set as 1. Two-tailed P values were calculated using standard t test.
The Intranuclear Distribution of Sp100 and Daxx in Parental HEp-2 and in Clone 1D2 PML−/− Cells.
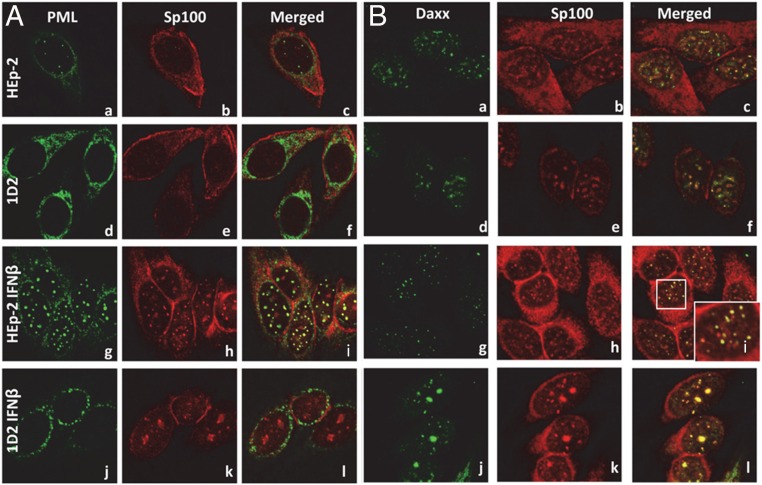
In this series of experiments, parental HEp-2 or 1D2 cells were grown on four-well slides in the presence of growth medium alone or medium supplemented with IFN-β (1,000 U/mL). After 24 h, the cells were fixed and reacted with monoclonal antibody to PML and polyclonal antibody to Sp100 (Fig. 3A) or monoclonal antibody to Daxx and polyclonal antibody to Sp100. The cells were examined with the aid of a Zeiss confocal microscope. The results may be summarized as follows. First, in the absence of IFN-β, the accumulation of Sp100 was weak and largely perinuclear (Fig. 3 A, b, c, e, f, and B, b, c, e, f). There were few ND10 structures in parental HEp-2 cells reacted with antibody to PML and Sp100 (Fig. 3 A, c), and these were more readily seen in cells reacted with antibodies against Daxx and Sp100 (Fig. 3 B, c). As expected, the monoclonal antibody to PML reacted with an antigen localized in the perinuclear space of 1D2 cells (Fig. 3 A, d and e) in close proximity to Sp100. The structures containing Sp100 and Daxx in parental HEp-2 cells could not be differentiated from those detected in nuclei of 1D2 cells.
Fig. 3.
Cellular localization of Sp100 and Daxx in HEp-2 and 1D2 cells. At 70% confluency, the medium of parental HEp-2 or 1D2 cells grown on four-well slides was replaced with fresh medium alone or medium supplemented with IFN-β (1,000 U/mL). After 24 h, the cells were fixed and reacted with polyclonal antibody to Sp100 (red) and either PML (green, A) or Daxx (green, B). (Magnification: 100×.)
Second, the number of ND10 bodies was consistently higher in HEp-2 cells treated with IFN-β (Fig. 3 A, i and B, i). As shown in the enlarged boxed area (Fig. 3 B, i, Inset), the intranuclear fluorescent structures were relatively uniform in size and contained both Sp100 and Daxx.
Finally, in 1D2 cells treated with IFN-β, the distribution of antigens reacting with anti-PML monoclonal antibody (Fig. 3 A, i) was similar to that observed in untreated cells (Fig. 3 A, a). In the IFN-β treated cells, Sp100 and Daxx colocalized in structures that varied in size (Fig. 3 A, k and, l and B, k and l), but on average, they were significantly larger than the ND10 bodies observed in parental HEp-2 treated with IFN-β (Fig. 3 A, l and B, l).
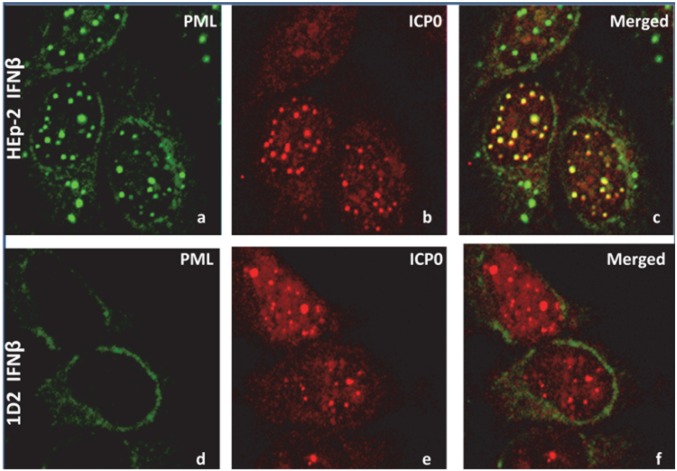
The results of the studies presented here suggest that the colocalization of Sp100 and Daxx is not dependent on the presence of PML. However, the variability of the structures that contain Sp100 and Daxx suggests that the relatively uniform sizes of ND10 structures in parental HEp-2 cells are defined by PML. To determine the role of ICP0 in defining the size of the aggregates early in infection, a monolayer culture of parental HEp-2 or 1D2 cells was exposed to 10 pfu HSV-1 per cell. After 2 h, the cultures were fixed and reacted with monoclonal antibody to PML and polyclonal antibody to ICP0. Typical images shown in Fig. 4 A–C indicate that the ND10 bodies containing both PML and ICP0 are uniform in size in HEp-2 cells. In contrast, ICP0 aggregates formed in infected 1D2 cells varied considerably in size (Fig. 4 D–F).
