Abstract
To compare lung adenocarcinoma (ADC) and lung squamous cell carcinoma (SqCC) and to identify new drivers of lung carcinogenesis, we examined exome sequences and copy number profiles of 660 lung ADC and 484 lung SqCC tumor/normal pairs. Recurrent alterations in lung SqCCs were more similar to other squamous carcinomas than to lung ADCs. Novel significantly mutated genes included PPP3CA, DOT1L, and FTSJD1 in lung ADC, RASA1 in lung SqCC, and KLF5, EP300, and CREBBP in both tumor types. Novel amplification peaks encompassed MIR21 in lung ADC, MIR205 in lung SqCC, and MAPK1 in both. Lung ADCs lacking receptor tyrosine kinase/Ras/Raf alterations revealed mutations in SOS1, VAV1, RASA1, and ARHGAP35. Regarding neoantigens, 47% of the lung ADC and 53% of the lung SqCC tumors had at least 5 predicted neoepitopes. While targeted therapies for lung ADC and lung SqCC are largely distinct, immunotherapies may aid in treatment for both subtypes.
INTRODUCTION
Lung cancer remains the leading cause of death from cancer around the world1. An estimated 221,000 new cases and 158,000 deaths from lung cancer occurred in the United States alone in 20152. The two major histological classes are non-small cell lung cancers (NSCLC) and small-cell lung cancers (SCLC). NSCLCs are mostly comprised of lung adenocarcinomas (ADC) and lung squamous cell carcinomas (SqCC). These two NSCLC subtypes have both unique and shared clinical and histopathological characteristics. For example, while smoking is the major risk factor for both subtypes, approximately 10–15% of lung ADCs are observed in never smokers3. Molecularly targeted therapies directed against receptor tyrosine kinases (RTKs) lead to dramatic responses in subsets of patients with lung ADCs harboring activating genomic alterations in the corresponding kinase genes, including EGFR, ALK, and ROS14. Other targeted therapies under current investigation are directed against activating alterations in the MET, RET, NTRK1, NTRK2, ERBB2, and BRAF kinases4,5.
Recent efforts have focused on comprehensively characterizing the changes found within the genome, epigenome, transcriptome, and proteome of lung ADCs and SqCCs in order to discover novel cancer driver genes that may be clinically actionable6–8. Identifying novel cancer genes can be challenging due to the large number of passenger mutations that can accumulate from prolonged exposure to tobacco carcinogens and from inherent mutagenic processes such as those caused by the aberrant activity of APOBEC cytidine deaminases9. Profiling larger numbers of samples within a tumor type and combining samples across tumor types can help overcome this problem, by providing the additional statistical power necessary to distinguish important genes mutated at a lower frequency from other genes with passenger mutations10. In addition, a comprehensive comparison of recurrently altered genes found in lung ADC and SqCC has not been performed. Such analyses may yield insights into the similarities and differences in carcinogenesis between the diseases and elucidate the degree to which common or distinct targeted and immunologic therapeutic strategies can be used to treat each cancer type.
RESULTS
Comparison of somatically altered genes
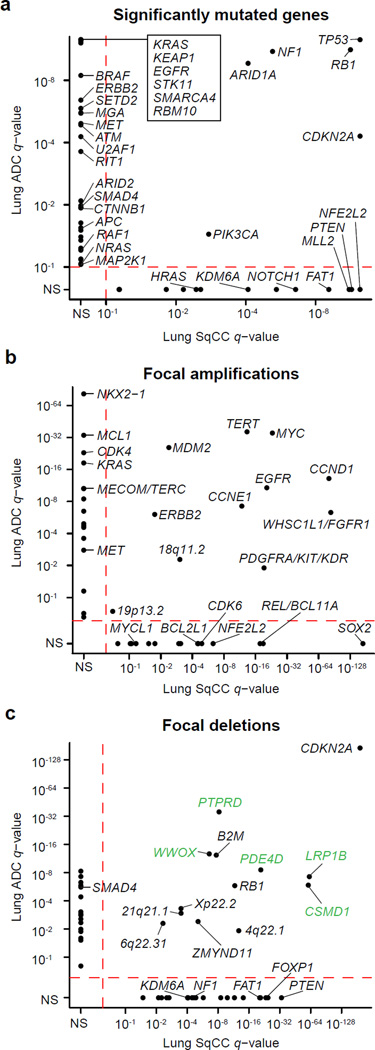
In order to compare the somatic profiles of lung ADC and lung SqCC and to identify novel genetic alterations, we studied 660 lung adenocarcinoma/normal paired exome sequences (including 274 previously unpublished cases, 227 previously described from The Cancer Genome Atlas (TCGA)6, and 159 cases from the Imielinski et al8 cohort) and 484 lung SqCC/normal paired exome sequences (including 308 previously unpublished cases and 176 previously described from TCGA7; Supplementary Tables 1–4). Similarly to previous studies6,7, we observed a median somatic mutation rate of 8.7/Mb and 9.7/Mb for lung ADCs and SqCCs, respectively. After excluding genes with lower median expression (6.16 log2 FPKM for ADCs and 6.27 log2 FPKM for SqCCs; Supplementary Fig. 1; Online Methods), we identified 38 genes as significantly mutated in ADC and 20 genes significantly mutated in SqCC using MutSig2CV10 (q-value < 0.1; Supplementary Table 5,6). Only 6 genes, TP53, RB1, ARID1A, CDKN2A, PIK3CA, and NF1, were significantly mutated in both tumor types and of these, the frequency of TP53, CDKN2A, and PIK3CA mutation was significantly higher in SqCC tumors (p < 0.01; Fisher’s exact test; Figure 1a). Likewise, only 11 out of 42 focal amplification peaks were identified as altered in both tumor types (Figure 1b), while 13 out of 50 focal deletion peaks were altered in both tumor types (Figure 1c). Interestingly, when compared to 19 other tumor types from TCGA10, the lists of significantly mutated genes for lung ADC and SqCC had a greater overlap with significantly mutated gene lists from other tumor types (>13% overlap; FDR q-value < 0.1) than to each other (12% overlap; p = 0.105; Supplementary Fig. 2), consistent with previous pan-cancer analyses11. Recurrently mutated and amplified genes in lung SqCC most closely resembled the alterations in head and neck squamous cell carcinoma (HNSC) and bladder cancer (BLCA), two other epithelial cancers with epidemiological associations with smoking (>25% overlap; Supplementary Fig. 2). Within these overlapping genes, TP53, CDKN2A, and FAT1 are specifically enriched in HPV- HNSC12. In contrast, the significantly mutated genes in lung ADC were most similar to glioblastoma (GBM) and colorectal cancer (CRC) (FDR q-value < 0.1). While lung ADC and lung SqCC did share several focal deletion peaks, five of these peaks were putative fragile sites (shown in green in Figure 1c). Taken together, these results suggest that the somatic drivers of carcinogenesis may be largely distinct between lung adenocarcinoma and lung squamous cell carcinoma.
Figure 1. Distinct somatic alterations in lung ADC and SqCC.
(a) The MutSig2CV algorithm10 was used to identify significantly mutated genes across 660 lung ADCs and 484 lung SqCCs. Genes with q-values < 0.1 were considered significant. The q-value for each gene in the lung ADC cohort is plotted against its respective q-value in the lung SqCC cohort. The majority of significantly mutated genes were unique to either tumor type. The GISTIC2.0 algorithm was used to identify significantly recurrent copy number gains and losses. The q-values for (b) amplifications and (c) deletions in the lung ADC cohort are plotted against the q-values in the lung SqCC cohort. Peaks with q-values < 0.25 were considered significant. Deletions located within putative fragile sites are indicated with green labels. Only points from previously characterized lung cancer genes are labeled. N.S. = Not Significant.