Fig. 4.
ICP0 formed aggregates in PML−/− cells. HEp-2 (A–C) and 1D2 cells (D–F) grown on four-well slides were treated with IFN-β, as earlier, and then exposed to 10 pfu HSV-(F) per cell. At 2 h after infection, the cells were fixed and reacted with polyclonal antibody to ICP0 (red) and monoclonal antibody to PML (green). (Magnification: 100×.)
The Role of PML in the Degradation of Sp100.
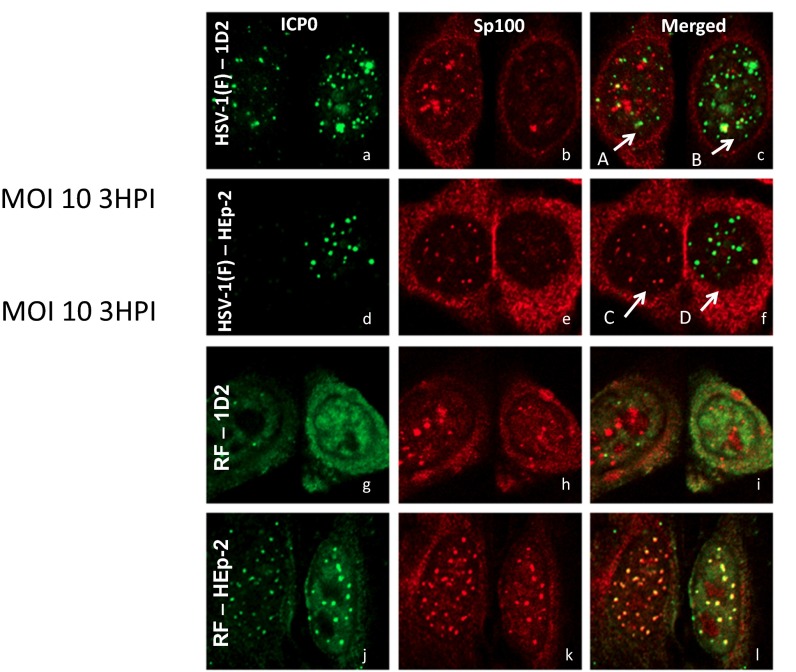
Studies published elsewhere have shown that ICP0 is an E3 ligase that, in conjunction with UbcH5a ubiquitin-conjugating enzyme, degrades PML and Sp100 (9–11). Mutations in the ring finger located in exon 2 of ICP0 abolish the ligase activity (11). The question posed here is whether the absence of PML hinders the degradation of Sp100. We report two series of experiments. In the first, parental HEp-2 cells or 1D2 cells grown on four-well slides were infected with 10 pfu HSV-1(F) or a ring finger (RF) mutant that is bereft of ubiquitin ligase activity. At 3 h after exposure to the virus, the cells were fixed and reacted with antibody to ICP0 and to Sp100, as described earlier.
The results may be summarized as follows: First, Fig. 5 A–C shows two 1D2 cells. The cell labeled A contains numerous aggregates of Sp100 differing in size and relatively few aggregates of ICP0. As shown in Fig. 5C, only a few of the aggregates of ICP0 (green) colocalized with the aggregates of Sp100 (red). Moreover, as apparent in Fig. 5C, the colocalized structures did not fully merge. In the cell labeled B, there are few prominent Sp100 bodies. The one prominent Sp100 body colocalized but did not merge with the ICP0 (green) aggregates.
Fig. 5.
(A–L) ICP0 and Sp100 aggregated with Daxx do not colocalize efficiently in 1D2 cells infected with wild-type or RF mutant viruses. Cells grown on four-well slide cultures and treated as earlier were exposed to 10 pfu wild-type or RF mutant virus per cell. At 3 h after infection, the cultures were fixed and reacted with monoclonal antibody to ICP0 and polyclonal antibody to Sp100. The images shown were acquired using a Zeiss confocal microscope. (Magnification: 100×.)
Second, Fig. 5 D–F shows two parental HEp-2 cells. The cell labeled C shows no signs of being infected. Thus, Sp100 aggregates are prominent, and no ICP0 can be seen. In contrast, there was little Sp100 and very prominent ICP0 aggregates in small uniform structures consistent with the size of ND10 bodies in the cell labeled D. The data are consistent with the hypothesis that Sp100 had been degraded.