Mutational signatures in lung cancer
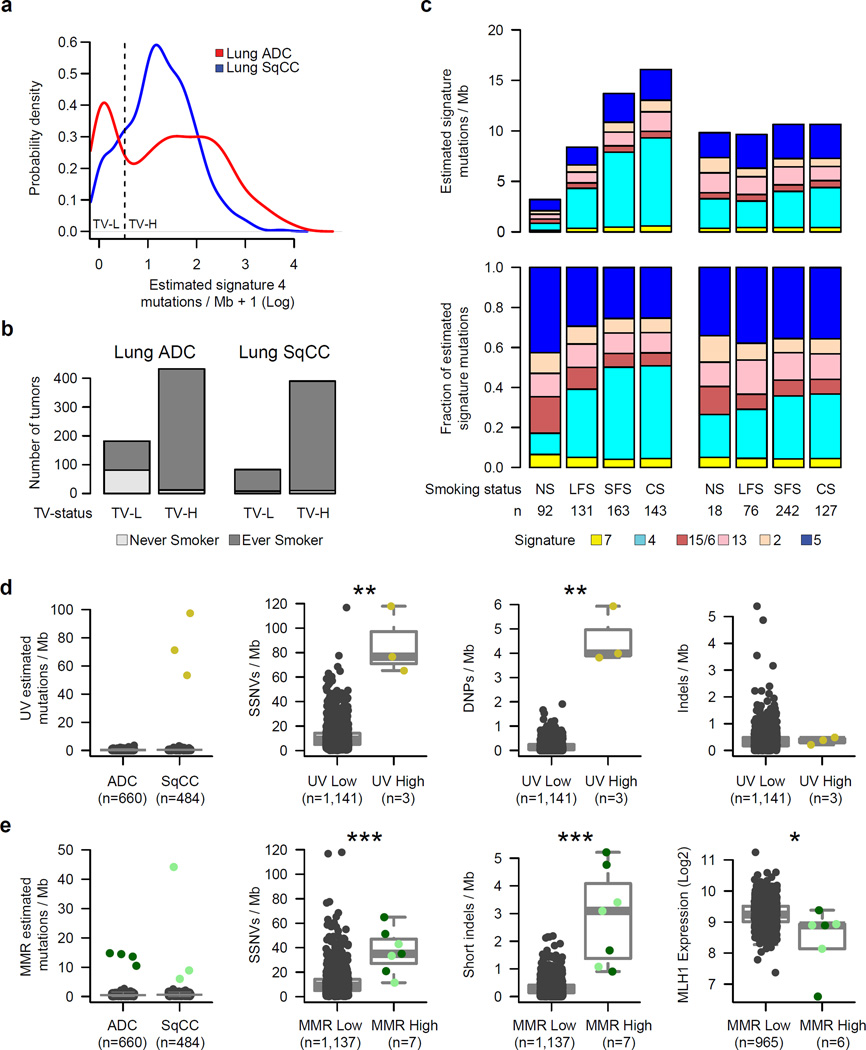
Various carcinogenic and cancer-related processes contribute to the mutational patterns observed in tumors13,14. Previous large-scale studies of lung cancer genomes have identified signatures associated with non-smoking and smoking cases6,8,15; here we extend these findings based on the improved statistical power of our larger sample set. Using non-negative matrix factorization13,16 (NMF, Online Methods), we identified six mutational signatures in this cohort, many of which are strongly correlated with previously defined signatures in the COSMIC database13,17 (Supplementary Figs. 3–5; Supplementary Table 7). These included a UV-related signature of C>T at TpCpC or CpCpC (COSMIC Signature 7, abbreviated SI7), a smoking-related signature of C>A transversions (SI4), a mismatch repair (MMR) signature of C>T at GpCpG (SI15/SI6), two APOBEC-related signatures of C>G or C>T at TpCpT or TpCpA (SI13 and SI2), and a final signature with a moderate correlation to COSMIC signature 5 (SI5) with putative “molecular clock” properties18 (Supplementary Fig. 5). In addition to identifying mutational signatures, NMF also estimates the number of mutations contributed by each signature within each tumor. The estimated number of SI4 (i.e. smoking-related) mutations per Mb displayed a bimodal pattern in lung ADC but not in lung SqCC (Figure 2a). Furthermore, the rate of SI4 mutations per Mb was able to classify tumors into those from never vs. ever smokers substantially better in lung ADCs (AUC=0.87; Supplementary Fig. 6) than in lung SqCCs (AUC=0.62) suggesting that the smoking statuses for the 18 never smokers with lung SqCC may be inaccurate. 87% of lung ADCs from never smokers were categorized as transversion-low (TV-L; ≤0.696 of SI4 per Mb; p = 8.5 × 10−37; Fisher’s exact test; Figure 2b; Supplementary Fig. 6). However, only 45% of transversion-low lung ADCs were from patients who were never smokers (Figure 2b). Within each tumor, we also derived the fraction of estimated mutations for a signature by dividing the number of estimated mutations for that signature by the sum of estimated mutations from all signatures. Lung SqCCs displayed significantly higher overall rates of SI5 mutations per Mb compared to all lung ADCs (p < 0.001; Wilcoxon rank-sum test). However, lung ADCs from never smokers displayed the highest fraction of estimated mutations from this signature on average (Figure 2c; Supplementary Fig. 7). In lung SqCC, we also observed moderate associations of tumor stage with SI5 activity and total mutation rate (p < 0.01; Supplementary Fig. 8).
Figure 2. Comparison of mutational signatures in lung cancer.
Six mutational signatures were identified using non-negative matrix factorization (NMF) on 192 distinct mutation types. (a) The estimated number of SI4 (smoking-related) mutations per Mb within each tumor displayed a bimodal pattern in lung ADC (red). (b) Lung ADCs categorized as transversion-low (TV-L) were enriched in clinically-annotated life-long never smokers (p = 8.5 × 10−37). (c) The estimated number of mutations for each signature per Mb (top) or the fraction of estimated mutations for each signature (bottom) was averaged across life-long never smokers (NS), longer-term former smokers (LFS), shorter-term former smokers (SFS), and current smokers (CS) for both lung ADCs and lung SqCCs (excluding UV-High and MMR-High tumors discussed below). (d) Three lung SqCCs had a high number of estimated mutations from a UV-associated signature commonly observed in melanoma. These tumors displayed a significantly higher overall rate of SSNVs and DNPs compared to all other lung tumors (p < 0.01). (e) Mutational profiles for another 7 tumors exhibited an MMR-like signature commonly observed in MSI colorectal carcinomas. These tumors had significantly higher rates of both SSNVs and short indels (p < 0.001), as well as lower levels of MHL1 expression (p = 0.011). Asterisks indicate significance level from a Wilcoxon rank-sum test (*p < 0.05, **p < 0.01, ***p < 0.001). Boxplots show median (middle bar), 1st quartile (bottom of box), 3rd quartile (top of box). Boxplot whiskers demark 1.5 times the interquartile range or minimum/maximum value.
The mutational profiles of three lung SqCCs (~1% of lung SqCCs) exhibited a pattern of UV-related mutations (SI7) commonly observed in melanoma and displayed a significantly higher mutation rate of somatic single nucleotide variants (SSNVs) and somatic dinucleotide polymorphisms (DNPs) compared to the other lung tumors (p < 0.01) but not higher rates of indels (p > 0.05; Figure 2d). One of these patients (TCGA-18–3409) had a previous history of basal cell carcinoma in the forehead raising the possibility that metastasis from the skin to the lung had occurred. The other two lung SqCCs with this signature may also represent squamous cell skin carcinomas metastatic to the lung. Mutational profiles for another 7 tumors (4 lung ADCs and 3 lung SqCCs) exhibited an MMR-like signature (SI15/SI6) commonly observed in microsatellite instable (MSI) colorectal carcinomas (Figure 2e)13. These tumors had significantly higher rates of both SSNVs and short indels (p < 0.00). They also displayed lower expression levels of the mismatch repair gene MLH1 (p = 0.011) suggesting a potential etiology for this signature in lung.
Novel significantly mutated genes
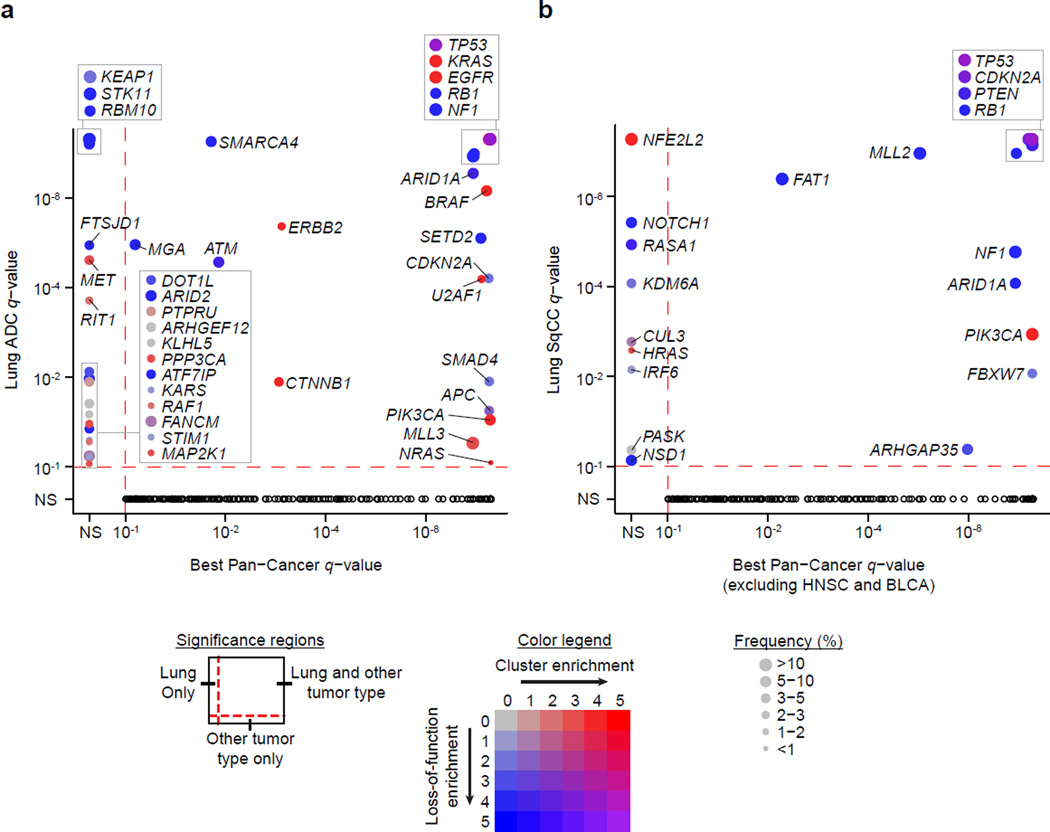
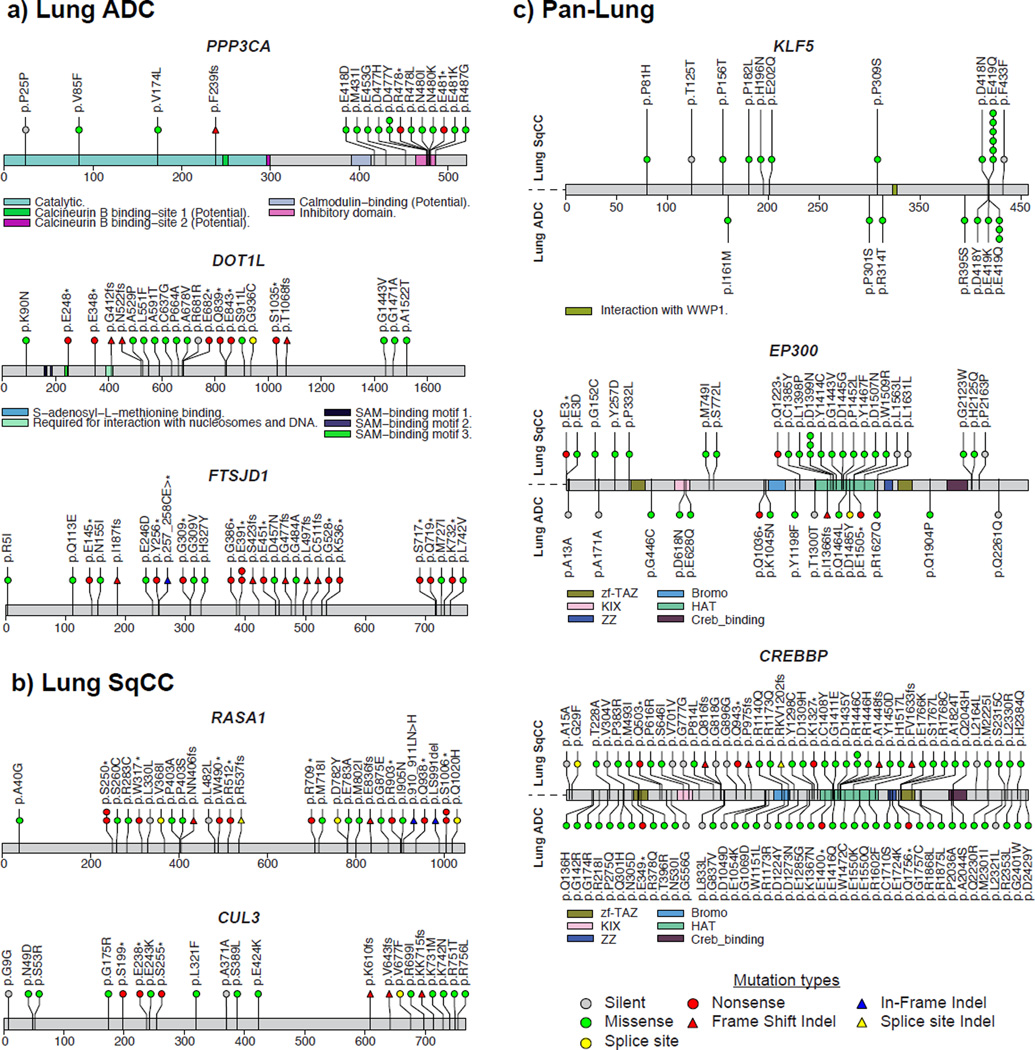
Comparing the significantly mutated genes to other tumor types from the TCGA Pan-Cancer study10 revealed that there were several genes significantly mutated exclusively in lung ADC including STK11, RBM10, KEAP1, RAF1, RIT1, and MET (MutSig2CV q-value < 0.1; Figure 3a; Supplementary Table 5). NFE2L2, KDM6A, RASA1, NOTCH1, and HRAS were significantly mutated in lung SqCC but not in other cancer types (excluding HNSC and BLCA) (Figure 3b; Supplementary Table 6). Genes that reached modest statistical significance in lung ADC that have been observed previously in lung cancer or in other tumor types include AKT1 with a recurrent mutation at p.E17K, CDK4 with a recurrent mutation at p.R24L, and DNMT3A (p < 0.005; Supplementary Table 5). Novel significantly mutated genes exclusive to lung ADC and which are absent in other tumor types include PPP3CA, which is the catalytic subunit for the calcium-dependent phosphatase, calcineurin. The mutations in PPP3CA clustered in the autoinhibitory domain near the C-terminus suggesting they may be gain-of-fuction alterations (Figure 4a). In addition, mutations in the autoinhibitory domain also tended to co-occur with activating KRAS mutations (p = 0.033) suggesting a potential relationship between K-Ras and calcineurin signaling pathways. Significantly mutated methyltransferase genes included MLL3 (KMT2C) and SETD2. A novel gene in this class was the H3K79 methyltransferase DOT1L, which was mutated in 3% of lung adenocarcinomas with enrichment for truncating mutations (Figure 4a). Recurrent mutations in lung adenocarcinoma have been previously reported in splicing factors such as U2AF1 and loss of function mutations in the RNA binding protein RBM108. In the current dataset, a cap methyltransferase, FTSJD1 (also known as CMTR2), was significantly mutated and enriched for frame shift mutations (Figure 4a). We also examined genes for other known proteins in this class and found recurrent mutations in SF3B119 and SNRPD3 (Supplementary Fig. 9). EGFR mutations were enriched in females, and SMARCA4 mutations were enriched in males (FDR q-value < 0.1; Supplementary Table 8). RBM10 mutations were modestly enriched in males as previously reported (q-value = 0.219)6. Novel significantly mutated genes in lung SqCC that were enriched for frame shift mutations (p < 0.001) included RASA1, whose protein product is p120GAP20 (Figure 4b). CUL3, whose protein product is a known interaction partner of KEAP1, also reached statistical significance in the lung SqCC cohort21 (Figure 4b). RB1 mutations were enriched in females, whereas PASK mutations were exclusive to males (FDR q-value < 0.1; Supplementary Table 9). We did not observe significant associations between mutation status and patient survival or tumor stage after correction for multiple hypothesis testing (Supplementary Tables 10–13). Controlling for tumor stage did not reveal additional significant associations between mutation status and survival.
Figure 3. Significantly mutated genes in lung cancer compared to other cancer types.
(a) The q-value for each significantly mutated gene in the lung ADC cohort is plotted against the best q-value for the same gene from 19 other tumor types from a Pan-Cancer study10. (b) The q-values from the lung SqCC cohort were similarly compared to the other tumor types excluding head and neck squamous cell (HNSC) and bladder urothelial carcinomas (BLCA). Size of the point is proportional to the frequency of mutations in the gene. The color of the point indicates enrichment for mutation clustering defined by MutSig2CV (−log10 pCL) and/or enrichment for loss-of-function mutations (−log10 p-value from a Fisher’s exact test, Online Methods). Black circles in the lower quadrant indicate genes significant in another cancer type but not in lung ADC and/or lung SqCC.
Figure 4. Novel significantly mutated genes in lung cancer.
Mutation profiles of novel genes specific to each lung tumor type include (a) PPP3CA, DOT1L, and FTSJD1 in lung ADC and (b) RASA1 and CUL3 in lung SqCC. (c) Combined analysis of both tumor types (Pan-Lung) revealed additional significantly mutated genes with hotspots including KLF5 and two paralogs, EP300 and CREBBP.