Finally, in parental (PML+/+) HEp-2 cells infected with RF mutant (Fig. 5 J–L), ICP0 and Sp100 aggregated in small nuclear structures relatively uniform in size indistinguishable from classical ND10 bodies. In contrast, in 1D2 cells, there was no evidence of large-scale colocalization of ICP0 and Sp100. Although Sp100 formed aggregates differing in size, ICP0 appeared to diffuse in the nucleus (Fig. 5 G–I).
The results presented here suggest that in PML−/− cells, the colocalization of ICP0 and Sp100 is a slow process and may not result in a total mixing of the two proteins. The paucity of Sp100 in Fig. 5C (cell labeled “B”) suggests Sp100 was degraded, possibly by massive accumulation of ICP0.
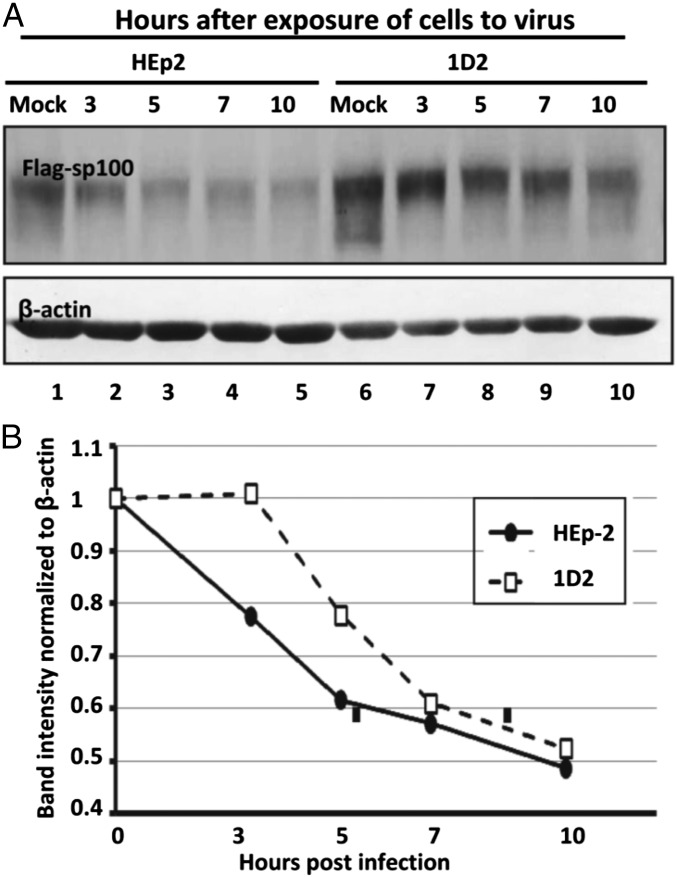
To quantify the pattern of decay of SP100 in the two cell lines, replicate cultures of HEp-2 or ID2 cells were transfected with FLAG-tagged SP100, as described in Materials and Methods. After 48 h, the cells were exposed to 10 pfu HSV-1(F) per cell. Cultures harvested at time of exposure to the virus (Mock) or at 3, 5, 7, or 10 h after infection were solubilized, subjected to electrophoresis in denaturing gels, and probed with antibody to Sp100 and β-actin. Fig. 6A shows the immunoblots of Sp100 and β-actin. Fig. 6B shows the results of a density scan normalized with respect to Sp100 present in mock-treated cells. The results show that under conditions used in these studies, the initiation of decay of Sp100 in 1D2 cells was delayed by at least 3 h, in contrast to the apparent initiation of decay of Sp100, which extrapolates to a time at or shortly after exposure of the cells to the virus.
Fig. 6.
Degradation of Sp100 by HSV-1(F) infection is delayed in PML−/− 1D2 cells. (A) HEp-2 and 1D2 cells were transfected with a plasmid-encoding Flag-tagged Sp100. At 48 h after transfection, the cells were exposed to 10 pfu wild-type virus per cell. At 0 (mock), 3, 5, 7, or 10 h after infection, the cells were harvested, solubilized, subjected to electrophoresis in denatured gels, and reacted with antibody to Sp100 and β-actin. (B) The relative amounts of Flag-tagged Sp100 were quantified and normalized to β-actin, and the amounts of Flag-tagged Sp100 in mock-treated cells (0 h after infection) with the aid of Image J. The degradation rates of FLAG-Sp100 were calculated as described in Materials and Methods.
The Replication of Wild-Type Virus Is Impaired in 1D2 Cells Infected at Low Ratios of Virus per Cell.