Previous studies have shown that joint analysis of different tumor types can yield additional statistical power to detect low-frequency events even if the tumor types are from vastly different tissues of origin and/or etiologies10. Additionally, although the individual drivers may be distinct between two tumor types, pathways such as MAP kinase are often altered similarly in both. We therefore hypothesized that combining the lung ADC and SqCC tumor cohorts (i.e. Pan-Lung) would reveal additional recurrent somatic pathway alterations common to both. We found 14 genes to be significantly mutated in the Pan-Lung cohort but not significantly mutated in either individual tumor type (q-value < 0.1; Supplementary Fig. 10; Supplementary Table 14). Many of these genes are involved in epigenetic regulation or immune-related pathways. KLF5, a transcription factor critical for lung development22 contained a novel recurrent mutation in the zinc finger domain, which was observed in both ADCs and SqCCs (Figure 4c). A regulator of KLF5, the E3 ubiquitin ligase FBXW723, was also significantly mutated in the lung SqCC and Pan-Lung cohorts but did not co-occur with KLF5 mutations. A super-enhancer duplication associated with increased KLF5 expression has also been recently reported in HNSC by our group24. The paralogs EP300 and CREBBP had a mutational hotspot region within the histone acetyltransferase (HAT) domain. All missense mutations in the HAT domains and other loss-of-function mutations outside this domain were non-overlapping within these two proteins. For sites with sufficient sequencing depth in the RNA-seq (power > 95%), we observed a somatic SNV validation rate of 88%.
Novel somatic copy number alterations
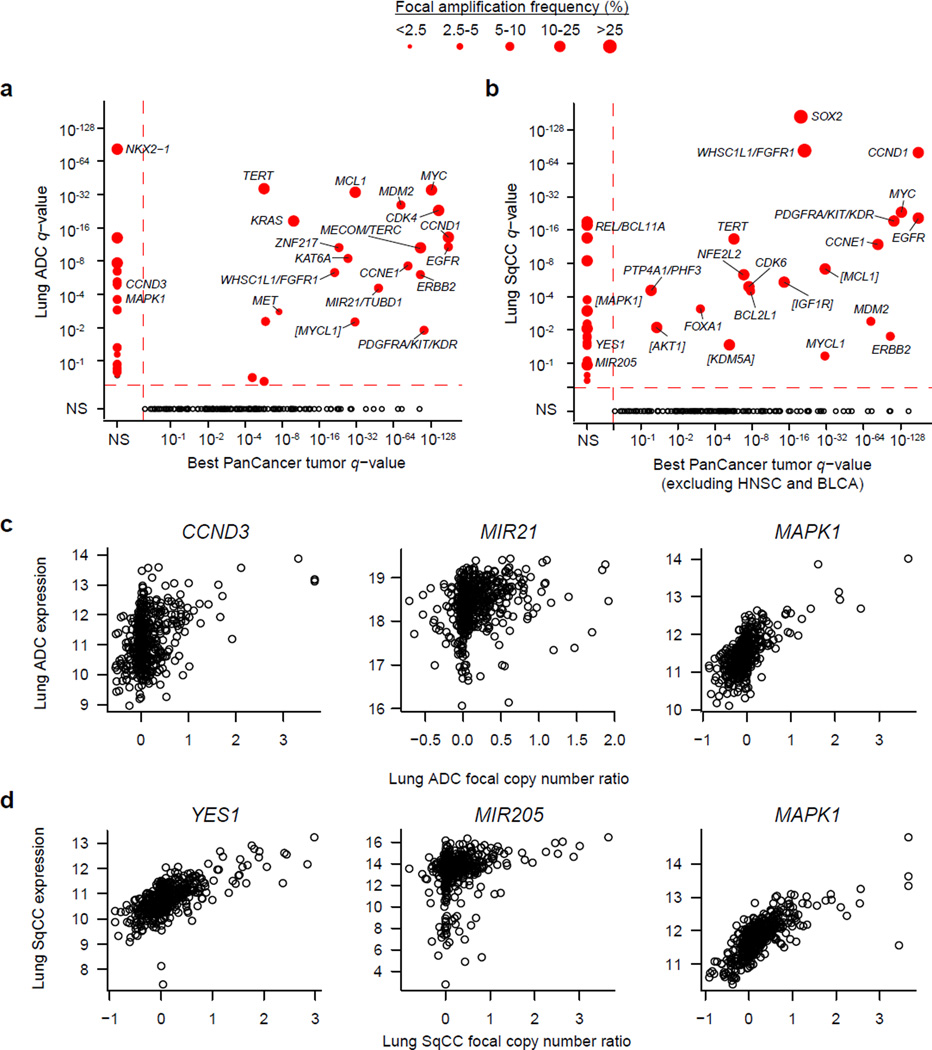
With a larger sample size, we had better resolution to detect novel copy number changes and ascertain the putative target genes of focal amplifications and deletions. For some peaks that still contained many genes, we inferred the most likely target gene by examining the same peak in a Pan-Cancer copy number analysis across 11 tumor types25 that included a subset of lung cancers from this set. The most significantly focally amplified genes in lung ADC were NKX2-1, MYC, TERT, MCL1, and MDM2 (Figure 5a; Supplementary Table 15), while peaks at SOX2, CCND1, WHSC1L1/FGFR1, MYC, and EGFR were among the top for lung SqCC (Figure 5b; Supplementary Table 16). Amplification peaks previously described in other tumor types but less characterized before in the lung tumors included KAT6A, ZNF217, and MYCL1 for lung ADC (Figure 5a) and IGF1R, KDM5A, PTP4A1/PHF3, and MYCL1 for lung SqCC (Figure 5b). CCND3 was specifically amplified in lung ADC while an amplification peak near MIR21/TUBD1 (Figure 5c) was also observed in breast cancer25. MIR21 expression has been shown to be a prognostic factor for early stage ADC26,27. Likewise, novel amplification peaks for lung SqCC included YES1, a Src family non-receptor protein kinase, and MIR205 (Figure 5d). Expression of MIR205 has been used to distinguish lung squamous cell carcinomas from other NSCLC types28 suggesting that amplification of this microRNA may represent a lineage specific alteration similar to that of SOX2 amplification. Finally, combined Pan-Lung copy number analysis revealed additional amplification peaks around MAPK1 (Figure 5a–d; Supplementary Table 17).
Figure 5. Significant amplifications in lung cancer.
(a) The q-value for amplifications in lung ADC are plotted against the best q-value for the same gene across 9 other non-lung tumor types25. (b) The q-values for amplifications in lung SqCC are compared against 7 other tumor types excluding HNSC and BLCA. Size of the point is proportional to the frequency of focal alterations. Brackets around gene names indicate that the most likely target gene was inferred from Pan-Cancer copy number analysis across 11 tumor types or from the combined Pan-Lung copy number analysis. Black circles in the lower quadrant indicate genes significantly altered in another cancer type but not in lung ADC and/or lung SqCC. Gene expression is plotted against focal copy number ratios for novel amplification peaks that include (c) CCND3, MIR21, and MAPK1 in lung ADC and (d) YES1, MIR205, and MAPK1 in lung SqCC.
Focal deletion peaks in lung ADC included the chromatin modifier genes SMARCA4 and ARID2 (Supplementary Fig. 11; Supplementary Table 18), which were also significantly mutated and enriched for loss-of-function mutations. Novel lung SqCC focal deletions observed in other tumor types included ZMYND11, CREBBP, ROBO1, USP22, and KDM6A (Supplementary Fig. 11; Supplementary Table 19). B2M (Beta2-microglobulin), a component of the MHC complex, was focally deleted in both tumor types, enriched for loss-of-function mutations in both tumor types (p < 0.01), and was significantly mutated in the Pan-Lung analysis (FDR q-value = 0.006). Combined Pan-Lung copy number analysis revealed another focal deletion peak around TRAF3 (Supplementary Table 20), which was also reported in HNSC12. In general, mRNA expression was significantly associated with copy number levels for target genes (Supplementary Figs. 12 and 13). We did not observe substantial batch effects within or across tumor types in both the mRNA expression and CNV data (Supplementary Figs. 14 and 15).