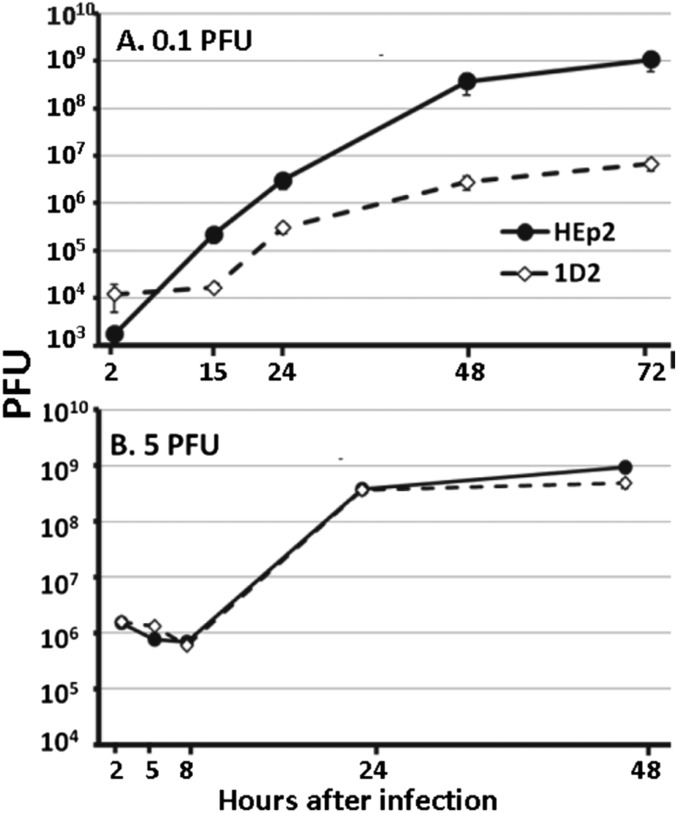
The degradation of PML mediated by ICP0 E3 ligase early after infection suggests that PML is inimical to the replication of HSV-1. To test this hypothesis, we exposed replicate cultures of parental HEp-2 or 1D2 cells to either 0.1 or 5 pfu HSV-1(F) per cell. The cells were harvested at intervals between 2 and 48 h in the case of cultures exposed to 5 pfu per cell, or between 2 and 72 h in the case of cultures exposed to 0.1 pfu virus per cell. As shown in Fig. 7B, the virus yields from cultures exposed to 5 pfu per cell were virtually identical. In contrast, the yields of virus obtained 48 and 72 h after infection of 1D2 cells with 0.1 pfu per cell were ∼100-fold lower than those obtained in parental HEp-2 cells exposed to the same ratio of virus per cell (Fig. 7A). The results suggest that PML may play both a supportive and inimical role in HSV-1 replication.
Fig. 7.
Growth of HSV-1(F) in PML−/− 1D2 cells is impaired in PML−/− cells exposed to low ratios of virus per cell. Replicate cultures of HEp-2 or 1D2 cells were exposed to 5 or 0.1 pfu virus per cell. At times shown, the cells were harvested and virus yields were titered in Vero cell.
The Accumulation of ICP0 Is Impaired in 1D2 Cells Infected at Low Ratios of Virus per Cell.
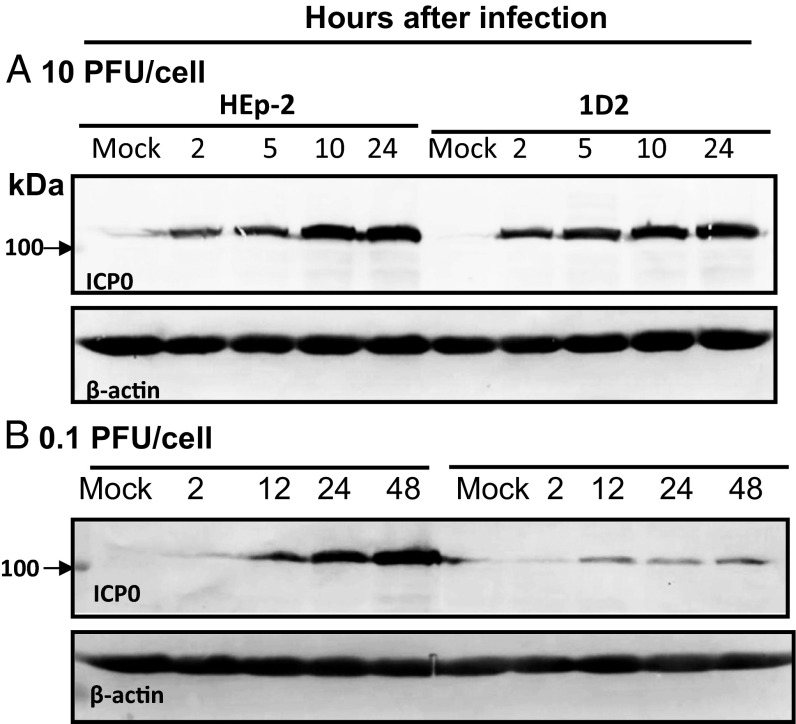
One hypothesis that could explain the reduced accumulation of virus in 1D2 cells at low multiplicities of infection is a reduction in the synthesis of ICP0. To test this hypothesis, replicate cultures of HEp-2 or 1D2 cells were infected with either 10 or 0.1 pfu virus per cell. At intervals after infection shown in Fig. 8, the cells were harvested, solubilized, subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose sheet, and probed with antibody to ICP0 and β-actin. The results shown in Fig. 8 indicate that the rates of accumulations of ICP0 in HEp-2 or 1D2 cells exposed to 10 pfu per cell were approximately equal. In contrast, the accumulation of ICP0 in 1D2 cells lagged significantly behind the accumulation of ICP0 observed in HEp-2 cells. The significance of the results rests on two considerations: Foremost, the synthesis and accumulation of ICP0 is dependent on the presence of PML, even though ICP0 ultimately degrades PML, and second, the delay in the degradation of Sp100, as shown in Fig. 5, is not a result of the lack of ICP0, as in those experiments, the cells were exposed to 10 pfu per cell.