Identifying Ras/Raf/RTK drivers in lung ADC
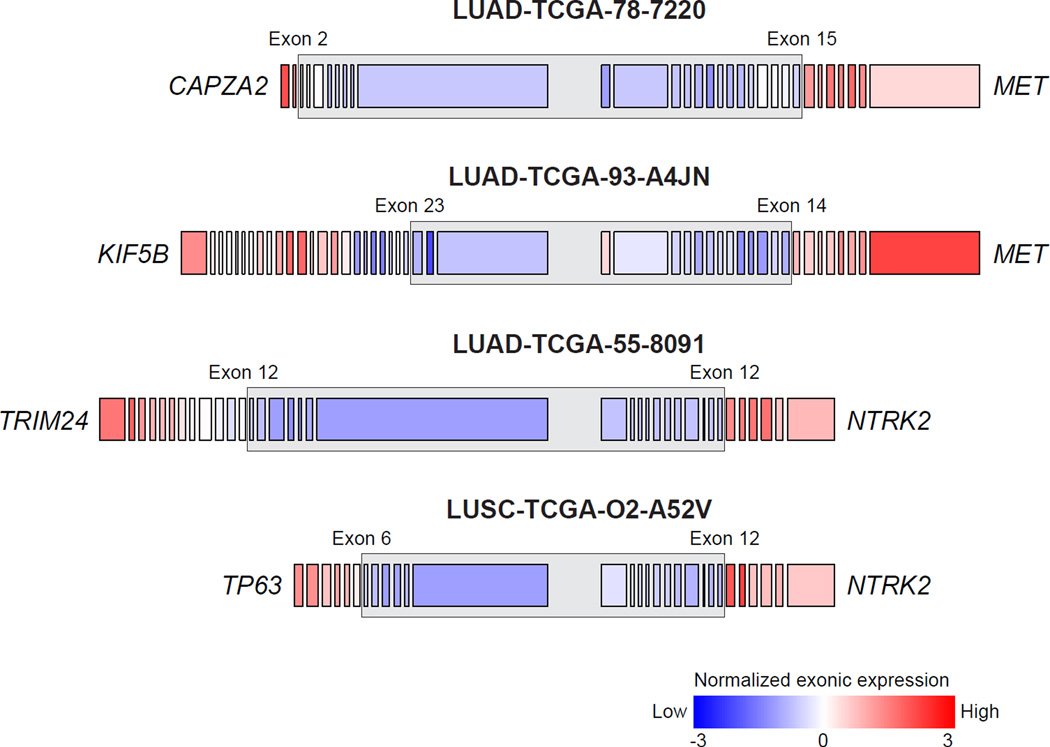
In lung ADC, mutually exclusive alterations have been characterized in members of the receptor tyrosine kinase (RTK)/Ras/Raf signaling pathways. These alterations are of particular interest because of the dramatic responses that have been observed in response to RTK inhibitors in clinical trials such as those for lung ADC patients harboring EGFR mutation or ALK or ROS1 translocations29. However, many lung ADCs do not exhibit a known activating mutation in the pathway raising the possibility that additional genes with low frequency somatic events are yet to be identified. To further understand the somatic landscape of this pathway, we first characterized alterations among the known pathway members and then identified novel genes with mutually exclusive alterations. Novel alterations in known pathway genes included a recurrent in-frame-insertion in MAP2K1 and a fusion of MET with its neighboring gene, CAPZA2 (Figure 6; Supplementary Table 21)30. Previously reported TRIM24-NTRK2 and KIF5B-MET fusions30 were observed in tumors without other known activating alterations. Interestingly, another NTRK2 fusion with TP63 was also found in a lung SqCC (Figure 6; Supplementary Table 21). As observed previously, high MET and ERBB2 amplifications were enriched in tumors without other known activating alterations in this pathway (p < 0.01; Supplementary Fig. 16)6. A single lung adenocarcinoma (TCGA-49–4512) contained an activating EGFR kinase domain duplication31. By manual review, we found additional canonical mutations in KRAS, EGFR, or ERBB2 in 17 tumors and complex indels in EGFR or MET in 11 tumors, some of which have been previously reported6,8,32 (Supplementary Table 22).
Figure 6. Fusions in MET and NTRK2.
Two fusions in MET were identified which retained the receptor tyrosine kinase domain including one with its neighboring gene, CAPZA2. This fusion mostly likely arose via tandem duplication resulting in the 3’ end of MET being fused with the 5’ end of CAPZA2. Previously reported TRIM24, NTRK2 and KIF5B-MET fusions30 were observed in lung ADCs without other known Ras/Raf/RTK activating alterations. Another NTRK2 fusion with TP63 was also found in a lung SqCC. The expression of exons retained in the putative fusion transcript was relatively higher than the expression of exons not in the putative fusion transcript (as indicated by the grey box).
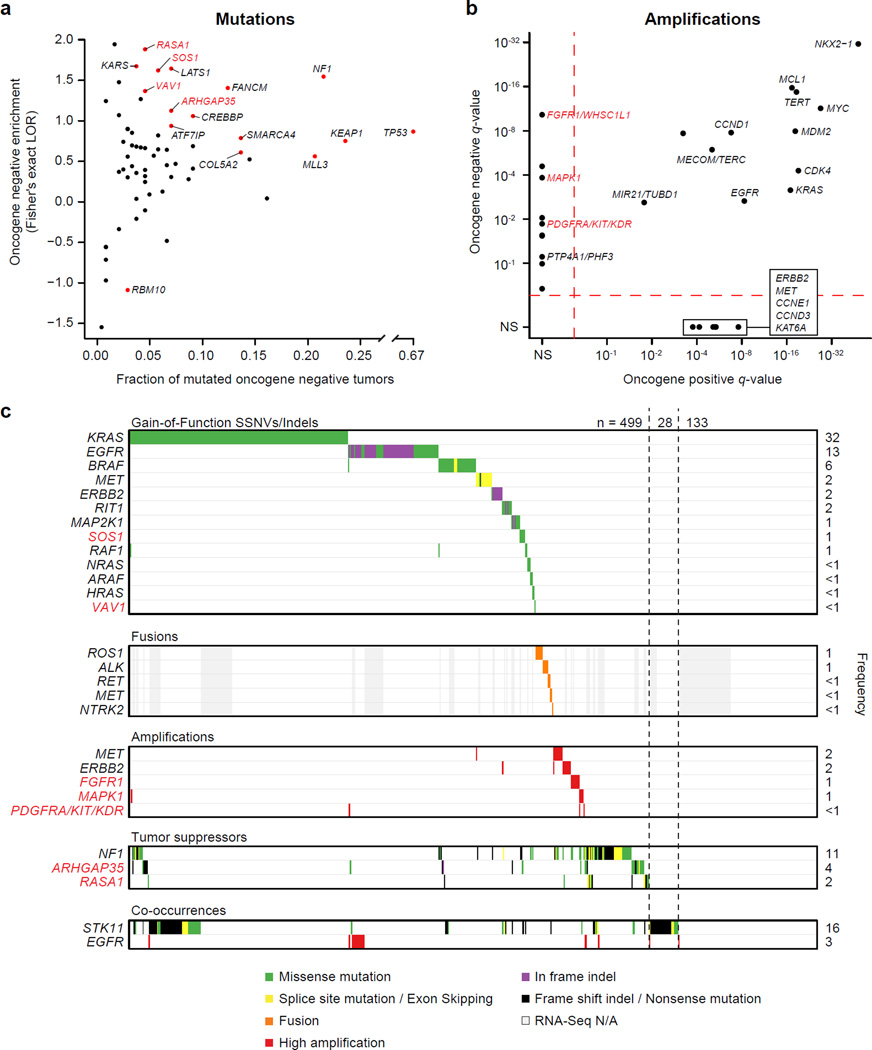
Lung ADCs that had an activating SSNV, indel, amplification or gene fusion in a known RTK/Ras/Raf driver6,33,34 were designated “oncogene positive” (n=418) while the remaining lung ADCs were considered “oncogene negative” (n=242). For the purposes of this analysis, we did not include NF1-altered tumors in the oncogene positive group as mutations in this gene are not entirely mutually exclusive with alterations in other Ras/Raf/RTK related genes. To identify additional potential drivers in this pathway, we determined if genes that are significantly mutated in any of the MutSig2CV analyses (Supplementary Tables 5, 6, and 14) or that are important in regulation of the Ras pathway35 were enriched in oncogene negative samples using a Fisher’s exact test. In total, 15 genes were significantly enriched among oncogene negative samples including known Ras pathway members SOS1 and RASA1, and Rho kinase pathway members VAV1 and ARHGAP35 (q-value < 0.1; Figure 7a,c; Supplementary Table 23). SOS1 is a guanine nucleotide exchange factor (GEF) bound to the RTK complex and assists in the activation of Ras proteins36. Recurrent p.N233Y mutations were observed in the autoinhibitory domain (DH) of SOS1 in 4 lung ADCs and the mutation p.D309Y in the same region has been reported in Noonan syndrome37,38 (Supplementary Fig. 17). Similarly, VAV1 is a GEF for the Rho family GTPases. Interactions between the calponin homology (CH), Acidic (Ac), and pleckstrin homology (PH) domains are important for autoinhibition of the catalytic Dbl homology domain39. The p.S67Y mutation is located near the interface of the CH, Ac and PH domains and mutagenesis at this site has been shown to increase overall GEF activity39 (Supplementary Fig. 17). RASA1 and ARHGAP35 (p190RhoGAP) are GTPase activating proteins (GAPs) for the Ras and Rho kinases, respectively, and are each enriched for loss-of-function mutations (p < 0.01). We also identified amplifications peaks near FGFR1/WHSC1L1 (8p11.21), PDGFRA/KIT/KDR (4q12), and MAPK1 (chr22q11) that were only significant in the oncogene negative tumor set (q-value < 0.25; Figure 7b,c). In total, 499 (76%) lung ADCs displayed an alteration in known or putative Ras/Raf/RTK driver genes (Figure 7c). Moreover, 193 out of 227 (85%) lung ADCs that previously underwent secondary expert pathological review and had RNA-seq data available for fusion analysis6 contained a predicted activating alteration in the RTK/Ras/Raf pathway.
Figure 7. Novel alterations in the Ras/Raf/Rho/RTK pathway in lung ADC.
Lung ADCs were classified as “oncogene positive” if they contained a known activating or recurrent alteration in previously characterized pathway members and classified as “oncogene negative” otherwise. (a) Mutations from 15 genes (red points) were significantly enriched among oncogene negative tumors (Fisher’s exact test; FDR q-value < 0.1; Supplementary Table 23). A log odds ratio (LOR) greater than zero indicates that the frequency of mutations was higher in the oncogene negative set. (B) Significant amplification peaks near FGFR1/WHSC1L1, PDGFRA/KIT/KDR, and MAPK1 were only found in the oncogene negative tumor set using GISTIC2.0 (q-value < 0.25). (c) Co-mutation plot for known and novel activators of this pathway. Tumors were considered to have high amplification for a given gene if they had a total log2 copy number ratio greater than 1. For genes with gain-of-function SSNVs or indels, only recurrently mutated sites or sites with previous experimental functional evidence are included. Novel oncogene negative enriched genes that are members of the Ras/Raf/Rho/RTK pathway are indicated with red labels in all panels.
Novel co-occurrences included MET amplifications and NF1 mutations (p = 0.019; Supplementary Figure 16). Additionally, high EGFR amplification significantly overlapped with activating EGFR mutations (p = 1.9 × 10−8)40,41 and STK11 mutations significantly overlapped with activating KRAS mutations (p = 1.1 × 10−6; Figure 7c)42,43. Furthermore, 28 lung ADCs that remain oncogene-negative for the RTK/Ras/Raf pathway harbor STK11 mutations (Figure 7c), suggesting the possibility of an additional hitherto-unrecognized KRAS-related genome alteration complementary to STK11 mutation in these cancer samples.
Assessment of neoantigen load and recurrence
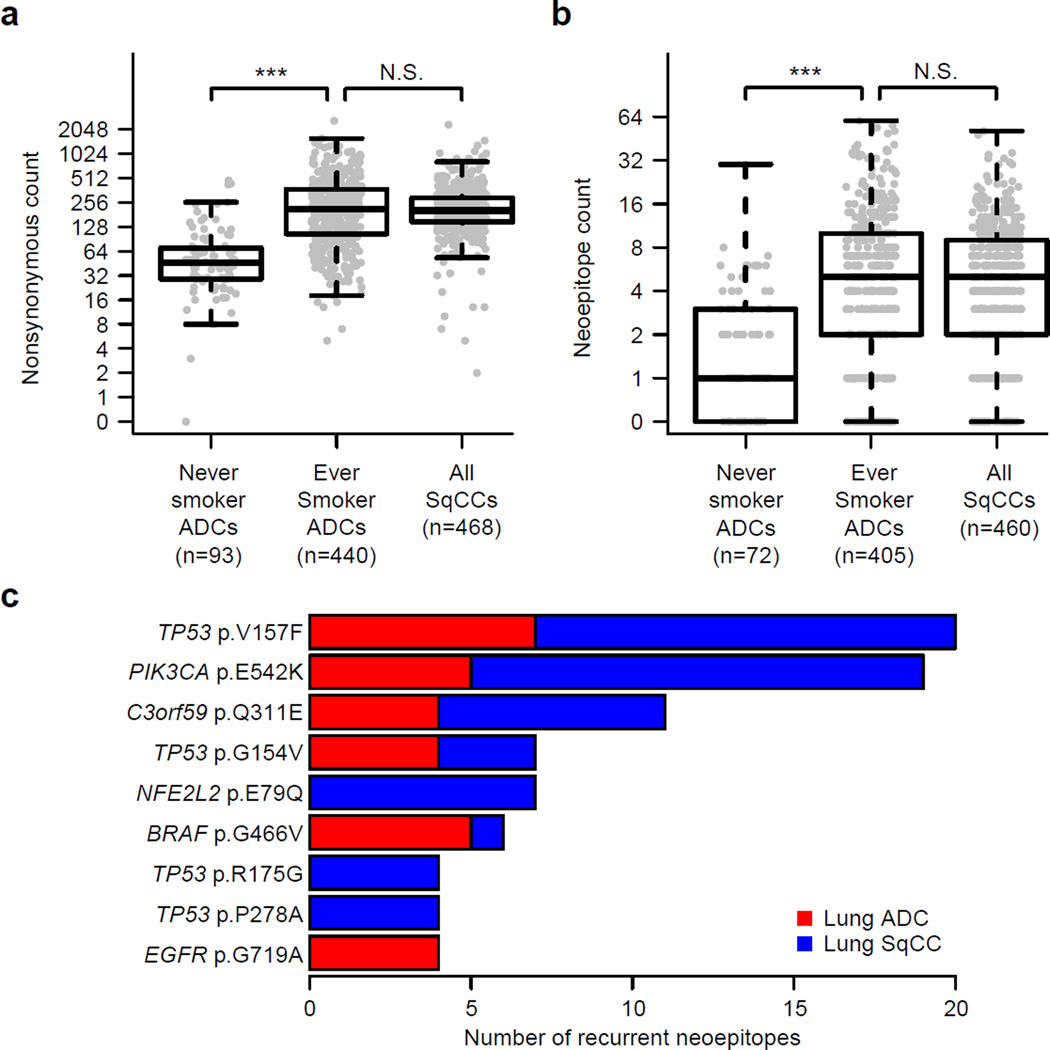
With increasing interest in the use of immune checkpoint inhibitors in lung cancer44,45, we comprehensively analyzed the potential immunogenic properties of the mutational landscape. Within each patient, we evaluated the ability of each somatic missense mutation to be processed and presented to immune cells by any one of the patient-specific HLA alleles46,47. We then assessed the association between the number of immunogenic mutations (i.e. neoepitopes or neoantigens) and clinical characteristics and identified the most common neoepitopes observed in lung cancer. Both nonsynonymous mutation and neoepitope counts were not significantly different between lung ADCs and lung SqCCs from ever smokers (Figure 8a,b). However, these counts were significantly lower in lung ADCs from never smokers compared to lung ADCs from ever smokers (p < 0.001; Wilcoxon rank-sum test; Figure 8a,b) and associated with overall smoking history in lung ADCs but not lung SqCCs (p < 0.001; Kruskal-Wallis test; Supplementary Fig. 18). Mutations predicted to be neoepitopes in at least 4 tumors included PIK3CA p.E542K, NFE2L2 p.E79Q, BRAF p.G466V, and EGFR p.G719A and several mutations in TP53, including p.V157F, p.G154V, p.R175G, and p.P278A (Figure 8c). A gene not previously implicated in lung cancer, C3orf59 (also known as MB21D2), contained a recurrent mutation at p.Q311E with predicted neoepitope properties (Figure 8c). Overall, 47% of lung ADC and 53% of lung SqCC samples had at least 5 predicted neoepitopes suggesting a great potential for immunotherapy.
Figure 8. Neoepitope load in lung cancer.
The immunogenicity of each missense mutation was predicted after inferring HLA alleles within each tumor with available RNA-seq data (n=971). (a) Nonsynonymous mutation counts and (b) neoepitope counts were not significantly different between ever smokers from lung ADCs and lung SqCCs (p > 0.05). However, these counts were significantly lower in lung ADCs from never smokers compared to lung ADCs from ever smokers (p < 0.001). (c) Some of the most common mutations predicted to be neoepitopes included TP53 p.V157F, PIK3CA p.E542K and C3orf59 p.Q311E.
DISCUSSION
We examined exome sequences and copy number profiles of 1,144 lung cancers to explore similarities and differences between lung adenocarcinoma and lung squamous cell carcinoma. Consistent with studies of gene expression11, this comparison showed that both mutated genes and recurrent SCNAs are largely distinct between the two lung cancer types. The similarity between lung SqCCs, head and neck squamous carcinomas, and a subset of bladder carcinomas was also observed when 12 tumor types were reclassified using clustering of five molecular data types48. These differences suggest that somatic alterations can have different oncogenic potential in different cellular contexts. Thus, cancers arising from developmentally similar cells of origin across different tissues will be more similar than cancers arising from different cells of origin within an anatomically defined tissue. As we had only one tumor sample per patient, we were not able to analyze intra-tumoral heterogeneity, as has been done in other studies49,50.
Several novel focal amplification peaks containing protein-coding genes, including MAPK1, YES1, and CCND3, were identified. Interestingly, we also found two peaks that contained or were near microRNAs (MIR21 in lung ADC and MIR205 in lung SqCC). We have also recently reported a non-coding super-enhancer duplication that results in increased MYC expression24. As the mutational analyses in this study focused on whole-exome sequencing of protein-coding genes, we were not able to examine mutations in non-coding genes or regulatory elements. Future studies examining large numbers of lung cancer whole-genomes may be better suited for discovery of other oncogenic alterations in non-coding genes or regulatory elements.