Fig. 8.
(A and B) Accumulation of ICP0 is reduced in PML−/−1D2 cells exposed to low ratios of virus per cell. Replicate cultures of HEp-2 or 1D2 cells were exposed to 10 or 0.1 pfu virus per cell. Cells were harvested at indicated points and were reacted with antibody to ICP0 and β-actin.
Effect of IFN-β on the Replication of HSV-1(F) in Parental HEp-2 and in 1D2 Cells.
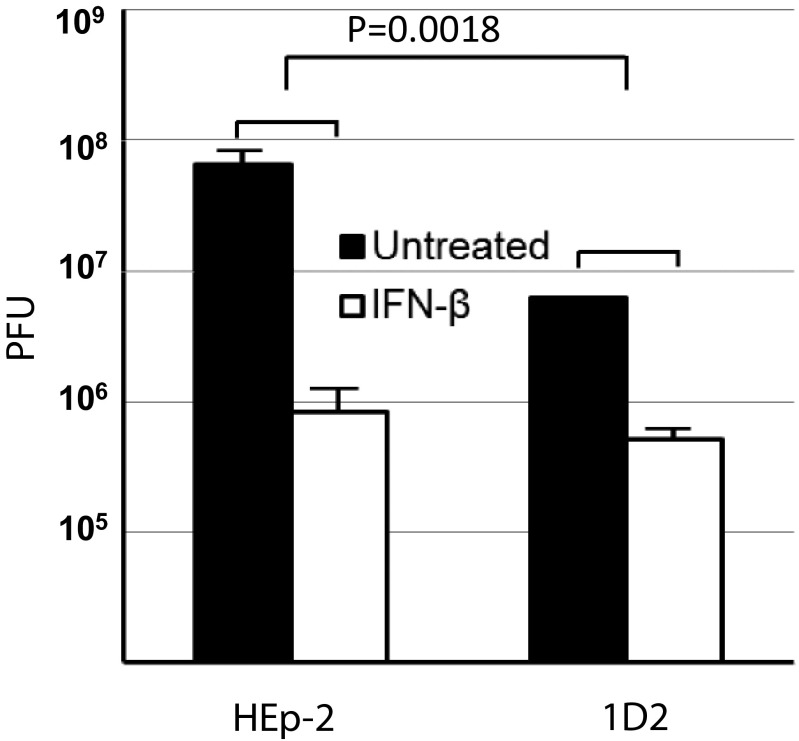
Earlier studies have shown that IFN-β is more effective in suppressing HSV-1 replication in primary murine fibroblast cultures than in sibling cells in which PML was knocked out. The question arose whether IFN-β had a similar effect in human cells, and in particular, in the parental HEp-2 and in PML−/− 1D2 clone cells used in the studies reported here. In the experiments reported, replicate cultures of the two cell lines were exposed to 0.1 pfu HSV-1 per cell and then either incubated for 24 h without further treatment or maintained in medium containing IFN. The results are shown in Fig. 9. As expected from the results in Fig. 7, the yields of untreated 1D2 cells were lower than those obtained in parental HEp-2 cells. In IFN-β–treated cells, the yield of virus at 15 h was reduced 100-fold compared with those obtained in untreated infected cells. In contrast, IFN-β decreased the yield of virus in 1D2 cells 10-fold compared with yields obtained in infected untreated 1D2 cells. In essence, the results obtained in a human cell line paralleled those obtained in murine cells.
Fig. 9.
The antiviral effects of IFN-β are diminished in cells in PML−/− cells. HEp-2 or PML−/− 1D2 cells were pretreated with IFN-β at 1,000 U/mL for 24 h and then exposed to 0.1 pfu virus per cell. The cells were harvested at 24 h after infection. Virus yields were titered in Vero cells. Antiviral effects of IFN-β treatment in HEp-2 or PML−/− 1D2 cells were represented by viral titer reduction posttreatment. P value was calculated by comparison of antiviral effects of IFN-β in HEp-2 and that in PML−/− 1D2 cells.