Our study has uncovered multiple significantly mutated genes in the RTK/Ras/Raf pathway, including newly identified genes such as RASA1, SOS1, and VAV1. Previous studies examining smaller numbers of lung tumors were not able to detect recurrent mutations in SOS18,38. The fact that we were able to detect these mutants further highlights the utility of increasing sample size to detect rare events. Since we did not have matching RNA-seq data for every tumor, we may be underestimating the rates of oncogenic fusions or MET exon 14 skipping events. As 15% to 25% of lung ADCs still do not contain a known detectable alteration in the RTK/Ras/Raf pathway, we may yet be underpowered to find additional rare recurrent mutations in known and novel pathway members. Similar considerations may be present for other pathways. For example, we identified new epigenetic modifier mutations in CREBBP and EP300, previously shown in small cell lung cancer51.
Finally, we examined the immunogenicity of individual missense mutations to understand more fully the association between neoepitope loads, overall nonsynonymous rates, and clinical variables such as smoking status. Some highly recurrent mutations were predicted to be neoepitopes. Future studies may further unravel the relationship between these candidates and clinical responses to immune checkpoint inhibitors and customized vaccine therapies.
ONLINE METHODS
Sample collection and pathology review
Sample collection and DNA sequencing were performed for the Imielinski et al and TCGA cohorts as previously described6–8. All specimens were obtained from patients with appropriate consent and with approval from the relevant institutional review boards. All patients were treatment-naïve with the exception of four patients with lung SqCC and three with lung ADC who received neoadjuvant treatment prior to resection (Supplementary Table 2). Initial pathological review was performed at the contributing tissue source sites (TSS) where each tumor was given an initial histological classification. After shipment of the frozen tissue to the Biospecimen Core Resource (BCR), one or two additional frozen sections were cut and stained with H&E to confirm the histological classification of the original TSS. 159 of the lung ADCs from Imielinski et al, 289 of the lung ADCs from TCGA, and 213 of the lung SqCCs from TCGA had also undergone additional histological review by an expert pathology committee led by Dr. William Travis (MSKCC) in previous studies6–8. Nucleic acid extraction and molecular quality control were performed at the BCR.
DNA-sequencing, alignment, and mutation calling
Exome capture was performed using the Agilent SureSelect Human All Exon 50MB kit followed by Illumina paired-end sequencing. Reads were processed using the Picard pipeline6. This pipeline utilizes BWA for read alignment, Picard tools for marking duplicates, and the Genome Analysis Tool Kit (GATK) for realignment around small insertions and deletions (indels) as well as base quality recalibration52. Contamination in tumor exomes was estimated using ContEst53. Only tumors with <5% contamination, an available SNP6.0 array for copy number analysis, and a valid ABSOLUTE54 solution were considered in the final analysis. The final sample set included 227 previously described lung ADCs from the TCGA6, 274 newly reported lung ADCs from the TCGA, and 159 lung ADCs from the Imielinski et al8 cohort, together with 176 previously described lung SqCCs from TCGA7, and 308 newly reported lung SqCCs from TCGA. Somatic single nucleotide variants (SSNVs) and indels were called using MuTect55 and Indelocator (www.broadinstitute.org/cancer/cga/indelocator), respectively. These algorithms compare the tumor to the matched normal in order to exclude germline variants. Somatic calls were excluded if found in a panel of over 2,900 normal exomes as previously described10. Coding mutation patterns can be viewed for individual genes at pubs.broadinstitute.org/panlung.
Identification of significantly mutated genes
Significantly mutated genes were identified using MutSig2CV which combines p-values from tests for high mutational frequency relative to the background mutation rate (pCV), clustering of mutations within the gene (pCL), and enrichment of mutations within evolutionarily conserved sites (pFN)10. For 660 lung adenocarcinomas, we had 100% power to detect genes mutated in 10% of patients and 73% power for genes mutated in 5% of patients assuming a mutation rate of 8.7/Mb10. For 484 lung squamous cell carcinomas, we had 100% power to detect genes mutated in 10% of patients and 41% power for genes mutated in 5% of patients assuming a mutation rate of 9.7/Mb10. In order to reduce the number of hypotheses tested in the MutSig2CV analysis, we excluded genes that exhibited low expression across tumors with relatively high purity. The median log2 FPKM value for each gene was obtained for 185 ADCs or 238 SqCCs which had a purity estimate from ABSOLUTE of >50% and available RNA-seq data (Supplementary Fig. 1). For each tumor type, a mixture model of two normal distributions was fit in R using the mclust package v4.2. Genes with 95% probability of belonging to the cluster with higher expression were considered in the multiple hypothesis correction of the MutSig2CV combined p-values. One gene, TRERF1, was excluded from the final results as closer inspection of its mutations revealed a recurrent frameshift deletion that was likely a false positive as all of these mutations had low allelic fractions (<1.5%) and had no supporting reads in matching RNA-seq data. A one-sided Fisher’s exact test was used to determine if the proportion of loss-of-function mutations (including nonsense, frameshift, and de novo start out-of-frame mutations) to other mutations for a given gene was significantly higher compared to the proportion of loss-of-function mutations to other mutations across all other genes.
Identification of recurrent copy number changes
DNA was hybridized onto Affymetrix SNP 6.0 arrays and normalized as previously described6. Segmentation was performed using Circular Binary Segmentation algorithm56 followed by Ziggurat Deconstruction to infer the length and amplitude each segment. Recurrent focal SCNA peaks were identified using GISTIC2.057. A peak was considered focally amplified or deleted within a tumor if the GISTIC2.0-estimated focal copy number ratio was greater than 0.1 or less than −0.1, respectively. Purity and ploidy were estimated using ABSOLUTE54. Two peaks were considered the same across tumor types if 1) the known target gene of each peak was the same or 2) the genomic location of the peaks overlapped +/− 1 Mb and each of the overlapping peaks had less than 25 genes and was smaller than 10 Mb.
RNA sequencing for expression and fusion analyses
Of the 1,144 tumors examined in this study, 495 lung ADCs and 476 lung SqCCs also had corresponding RNA-seq data from TCGA. RNA reads were generated, aligned to the hg19 genome assembly with Mapsplice58, and normalized with RSEM59 to Fragment per Kilobase per Million (FPKM) expression estimates as previously described6. Expression values less than 1 FPKM were set to 1 and all data were log2 transformed. Exon skipping of MET exon 14 was identified with juncBASE60 as previously described6. Lists of fusions were obtained from previous studies6,3061. Fusions for additional tumors were identified with the PRADA pipeline62. For plotting of exonic expression of fusion transcripts, exon expression levels were counted and normalized to reads per kilobase per million (RPKM) as previously described6. Expression for an individual exon was first Z-score transformed across all tumors within each tumor type. Subsequently, all exons for a gene were Z-score transformed again within each tumor. Transcript annotations used for this analysis included ENST00000397752 for MET, ENST00000361183 for CAPZA2, ENST00000302418 for KIF5B, ENST00000323115 for NTRK2, ENST00000343526 for TRIM24, and ENST00000354600 for TP63.
Identification of mutational signatures
Non-negative matrix factorization (NMF) was used to deconvolute a K × G matrix of mutation catalogues into a K × N matrix of mutational processes and an N × G matrix of mutational exposures (where G is the number of lung exomes, K is the number of mutational states, and N is the number of estimated mutational processes)16. Code to run NMF was obtained from MATLAB Central (see URLs) and run using the nnmf function from the MATLAB Statistics Toolbox. We used 6 mutation types with 16 different trinucleotide contexts and 2 transcriptional strands for a total of 192 mutational states. The number of possible signatures was varied from 1 to 10 and signature stability was assessed via bootstrapping as previously described16. Within each tumor, the fraction of estimated mutations for a signature was derived by dividing the number of estimated mutations for that signature by the sum of estimated mutations from all signatures.
Predicting Immunogenicity
HLA alleles were called with POLYSOLVER46 for all lung cancer exomes. Within each tumor, epitope predictions were made between confidently called alleles and single amino acid missense mutations. Separate lists were made consisting of wildtype and mutant peptides of length 8, 9, 10 and 11 amino acids since these are the possible peptide lengths known to be presented by human MHC class I molecules63. We then predicted MHC binding affinity for each of the peptide as described previously47. First, the proteasome processing score was calculated using the NetChop program64. Then, we used the NetMHC65, NetMHCpan66, SMM67, and SMMPMBEC68 methods to predict the MHC binding affinity values for each peptide and used the median affinity value across all algorithms as a composite measure of binding strength. We also defined the neoepitope ratio for each mutant-wildtype peptide pair as the mutant median affinity value divided by wildtype median affinity value. This value was found to be a reliable comparator of the relative immunogenicities of the mutant versus wildtype peptide sequences47. Peptide pairs were further considered if the mutant peptide displayed a processing score ≥0.7, a median affinity value ≥0.01, a neoepitope ratio ≥1, and the mRNA transcript of the gene was expressed in the RNA-seq data for that tumor (top 15,000 expressed genes within each tumor). Since epitope binding is HLA-dependent, the previous steps were performed for each of the called MHC-I alleles. After that, only those peptides predicted to be the best epitopes for each mutation were considered.