Discussion
The function, and ultimately the degradation, of PML in HSV-1-infected cells has been the subject of numerous studies. The preponderance of evidence supports the hypothesis that PML is inimical to virus replication. Thus, the accumulation of PML is enhanced after exposure of cells to IFN-β (26). In addition, as reinforced in this study, the antiviral effects of IFN-β are drastically reduced in the absence of PML (27). Finally, one of the first and most prominent actions of ICP0, a product made immediately after infection, is to degrade PML and Sp100, another key component of ND10 bodies. The enhancement of synthesis of PML and Sp100 by IFN-β and the targeting of these two components of ND10 bodies for degradation may be selective. Thus, Daxx, an ND10 constituent that colocalizes with Sp100, is neither enhanced by IFN-β nor degraded by ICP0. PML appears to be an effector of INF-β, and one expectation of our studies was that the absence of PML would be beneficial to HSV replication. This is, in fact, not the case. Our findings are as follows: In PML−/− cells, the typical ND10 bodies were replaced with bodies containing at least Daxx and Sp100. These bodies vary in size, but on average, are larger than the typical ND10 bodies present in parental PML+/+ cells. In PML−/− cells, ICP0 formed aggregates that in some cells partially colocalized but did not merge with the SP100/Daxx bodies. Consistent with the evidence that colocalization of ICP0 and Sp100/Daxx bodies was not a common event, Sp100 was degraded, but only after a delay of several hours. The delay in the degradation of Sp100 was not a result of a lack of ICP0 inasmuch as, under conditions tested, cells infected with 10 pfu virus per cell, the two cell lines accumulated equivalent amounts of ICP0. We conclude that some constituent of ND10 bodies aggregate in the absence of PML. PML appears to confer typical morphology to ND10 bodies, and the aggregation of ICP0 near ND10 bodies ultimately leads to the degradation of SP100. In addition, at high multiplicities of infection, the pattern of accumulation of infectious virus in PML−/− cells is not significantly different from that in parental PML+/+. In contrast, in cells infected with 0.1 pfu per cell, the yields were 100-fold lower than in PML+/+ cells exposed to the same ratio of virus per cell. Equally unexpected, the amounts of ICP0 that accumulated in PML−/− cells exposed to 0.1 pfu per cell was significantly lower than that produced in PML+/+ cells exposed to the same ratio of virus per cell.
The data presented in this report do not support the current consensus on the events taking place in the infected cells immediately after entry of viral DNA into the nucleus. Thus, the current consensus is that on entry of viral DNA into the nucleus, numerous cellular proteins attempt to transcriptionally silence viral DNA (1, 7, 8, 35–37). Concurrently, VP16, a key viral transcriptional factor, recruits cellular protein to transcribe and enable the synthesis of α proteins (38–41). In addition, concurrently or sequentially, a ND10 body assembles at viral DNA (7, 8), and one α protein, ICP0, interacts with PML, gains entry, and diffuses throughout the ND10 body (12). Finally, ICP0 associated with the ND10 body degrades PML and Sp100 (9, 10, 42). According to this sequence of events, the synthesis of α proteins is independent of the interaction of virion components with PML. The data presented here suggest that optimal accumulation of viral α proteins, and ultimately the accumulation of viral proteins, is enhanced by PML.
An intriguing question is why the parental HEp-2 cells infected at high or very low ratios of pfu per cell produced identical yields, whereas 1D2 cells infected at low ratios of virus per cell produced significantly lower yields than cells infected at high ratios of virus per cell. Several nonexclusive mechanisms may be responsible. Specifically, in principle, in cultures infected at low ratios of virus per cell, the final yield is the sum of virus produced during multiple cycles of infection, replication, and spread to uninfected cells. The inherent assumption has been that the yield of virus from the first cycle is identical to that of the last cycle. This concept has been challenged by the observation that cells export along with infectious particles exosomes containing micro-RNAs that may affect the synthesis of virus gene products and inimical cellular innate immune factors (e.g., STING) (43, 44). It is conceivable that the negative effect of exosomes is much higher on late cycles than on earlier ones.
In addition, the effect on viral replication in cells contracting concurrently both virus and the contents of exosomes bearing antiviral factors may be quite different from that taking place in cells in which entry of antiviral factors born by exosomes precedes virus entry by several hours. The delay in the degradation of SP100 in cells exposed to 10 pfu per cell suggests that assembly of virus factories required for optimal accumulation of gene products in cells infected at lower multiplicities may be even more delayed.
Last, two highly significant studies showed that in infected cells, the number of virus factories or assemblons initiated by single genomes is less than 10, irrespective of the multiplicity of infection (45, 46). Implicit in this finding is the notion that either a critical component necessary for the assembly of virus factories is limited in supply, or that there is a gradual accumulation of innate immune factors that curtail the formation of virus factories once a critical number is formed. A convenient but unsupported hypothesis that could explain our data is that in the absence of PML, the resistance to formation of virus factories increases.
Irrespective of whether the hypotheses are ultimately borne out by further studies, the results of this report indicate that HSV-1 evolved the means to take advantage of an inimical cellular defense mechanism to degrade it, and at the same time use it to gain access to a nuclear domain essential for efficient replication.
Materials and Methods
Cells and Viruses.
The sources and maintenance of HEP-2 and Vero cells were reported elsewhere (47). HSV-1(F) is the prototype strain used in our laboratory (48). RF-HSV (F) was a recombinant virus containing ring-finger domain deletion of both copies of ICP0 protein described elsewhere (12). All viruses were amplified in HEp-2 cells. Plaque assays were performed in Vero cells.
RNA Isolation and Real-Time PCR.
RNA extraction and assays were done as previously described (49). mRNA level of Sp100 and Daxx were measured by gene-specific primers: Sp100 forward primer, 5′-GCTCAGGACCCCAGATTGTAC-3′, and reverse primer, 5′-CTAATCTTCTTTACCTGACCC-3′, and Daxx forward primer, 5′-GACGGACATTTCCTCTTCCA-3′, and reverse primer, 5′-CGCCTCCATTGAAGGAAGTA-3′.
Transfection Plus Superinfection.
Parental HEp-2 and PML−/− clone 1D2 cells were grown to 70–90% confluency in T150 flasks. Each flask was transfected with 18 μg of a plasmid DNA containing a CMV promoter-driven FLAG-tagged Sp100 ORF, using Lipofectamine LTX with Plus Reagent (Life Technologies, 15338100), according to manufacturer’s instructions. At 24 h after transfection, the cells contained in T150 flasks were split into T25 flasks. At 24 h after seeding, the mock-transfected and FLAG-tagged Sp100-transfected cells were mock infected or exposed to 10 pfu HSV-1(F) per cell. The cells were harvested and processed as described in Results.
Generation of PML−/− Cell Line Using CRISPR.
A CRISPR kit targeting the first exon of PML gene was obtained from Origene (cat. no. KN200700). Parental HEp-2 cells were cotransfected with a gRNA vector plasmid targeting sequences 5′-CTCGGAGATCGGGCGGGTGC-3′ (150th–169th bp in exon 1; 4th–10th amino acid from the start codon) or 5′-GGCCTTCAGAGGGGGTCTCG-3′ (218th–237th bp in exon1; 26th–32nd amino acid from the start codon), and a donor vector containing a selection cassette expressing GFP–puromycin was inserted into the targeted site in human PML gene. Selected cells (GFP+ and puromycin resistant) were serially diluted to form single cell-derived colonies. The colonies were further screened for expression of PML by immunoblotting and immunofluorescence.
Immunoblot Analyses.
At indicated times, the cells were lysed, heat denatured, electrophoretically separated in denaturing gels, transferred to polyvinylidene difluoride membranes, and probed with the appropriate primary antibody, as previously described (49). The antibodies included monoclonal anti-PML (Santa Cruz, sc-966) (50), rabbit polyclonal anti-PML (Abcam, 79466), mouse monoclonal anti–β-actin (Thermo Fisher, MA5-15739), rabbit polyclonal anti-ICP0 (Lab stock), and mouse monoclonal anti-FLAG (Sigma, F1804). The membranes were then incubated with alkaline phosphatase-conjugated goat anti-mouse or goat anti-rabbit secondary antibody (Sigma) and visualized with BCIP/NBT Western Blotting Detection Reagent (GE Healthcare).
ImageJ Quantification and Statistics.
Image J software was used to quantify band intensity. FLAG-Sp100 levels were first normalized to β-actin. Degradation rates of FLAG-Sp100 were calculated by normalization to Sp100 in mock-treated cells. Error bars were calculated from the results of three independent quantifications of band intensities. Two-tailed P values were calculated using standard t test. To compare ICP0 expression level, the band intensity of ICP0 was first normalized to β-actin. For high multiplicity of infection, ICP0 expression level of all points examined was then normalized to that in HEp-2 at 24 h postinfection; for low multiplicity of infection, ICP0 expression level of all points examined was then normalized to that in HEp-2 at 48 h postinfection.
Immunofluorescence Staining and Confocal Microscopy.
Cells in four-welled slides were fixed and permeabilized, blocked, and then incubated with primary antibodies, as previously described (47). Those included antibodies anti-PML, as described earlier, rabbit anti-Sp100 (Abcam, ab43151); mouse anti-Daxx (Abcam, ab9091); and rabbit anti-ICP0 or mouse anti-ICP0, described elsewhere (47), at 4 °C overnight, rinsed three times with blocking solution, and subjected to reaction with fluorescein isothiocyanate-conjugated goat anti-mouse (Invitrogen) or/and Texas Red-conjugated goat anti-rabbit (Sigma) secondary antibodies. Images were taken at 100× magnification with a Zeiss confocal microscopy.