Statistical comparisons
Nonparametric tests such as the Wilcoxon rank-sum test (comparison between 2 groups) or the Kruskal-Wallis test (comparison between more than two groups) were used for continuous variables unless otherwise noted. The Fisher’s exact test was used when comparing 2 categorical variables. In total, longitudinal data on survival were available for 481 patients with lung ADC and 473 patients with lung SqCC from TCGA. The Cox proportional hazards model was used to examine associations between patient survival and mutation status, with and without controlling for stage. Correction for multiple hypothesis testing was performed with the Benjamini-Hochberg procedure.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institute as part of The Cancer Genome Atlas project: U24CA126546, U24CA143867, U24CA143845, U24CA126544, and U24CA143883. Additionally, this work was also funded by the National Cancer Institute K08 CA163677 (P.S.H.), Government of Russian Federation Grant 074-U01 (A.A.), the Department of Defense W81XWH-12-1-0269 (M.M.), the American Cancer Society Research Professor Award (M.M.), and the National Cancer Institute R35CA197568 (M.M.).
Competing Financial Interest
Research support from Bayer Pharmaceuticals (C.S.P., B.A.M., A.D.C., M.M.).
Collaborators
Jean C. Zenklusen, Jiashan Zhang, Ina Felau, John A. Demchok, Liming Yang, Zhining Wang, Martin L. Ferguson, Roy Tarnuzzer, Carolyn M. Hutter, Heidi J. Sofia, Todd Pihl, Yunhu Wan, Sudha Chudamani, Jia Liu, Charlie Sun, Rashi Naresh, Laxmi Lolla, Ye Wu, Chad J. Creighton, W. Kimryn Rathmell, J. Todd Auman, Saianand Balu, Tom Bodenheimer, D. Neil Hayes, Katherine A. Hoadley, Alan P. Hoyle, Corbin D. Jones, Stuart R. Jefferys, Shaowu Meng, Piotr A. Mieczkowski, Lisle E. Mose, Charles M. Perou, Jeffrey Roach, Yan Shi, Janae V. Simons, Tara Skelly, Matthew G. Soloway, Donghui Tan, Junyuan Wu, Umadevi Veluvolu, Joel S. Parker, Matthew D. Wilkerson, Lori Boice, Mei Huang, Leigh B. Thorne, Gad Getz, Michael S. Noble, Hailei Zhang, David I. Heiman, Juok Cho, Nils Gehlenborg, Gordon Saksena, Doug Voet, Pei Lin, Scott Frazer, Jaegil Kim, Michael S. Lawrence, Lynda Chin, Ming-Sound Tsao, Frances Allison, Dianne Chadwick, Thomas Muley, Michael Meister, Hendrik Dienemann, Raju Kucherlapati, Peter Park, Jay Bowen, Julie M. Gastier-Foster, Mark Gerken, Kristen M. Leraas, Tara M. Lichtenberg, Nilsa C. Ramirez, Lisa Wise, Erik Zmuda, Josh Stuart, Eric Collisson, Martin Peifer, David Kwiatkowski, Joshua D. Campbell, Bradley A. Murray, Andrew D. Cherniack, Alice H. Berger, Carrie Sougnez, Gordon Saksena, Steven E. Schumacher, Juliann Shih, Rameen Beroukhim, Travis I. Zack, Stacey B. Gabriel, Matthew Meyerson, Lauren A. Byers, Tanja Davidsen, Peter W. Laird, Daniel J. Weisenberger, David J. Van Den Berg, Moiz S. Bootwalla, Phillip H. Lai, Dennis T. Maglinte, Stephen B. Baylin, James G. Herman, Ludmila Danilova, Leslie Cope, Daniel J. Crain, Erin Curley, Johanna Gardner, Kevin Lau, David Mallery, Scott Morris, Joeseph Paulauskis, Robert Penny, Candace Shelton, Troy Shelton, Mark Sherman, Peggy Yena
Footnotes
URLs
Picard tools: http://broadinstitute.github.io/picard/,
MutSig algorithm: www.broadinstitute.org/cancer/cga/MutSig,
Indelocator: www.broadinstitute.org/cancer/cga/indelocator,
Broad Institute Firehose pipeline: http://www.broadinstitute.org/cancer/cga/,
Oncotator: http://www.broadinstitute.org/oncotator/
Power calculations: http://www.tumorportal.org/,
PRADA fusions: http://www.tumorfusions.org,
Mutational signatures: http://www.mathworks.com/matlabcentral/fileexchange/38724,
Accession codes
Clinical and molecular data from Imielinski et al8 are available in dbGAP under the accession phs000488.v1.p1. Binary alignment (BAM) files for all TCGA samples6,7 can be downloaded from the University of California Santa Cruz Cancer Genomics Hub (cghub.ucsc.edu) using the UUIDs in Supplementary Table 2. Additional clinical and molecular data for TCGA samples can be accessed via TCGA Data Portal (tcga-data.nci.nih.gov/tcga/). Mutational patterns for individual genes can be viewed via the Pan-Lung Tumor Portal (pubs.broadinstitute.org/panlung).
Author Contributions
J.D.C. performed sample quality control, mutation calling and review, ABSOLUTE analysis of tumors from the Imielinski et al cohort, identification and comparison of recurrently altered genes, mutational signature identification and characterization, identification of EGFR complex indels, and wrote the manuscript; A.A., M.N.A., and R.S. generated neoantigen calls; J.K. contributed to mutational signature analyses; J.W. contributed to EGFR complex indel characterization; A.H.B. contributed to oncogene negative analysis and manuscript preparation; C.S.P. generated the Pan-Lung portal; A.N.B. identified MET exon 14 skipping events using RNA-seq; X.H. and R.G.W.V. generated fusion calls; S.L. and R.A. performed batch effect analyses; G.Guo contributed to MET exon 14 complex indel identification; M.R., M.I., M.S.L., and G.Getz contributed algorithms for mutation calling and analyses; B.A.M. and A.D.C. contributed to copy number and ABSOLUTE analyses; S.A.S. and C.J.W. performed HLA genotyping; C.C. contributed to sample coordination and quality control; A.R., A.D.C., E.A.C., J.N.W., P.S.H., and D.J.K. contributed to manuscript preparation; R.G. and M.M. conceived and designed the study and wrote the manuscript.
References
- 1.Stewart BW, Wild CP, editors. World Cancer Report 2014. International Agency for Research on Cancer; 2014. [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Samet JM, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15:5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardarella S, Johnson BE. The impact of genomic changes on treatment of lung cancer. Am J Respir Crit Care Med. 2013;188:770–775. doi: 10.1164/rccm.201305-0843PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaishnavi A, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 20.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 21.Hast BE, et al. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res. 2014;74:808–817. doi: 10.1158/0008-5472.CAN-13-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan H, et al. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2015 doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akagi I, et al. Combination of protein coding and noncoding gene expression as a robust prognostic classifier in stage I lung adenocarcinoma. Cancer Res. 2013;73:3821–3832. doi: 10.1158/0008-5472.CAN-13-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito M, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebanony D, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 29.Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;31:1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallant JN, et al. EGFR Kinase Domain Duplication (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov. 2015;5:1155–1163. doi: 10.1158/2159-8290.CD-15-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye K, et al. Systematic discovery of complex insertions and deletions in human cancers. Nat Med. 2016;22:97–104. doi: 10.1038/nm.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 34.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;18:349–351. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- 35.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007;1773:1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Lepri F, et al. SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype-phenotype correlations. Hum Mutat. 2011;32:760–772. doi: 10.1002/humu.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson KD, et al. SOS1 mutations are rare in human malignancies: implications for Noonan Syndrome patients. Genes Chromosomes Cancer. 2008;47:253–259. doi: 10.1002/gcc.20527. [DOI] [PubMed] [Google Scholar]
- 39.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–256. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan L, et al. Concurrence of EGFR amplification and sensitizing mutations indicate a better survival benefit from EGFR-TKI therapy in lung adenocarcinoma patients. Lung Cancer. 2015;89:337–342. doi: 10.1016/j.lungcan.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Sholl LM, et al. Lung adenocarcinoma with EGFR amplification has distinct clinicopathologic and molecular features in never-smokers. Cancer Res. 2009;69:8341–8348. doi: 10.1158/0008-5472.CAN-09-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;3:870–879. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HS, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155:552–566. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brahmer J, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shukla SA, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 52.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 57.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang K, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brooks AN, et al. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 2011;21:193–202. doi: 10.1101/gr.108662.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshihara K, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2014 doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torres-Garcia W, et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30:2224–2226. doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alberts B. Molecular biology of the cell. New York: Garland Science; 2002. p. 1548. xxxiv. [Google Scholar]
- 64.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 65.Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim Y, Sidney J, Pinilla C, Sette A, Peters B. Derivation of an amino acid similarity matrix for peptide: MHC binding and its application as a Bayesian prior. BMC Bioinformatics. 2009;10:394. doi: 10.1186/1471-2105-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.