Acknowledgments
We thank Lindsay Smith for invaluable assistance; and Guoying Zhou, Rozanne Sandri-Goldin, and David Bloom for careful reading of the article and useful comments and suggestions. These studies were aided by a grant from the Joseph Regenstein Foundation.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104(43):17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knipe DM, et al. Snapshots: Chromatin control of viral infection. Virology. 2013;435(1):141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conn KL, Schang LM. Chromatin dynamics during lytic infection with herpes simplex virus 1. Viruses. 2013;5(7):1758–1786. doi: 10.3390/v5071758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson Z, Dhummakupt A, Messer H, Phelan D, Bloom D. Role of polycomb proteins in regulating HSV-1 latency. Viruses. 2013;5(7):1740–1757. doi: 10.3390/v5071740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guise AJ, Budayeva HG, Diner BA, Cristea IM. Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses. 2013;5(7):1607–1632. doi: 10.3390/v5071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel JL, Kristie TM. The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection. Viruses. 2013;5(5):1272–1291. doi: 10.3390/v5051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79(8):5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett RD, Murray J, Orr A, Preston CM. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol. 2007;81(20):10991–11004. doi: 10.1128/JVI.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100(15):8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelbi-Alix MK, de Thé H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18(4):935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 11.Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76(2):841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu H, Zheng Y, Roizman B. Interaction of herpes simplex virus ICP0 with ND10 bodies: A sequential process of adhesion, fusion, and retention. J Virol. 2013;87(18):10244–10254. doi: 10.1128/JVI.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkham J, Coen DM, Hwang CB, Weller SK. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J Virol. 2001;75(5):2353–2367. doi: 10.1128/JVI.75.5.2353-2367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett RD, Sourvinos G, Orr A. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J Virol. 2003;77(6):3680–3689. doi: 10.1128/JVI.77.6.3680-3689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukonis CJ, Burkham J, Weller SK. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: Only a subset are likely to be precursors to replication compartments. J Virol. 1997;71(6):4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 17.Dellaire G, Bazett-Jones DP. PML nuclear bodies: Dynamic sensors of DNA damage and cellular stress. BioEssays. 2004;26(9):963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 18.Ishov AM, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147(2):221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong S, et al. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95(9):2748–2752. [PubMed] [Google Scholar]
- 20.Szostecki C, Guldner HH, Netter HJ, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145(12):4338–4347. [PubMed] [Google Scholar]
- 21.Dyck JA, et al. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76(2):333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 22.Li H, et al. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20(5):1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koken MH, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13(5):1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisole S, Stoye JP, Saïb A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3(10):799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 25.Dror N, et al. Interferon regulatory factor-8 is indispensable for the expression of promyelocytic leukemia and the formation of nuclear bodies in myeloid cells. J Biol Chem. 2007;282(8):5633–5640. doi: 10.1074/jbc.M607825200. [DOI] [PubMed] [Google Scholar]
- 26.Stadler M, et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11(12):2565–2573. [PubMed] [Google Scholar]
- 27.Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77(12):7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regad T, et al. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 2001;20(13):3495–3505. doi: 10.1093/emboj/20.13.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20(49):7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida H, et al. PML-retinoic acid receptor alpha inhibits PML IV enhancement of PU.1-induced C/EBPepsilon expression in myeloid differentiation. Mol Cell Biol. 2007;27(16):5819–5834. doi: 10.1128/MCB.02422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Condemine W, et al. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66(12):6192–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- 32.Gratz SJ, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Zhang L, Huang X. Genome modification by CRISPR/Cas9. FEBS J. 2014;281(23):5186–5193. doi: 10.1111/febs.13110. [DOI] [PubMed] [Google Scholar]
- 35.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15(11):1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuchet-Lourenço D, et al. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 2011;7(7):e1002123. doi: 10.1371/journal.ppat.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6(3):211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 38.McKnight JL, Kristie TM, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc Natl Acad Sci USA. 1987;84(20):7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristie TM, Roizman B. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc Natl Acad Sci USA. 1987;84(1):71–75. doi: 10.1073/pnas.84.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKnight JL, Pellett PE, Jenkins FJ, Roizman B. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate alpha-trans-inducing factor-dependent activation of alpha genes. J Virol. 1987;61(4):992–1001. doi: 10.1128/jvi.61.4.992-1001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kristie TM, LeBowitz JH, Sharp PA. The octamer-binding proteins form multi-protein–DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989;8(13):4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13(21):5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA. 2014;111(46):E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Z, et al. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci USA. 2016;113(7):E894–E901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobiler O, Lipman Y, Therkelsen K, Daubechies I, Enquist LW. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat Commun. 2010;1:146. doi: 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobiler O, Brodersen P, Taylor MP, Ludmir EB, Enquist LW. Herpesvirus replication compartments originate with single incoming viral genomes. MBio. 2011;2(6):e00278-11. doi: 10.1128/mBio.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalamvoki M, Roizman B. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 2008;105(51):20488–20493. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA. 2005;102(21):7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du T, Zhou G, Roizman B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci USA. 2011;108(46):18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flenghi L, et al. Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: Immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells, and epithelia. Blood. 1995;85(7):1871–1880. [PubMed] [Google Scholar]